Abstract
Plants are subjected to abiotic stresses throughout their developmental period. Abiotic stresses include drought, salt, heat, cold, heavy metals, nutritional elements, and oxidative stresses. Improving plant responses to various environmental stresses is critical for plant survival and perpetuation. WRKY transcription factors have special structures (WRKY structural domains), which enable the WRKY transcription factors to have different transcriptional regulatory functions. WRKY transcription factors can not only regulate abiotic stress responses and plant growth and development by regulating phytohormone signalling pathways but also promote or suppress the expression of downstream genes by binding to the W-box [TGACCA/TGACCT] in the promoters of their target genes. In addition, WRKY transcription factors not only interact with other families of transcription factors to regulate plant defence responses to abiotic stresses but also self-regulate by recognising and binding to W-boxes in their own target genes to regulate their defence responses to abiotic stresses. However, in recent years, research reviews on the regulatory roles of WRKY transcription factors in higher plants have been scarce and shallow. In this review, we focus on the structure and classification of WRKY transcription factors, as well as the identification of their downstream target genes and molecular mechanisms involved in the response to abiotic stresses, which can improve the tolerance ability of plants under abiotic stress, and we also look forward to their future research directions, with a view of providing theoretical support for the genetic improvement of crop abiotic stress tolerance.
1. Introduction
Crops are an indispensable and important resource in human life, carrying people’s survival and development. From planting to harvesting, from processing to consumption, food has accompanied the historical process of humankind and profoundly influenced the way of life and social structure of humankind [1]. Crops are able to provide humans with an abundance of carbohydrates, proteins, and multivitamins to maintain the normal physiological functions of the human body [2]. In conclusion, ensuring the normal development of crops is an important guarantee for the sustainable and stable development of human society [3].
Plants are subjected to a variety of stresses during growth and development, such as drought, temperature extremes, pests, diseases, etc., of which abiotic stresses are the major threats affecting the growth of food crops [4,5,6,7]. Currently, numerous studies have been conducted on the response of plants to abiotic stresses, for example, the response to cold temperature stress, drought stress, salt stress, heavy metal stress, etc. In order to survive, plants need to make a series of morphological and physiological-biochemical metabolic adjustments to adapt to adversity when subjected to abiotic stress [8,9,10]. First, when the outer cell membrane is stimulated by abiotic stress, second signalling molecules such as reactive oxygen species (ROS) are produced. The second signalling molecules then stimulate the intracellular membrane, which, by regulating intracellular Ca2+ levels, initiates a protein phosphorylation cascade reaction and produces phosphorylated protein molecules that are involved in the cytoprotection of proteins or transcription factors that regulate specific stress-regulated genes. The products of some of these genes can participate in the production of abscisic acid (ABA) and ethylene (ETH), which activates the expression of transcription factors. The transcription factors are able to bind specifically to their downstream target gene promoter sequence portions, thereby regulating the expression of downstream functional genes and ultimately improving the plant’s ability to cope with abiotic stresses [11,12]. Studies have been deepened to the level of molecular mechanisms, and WRKY transcription factors, which are one of the largest families of transcription factors in plants, have gained great attention. Nowadays, a large number of WRKY transcription factor genes have been identified, and they are widely involved in the regulation of plant secondary metabolism, abiotic stress, growth, and development [13,14,15]. Wang et al. found a WRKY transcription factor, ZmWRKY40, was induced by drought, high salt, high temperature, and ABA. ZmWRKY40 overexpression increased drought tolerance in transgenic Arabidopsis thaliana due to the modulation of the expression of downstream abiotic stress genes and reduced the ROS content of the transgenic lines through the enhancement of peroxide dismutase (POD) and catalase (CAT) activities under drought stress [16]. Yin et al. found a WRKY transcription factor, PcWRKY33, specifically binds to the W-box in the promoters of downstream target genes to regulate their expression. The expression of PcWRKY33 can be induced by various abiotic stresses. Overexpression of PcWRKY33 reduced tolerance to salt stress in Arabidopsis. In transgenic plants, the expression of stress-related genes was decreased, the ability to maintain Na+/K+ homeostasis was weakened, the activity of ROS scavenging enzymes was decreased, and the accumulation of ROS was increased after salt stress [17]. Zhang et al. identified a WRKY transcription factor, OsWRKY63, negatively regulates cold tolerance in Oryza sativa. Overexpression of OsWRKY63 lines was more sensitive to cold stress, and the knockout mutant line showed higher cold tolerance. OsWRKY63 was able to repress the expression of OsWRKY76, and the OsWRKY76-knockout mutant lines showed significantly lower cold tolerance and suppressed cold-induced expression of the five OsDREB1 genes [18]. Ma et al. found that the tomato WRKY transcription factor SlWRKY57 acts as a negative regulator in the salt stress response by directly attenuating the transcription of salt-responsive genes (SlRD29B and SlDREB2) and an ion homeostasis gene (SlSOS1) [19]. Devaiah et al. found WRKY75 was located in the nucleus and was differentially induced in plants with phosphate (Pi) deficiency. When WRKY75 expression was inhibited, the expression of several genes involved in the Pi starvation response decreased, and the lateral root length and number as well as the number of root hairs significantly increased [20]. Therefore, the important role of WRKY transcription factors in the regulation of abiotic stress and plant growth and development has made it become a popular gene family for plant stress breeding research.
This paper reviews the structural domains and classification of WRKY transcription factors and which transcriptional regulation downstream genes are involved in the response to abiotic stress. It also discusses the research progress on the role of WRKY transcription factors in regulating the response of plants to abiotic stress, which will provide a theoretical reference for the future study of the ability of WRKY transcription factors to improve the ability of plants to cope with abiotic stresses.
