Pathogenesis-Related 1 (PR1) Protein Family Genes Involved in Sugarcane Responses to Ustilago scitaminea Stress
Abstract
1. Introduction
2. Results
2.1. Identification, Physio-Chemical Characterization, and Phylogeny of SsnpPR1 Genes
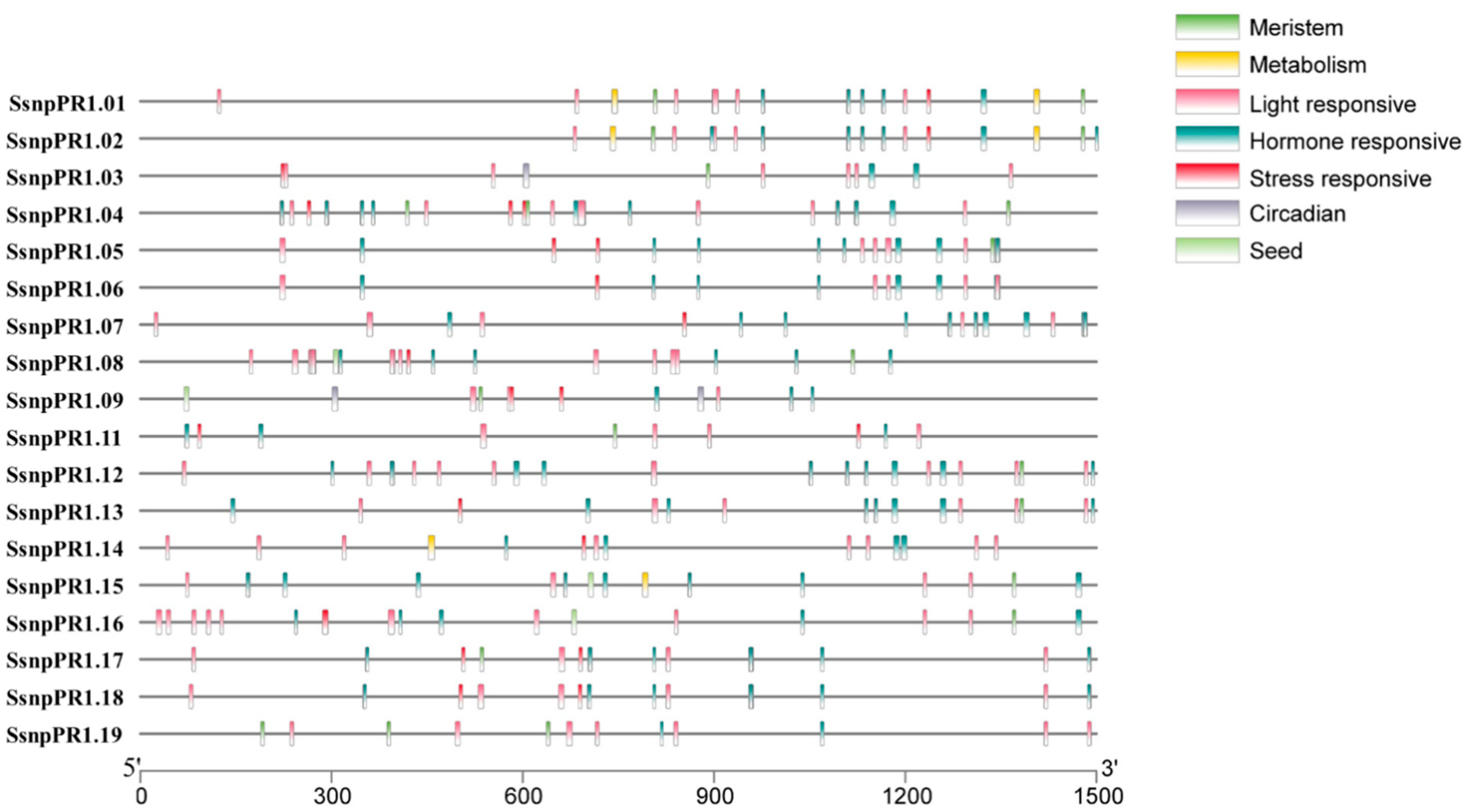
2.2. Gene Structure and cis-Regulatory Elements of SsnpPR1 Genes
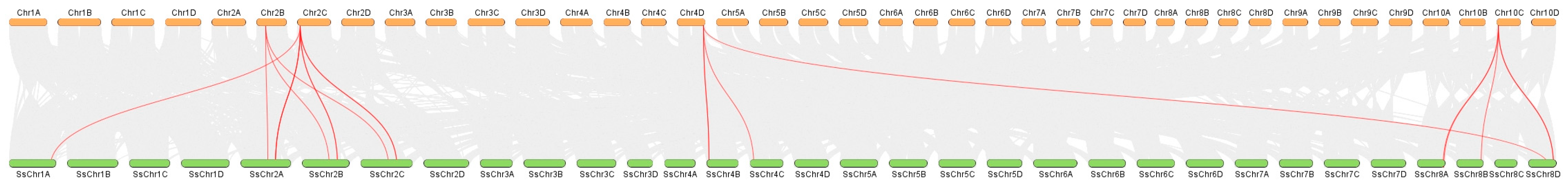
2.3. Chromosomal Mapping, Gene Duplication, and Collinearity Analysis of SsnpPR1 Genes
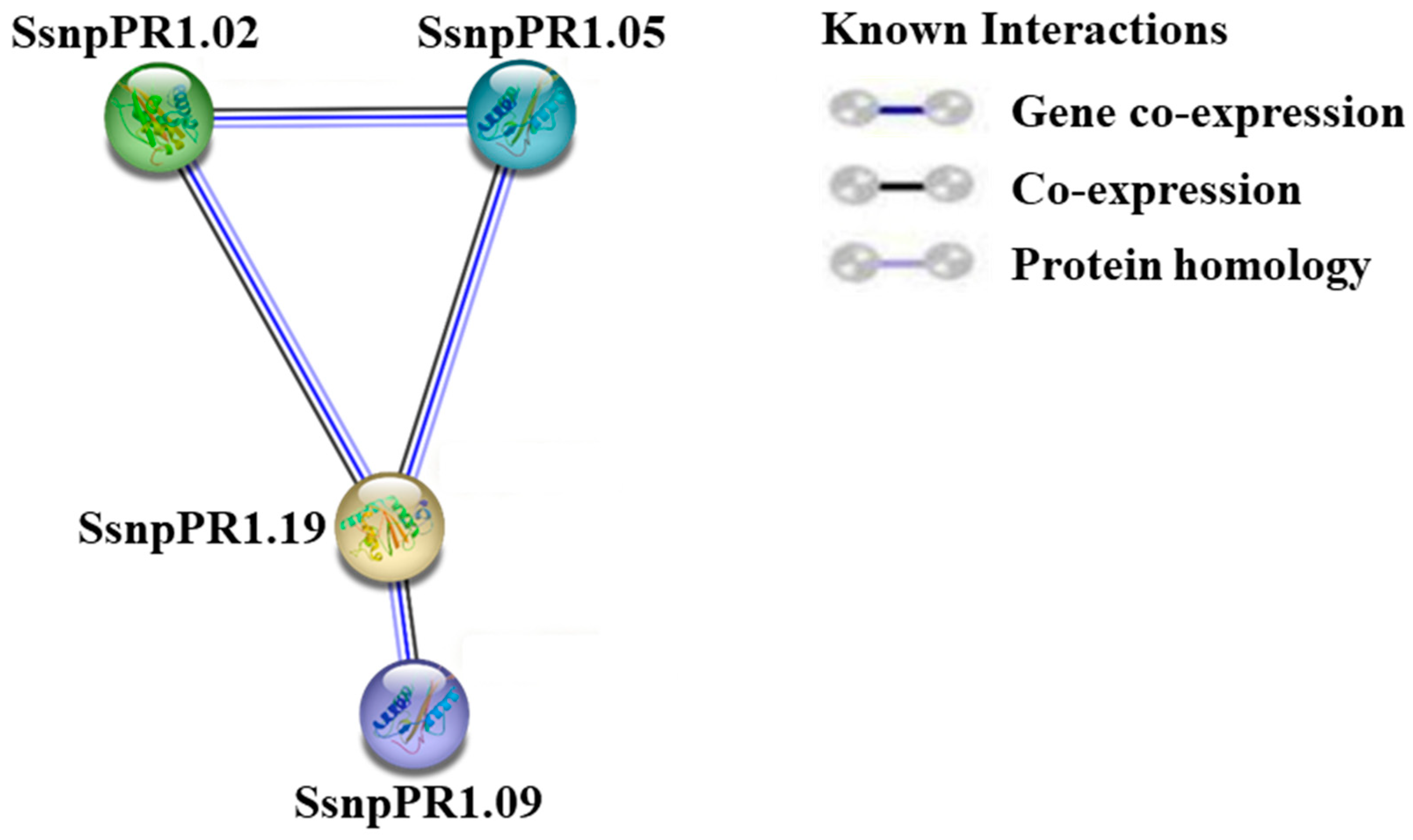
2.4. Protein–Protein Interaction Analysis
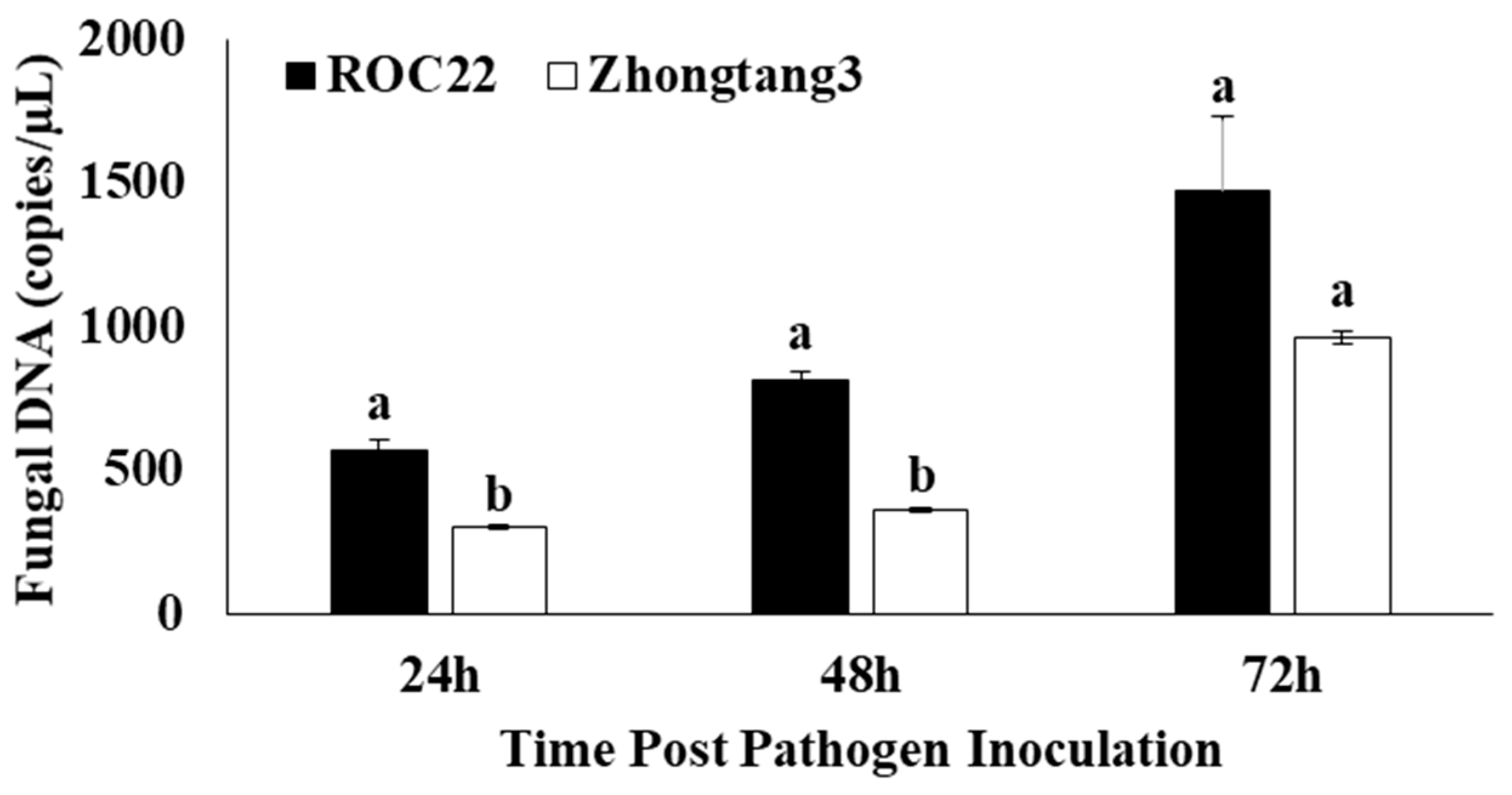
2.5. Sugarcane Cultivar Responses to Pathogen Infection
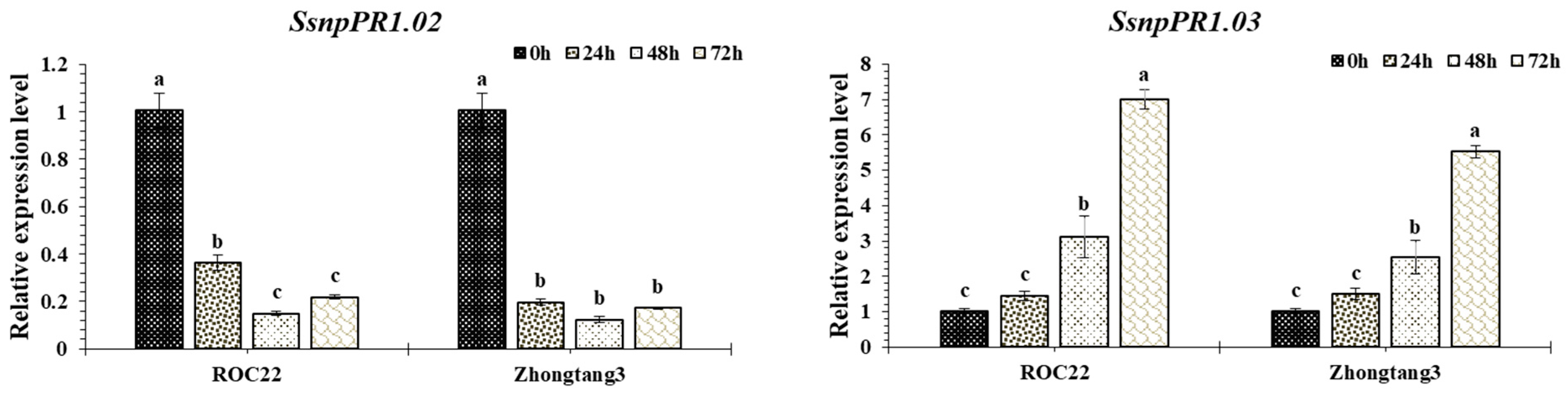
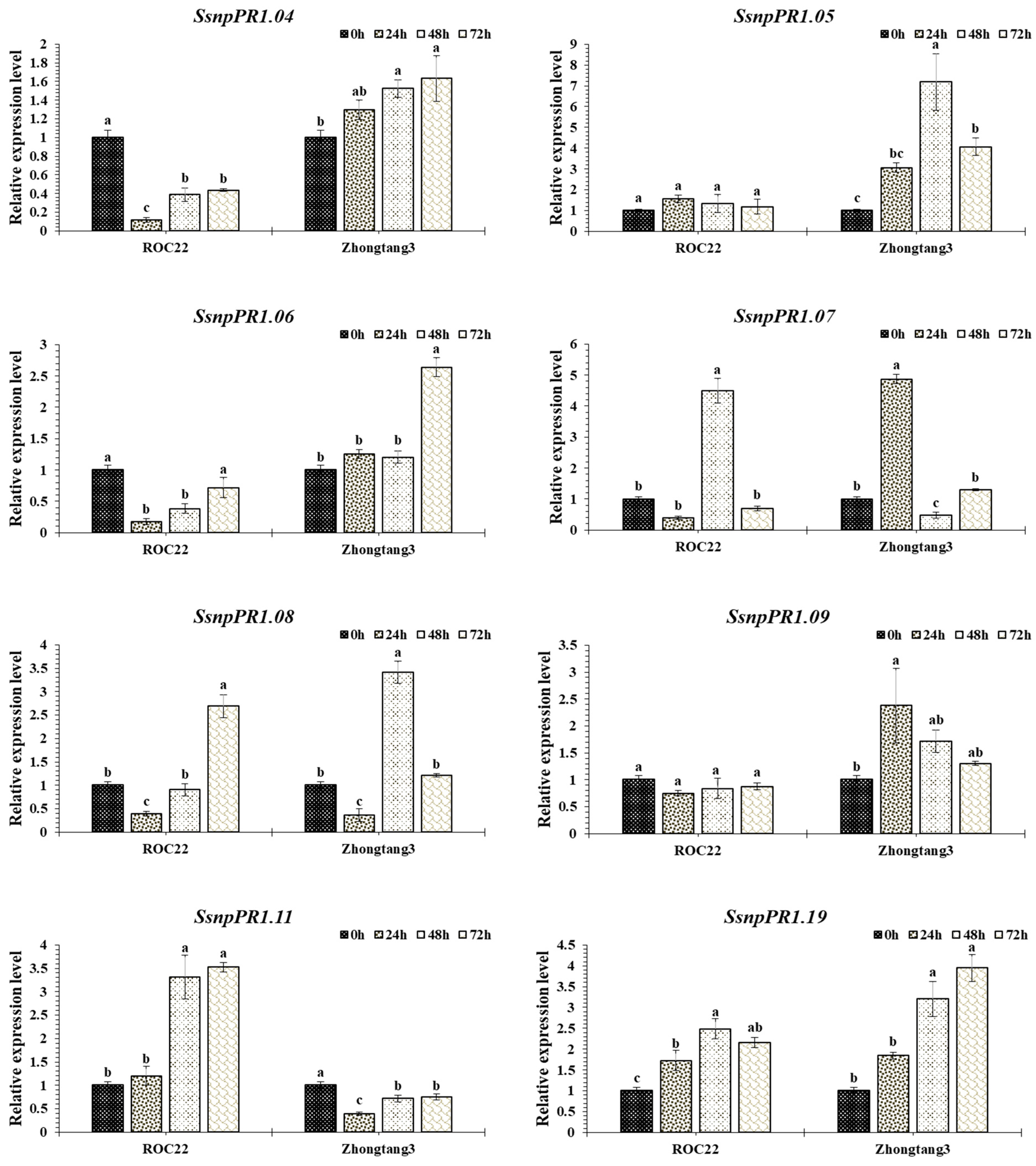
2.6. Transcript Profiling of SsnpPR1 Genes in Response to Pathogen Infection
3. Discussion
3.1. Genetic Diversity of SsnpPR1 Genes
3.2. Functional Divergence of SsnpPR1 Genes
4. Materials and Methods
4.1. Identification and Physio-Chemical Characterization of PR1 Family Members
4.2. Phylogenetic and Gene Structure Analysis
4.3. Chromosomal Localization, Gene Duplication, and Collinearity Analysis
4.4. Protein Interaction Network and cis-Regulatory Elements
4.5. Crop Husbandry, Pathogen Inoculation, and Leaf Sampling
4.6. Determination of Fungal Population Size by qPCR Assay
4.7. Expression Profiling of SsnpPR1s by RT-qPCR Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kummu, M.; Heino, M.; Taka, M.; Varis, O.; Viviroli, D. Climate change risks pushing one-third of global food production outside the safe climatic space. One Earth 2021, 4, 720–729. [Google Scholar] [CrossRef]
- Yuan, M.; Ngou, B.P.M.; Ding, P.; Xin, X.F. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Boil. 2021, 62, 102030. [Google Scholar] [CrossRef]
- Javed, T.; Gao, S.J. WRKY transcription factors in plant defense. Trends Genet. 2023, 39, 787–801. [Google Scholar] [CrossRef]
