Myogenic microRNAs as Therapeutic Targets for Skeletal Muscle Mass Wasting in Breast Cancer Models
Abstract
1. Introduction
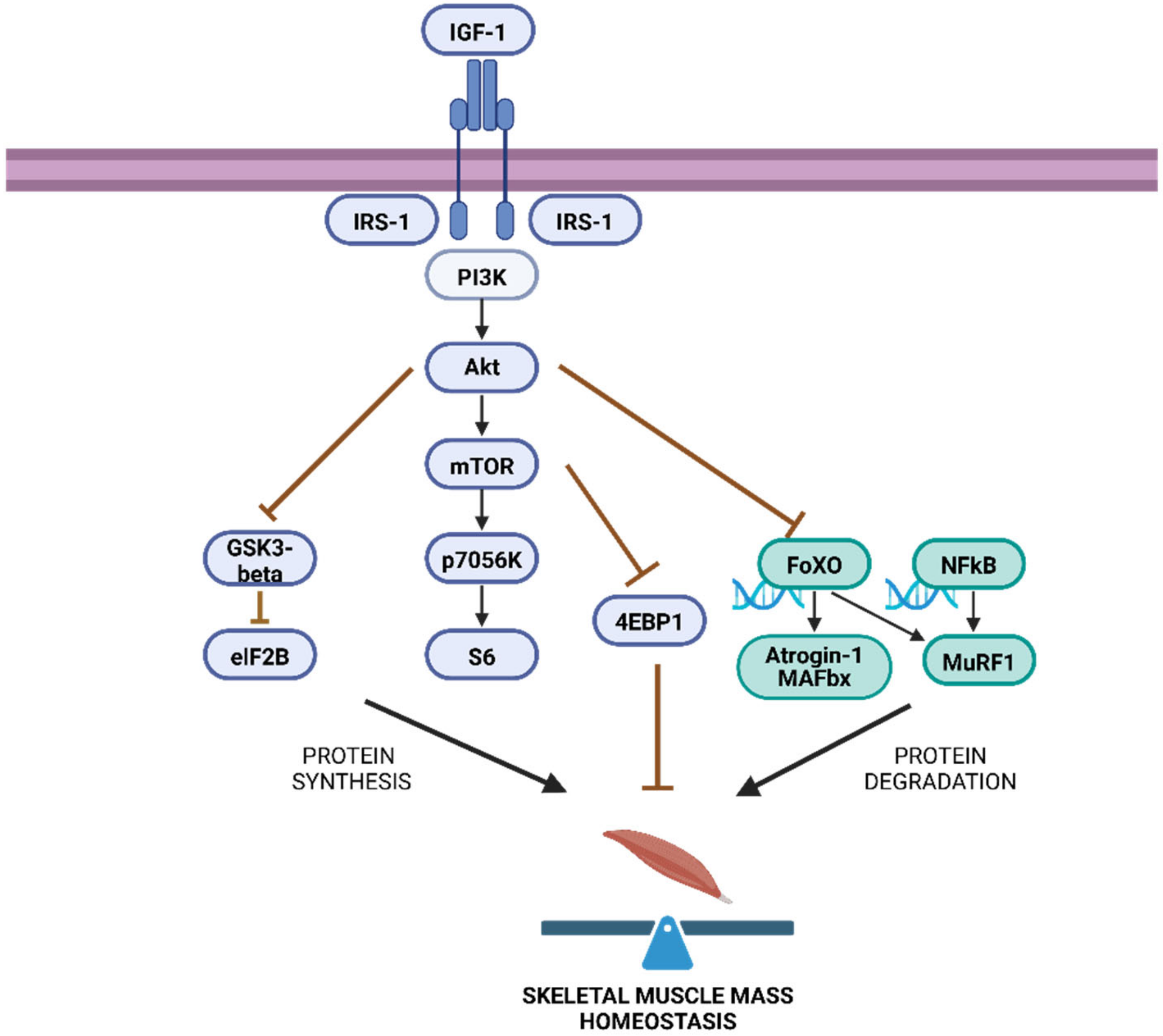
2. Pathways Involved in the Regulation of Skeletal Muscle Mass under Homeostasis
3. Skeletal Muscle Mass Loss in Breast Cancer Patients
4. Role of miRNAs in Maintaining Skeletal Muscle Mass Homeostasis
5. Search on Skeletal Muscle Atrophy miRNAs in Breast Cancer
6. miRNAs as Potential Therapeutic Targets of Resistance Exercise Training in a Breast Cancer
7. Discussion and Future Directions
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Demark-Wahnefried, W.; Peterson, B.L.; Winer, E.P.; Marks, L.; Aziz, N.; Marcom, P.K.; Blackwell, K.; Rimer, B.K. Changes in Weight, Body Composition, and Factors Influencing Energy Balance Among Premenopausal Breast Cancer Patients Receiving Adjuvant Chemotherapy. J. Clin. Oncol. 2001, 19, 2381–2389. [Google Scholar] [CrossRef] [PubMed]
- Freedman, R.J.; Aziz, N.; Albanes, D.; Hartman, T.; Danforth, D.; Hill, S.; Sebring, N.; Reynolds, J.C.; Yanovski, J.A. Weight and Body Composition Changes during and after Adjuvant Chemotherapy in Women with Breast Cancer. J. Clin. Endocrinol. Metab. 2004, 89, 2248–2253. [Google Scholar] [CrossRef] [PubMed]
- Prado, C.M.M.; Baracos, V.E.; McCargar, L.J.; Reiman, T.; Mourtzakis, M.; Tonkin, K.; Mackey, J.R.; Koski, S.; Pituskin, E.; Sawyer, M.B. Sarcopenia as a Determinant of Chemotherapy Toxicity and Time to Tumor Progression in Metastatic Breast Cancer Patients Receiving Capecitabine Treatment. Clin. Cancer Res. 2009, 15, 2920–2926. [Google Scholar] [CrossRef] [PubMed]
- Roberto, M.; Barchiesi, G.; Resuli, B.; Verrico, M.; Speranza, I.; Cristofani, L.; Pediconi, F.; Tomao, F.; Botticelli, A.; Santini, D. Sarcopenia in Breast Cancer Patients: A Systematic Review and Meta-Analysis. Cancers 2024, 16, 596. [Google Scholar] [CrossRef] [PubMed]
- Baracos, V.E.; Martin, L.; Korc, M.; Guttridge, D.C.; Fearon, K.C.H. Cancer-associated cachexia. Nat. Rev. Dis. Primers 2018, 4, 17105. [Google Scholar] [CrossRef] [PubMed]
- Smoot, B.; Wampler, M.; Topp, K.S. Breast Cancer Treatments and Complications: Implications for Rehabilitation. Rehabil. Oncol. 2009, 27, 16–26. [Google Scholar] [CrossRef]
- Zhang, X.M.; Dou, Q.L.; Zeng, Y.; Yang, Y.; Cheng, A.S.K.; Zhang, W.W. Sarcopenia as a predictor of mortality in women with breast cancer: A meta-analysis and systematic review. BMC Cancer 2020, 20, 172. [Google Scholar] [CrossRef] [PubMed]
- Kubo, Y.; Naito, T.; Mori, K.; Osawa, G.; Aruga, E. Skeletal muscle loss and prognosis of breast cancer patients. Support. Care Cancer 2017, 25, 2221–2227. [Google Scholar] [CrossRef]
- Lee, B.M.; Cho, Y.; Kim, J.W.; Ahn, S.G.; Kim, J.H.; Jeung, H.C.; Jeong, J.; Lee, I.J. Association between Skeletal Muscle Loss and the Response to Neoadjuvant Chemotherapy for Breast Cancer. Cancers 2021, 13, 1806. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Nathan, J.A.; Goldberg, A.L. Muscle wasting in disease: Molecular mechanisms and promising therapies. Nat. Rev. Drug Discov. 2015, 14, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Mourtzakis, M.; Bedbrook, M. Muscle atrophy in cancer: A role for nutrition and exercise. Appl. Physiol. Nutr. Metab. 2009, 34, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, S.Y.; Holloway, T.M.; Phillips, S.M. The Impact of Step Reduction on Muscle Health in Aging: Protein and Exercise as Countermeasures. Front. Nutr. 2019, 6, 453801. [Google Scholar] [CrossRef] [PubMed]