1.1. Structural Domains and Classification of WRKY Transcription Factors
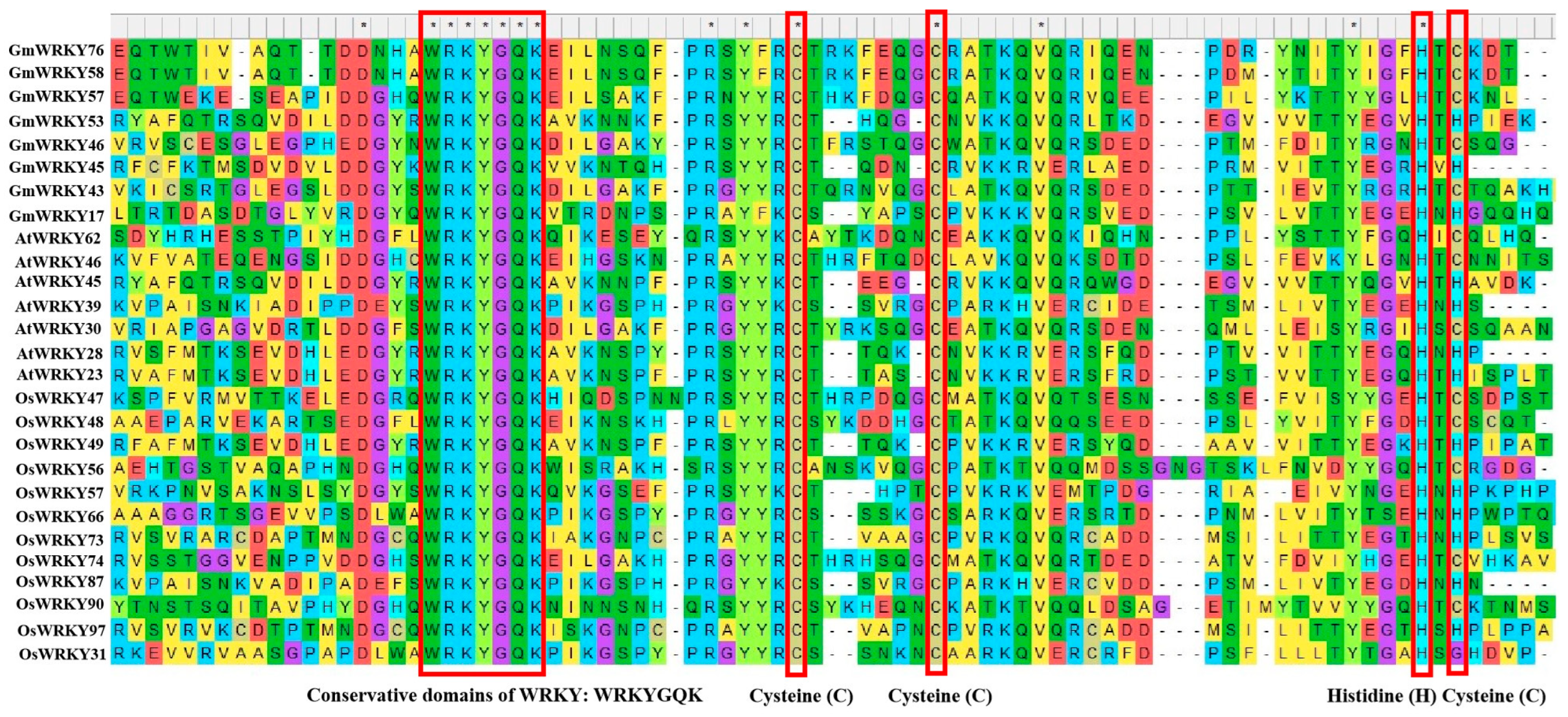
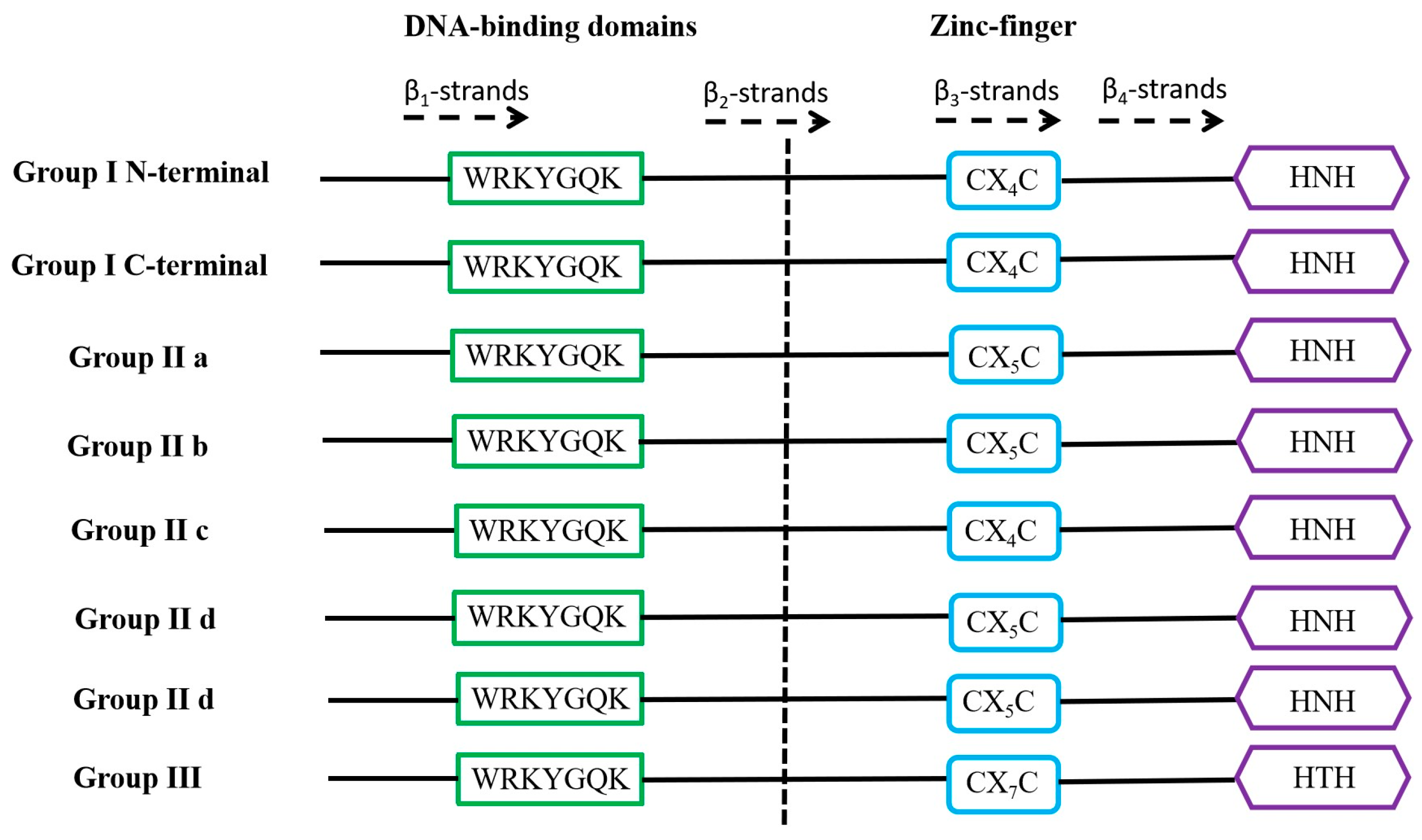
WRKY is a family of transcription factors unique to plants and defined as WRKY transcription factors because their protein sequences contain a DNA-binding domain with the highly conserved amino acid sequence WRKYGQK at the N-terminus of the domain [21]. However, in a few WRKY proteins, there are different types of mutations in the WRKYGQK amino acid sequence, mainly WRKYGEK, WRKYGMK, WRKYGKK, WSKYEQK, or WIKYGEN (Figure 1). The WRKY transcription factor proteins are usually composed of 60 amino acids, and this sequence recognises the W-box [TGACCA/TGACCT] and some similar W-boxes containing the TGAC core structure in the homoeopathic element and specifically binds to them to regulate the downstream target genes [22,23]. The neighbouring sequences of the TGAC core structure determine the priority of the WRKY transcription factor binding sites [24]. A large number of previous studies have shown that most of the promoters of genes related to abiotic stresses contain more than one W-box sequence, which explains the ability of the WRKY family of transcription factors to be widely involved in the regulation of the expression of many plants’ abiotic stress-related genes [25] (Table 1). Typically, WRKY transcription factors contain at least one WRKY structural domain at the N-terminal end and an atypical zinc-finger structure at the C-terminal end [26]. Based on the number of WRKY-binding domains and the characteristics of the zinc finger-like motifs, they can be classified into three groups: Group I contains two WRKY-binding domains with C2-H2 motifs (C-X4-5-CX22-23-H-X1-H); Group II contains one WRKY-binding domain with C2-H2 motifs. Group II can be generally divided into five subgroups (II a, II b, II c, II d, and II e). Group III contains a WRKY-binding domain and a different zinc finger-like motif, C2-H-C (C-X7-C-X23-H-X1-C). Analyses based on phylogenetic data indicate that WRKY families in higher plants are more accurately classified into groups I, II a + II b, II c, II d + II e, and III [27,28] (Figure 2).