- Javed, T.; Shabbir, R.; Ali, A.; Afzal, I.; Zaheer, U.; Gao, S.J. Transcription factors in plant stress responses: Challenges and potential for sugarcane improvement. Plants 2020, 9, 491. [Google Scholar] [CrossRef] [PubMed]
- Chu, N.; Zhou, J.R.; Rott, P.C.; Li, J.; Fu, H.Y.; Huang, M.T.; Zhang, H.L.; Gao, S.J. ScPR1 plays a positive role in the regulation of resistance to diverse stresses in sugarcane (Saccharum spp.) and Arabidopsis thaliana. Ind. Crop. Prod. 2022, 180, 114736. [Google Scholar] [CrossRef]
- Islam, M.M.; El-Sappah, A.H.; Ali, H.M.; Zandi, P.; Huang, Q.; Soaud, S.A.; Alazizi, E.M.; Wafa, H.A.; Hossain, M.A.; Liang, Y. Pathogenesis-related proteins (PRs) countering environmental stress in plants: A review. S. Afr. J. Bot. 2023, 160, 414–427. [Google Scholar] [CrossRef]
- Javed, T.; Wang, W.; Yang, B.; Shen, L.; Sun, T.; Gao, S.J.; Zhang, S. Pathogenesis related-1 proteins in plant defense: Regulation and functional diversity. Crit. Rev. Biotechnol. 2024, 1–9. [Google Scholar] [CrossRef]
- Yin, W.; Bai, Y.; Wang, S.; Xu, K.; Liang, J.; Shang, Q.; Sa, W.; Wang, L. Genome-wide analysis of pathogenesis-related protein-1 (PR-1) genes from Qingke (Hordeum vulgare L. var. nudum) reveals their roles in stress responses. Heliyon 2023, 9, e14899. [Google Scholar]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Xiong, D.; Schneiter, R.; Tian, C. The function of plant PR1 and other members of the CAP protein superfamily in plant–pathogen interactions. Mol. Plant Pathol. 2023, 24, 651–668. [Google Scholar] [CrossRef]
- Li, J.J.; Luo, C.; Yang, X.Z.; Peng, L.H.; Lu, T.T.; Yang, J.H.; Zhang, X.J.; Xie, Y.Q.; Yang, Z.Y.; Xu, F.; et al. Genome-wide identification of the mango pathogenesis-related 1 (PR1) gene family and functional analysis of MiPR1A genes in transgenic Arabidopsis. Sci. Hortic. 2023, 321, 112254. [Google Scholar] [CrossRef]