- Atherton, P.J.; Greenhaff, P.L.; Phillips, S.M.; Bodine, S.C.; Adams, C.M.; Lang, C.H. Control of skeletal muscle atrophy in response to disuse: Clinical/preclinical contentions and fallacies of evidence. Am. J. Physiol.-Endocrinol. Metab. 2016, 311, E594–E604. [Google Scholar] [CrossRef] [PubMed]
- Kalyani, R.R.; Corriere, M.; Ferrucci, L. Age-related and disease-related muscle loss: The effect of diabetes, obesity, and other diseases. Lancet Diabetes Endocrinol. 2014, 2, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Nunes, E.A.; Stokes, T.; McKendry, J.; Currier, B.S.; Phillips, S.M. Disuse-induced skeletal muscle atrophy in disease and nondisease states in humans: Mechanisms, prevention, and recovery strategies. Am. J. Physiol.-Cell Physiol. 2022, 322, C1068–C1084. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wu, Q.; Li, Z.; Wang, Z.; Tu, Y.; Chen, C.; Sun, S.; Sun, S. Exosomal microRNAs in cancer-related sarcopenia: Tumor-derived exosomal microRNAs in muscle atrophy. Exp. Biol. Med. 2021, 246, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Deng, J.; Qiu, Y.; Gao, J.; Li, J.; Guan, L.; Lee, H.; Zhou, Q.; Xiao, J. Non-coding RNA basis of muscle atrophy. Mol. Ther. Nucleic Acids 2021, 26, 1066–1078. [Google Scholar] [CrossRef]
- Suzuki, T.; Springer, J. MicroRNAs in muscle wasting. J. Cachexia Sarcopenia Muscle 2018, 9, 1209–1212. [Google Scholar] [CrossRef] [PubMed]
- Siracusa, J.; Koulmann, N.; Banzet, S. Circulating myomiRs: A new class of biomarkers to monitor skeletal muscle in physiology and medicine. J. Cachexia Sarcopenia Muscle 2018, 9, 20–27. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Xhuti, D.; Nilsson, M.I.; Manta, K.; Tarnopolsky, M.A.; Nederveen, J.P. Circulating exosome-like vesicle and skeletal muscle microRNAs are altered with age and resistance training. J. Physiol. 2023, 601, 5051–5073. [Google Scholar] [CrossRef]
- Gazova, A.; Samakova, A.; Laczo, E.; Hamar, D.; Polakovicova, M.; Jurikova, M.; Kyselovic, J. Clinical Utility of miRNA-1, miRNA-29g and miRNA-133s Plasma Levels in Prostate Cancer Patients with High-Intensity Training After Androgen-Deprivation Therapy. Physiol. Res. 2019, 68 (Suppl. S2), S139–S147. [Google Scholar] [CrossRef] [PubMed]
- Glass, D.J. PI3 kinase regulation of skeletal muscle hypertrophy and atrophy. Curr. Top. Microbiol. Immunol. 2010, 346, 267–278. [Google Scholar] [PubMed]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt Promotes Cell Survival by Phosphorylating and Inhibiting a Forkhead Transcription Factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Sacheck, J.M.; Ohtsuka, A.; McLary, S.C.; Goldberg, A.L. IGF-I stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and MuRF1. Am. J. Physiol.-Endocrinol. Metab. 2004, 287, E591–E601. [Google Scholar] [CrossRef] [PubMed]
- Bodine, S.C.; Latres, E.; Baumhueter, S.; Lai, V.K.M.; Nunez, L.; Clarke, B.A.; Poueymirou, W.T.; Panaro, F.J.; Na, E.; Dharmarajan, K.; et al. Identification of Ubiquitin Ligases Required for Skeletal Muscle Atrophy. Science 2001, 294, 1704–1708. [Google Scholar] [CrossRef] [PubMed]
- Gomes, M.D.; Lecker, S.H.; Jagoe, R.T.; Navon, A.; Goldberg, A.L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA 2001, 98, 14440–14445. [Google Scholar] [CrossRef] [PubMed]
- Sandri, M. Autophagy in skeletal muscle. FEBS Lett. 2010, 584, 1411–1416. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.S.; Ghosh, S. NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev. 2012, 26, 203–234. [Google Scholar] [CrossRef] [PubMed]