Figure 1.
Conserved domains of WRKY family transcription factors in Glycine max, Arabidopsis thaliana, and Oryza sativa. These asterisks “*” and the red frames above the maps indicate the domains of conservative cysteine (C), cysteine (C), histidine (H), and cysteine (C), respectively.

Table 1.
The WRKY genes’ total numbers in different plants.
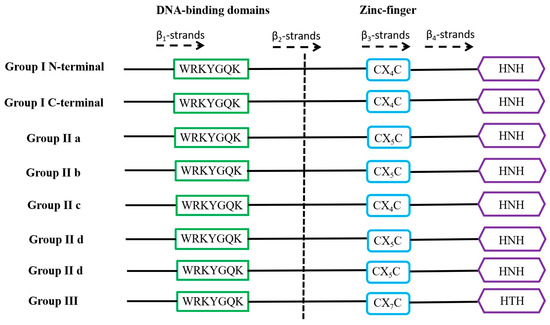
Figure 2.
Domain structures of different WRKY subfamilies in higher plants. The WRKY motif, the cysteines, and the histidines that form the zinc finger are shown in boxes. The 4 β-strands are shown with dashed arrows.
By summarising the localisation, self-activating activity, and binding elements of published WRKY family transcription factors, we have found that most of the identified WRKY family transcription factors are localised in the nucleus, have self-activating activity, and are able to bind to W-box elements. (Refs. [46,47,48,49,50,51,52] are cited in Supplemental Table S1).
1.2. W-box cis-Element in the Promoter Region of WRKY Downstream Genes
It has been shown that WRKY transcription factors can specifically bind to DNA through the cis-acting element W-box (5′-[TGACCA/TGACCT]-3′) in the promoters of target genes to regulate the expression of related genes and affect plants under abiotic stresses, plant growth, and development. The core sequence of the W-box is “TGAC”, so the W-box can be used for the prediction of the target genes of WRKY transcription factors [21]. Different WRKY transcription factors bind to different sequences near the W-box, which affects the selectivity and strength of WRKY transcription factor binding [53]. Mutations in the conserved sequence WRKYGQK in the WRKY structural domain or changes in any nucleotide in the W-box reduce the binding activity of WRKY transcription factors to DNA, whereas the substitution of conserved cysteine (Cys) and histidine (His) residues in the C-terminal zinc-finger structure removes their DNA-binding activity [54].
2. Response and Tolerance of WRKY Transcription Factor Family to Abiotic Stresses in Plants
Plants may encounter the effects of a wide range of abiotic stresses during growth and development, including drought, temperature extremes (high temperature and low temperature), salinity, heavy metal stresses, and nutrient deficiencies. With climate change and increasing weather extremes, the impact of abiotic stresses on crop production is increasing, leading to growth retardation, quality deterioration, and yield reduction [55,56]. Under abiotic stress conditions, WRKY transcription factors activate or suppress the transcription of downstream genes to regulate the expression of abiotic stress-responsive genes or the direct regulation of abiotic stress-responsive gene expression, activate the defence mechanism against abiotic stresses in crops, and improve crops’ resilience to ensure grain yield under abiotic stresses [57,58].
2.1. Molecular Mechanisms of WRKY Transcription Factors Associated with Drought Stress
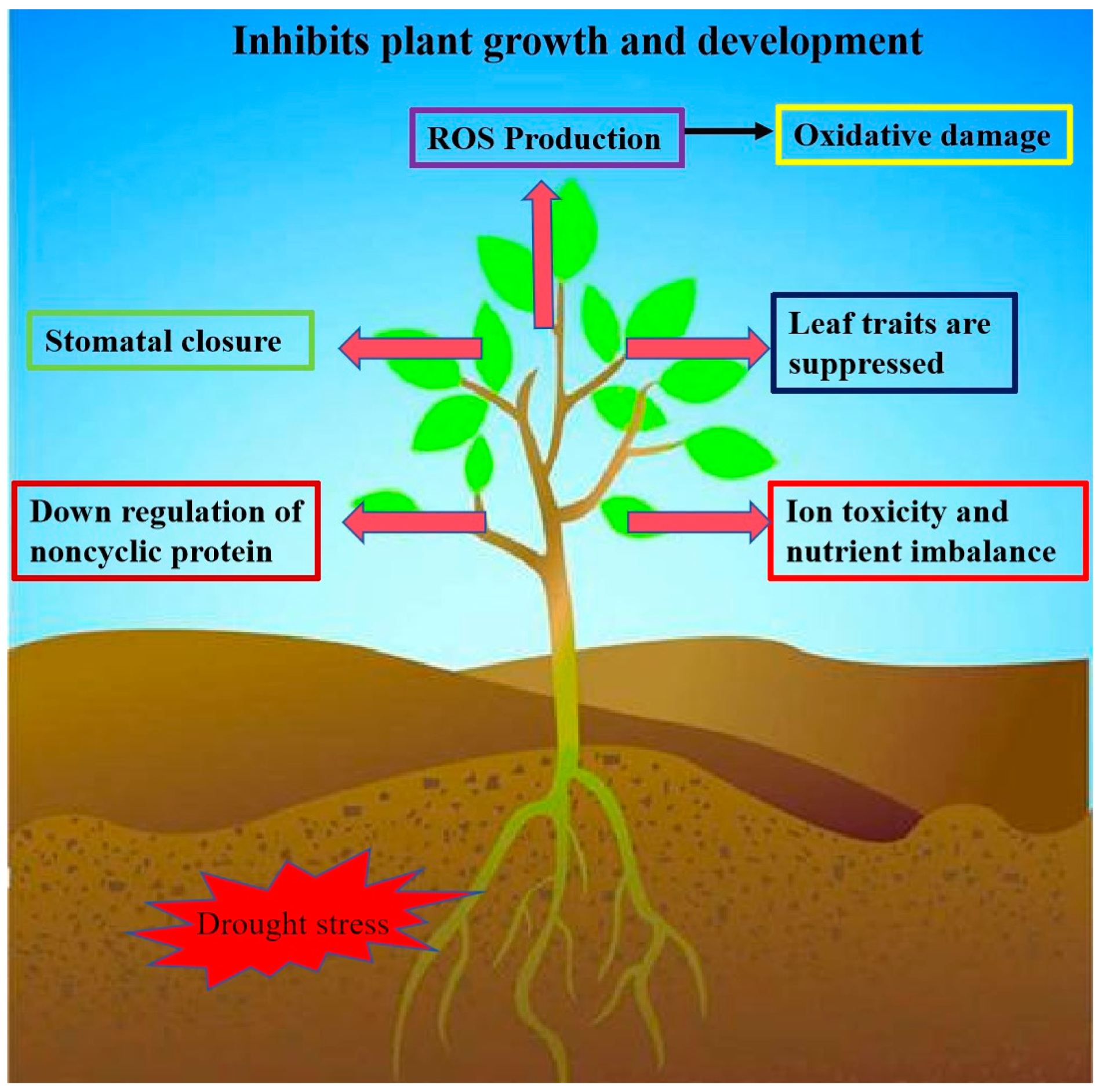
Drought is one of the major environmental factors affecting plant growth and development and crop yield (Figure 3). Many WRKY transcription factor genes have been identified to regulate drought tolerance in plants, and overexpression lines or knockdown lines of these WRKY genes have improved drought tolerance and even seed yield in Arabidopsis thaliana, Nicotiana tabacum, Glycine max, Oryza sativa, Triticum, and cotton [55,56,57,58]. It has been shown that WRKYs can improve plants’ tolerance to drought stress by reducing their H2O2 content through the ROS-scavenging system. For example, overexpression of MdWRKY70L in Nicotiana tabacum reduced H2O2 and O2− accumulation and enhanced drought tolerance in transgenic plants [59]. Overexpression of GmWRKY17 enhances drought tolerance in soybean, and it positively regulates drought tolerance in soybean by activating the expression of the drought-inducible gene GmDREB1D and the ABA-associated gene GmABA2 by combining their promoters [49]. The expression of ZmWRKY106 was significantly induced by drought, high temperature, and exogenous ABA. ZmWRKY106 overexpression lines in Arabidopsis showed increased tolerance to drought and high temperature. Under drought stress, ZmWRKY106 reduced the ROS content in the transgenic lines by enhancing superoxide dismutase (SOD), POD, and CAT activities [60]. Duan et al. found that the transcript abundance of MdWRKY56 was upregulated under drought stress. A MdWRKY56 overexpression line exhibited lower electrolyte leakage, malondialdehyde (MDA) content, ROS accumulation, proline content, and antioxidant enzyme activities [61]. Zhang et al. found that ChaWRKY40 may enhance hazelnuts’ drought tolerance by positively regulating the ChaP5CS gene’s expression to increase the proline content. In the wild type, the expression of