- Zribi, I.; Ghorbel, M.; Haddaji, N.; Besbes, M.; Brini, F. Genome-wide identification and expression profiling of pathogenesis-related protein 1 (PR-1) genes in durum wheat (Triticum durum Desf.). Plants 2023, 12, 1998. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Lu, J.; Xing, J.; Xue, L.; Wu, Y.; Zhang, L. Characterization and functional analyses of wheat TaPR1 genes in response to stripe rust fungal infection. Sci. Rep. 2023, 13, 3362. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Shao, Q.; Xiong, Z. Isolation and characterization of a pathogenesis-related protein 1 (SlPR1) gene with induced expression in tomato (Solanum lycopersicum) during Ralstonia solanacearum infection. Gene 2023, 855, 147105. [Google Scholar] [CrossRef] [PubMed]
- Javed, T.; Zhou, J.R.; Li, J.; Hu, Z.T.; Wang, Q.N.; Gao, S.J. Identification and expression profiling of WRKY family genes in sugarcane in response to bacterial pathogen infection and nitrogen implantation dosage. Front. Plant Sci. 2022, 13, 917953. [Google Scholar] [CrossRef]
- Ali, A.; Khan, M.; Sharif, R.; Mujtaba, M.; Gao, S.J. Sugarcane omics: An update on the current status of research and crop improvement. Plants 2019, 8, 344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Qi, Y.; Pan, H.; Tang, H.; Wang, G.; Hua, X.; Wang, Y.; Lin, L.; Li, Z.; Li, Y.; et al. Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum. Nat. Genet. 2022, 54, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Piperidis, N.; D’Hont, A. Sugarcane genome architecture decrypted with chromosome-specific oligo probes. Plant J. 2020, 103, 2039–2051. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, X.; Tang, H.; Zhang, Q.; Hua, X.; Ma, X.; Zhu, F.; Jones, T.; Zhu, X.; Bowers, J.; et al. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 2018, 50, 1565–1573. [Google Scholar] [CrossRef]
- Almeida-Silva, F.; Venancio, T.M. Pathogenesis-related protein 1 (PR-1) genes in soybean: Genome-wide identification, structural analysis and expression profiling under multiple biotic and abiotic stresses. Gene 2022, 809, 146013. [Google Scholar] [CrossRef]