- Bonaldo, P.; Sandri, M. Cellular and molecular mechanisms of muscle atrophy. Dis. Model. Mech. 2013, 6, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Wilson, H.E.; Stanton, D.A.; Rellick, S.; Geldenhuys, W.; Pistilli, E.E. Breast cancer-associated skeletal muscle mitochondrial dysfunction and lipid accumulation is reversed by PPARG. Am. J. Physiol.-Cell Physiol. 2021, 320, C577–C590. [Google Scholar] [CrossRef] [PubMed]
- Bohlen, J.; McLaughlin, S.L.; Hazard-Jenkins, H.; Infante, A.M.; Montgomery, C.; Davis, M.; Pistilli, E.E. Dysregulation of metabolic-associated pathways in muscle of breast cancer patients: Preclinical evaluation of interleukin-15 targeting fatigue. J. Cachexia Sarcopenia Muscle 2018, 9, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Mijwel, S.; Cardinale, D.A.; Norrbom, J.; Chapman, M.; Ivarsson, N.; Wengström, Y.; Sundberg, C.J.; Rundqvist, H. Exercise training during chemotherapy preserves skeletal muscle fiber area, capillarization, and mitochondrial content in patients with breast cancer. FASEB J. 2018, 32, 5495–5505. [Google Scholar] [CrossRef] [PubMed]
- Kawanishi, N.; Funakoshi, T.; Machida, S. Time-course study of macrophage infiltration and inflammation in cast immobilization–induced atrophied muscle of mice. Muscle Nerve 2018, 57, 1006–1013. [Google Scholar] [CrossRef]
- Hanson, A.M.; Harrison, B.C.; Young, M.H.; Stodieck, L.S.; Ferguson, V.L. Longitudinal characterization of functional, morphologic, and biochemical adaptations in mouse skeletal muscle with hindlimb suspension. Muscle Nerve 2013, 48, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.; Neuparth, M.; Vitorino, R.; Appell, H.; Amado, F.; Duarte, J. Evidences of apoptosis during the early phases of soleus muscle atrophy in hindlimb suspended mice. Physiol. Res. 2008, 57, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Møller, A.B.; Lønbro, S.; Farup, J.; Voss, T.S.; Rittig, N.; Wang, J.; Højris, I.; Mikkelsen, U.R.; Jessen, N. Molecular and cellular adaptations to exercise training in skeletal muscle from cancer patients treated with chemotherapy. J. Cancer Res. Clin. Oncol. 2019, 145, 1449–1460. [Google Scholar] [CrossRef] [PubMed]
- Lønbro, S.; Farup, J.; Bentsen, S.; Voss, T.; Rittig, N.; Wang, J.; Ørskov, M.; Højris, I.; Mikkelsen, U.R. Lean body mass, muscle fibre size and muscle function in cancer patients during chemotherapy and 10 weeks exercise. JCSM Clin. Rep. 2017, 2, 1–15. [Google Scholar] [CrossRef][Green Version]
- Hiensch, A.E.; Bolam, K.A.; Mijwel, S.; Jeneson, J.A.L.; Huitema, A.D.R.; Kranenburg, O.; van der Wall, E.; Rundqvist, H.; Wengstrom, Y.; May, A.M. Doxorubicin-induced skeletal muscle atrophy: Elucidating the underlying molecular pathways. Acta Physiol. 2020, 229, e13400. [Google Scholar] [CrossRef] [PubMed]
- Temparis, S.; Asensi, M.; Taillandier, D.; Aurousseau, E.; Larbaud, D.; Obled, A.; Béchet, D.; Ferrara, M.; Estrela, J.M.; Attaix, D. Increased ATP-ubiquitin-dependent proteolysis in skeletal muscles of tumor-bearing rats. Cancer Res. 1994, 54, 5568–5573. [Google Scholar] [PubMed]
- Schmitt, T.L.; Martignoni, M.E.; Bachmann, J.; Fechtner, K.; Friess, H.; Kinscherf, R.; Hildebrandt, W. Activity of the Akt-dependent anabolic and catabolic pathways in muscle and liver samples in cancer-related cachexia. J. Mol. Med. 2007, 85, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Murton, A.J.; Maddocks, M.; Stephens, F.B.; Marimuthu, K.; England, R.; Wilcock, A. Consequences of Late-Stage Non–Small-Cell Lung Cancer Cachexia on Muscle Metabolic Processes. Clin. Lung Cancer 2017, 18, e1–e11. [Google Scholar] [CrossRef]