ChaWRKY40 and ChaP5CS increased with the increase in the PEG-6000 concentration in the leaves and the gradual decrease in the relative water content in the leaves [62]. Wang et al. found that the WRKY transcription factor EjWRKY17 was identified in Eriobotrya japonicaloqua, which was significantly upregulated in leaves by melatonin treatment during drought stress. The EjWRKY17 overexpression line was able to increase drought tolerance in plants, which had low water loss, limited electrolyte leakage, and lower levels of ROS and MDA compared with the wild type [63]. Huang et al. found that MfWRKY40 promoted primordial root length elongation, increased water uptake, and reduced water loss under stress, and the antioxidant capacity of the overexpression lines was also significantly enhanced, as evidenced by the higher chlorophyll content and antioxidant enzyme activities and less malondialdehyde and ROS accumulation [64].
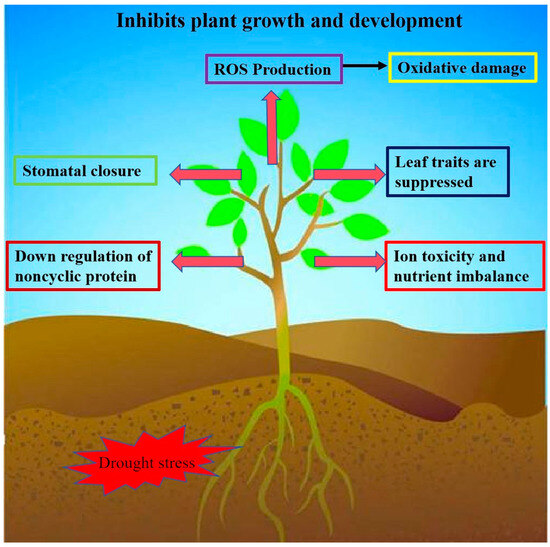
Figure 3.
Impacts of drought stress on plants. Overexpression of WRKY transcription factor genes in plants can reduce ROS production, stomatal closure, and downregulation of noncyclic proteins, and leaf traits, ion toxicity, and nutrient imbalance are suppressed. This allows plants to improve their water retention capacity to cope with plant survival rate under drought stress.
Drought is one of the major abiotic stresses that limit plant growth and development and reduce crop yield. Many members of the WRKY transcription factor family have been found to be able to respond to drought stress by binding downstream to the W-box element in the promoter region of drought stress-related genes and regulating the expression of these genes, which ultimately improves the ability of plants to cope with drought stress.
2.2. WRKY Transcription Factors Involved in Response to Temperature Stress
Temperature is considered the major abiotic stress for plants. Extreme high or low temperatures can lead to severe effects on plants, resulting in plant mortality and extensive agricultural economic losses. It is, therefore, important to increase the tolerance of plant cells to drastic changes in temperature and necessary to protect food production. WRKY transcription factors help plants resist temperature changes by regulating the expression of genes involved in temperature stress, and they also respond to extreme temperatures by regulating the expression of genes involved in the ABA response [65,66,67].
2.2.1. WRKY Transcription Factors and High-Temperature Stress
When the temperature exceeds the maximum upper limit of the temperature to which a plant can adapt, it has an injurious effect on the plant and stunts its growth and development. High temperatures can also weaken photosynthesis and enhance respiration, causing plants to over-consume their own energy and causing them to die from long-term starvation. High temperatures can also disrupt the water balance of plants, leading to the accumulation of harmful metabolites in the body. Therefore, high temperature is a significant stress factor that limits the normal growth and development of plants. Nowadays, more and more researchers are focusing on high-temperature stress in their studies [68,69]. He et al. found that the WRKY transcription factors TaWRKY1 and TaWRKY33 were identified in Triticum aestivum L. TaWRKY33 overexpression lines showed enhanced heat stress tolerance. TaWRKY1 was slightly upregulated by high temperatures, and ABA was downregulated by low temperatures. TaWRKY33 was involved in the response to high and low temperatures. The overexpression of TaWRKY1 and TaWRKY33 activated several stress-related downstream genes and promoted root growth in Arabidopsis under various stresses [70]. Wang et al. identified a WRKY family transcription factor SlWRKY3 that was induced and upregulated under heat stress, whereas a knockout strain of wrky3 resulted in reduced heat stress tolerance. The overexpression of SlWRKY3 accumulated less ROS, whereas wrky3-knockout lines accumulated more ROS under heat stress. They concluded that SlWRKY3 activated the expression of a range of abiotic stress-responsive genes involved in ROS scavenging [71]. Wu et al. performed transcriptome analysis on lily (Lilium longiflorum) and identified the WRKY family transcription factor gene LlWRKY22, whose expression was activated at high temperatures, and the overexpression of LlWRKY22 in Lilium longiflorum increased its heat tolerance and activated the expression of heat-related LlDREB2B genes, which play a positive role in the regulation of heat tolerance [72]. Balfagón et al. found that the WRKY family transcription factor AtWRKY48 was able to negatively control Arabidopsis acclimation to a combination of high light and heat stress and that the AtWRKY48 gene’s expression was reduced by jasmonic acid (JA) under these conditions [73].