- Zaynab, M.; Peng, J.; Sharif, Y.; Al-Yahyai, R.; Jamil, A.; Hussain, A.; Khan, K.A.; Alotaibi, S.S.; Li, S. Expression profiling of pathogenesis-related Protein-1 (PR-1) genes from Solanum tuberosum reveals its critical role in Phytophthora infestans infection. Microb. Pathog. 2021, 161, 105290. [Google Scholar] [CrossRef]
- Kattupalli, D.; Srinivasan, A.; Soniya, E.V. A genome-wide analysis of pathogenesis-related protein-1 (PR-1) genes from Piper nigrum reveals its critical role during Phytophthora capsici infection. Genes 2021, 12, 1007. [Google Scholar] [CrossRef] [PubMed]
- Anuradha, C.; Chandrasekar, A.; Backiyarani, S.; Thangavelu, R.; Giribabu, P.; Uma, S. Genome-wide analysis of pathogenesis-related protein 1 (PR-1) gene family from Musa spp. and its role in defense response during stresses. Gene 2022, 821, 146334. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, O.K.; Shchennikova, A.V.; Kochieva, E.Z.; Filyushin, M.A. Pathogenesis-related genes of PR1, PR2, PR4, and PR5 families are involved in the response to Fusarium infection in garlic (Allium sativum L.). Int. J. Mol. Sci. 2021, 22, 6688. [Google Scholar] [CrossRef]
- Zhang, Q.; Guo, N.; Zhang, Y.; Yu, Y.; Liu, S. Genome-Wide characterization and expression analysis of pathogenesis-related 1 (PR-1) gene family in tea plant (Camellia sinensis (L.) O. Kuntze) in response to blister-blight disease stress. Int. J. Mol. Sci. 2022, 23, 1292. [Google Scholar] [CrossRef]
- Ali, A.; Javed, T.; Zaheer, U.; Zhou, J.R.; Huang, M.T.; Fu, H.Y.; Gao, S.J. Genome-wide identification and expression profiling of the bHLH transcription factor gene family in Saccharum spontaneum under bacterial pathogen stimuli. Trop. Plant Biol. 2021, 14, 283–294. [Google Scholar] [CrossRef]
- Ali, A.; Chu, N.; Ma, P.; Javed, T.; Zaheer, U.; Huang, M.T.; Fu, H.Y.; Gao, S.J. Genome-wide analysis of mitogen-activated protein (MAP) kinase gene family expression in response to biotic and abiotic stresses in sugarcane. Physiol. Plant. 2021, 171, 86–107. [Google Scholar] [CrossRef]