- Wilson, H.E.; Rhodes, K.K.; Rodriguez, D.; Chahal, I.; Stanton, D.A.; Bohlen, J.; Davis, M.; Infante, A.M.; Hazard-Jenkins, H.; Klinke, D.J.; et al. Human Breast Cancer Xenograft Model Implicates Peroxisome Proliferator–activated Receptor Signaling as Driver of Cancer-induced Muscle Fatigue. Clin. Cancer Res. 2019, 25, 2336–2347. [Google Scholar] [CrossRef] [PubMed]
- Hussain, N.; Said, A.S.A.; Khan, Z. Safety Assessment of Neoadjuvant Pertuzumab Combined with Trastuzumab in Nonmetastatic HER2-Positive Breast Cancer in Postmenopausal Elderly Women of South Asia. Int. J. Breast Cancer 2018, 2018, 6106041. [Google Scholar] [CrossRef] [PubMed]
- Nyrop, K.A.; Deal, A.M.; Shachar, S.S.; Basch, E.; Reeve, B.B.; Choi, S.K.; Lee, J.T.; Wood, W.A.; Anders, C.K.; Carey, L.A.; et al. Patient-Reported Toxicities During Chemotherapy Regimens in Current Clinical Practice for Early Breast Cancer. Oncologist 2019, 24, 762–771. [Google Scholar] [CrossRef] [PubMed]
- De Groef, A.; Geraerts, I.; Demeyer, H.; Van der Gucht, E.; Dams, L.; de Kinkelder, C.; Dukers-van Althuis, S.; Van Kampen, M.; Devoogdt, N. Physical activity levels after treatment for breast cancer: Two-year follow-up. Breast 2018, 40, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Yildiz Kabak, V.; Gursen, C.; Aytar, A.; Akbayrak, T.; Duger, T. Physical activity level, exercise behavior, barriers, and preferences of patients with breast cancer–related lymphedema. Support. Care Cancer 2021, 29, 3593–3602. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, K.M.; Haddad, F.; Pandorf, C.E.; Roy, R.R.; Edgerton, V.R. Alterations in muscle mass and contractile phenotype in response to unloading models: Role of transcriptional/pretranslational mechanisms. Front. Physiol. 2013, 4, 284. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, D.M.d.R.; das Neves, R.X.; de Matos-Neto, E.M.; Camargo, R.G.; Lima, J.D.C.C.; Radloff, K.; Alves, M.J.; Costa, R.G.F.; Tokeshi, F.; Otoch, J.P.; et al. Plasma Lipid Profile and Systemic Inflammation in Patients With Cancer Cachexia. Front. Nutr. 2020, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Scott, H.; McMillan, D.; Crilly, A.; McArdle, C.; Milroy, R. The relationship between weight loss and interleukin 6 in non-small-cell lung cancer. Br. J. Cancer 1996, 73, 1560–1562. [Google Scholar] [CrossRef] [PubMed]
- Moses, A.G.; Maingay, J.; Sangster, K.; Fearon, K.C.; Ross, J.A. Pro-inflammatory cytokine release by peripheral blood mononuclear cells from patients with advanced pancreatic cancer: Relationship to acute phase response and survival. Oncol. Rep. 2009, 21, 1091–1095. [Google Scholar]
- Vainshtein, A.; Sandri, M. Signaling Pathways That Control Muscle Mass. Int. J. Mol. Sci. 2020, 21, 4759. [Google Scholar] [CrossRef]
- Durhan, G.; Erdemir, A.; Konan, A. Evaluation of Latissimus Dorsi Muscle Atrophy via Computed Tomography in Patients with Breast Cancer After Axillary Lymph Node Dissection. Indian J. Surg. 2022, 84, 294–298. [Google Scholar] [CrossRef]
- Jurdana, M. Radiation effects on skeletal muscle. Radiol. Oncol. 2008, 42, 15–22. [Google Scholar] [CrossRef]
- Johns, N.; Stephens, N.A.; Fearon, K.C.H. Muscle wasting in cancer. Int. J. Biochem. Cell Biol. 2013, 45, 2215–2229. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.N.; Gradishar, W.J. Adjuvant Anthracyclines in Breast Cancer: What Is Their Role? Oncologist 2018, 23, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Padilha, C.S.; Marinello, P.C.; Galvão, D.A.; Newton, R.U.; Borges, F.H.; Frajacomo, F.; Deminice, R. Evaluation of resistance training to improve muscular strength and body composition in cancer patients undergoing neoadjuvant and adjuvant therapy: A meta-analysis. J. Cancer Surviv. 