2.2.2. WRKY Transcription Factors and Low-Temperature Stress
Temperature is one of the main environmental factors affecting the normal growth of plants and is necessary to ensure the normal growth of plants. Too low a temperature can cause stress to plants, affecting the growth and development of the plants. With the change in the global climate, low temperature has become an agrometeorological disaster. When the temperature drops to the lowest limit that plants can tolerate, it causes crop growth obstacles and damage to the fruiting organs and ultimately leads to the inability for normal growth and fruiting, which results in a substantial reduction in crop yields. Therefore, the study of the molecular mechanisms of plants’ responses to low-temperature stress is of great scientific significance in improving the cold tolerance of crops [74,75]. Mi et al. identified a WRKY family transcription factor, CsWRKY21, in tea tree. CsWRKY21 was induced by low temperatures and expressed six times more compared with the control [76]. Yu et al. identified 42 PgWRKY genes of seven subclasses in the genome of Platycodon grandiflorus. Among them, the expression of PgWRKY26 significantly increased after 6 h under cold stress [77]. Wang et al. identified a cold-inducible WRKY gene, PmWRKY57, which was cloned in P. mume. A PmWRKY57 overexpression line increased cold tolerance in Arabidopsis. Under cold treatment, the transgenic lines had significantly lower malondialdehyde contents and significantly higher superoxide dismutase activity, peroxidase activity, and proline contents in leaves than in wild-type plants. The expression levels of cold-responsive genes such as AtCOR6.6, AtCOR47, AtKIN1, and AtRCI2A were upregulated in transgenic Arabidopsis thaliana leaves compared with the wild type [78]. Liu et al. isolated an uncharacterised WRKY family transcription factor, VvWRKY28, in Beichun (V. vinifera x V.amurensis). Cold treatments can induce the high expression of VvWRKY28. VvWRKY28 overexpression lines improved the tolerance to low temperatures in Arabidopsis. Among them, MDA contents decreased, chlorophyll and proline contents increased, and SOD, POD, and CAT activities increased in the VvWRKY28 overexpression lines. In addition, WRKY28 may be associated with the regulation of the expression of downstream genes associated with cold stress (RAB18, COR15A, ERD10, PIF4, COR47, and ICS1) [79]. Wang et al. identified a new WRKY family transcription factor, SlWRKY50, in Solanum lycopersicum. SlWRKY50 responds to cold stimuli and plays a key role in JA biosynthesis. SlWRKY50 overexpression lines increased cold resistance in tomato, leading to higher levels of Fv/Fm, antioxidative enzymes, allene oxide synthase expression, and JA accumulation [80].
2.3. WRKY Transcription Factors in Response to Salt Stress
When the salt concentration in the soil is too high, it leads to the dehydration of plant cells, affecting nutrient absorption and thus inhibiting normal root growth. Salt is also capable of causing ionic toxicity in plants, leading to an ionic imbalance in the plants, affecting the osmotic pressure balance inside and outside the root cells, and leading to cell swelling or shrinkage and, in severe cases, cell death. Salt, therefore, plays an important role in plant growth and development [81,82,83]. Huang et al. isolated a WRKY family transcription factor, OsWRKY50, and OsWRKY50 overexpression lines enhanced salt stress tolerance in plants. OsWRKY50 transcription was repressed under salt stress conditions but was activated after ABA treatment. OsWRKY50 was able to bind to the promoter of OsNCED5 and repress its transcription [84]. Huang et al. identified a new WRKY family transcription factor, OsWRKY54. Salt stress resulted in a rapid induction in OsWRKY54 expression in roots. The wrky54 mutant led to greater sodium accumulation in shoots and enhanced the sensitivity of rice plants to salt stress. OsWRKY54 regulates the expression of OsHKT1;5; this is an essential gene related to salt tolerance. OsWRKY54 regulates the expression of OsHKT1;5 by directly binding to the W-box motif in its promoter [85]. Fang et al. found a new transcription factor gene, ZmWRKY86, in Zea mays L., whose expression was upregulated by salt stress. The wrky86 mutant enhanced plants’ tolerance to salt stress, with higher viability, catalase activity, and K+ contents, and lower malondialdehyde accumulation and Na+ contents under salt stress conditions [86]. Yu et al. found that TaWRKY17 expression was upregulated by salt, drought, hydrogen peroxide (H2O2), and ABA treatments in wheat. Overexpression of TaWRKY17 in Arabidopsis thaliana and wheat resulted in a significant increase in the plants’ salt stress tolerance. Among the TaWRKY17 overexpression plants, SOD, POD, and CAT activities were elevated, whereas H2O2 and MDA accumulation were reduced [87].
2.4. Role of WRKY Transcription Factors in Plants’ Response to Heavy Metal Stress
Heavy metal levels in plants that exceed thresholds can affect produce quality and food safety. Heavy metals can also inhibit the growth of plants, resulting in short plants, the loss of green leaves, poor root development, and other phenomena. Heavy metals can interfere with the normal physiological and metabolic processes of plants, affecting photosynthesis and respiration, leading to poor nutrient uptake and ultimately causing the death of plants and affecting plant yields. If heavy metals excessively accumulate in food crops, human consumption of such crops will cause great harm to human health. Therefore, controlling heavy metal levels is essential for human food security [88,89,90,91,92]. Gu et al. identified that the WRKY transcription factor gene ZmWRKY64 is enhanced in maize roots and leaves under cadmium stress and that knocking down the expression of ZmWRKY64 leads to excessive cadmium accumulation in leaf and root cells, resulting in a cadmium-sensitive phenotype. ZmSRG7 is a key gene that regulates ROS homeostasis under abiotic stress, and ZmWRKY64 directly enhances the transcription of this gene, thereby regulating maize tolerance in response to cadmium stress [93]. Jia et al. identified a WRKY transcription factor, TaWRKY70, that regulates the tolerance to the heavy metal cadmium in wheat. Cadmium accumulated in TaWRKY70-overexpressing Arabidopsis roots but not in leaf tissues. When TaWRKY70 was expressed, the net influx of Cd2+ into Arabidopsis roots was reduced. The overexpression of TaWRKY70 in Arabidopsis thaliana showed lower electrolyte leakage and malondialdehyde and hydrogen peroxide contents than in the wild type and higher antioxidant enzyme activities than in the wild type. TaWRKY70 directly binds to and regulates the expression of the TaCAT5 promoter, which, in turn, regulates the tolerance of plants to cadmium stress [94]. Xian et al. identified a WRKY transcription factor gene, GmWRKY172, whose expression was significantly upregulated under cadmium stress. GmWRKY172 overexpression lines exhibited an enhanced cadmium tolerance and reduced cadmium content in shoots. Under cadmium stress, transgenic soybean accumulated less MDA and H2O2 and had higher flavonoid contents, lignin contents, and POD activity than the wild type [95]. Under cadmium stress, the StWRKY6 overexpression strain had significantly higher soil and plant analysis development values and reactive oxygen-scavenging enzyme contents than the wild type. The ability of cadmium to induce StWRKY6 transcription factors upregulated the expression of a number of potential genes, including those involved in cadmium chelation, such as APR2 and DFRA; plant defences, such as VSP2 and PDF1.4; toxic substance efflux, such as ABCG1; light morphological development, such as BBX20; and auxin signalling, such as SAUR64/67. He et al. demonstrated that these genes coordinate the regulation of cadmium tolerance in StWRKY6 overexpression lines [96]. A new WRKY transcription factor, GmWRKY142, positively regulating cadmium stress, was identified by Cai et al. GmWRKY142 was highly expressed in roots, and the expression of this gene was significantly upregulated under cadmium stress, and the overexpression of GmWRKY142 in Arabidopsis and soybean hairy roots significantly enhanced cadmium tolerance. ATCDT1, GmCDT1-1, and GmCDT1-2, encoding cadmium tolerance 1, were induced in the GmWRKY142 overexpression lines [97].