- Zhou, J.R.; Li, J.; Lin, J.X.; Xu, H.M.; Chu, N.; Wang, Q.N.; Gao, S.J. Genome-wide characterization of cys-tathionine-β-synthase domain-containing proteins in sugarcane reveals their role in defense responses under multiple stressors. Front. Plant Sci. 2022, 13, 985653. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, J.Y.; Shi, Y.; Fu, H.Y.; Huang, M.T.; Meng, J.Y.; Gao, S.J. Systematic and functional analysis of non-specific lipid transfer protein family genes in sugarcane under Xanthomonas albilineans infection and salicylic acid treatment. Front. Plant Sci. 2022, 13, 1014266. [Google Scholar] [CrossRef]
- Kim, S.-J.; Park, J.-S.; Shin, Y.-H.; Park, Y.-D. Identification and validation of genetic variations in transgenic chinese cabbage plants (Brassica rapa ssp. pekinensis) by next-generation sequencing. Genes 2021, 12, 621. [Google Scholar]
- Wang, P.; Zhou, J.; Sun, W.; Li, H.; Li, D.; Zhuge, Q. Characteristics and function of the pathogenesis-related protein 1 gene family in poplar. Plant Sci. 2023, 336, 111857. [Google Scholar] [CrossRef] [PubMed]
- Aboulila, A.A. Efficiency of plant growth regulators as inducers for improve systemic acquired resistance against stripe rust disease caused by Puccinia striiformis f. sp. tritici in wheat through up-regulation of PR-1 and PR-4 genes expression. Physiol. Mol. Plant Pathol. 2022, 121, 101882. [Google Scholar]
- Zhao, T.; Zhang, Y.; Wang, F.; Zhang, B.; Chen, Q.; Liu, L.; Yan, L.; Yang, Y.; Meng, Q.; Huang, J.; et al. Transcriptome mapping related genes encoding PR1 protein involved in necrotic symptoms to soybean mosaic virus infection. Mol. Breed. 2023, 43, 7. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Tian, T.; Feng, L.; Yang, X.; Li, L.; Tan, X.; Wu, W.; Li, Z.; Treves, H.; Serneels, F.; et al. Pathogenesis-related protein 1 suppresses oomycete pathogen by targeting against AMPK kinase complex. J. Adv. Res. 2023, 43, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Bai, Y.; Wei, Y.; Dong, Y.; Zeng, H.; Reiter, R.J.; Shi, H. Fine-tuning of pathogenesis-related protein 1 (PR1) activity by the melatonin biosynthetic enzyme ASMT2 in defense response to cassava bacterial blight. J. Pineal Res. 2022, 72, e12784. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Shen, S.; Zhao, C.; Cui, Z.; Meng, L.; Wu, W.; Liu, D.; Wang, H. TaPR1 interacts with TaTLP1 via the αIV helix to be involved in wheat defense to Puccinia triticina through the CAPE1 motif. Front. Plant Sci. 2022, 13, 874654. [Google Scholar] [CrossRef]
- Chien, P.S.; Nam, H.G.; Chen, Y.R. A salt-regulated peptide derived from the CAP superfamily protein negatively regulates salt-stress tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 5301–5313. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Su, Y.; Wang, S.; Guo, J.; Xue, B.; Xu, L.; Que, Y. A TaqMan real-time PCR assay for detection and quantification of Sporisorium scitamineum in sugarcane. Sci. World J. 2013, 2023, 942682. [Google Scholar]



| Gene ID | Nomenclature | Length of Amino Acids | Molecular Weight | Isoelectric Point | Instability Index | Aliphatic Index | GRAVY |
|---|---|---|---|---|---|---|---|
| Npp.04C028570.1 | SsnpPR1.01 | 173 | 18,732 | 7.5 | 58.9 | 69.5 | −0.18 |
| Npp.04D024970.1 | SsnpPR1.02 | 173 | 18,732 | 7.5 | 58.9 | 69.5 | −0.18 |
| Npp.02D002590.1 | SsnpPR1.03 | 155 | 16,428 | 6.8 | 40.1 | 59.2 | −0.23 |
| Npp.02C003580.1 | SsnpPR1.04 | 155 | 16,428 | 6.8 | 40.1 | 59.2 | −0.23 |
| Npp.02C003610.1 | SsnpPR1.05 | 167 | 17,449 | 6.2 | 26.7 | 76.5 | −0.05 |
| Npp.02D002640.1 | SsnpPR1.06 | 167 | 17,521 | 6.1 | 26.5 | 77.1 | −0.06 |
| Npp.02C003670.1 | SsnpPR1.07 | 273 | 28,694 | 8.9 | 38.5 | 76.5 | −0.17 |
| Npp.02C003650.1 | SsnpPR1.08 | 271 | 27,796 | 8.5 | 35.9 | 79.5 | −0.02 |
| Npp.02C003630.1 | SsnpPR1.09 | 216 | 23,213 | 8.8 | 56.1 | 82.6 | −0.07 |
| Npp.01C028770.1 | SsnpPR1.11 | 291 | 31,117 | 10.1 | 54.7 | 66.2 | −0.29 |
| Npp.02A009930.1 | SsnpPR1.12 | 171 | 18,146 | 6.3 | 34.7 | 66.8 | −0.11 |