2017, 11, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Mok, G.F.; Lozano-Velasco, E.; Münsterberg, A. microRNAs in skeletal muscle development. Semin. Cell Dev. Biol. 2017, 72, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Pasquinelli, A.E. MicroRNAs and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012, 13, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, M.S.; Morris, K.V. Transcriptional gene silencing in humans. Nucleic Acids Res. 2016, 44, 6505–6517. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Shih I hung Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of Mammalian MicroRNA Targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Williams, A.H.; Liu, N.; van Rooij, E.; Olson, E.N. MicroRNA control of muscle development and disease. Curr. Opin. Cell Biol. 2009, 21, 461–469. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, J.J.; Esser, K.A. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J. Appl. Physiol. 2007, 102, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Drummond, M.J.; Dreyer, H.C.; Pennings, B.; Fry, C.S.; Dhanani, S.; Dillon, E.L.; Sheffield-Moore, M.; Volpi, E.; Rasmussen, B.B. Skeletal muscle protein anabolic response to resistance exercise and essential amino acids is delayed with aging. J. Appl. Physiol. 2008, 104, 1452–1461. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Zhao, L.; Guo, J.; Zhao, J. miR-206 regulates the growth of the teleost tilapia (Oreochromis niloticus) through the modulation of IGF-1 gene expression. J. Exp. Biol. 2013, 216, 1265–1269. [Google Scholar] [CrossRef] [PubMed]
- Winbanks, C.E.; Beyer, C.; Hagg, A.; Qian, H.; Sepulveda, P.V.; Gregorevic, P. miR-206 Represses Hypertrophy of Myogenic Cells but Not Muscle Fibers via Inhibition of HDAC4. PLoS ONE 2013, 8, e73589. [Google Scholar] [CrossRef]
- Blanco-Aparicio, C.; Renner, O.; Leal, J.F.M.; Carnero, A. PTEN, more than the AKT pathway. Carcinogenesis 2007, 28, 1379–1386. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, R.; Workeneh, B.; Dong, Y.; Wang, X.; Hu, Z. Transcription factor FoxO1, the dominant mediator of muscle wasting in chronic kidney disease, is inhibited by microRNA-486. Kidney Int. 2012, 82, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Hitachi, K.; Nakatani, M.; Tsuchida, K. Myostatin signaling regulates Akt activity via the regulation of miR-486 expression. Int. J. Biochem. Cell Biol. 2014, 47, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, I.; Eran, A.; Nishino, I.; Moggio, M.; Lamperti, C.; Amato, A.A.; Lidov, H.G.; Kang, P.B.; North, K.N.; Mitrani-Rosenbaum, S.; et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc. Natl. Acad. Sci. USA 2007, 104, 17016–17021. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Kumar, B.; Doud, E.H.; Mosley, A.L.; Alexander, M.S.; Kunkel, L.M.; Nakshatri, H. Skeletal muscle-specific overexpression of miR-486 limits mammary tumor-induced skeletal muscle functional limitations. Mol. Ther. Nucleic Acids 2022, 28, 231–248. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Goswami, C.P.; Burnett, R.M.; Anjanappa, M.; Bhat-Nakshatri, P.; Muller, W.; Nakshatri, H. Cancer Affects microRNA Expression, Release, and Function in Cardiac and Skeletal Muscle. Cancer Res. 2014, 74, 4270–4281. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Cao, M.; Ruan, X.; Jiang, L.; Lee, S.; Lemanek, A.; Ghassemian, M.; Pizzo, D.P.; Wan, Y.; Qiao, Y.; et al. Cancer-cell-secreted miR-122 suppresses O-GlcNAcylation to promote skeletal muscle proteolysis. Nat. Cell Biol. 2022, 24, 793–804. [Google Scholar] [CrossRef]