2.5. WRKY Transcription Factors Involved in Plant Response to Nutritional Element Stress
Nitrogen (N) is one of the most important nutrients in the growth and development of plants and is a component of organic compounds such as proteins, chlorophyll, nucleic acids, and various biological enzymes. Plants mainly obtain inorganic nitrogen nutrients in the form of nitrate nitrogen (NO3−) and ammonium nitrogen (NH4+) in the soil through the root system. The inorganic nitrogen in the soil that can be directly used by plants is very little, and inorganic nitrogen is easy to leach and volatilise but also be fixed by the organic matter in the soil, so the effective nitrogen in the soil is far from enough for the normal growth of plants [98,99,100]. Javed et al. found four genes, ShWRKY13-2, ShWRKY39-1, ShWRKY49-3 and ShWRKY125-3, that exhibited significant upregulation in resistance to leaf scald LCP85-384 in two Saccharum spp. varieties triggered by Xanthomonas albilineans (Xa). In particular, ShWRKY22-1, ShWRKY49-3, and ShWRKY52-1 acted as negative regulators in both Saccharum spp. varieties in response to a range of N injection doses [101]. To assess whether the nitrogen form affects the synthesis of the high-value terpene metabolite steviol glycosides (SGs) in stevia (Stevia rebaudiana), Sun et al. utilised stevia plants at the same nitrogen level with NO3− or NH4+, and they found that the nitrogen form had no significant effect on the stevia leaf biomass or total nitrogen content, but NO3− increased the leaf SG content. Combined transcriptome analysis identified 397 genes that were differentially expressed (DEGs) between the NO3− and NH4+ treatments. It was concluded that NO3− could promote leaf SG synthesis through the NO3−-MYB/WRKY-GGPPS/CPS module [102]. Betalain is a water-soluble nitrogenous pigment. Zhang et al. identified a novel WRKY transcription factor, HmoWRKY40, in Hylocereus monacanthus. The betalain content and HmoWRKY40 expression rapidly increased during dragon fruit colouring, and the silencing of the HmoWRKY40 gene led to significant reductions in the betacyanin content. HmoWRKY40 binds to the promoter of HmoCYP76AD1 and activates its expression, thereby regulating betalain biosynthesis in Hylocereus monacanthus fruit [103].
Phosphorus (P) is one of the essential nutrients for plant growth and development, and phosphorus deficiency leads to morphological and physiological changes in plants. Phosphorus deficiency has an effect on photosynthesis, respiration, and biosynthetic processes in plants. Phosphorus is also an integral part of plant cells and is closely linked to all plant life activities, playing a vital role in plant growth and development [104,105,106,107]. Wang et al. identified a WRKY family of transcription factors, including OsWRKY108 and OsWRKY21, in rice, and the overexpression of these two genes resulted in the upregulation of Pi transporter protein genes, thereby enhancing Pi accumulation [108]. Wang et al. identified the FtWRKY29 transcription factor in Fagopyrum tataricum Gaertn. FtWRKY29 regulates the ability to tolerate phosphorus deficiency. Overexpression of FtWRKY29 in Arabidopsis produced transgenic lines that increased phosphorus uptake, regulated anthocyanin accumulation, and were less sensitive to low-phosphorus-induced stress. The low-phosphorus-responsive genes PHT1;1, PHT1;4, and PHO1 were significantly upregulated in these lines [109]. Liu et al. found that the GmWRKY46 gene is involved in the regulation of phosphorus deficiency tolerance in soybean. The expression of GmWRKY46 was significantly higher in low-phosphorus-sensitive soybean varieties than in phosphorus-tolerant soybean varieties, and the gene was strongly induced by phosphorus deficiency. The expression patterns of many P-responsive genes, for example, GmPht1;1, GmPht1;4, GmPTF1, GmACP1, GmPAP21, and GmExpansin-A7, were altered in the GmWRKY46 overexpression lines and GmWRKY46-silenced lines [47]. Zhang et al. found that the overexpression of OsWRKY21 or OsWRKY108 caused an increase in Pi accumulation due to the elevated expression of phytophosphate transporter protein 1 (PHT1). Oswrky21 and Oswrky108 double mutants showed decreased Pi accumulation and OsPHT1;1 expression in a Pi-dependent manner. Their results demonstrate that rice WRKY transcription factors function redundantly to promote Pi uptake by activating OsPHT1;1 expression under Pi sufficiency conditions [110]. Knockdown of OsWRKY10 results in increased Pi uptake and accumulation under conditions of Pi sufficiency. OsPHT1;2 results in increased Pi accumulation in oswrky10. OsWRKY10 is a transcriptional repressor, induced by Pi transcription, and it is upregulated by a subset of the PHT1 gene upon its mutation. Under Pi starvation, the OsWRKY10 protein is degraded via the 26S proteasome. These results demonstrate that the OsWRKY10-OsPHT1;2 module inhibits Pi uptake only in the presence of sufficient Pi [111].
2.6. WRKY Transcription Factors and Oxidative Stress
Oxidative stress is one of the most severe stresses caused by various other stresses. There are four main types of ROS in plants: singlet oxygen (1O2), superoxide (O2−), hydroxyl radical (OH), and hydrogen peroxide (H2O2). The induction of ROS accumulation in Arabidopsis with salt stress or by treating plants with H2O2 or methyl viologen (MV) induces the expression of multiple genes encoding WRKY family transcription factors [112,113,114,115]. Jia et al. identified a WRKY family of transcription factors, GhWRKY68. Promoter-driven β-glucuronidase activity is enhanced after exposure to drought, salt, ABA, and H2O2. GhWRKY68 overexpression lines showed reduced resistance to drought and salt and reduced tolerance to oxidative stress, which was associated with the accumulation of ROS, reduced enzyme activity, increased MDA contents, and an altered expression of ROS-related genes [116]. Sun et al. found that AtWRKY53 overexpression lines were highly sensitive to drought stress compared with Col-0 plants. The activated expression of AtWRKY53 inhibited stomatal closure by reducing the H2O2 content in guard cells. AtWRKY53 could bind directly to the qua-quine starch (QQS, AT3G30720) gene promoter sequence, leading to enhanced starch metabolism [117].