| Npp.02B011150.1 | SsnpPR1.13 | 171 | 18,054 | 7.6 | 30.2 | 71.9 | 0.02 |
| Npp.02C003600.1 | SsnpPR1.14 | 170 | 17,910 | 4.4 | 38.7 | 72.4 | −0.09 |
| Npp.10C002330.1 | SsnpPR1.15 | 167 | 18,189 | 8.6 | 31.9 | 68.4 | −0.27 |
| Npp.10C002360.1 | SsnpPR1.16 | 167 | 18,189 | 8.6 | 31.9 | 68.4 | −0.27 |
| Npp.04C028540.1 | SsnpPR1.17 | 181 | 19,163 | 9.9 | 50.8 | 69.7 | −0.07 |
| Npp.04D024890.1 | SsnpPR1.18 | 181 | 19,163 | 9.9 | 50.8 | 69.7 | −0.07 |
| Npp.04A034860.1 | SsnpPR1.19 | 181 | 19,226 | 9.8 | 55.9 | 66.5 | −0.11 |
| Chr2B | Npp.02B011150.1.t1 | == | SsChr2A | Sspon.02G0020390-1A |
| Chr2B | Npp.02B011150.1.t1 | == | SsChr2B | Sspon.02G0020390-2B |
| Chr2B | Npp.02B011150.1.t1 | == | SsChr2C | Sspon.02G0020390-3C |
| Chr2C | Npp.02C003600.1.t1 | == | SsChr1A | Sspon.01G0029040-1A |
| Chr2C | Npp.02C003600.1.t1 | == | SsChr2A | Sspon.01G0029040-1P |
| Chr2C | Npp.02C003610.1.t1 | == | SsChr2A | Sspon.02G0024520-1A |
| Chr2C | Npp.02C003630.1.t1 | == | SsChr2A | Sspon.02G0024500-1A |
| Chr2C | Npp.02C003610.1.t1 | == | SsChr2B | Sspon.02G0024520-2B |
| Chr2C | Npp.02C003630.1.t1 | == | SsChr2B | Sspon.02G0024500-1P |
| Chr2C | Npp.02C003610.1.t1 | == | SsChr2C | Sspon.02G0024530-3C |
| Chr2C | Npp.02C003600.1.t1 | == | SsChr2C | Sspon.01G0029040-3C |
| Chr4D | Npp.04D024890.1.t1 | == | SsChr4B | Sspon.04G0002650-2B |
| Chr4D | Npp.04D024970.1.t1 | == | SsChr4B | Sspon.04G0002610-2B |
| Chr4D | Npp.04D024890.1.t1 | == | SsChr4C | Sspon.04G0002650-1P |
| Chr4D | Npp.04D024970.1.t1 | == | SsChr8D | Sspon.04G0002610-3D |
| Chr10C | Npp.10C002360.1.t1 | == | SsChr8A | Sspon.08G0016170-1A |
| Chr10C | Npp.10C002330.1.t1 | == | SsChr8A | Sspon.08G0016290-1A |
| Chr10C | Npp.10C002330.1.t1 | == | SsChr8B | Sspon.08G0016290-2B |
| Chr10C | Npp.10C002360.1.t1 | == | SsChr8D | Sspon.08G0016170-3D |
| Chr10C | Npp.10C002330.1.t1 | == | SsChr8D | Sspon.08G0016290-3D |
| Gene Name | Primer/Probe Name | Forward Primer (5′→3′) | Reverse Primer (3′→5′) |
|---|---|---|---|