- Gomes, J.L.P.; Tobias, G.C.; Fernandes, T.; Silveira, A.C.; Negrão, C.E.; Chammas, R.; Brum, P.C.; Oliveira, E.M. Effects of Aerobic Exercise Training on MyomiRs Expression in Cachectic and Non-Cachectic Cancer Mice. Cancers 2021, 13, 5728. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Sun, S.; Li, Z.; Yang, Q.; Li, B.; Zhu, S.; Wang, L.; Wu, J.; Yuan, J.; Wang, C.; et al. Breast cancer-released exosomes trigger cancer-associated cachexia to promote tumor progression. Adipocyte 2019, 8, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Small, E.M.; O’Rourke, J.R.; Moresi, V.; Sutherland, L.B.; McAnally, J.; Gerard, R.D.; Richardson, J.A.; Olson, E.N. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci. USA 2010, 107, 4218–4223. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.S.; Casar, J.C.; Motohashi, N.; Vieira, N.M.; Eisenberg, I.; Marshall, J.L.; Gasperini, M.J.; Leque, A.; Myers, J.A.; Estrella, E.A.; et al. MicroRNA-486–dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy–associated symptoms. J. Clin. Investig. 2014, 124, 2651–2667. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Li, X.; Quan, X.; Yang, X.; Zheng, C.; Hao, X.; Qu, R.; Zhou, B. MiR-486 as an effective biomarker in cancer diagnosis and prognosis: A systematic review and meta-analysis. Oncotarget 2018, 9, 13948–13958. [Google Scholar] [CrossRef] [PubMed]
- Rask, L.; Balslev, E.; Søkilde, R.; Høgdall, E.; Flyger, H.; Eriksen, J.; Litman, T. Differential expression of miR-139, miR-486 and miR-21 in breast cancer patients sub-classified according to lymph node status. Cell. Oncol. 2014, 37, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Guescini, M.; Canonico, B.; Lucertini, F.; Maggio, S.; Annibalini, G.; Barbieri, E.; Luchetti, F.; Papa, S.; Stocchi, V. Muscle Releases Alpha-Sarcoglycan Positive Extracellular Vesicles Carrying miRNAs in the Bloodstream. PLoS ONE 2015, 10, e0125094. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Lin, J.; Sun, Y.; Cong, S.; Liu, S.; Zhang, Y.; Chen, Q.; Chen, J. miR-122-5p negatively regulates the transforming growth factor-β/Smad signaling pathway in skeletal muscle myogenesis. Cell Biochem. Funct. 2020, 38, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Santagnello, S.B.; Martins, F.M.; de Oliveira Junior, G.N.; de Freitas Rodrigues de Sousa, J.; Nomelini, R.S.; Murta, E.F.C.; Orsatti, F.L. Improvements in muscle strength, power, and size and self-reported fatigue as mediators of the effect of resistance exercise on physical performance breast cancer survivor women: A randomized controlled trial. Support. Care Cancer 2020, 28, 6075–6084. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, M.G. Effects of Exercise Interventions on Breast Cancer Patients During Adjuvant Therapy. Cancer Nurs. 2020, 43, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Soares Falcetta, F.; de Araújo Vianna Träsel, H.; de Almeida, F.K.; Rangel Ribeiro Falcetta, M.; Falavigna, M.; Dornelles Rosa, D. Effects of physical exercise after treatment of early breast cancer: Systematic review and meta-analysis. Breast Cancer Res. Treat. 2018, 170, 455–476. [Google Scholar] [CrossRef] [PubMed]
- Asselain, B.; Barlow, W.; Bartlett, J.; Bergh, J.; Bergsten-Nordström, E.; Bliss, J.; Boccardo, F.; Boddington, C.; Bogaerts, J.; Bonadonna, G.; et al. Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: Meta-analysis of individual patient data from ten randomised trials. Lancet Oncol. 