3. Conclusions and Prospects
Due to the continuous changes in the global climatic environment, plants often suffer from different forms of biotic and abiotic stress during their growth and development. In order to cope with these adverse conditions, plants have developed a complete coping system in the process of evolution. Plant adversities are dealt with through the protein regulation of the expression levels of downstream genes or interactions between proteins. Because transcription factors contain different gene-binding sites, they can often regulate the expression of multiple downstream genes. Therefore, focusing on the functions of transcription factors has become a major issue in crops. As a large class of important transcription factors in plants, WRKY transcription factor genes play a key role throughout a plant’s life cycle. With the continuous development of gene editing and third-generation sequencing technology, many researchers have verified the functions of WRKY family members in different types of plants and have found and proved that WRKY genes play a key role in plant growth and development and biotic and abiotic stresses [118,119,120,121,122]. In order to investigate the phylogeny and evolutionary relationships between WRKY family genes in different species, we constructed phylogenetic trees of the amino acid sequences of some WRKY family transcription factors in Glycine max, Arabidopsis thaliana Oryza sativa, and other plants using the MEGA 11.0 software [123]. Among them, we found that OsWRKY97 and SgWRKY11 were highly related; HvWRKY72 and TaWRKY72 were highly related; SbWRKY72 and ZmWRKY61 were highly related; OsWRKY90 and PhWRKY63 were highly related; GmWRKY58 and GmWRKY76 were highly related; PbWRKY45 and PdWRKY22-like were highly related; ZmWRKY22, SbWRKY22, PaWRKY22-like, DoWRKY22, and SiWRKY22 were highly related; and OsWRKY87 and AtWRKY39 were highly related, which indicated that the WRKY family of transcription factors are highly related in various species and have close affinities between them, and it is possible that WRKY family transcription factors have similar functions in different species (Figure S1).
Future research on WRKY transcription factors can start from the following aspects, such as the use of CRISPR/Cas9 technology to knock out the WRKY family transcription factor genes and the overexpression of genes and other technologies to cultivate new varieties of resilient and excellent crops and to promote the sustainable development of agriculture; to further investigate the upstream and downstream regulators and target genes of WRKY family transcription factor genes by using existing technologies; to analyse their regulatory networks in response to adversity stress; and to elucidate the molecular mechanisms of WRKY and plants’ responses to abiotic stresses.
Nowadays, scientists mainly focus on the response of WRKY transcription factors to common abiotic stresses such as drought, cold, salt, and heavy metals (Table 2). Meanwhile, the molecular mechanisms of WRKY transcription factors in response to chemical agent stresses are less reported. In today’s increasingly developed industry, chemical pollutants such as car exhaust, haze, and pesticides are extremely harmful to crops. The main component of automobile exhaust and haze is sulphide, and SO2 in the atmosphere generates SO3 under the action of sunlight, water vapour, drifting dust, and so on. Firstly, SO3 falls to the ground in the form of rainfall and drenches plants, damaging the waxy protective layer of the epidermis of the plant leaves and impairing the normal transpiration and gas exchange process. Secondly, acid rain also destroys a class of alkaline nutrients such as potassium, calcium, and phosphorus in the soil, leading to the withering and death of plants that cannot absorb nutrients in soils with insufficient fertility. The excessive use of pesticides inhibits the growth and development of plants, resulting in short plants, a small leaf area, and the yellowing of the leaf colour. The toxicity of pesticides can also lead to cell membrane rupture and cytoplasmic leakage and ultimately affects the normal growth and development of plants. Today, people are more and more concerned about crop security. If we pay more attention to the molecular mechanisms of WRKY transcription factors and chemical reagents in the future, this will provide a guarantee for crop security.

Table 2.
Abiotic stress-responsive WRKY transcription factors in plants.
In summary, the WRKY family of transcription factors is critical for plant growth and development as well as regulation in plants’ response to abiotic stresses. In the future, we will pay more attention to the mechanisms by which WRKY transcription factors can regulate downstream target genes in response to abiotic stresses, which can provide a more theoretical basis for the improvement of food safety.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms25136845/s1.
Author Contributions
All the authors contributed to the present form of this manuscript. Z.M. collected the data and drafted this manuscript; Z.M. edited this manuscript; Z.M. and L.H. created the figures and tables; Z.M. and L.H. supervised; Z.M. and L.H. finalised and approved the final version of this manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 32201695). Open-access funding was provided by the Max Planck Society.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no competing interests.
References
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Liu, B.; Yao, Y.; Guo, Z.; Jia, H.; Kong, L.; Zhang, A.; Ma, W.; Ni, Z.; Xu, S.; et al. Wheat genomic study for genetic improvement of traits in China. Sci. China Life Sci. 2022, 65, 1718–1775. [Google Scholar] [CrossRef] [PubMed]
- Valliyodan, B.; Ye, H.; Song, L.; Murphy, M.; Shannon, J.G.; Nguyen, H.T. Genetic diversity and genomic strategies for improving drought and waterlogging tolerance in soybeans. J. Exp. Bot. 2017, 68, 1835–1849. [Google Scholar] [CrossRef] [PubMed]
- Thudi, M.; Palakurthi, R.; Schnable, J.C.; Chitikineni, A.; Dreisigacker, S.; Mace, E.; Srivastava, R.K.; Satyavathi, C.T.; Odeny, D.; Tiwari, V.K.; et al. Genomic resources in plant breeding for sustainable agriculture. J. Plant Physiol. 2021, 257, 153351. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Jin, Y.M.; Wu, T.; Hu, L.; Zhang, Y.; Jiang, W.; Du, X. OsDREB2B, an AP2/ERF transcription factor, negatively regulates plant height by conferring GA metabolism in rice. Front. Plant Sci. 2022, 13, 1007811. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Wu, T.; Huang, K.; Jin, Y.M.; Li, Z.; Chen, M.; Yun, S.; Zhang, H.; Yang, X.; Chen, H.; et al. A Novel AP2/ERF Transcription Factor, OsRPH1, Negatively Regulates Plant Height in Rice. Front. Plant Sci. 2020, 11, 709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, Y.; Zhu, J.K. Thriving under Stress: How Plants Balance Growth and the Stress Response. Dev. Cell 2020, 55, 529–543. [Google Scholar] [CrossRef]