| PR1.02 | q-PR1.02-F/R | GCTCAAGTACACGGAGGAGA | AGTTGGAGCCGTAGTCGTAG |
| PR1.03 | q-PR1.03-F/R | ACGGTGAGAACCTCTTCTGG | GGTCGTAGTACTGCCTCTCC |
| PR1.04 | q-PR1.04-F/R | CCGAGAAGCAGTTCTACGAC | AGTTGCAGGTGATGAAGACG |
| PR1.05 | q-PR1.05-F/R | CTGTGGCCTTAGCAACCATC | ATACGTGGCTAGGCTCTCG |
| PR1.06 | q-PR1.06-F/R | CTGTGGCCTTAGCAACCATC | AGGCTCTCATTCCACGACAC |
| PR1.07 | q-PR1.07-F/R | TCGAGCTTCACAGACTGGTT | CGCCTTGTTGTGCAAGTCTA |
| PR1.08 | q-PR1.08-F/R | TCGTCAACTTGCACAACGAG | AGAAGAGGTTCTCGCCGTAG |
| PR1.09 | q-PR1.09-F/R | TACGACCACGCCACTAACA | GAGTCTAGTACGGGCTCTGG |
| PR1.11 | q-PR1.11-F/R | CCACTATGCCTCACCCATCA | CTGAGGTGGTGCTTGCTATG |
| PR1.19 | q-PR1.19-F/R | CTACGACAGGTCCACCAACT | TCAGTAGGGCCTCATCCC |
| GAPDH | GAPDH-F/R | CACGGCCACTGGAAGCA | TCCTCAGGGTTCCTGATGCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Javed, T.; Wang, W.; Sun, T.; Shen, L.; Feng, X.; Huang, J.; Zhang, S. Pathogenesis-Related 1 (PR1) Protein Family Genes Involved in Sugarcane Responses to Ustilago scitaminea Stress. Int. J. Mol. Sci. 2024, 25, 6463. https://doi.org/10.3390/ijms25126463
Javed T, Wang W, Sun T, Shen L, Feng X, Huang J, Zhang S. Pathogenesis-Related 1 (PR1) Protein Family Genes Involved in Sugarcane Responses to Ustilago scitaminea Stress. International Journal of Molecular Sciences. 2024; 25(12):6463. https://doi.org/10.3390/ijms25126463
Chicago/Turabian StyleJaved, Talha, Wenzhi Wang, Tingting Sun, Linbo Shen, Xiaoyan Feng, Jiayan Huang, and Shuzhen Zhang. 2024. "Pathogenesis-Related 1 (PR1) Protein Family Genes Involved in Sugarcane Responses to Ustilago scitaminea Stress" International Journal of Molecular Sciences 25, no. 12: 6463. https://doi.org/10.3390/ijms25126463
APA StyleJaved, T., Wang, W., Sun, T., Shen, L., Feng, X., Huang, J., & Zhang, S. (2024). Pathogenesis-Related 1 (PR1) Protein Family Genes Involved in Sugarcane Responses to Ustilago scitaminea Stress. International Journal of Molecular Sciences, 25(12), 6463. https://doi.org/10.3390/ijms25126463