2018, 19, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.J.; Chen, S.L. Prevention and Management of Lymphedema after Breast Cancer Treatment. Breast J. 2015, 21, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Hagstrom, A.; Denham, J. microRNAs in High and Low Responders to Resistance Training in Breast Cancer Survivors. Int. J. Sports Med. 2018, 39, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.C.; Tavares, P.; Neves, B.; Oliveira, P.F.; Vitorino, R.; Moreira-Gonçalves, D.; Ferreira, R. Exploiting the therapeutic potential of contracting skeletal muscle-released extracellular vesicles in cancer: Current insights and future directions. J. Mol. Med. 2024, 102, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Schild, M.; Eichner, G.; Beiter, T.; Zügel, M.; Krumholz-Wagner, I.; Hudemann, J.; Pilat, C.; Krüger, K.; Niess, A.M.; Steinacker, J.M.; et al. Effects of Acute Endurance Exercise on Plasma Protein Profiles of Endurance-Trained and Untrained Individuals over Time. Mediat. Inflamm. 2016, 2016, 4851935. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.; McClean, C.M.; Davison, G.W.; Brown, J.C.W.; Murphy, M.H. The acute effects of walking exercise intensity on systemic cytokines and oxidative stress. Eur. J. Appl. Physiol. 2018, 118, 2111–2120. [Google Scholar] [CrossRef] [PubMed]
- Nair, V.D.; Ge, Y.; Li, S.; Pincas, H.; Jain, N.; Seenarine, N.; Amper, M.A.S.; Goodpaster, B.H.; Walsh, M.J.; Coen, P.M.; et al. Sedentary and Trained Older Men Have Distinct Circulating Exosomal microRNA Profiles at Baseline and in Response to Acute Exercise. Front. Physiol. 2020, 11, 605. [Google Scholar] [CrossRef] [PubMed]
- Jacques, M.; Hiam, D.; Craig, J.; Barrès, R.; Eynon, N.; Voisin, S. Epigenetic changes in healthy human skeletal muscle following exercise—A systematic review. Epigenetics 2019, 14, 633–648. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Ridinger, J.; Rupp, A.K.; Janssen, J.W.; Altevogt, P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011, 9, 86. [Google Scholar] [CrossRef]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef] [PubMed]
- Tarnopolsky, M.A.; Pearce, E.; Smith, K.; Lach, B. Suction-modified Bergström muscle biopsy technique: Experience with 13,500 procedures. Muscle Nerve 2011, 43, 716–725. [Google Scholar] [CrossRef]
- Walters, J.; Baborie, A. Muscle biopsy: What and why and when? Pract. Neurol. 2020, 20, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Dengler, J.; Linke, P.; Gdynia, H.J.; Wolf, S.; Ludolph, A.C.; Vajkoczy, P.; Meyer, T. Differences in pain perception during open muscle biopsy and Bergstroem needle muscle biopsy. J. Pain Res. 2014, 7, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S.S. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2021, 28, 127–138. [Google Scholar] [CrossRef] [PubMed]


| Study | Country | Aim | Population | miRNA | Function |
|---|---|---|---|---|---|
| Wang R (2022) [76]. | United States | To investigate whether the overexpression of miR-486 can overcome skeletal muscle defects in a breast cancer model. | Neu/miR-186+ female transgenic mice representing HER2 breast cancer subtype and miR-186 overexpression. | miRNA-486 | It plays an integral role in the myogenesis signalling network involving Pax7, MyoD, myostatin, and NF-κB. It blocks PTEN which upregulates PI3K/Akt. |
| Chen D (2014) [77]. | United States | To identify miRNAs that are present at a lower level in circulation in breast cancer models and their role in skeletal muscle. | Female MMTV-PyMT and MMTV-HER2 transgenic mice representing two subtypes of breast cancer. | miRNA-486 | It downregulates PTEN and FOXO1, which consequently activates the PI3K/Akt pathway in cardiac and skeletal muscle. |
| Yan W (2022) [78]. | United States | To determine whether miR-122 secreted by cancer cells suppresses O-GlcNAcylation (OGT) to promote skeletal muscle proteolysis. | Female mice (different types of breast cancer models MDA-MB-231). | miRNA-122 | It regulates OGT, which controls OGT of the RYR1 receptor, releasing cytosolic Ca+2 in skeletal muscle, which is why it is involved in muscle homeostasis. |
| Gomes JLP (2021) [79]. | Brazil | To investigate the profile of miRNAs that regulate muscle mass in cachectic CT26 mice and non-cachectic MMTV-PyMT mice. | Cachectic female mice with colon cancer (CT26) and non-cachectic female mice with breast cancer (MMTV-PyMT). | miRNA-486 | It regulates PTEN mRNA, a key protein that controls the PI3K/Akt/mTOR pathway. |
| miRNA-206 | Skeletal muscle-specific miRNA directly and inversely regulates the expression of the IGF1 gene. | ||||
| Wu Q (2019) [80]. | China | To identify breast cell-specific miRNAs involved in cancer cachexia. | Human breast cancer cell lines MCF-7 and MDA-MB-231, C2C12, and HEK 293T. | miRNA-155 | It modulates PPARγ, whose main function is to regulate the transcription of genes related to lipid and carbohydrate metabolism and inflammation. |
| Author | Sample | Method Used to Identify miRNAs | miRNAs Differentially Expressed in Muscle Atrophy | The Role of Dysregulated miRNAs in Muscle Atrophy |
|---|---|---|---|---|
| Wang R (2022) [76]. | Plasma and tibialis anterior muscle tissue—gastrocnemius of the hind legs of mouse model. | RT-qPCR | ↓ miR-486 | ↓ Levels of p-AKT, p53, and p-LAMA2. ↓ Muscle strength and performance. ↑ Inflammatory cytokines (IL-6, IL-4, TNF-a, and CCL4). |
| Chen D (2014) [77]. | Serum and C2C12 cells from mouse myoblasts. | RT-qPCR Microarray | ↓ miR-486 | ↓ PI3K/AKT pathway signal. ↑ Expression of PTEN and FOXO1 genes. |
| Yan W (2022) [78]. | Breast cancer cells and mouse EDL muscle tissue. | RT-qPCR | ↑ miR-122 | ↑ Muscle fibre degradation. It suppresses the OGT protein and increases RYR1, which favours the cytosolic transport of Ca+2 and induces myofibrillar destruction mediated by the UPS system in skeletal muscle. |
| Gomes JLP (2021) [79]. | Mouse tibialis anterior muscle tissue and serum. | RT-qPCR | Cachectic model ↓ miR-486 ↓ miR-206 | ↓ PI3K and mTOR protein levels. |
| Non-cachectic model ↓ miR-486 ↑ miR-206 | ↓ PI3K, p-AKT, and mTOR protein levels. ↑ miR-206 is associated with ↓ skeletal muscle mass (IGF1 reverse regulation). | |||
| Wu Q (2019) [80]. | Human breast cancer cells. | RT-qPCR Microarray | ↑ miR-155 | ↓ PPARγ expression alters the energy metabolism of adipocytes and muscle cells. ↑ Adipocyte lipolysis and skeletal muscle cell apoptosis. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Artigas-Arias, M.; Curi, R.; Marzuca-Nassr, G.N. Myogenic microRNAs as Therapeutic Targets for Skeletal Muscle Mass Wasting in Breast Cancer Models. Int. J. Mol. Sci. 2024, 25, 6714. https://doi.org/10.3390/ijms25126714
Artigas-Arias M, Curi R, Marzuca-Nassr GN. Myogenic microRNAs as Therapeutic Targets for Skeletal Muscle Mass Wasting in Breast Cancer Models. International Journal of Molecular Sciences. 2024; 25(12):6714. https://doi.org/10.3390/ijms25126714
Chicago/Turabian StyleArtigas-Arias, Macarena, Rui Curi, and Gabriel Nasri Marzuca-Nassr. 2024. "Myogenic microRNAs as Therapeutic Targets for Skeletal Muscle Mass Wasting in Breast Cancer Models" International Journal of Molecular Sciences 25, no. 12: 6714. https://doi.org/10.3390/ijms25126714
APA StyleArtigas-Arias, M., Curi, R., & Marzuca-Nassr, G. N. (2024). Myogenic microRNAs as Therapeutic Targets for Skeletal Muscle Mass Wasting in Breast Cancer Models. International Journal of Molecular Sciences, 25(12), 6714. https://doi.org/10.3390/ijms25126714