- Huang, S.; Ma, Z.; Hu, L.; Huang, K.; Zhang, M.; Zhang, S.; Jiang, W.; Wu, T.; Du, X. Involvement of rice transcription factor OsERF19 in response to ABA and salt stress responses. Plant Physiol. Biochem. 2021, 167, 22–30. [Google Scholar] [CrossRef]
- Ma, Z.; Hu, L. MicroRNA: A Dynamic Player from Signalling to Abiotic Tolerance in Plants. Int. J. Mol. Sci. 2023, 24, 11364. [Google Scholar] [CrossRef]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Hu, L.; Jiang, W. Understanding AP2/ERF Transcription Factor Responses and Tolerance to Various Abiotic Stresses in Plants: A Comprehensive Review. Int. J. Mol. Sci. 2024, 25, 893. [Google Scholar] [CrossRef]
- Yamada, Y.; Sato, F. Transcription factors in alkaloid biosynthesis. Int. Rev. Cell Mol. Biol. 2013, 305, 339–382. [Google Scholar]
- Wang, H.; Chen, W.; Xu, Z.; Chen, M.; Yu, D. Functions of WRKYs in plant growth and development. Trends Plant Sci. 2023, 28, 630–645. [Google Scholar] [CrossRef]
- Guo, X.; Ullah, A.; Siuta, D.; Kukfisz, B.; Iqbal, S. Role of WRKY Transcription Factors in Regulation of Abiotic Stress Responses in Cotton. Life 2022, 12, 1410. [Google Scholar] [CrossRef]
- Su, M.; Zuo, W.; Wang, Y.; Liu, W.; Zhang, Z.; Wang, N.; Chen, X. The WKRY transcription factor MdWRKY75 regulates anthocyanins accumulation in apples (Malus domestica). Funct. Plant Biol. 2022, 49, 799–809. [Google Scholar] [CrossRef]
- Yin, Y.; Fu, H.; Mi, F.; Yang, Y.; Wang, Y.; Li, Z.; He, Y.; Yue, Z. Genomic characterization of WRKY transcription factors related to secoiridoid biosynthesis in Gentiana macrophylla. BMC Plant Biol. 2024, 24, 66. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, R.; Huang, K.; Huang, S.; Wang, H.; Wei, Z.; Li, Z.; Bian, M.; Jiang, W.; Wu, T.; et al. The OsWRKY63-OsWRKY76-OsDREB1B module regulates chilling tolerance in rice. Plant J. 2022, 112, 383–398. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, C.; Sun, L.; Ma, X.; Qiao, H.; Zhao, W.; Yang, R.; Song, S.; Wang, S.; Huang, H. The SlWRKY57-SlVQ21/SlVQ16 module regulates salt stress in tomato. J. Integr. Plant Biol. 2023, 65, 2437–2455. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, N.; Yoshioka, H. Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 2012, 15, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Aamir, M.; Singh, V.K.; Meena, M.; Upadhyay, R.S.; Gupta, V.K.; Singh, S. Structural and Functional Insights into WRKY3 and WRKY4 Transcription Factors to Unravel the WRKY-DNA (W-Box) Complex Interaction in Tomato (Solanum lycopersicum L.). A Computational Approach. Front Plant Sci. 2017, 8, 819. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Grzechowiak, M.; Ruszkowska, A.; Sliwiak, J.; Urbanowicz, A.; Jaskolski, M.; Ruszkowski, M. New aspects of DNA recognition by group II WRKY transcription factor revealed by structural and functional study of AtWRKY18 DNA binding domain. Int. J. Biol. Macromol. 2022, 213, 589–601. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef]
- Li, W.; Pang, S.; Lu, Z.; Jin, B. Function and Mechanism of WRKY Transcription Factors in Abiotic Stress Responses of Plants. Plants 2020, 9, 1515. [Google Scholar] [CrossRef]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef]
- Wu, K.L.; Guo, Z.J.; Wang, H.H.; Li, J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005, 12, 9–26. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Mangelsen, E.; Kilian, J.; Berendzen, K.W.; Kolukisaoglu, U.H.; Harter, K.; Jansson, C.; Wanke, D. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genom. 2008, 9, 194. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 471. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Yu, E.; Hou, L.; Hua, P.; Zhao, M.; Wang, Y.; Hu, J.; Zhang, M.; Wang, K.; Wang, Y. Transcriptome-Based Identification, Characterization, Evolutionary Analysis, and Expression Pattern Analysis of the WRKY Gene Family and Salt Stress Response in Panax ginseng. Horticulturae 2022, 8, 756. [Google Scholar] [CrossRef]
- De Silva Matos, M.K.; Benko-Iseppon, A.M.; Bezerra-Neto, J.P.; Ferreira-Neto, J.R.C.; Wang, Y.; Liu, H.; Pandolfi, V.; Amorim, L.L.B.; Willadino, L.; do Vale Amorim, T.C.; et al. The WRKY transcription factor family in cowpea: Genomic characterization and transcriptomic profiling under root dehydration. Gene 2022, 823, 146377. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Zhang, Z.; Tong, T.; Fang, Y.; Zhang, X.; Niu, C.; Li, J.; Wu, Y.; Xue, D.; Zhang, X. Genome-Wide Identification of WRKY Gene Family and Expression Analysis under Abiotic Stress in Barley. Agronomy 2021, 11, 521. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, C.; Guo, F.; Sun, Q.; Yu, J.; Dong, T.; Wang, X.; Song, W.; Li, Z.; Meng, X.; et al. A systematical genome-wide analysis and screening of WRKY transcription factor family engaged in abiotic stress response in sweetpotato. BMC Plant Biol. 2022, 22, 616. [Google Scholar] [CrossRef]
- Yao, H.; Yang, T.; Qian, J.; Deng, X.; Dong, L. Genome-Wide Analysis and Exploration of WRKY Transcription Factor Family Involved in the Regulation of Shoot Branching in Petunia. Genes 2022, 13, 855. [Google Scholar] [CrossRef]
- Cheng, Y.; Luo, J.; Li, H.; Wei, F.; Zhang, Y.; Jiang, H.; Peng, X. Identification of the WRKY Gene Family and Characterization of Stress-Responsive Genes in Taraxacum kok-saghyz Rodin. Int. J. Mol. Sci. 2022, 23, 10270. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, W.; Wang, D.; Hu, S.; Zhang, Q.; Wang, Z.; Cui, L. Genome-Wide Identification and Characterization of the WRKY Gene Family in Scutellaria baicalensis Georgi under Diverse Abiotic Stress. Int. J. Mol. Sci. 2022, 23, 4225. [Google Scholar] [CrossRef]
- Chen, C.; Xie, F.; Shah, K.; Hua, Q.; Chen, J.; Zhang, Z.; Zhao, J.; Hu, G.; Qin, Y. Genome-Wide Identification of WRKY Gene Family in Pitaya Reveals the Involvement of HmoWRKY42 in Betalain Biosynthesis. Int. J. Mol. Sci. 2022, 23, 10568. [Google Scholar] [CrossRef]
- Nan, H.; Gao, L.Z. Genome-Wide Analysis of WRKY Genes and Their Response to Hormone and Mechanic Stresses in Carrot. Front. Genet. 2019, 10, 363. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Saiyinduleng; Chang, Q.; Cheng, C.; Zheng, Z.; Yu, S. Identification of yellowhorn (Xanthoceras sorbifolium) WRKY transcription factor family and analysis of abiotic stress response model. J. For. Res. 2021, 32, 987–1004. [Google Scholar]
- Du, Z.; You, S.; Zhao, X.; Xiong, L.; Li, J. Genome-Wide Identification of WRKY Genes and Their Responses to Chilling Stress in Kandelia obovata. Front. Genet. 2022, 13, 875316. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chi, Y.; Wang, Z.; Zhou, Y.; Fan, B.; Chen, Z. Functional analysis of structurally related soybean GmWRKY58 and GmWRKY76 in plant growth and development. J. Exp. Bot. 2016, 67, 4727–4742. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, Y.; Wang, R.; Cui, R.; Xu, H.; Sun, C.; Wang, J.; Zhang, H.; Chen, H.; Zhang, D. GmWRKY46, a WRKY transcription factor, negatively regulates phosphorus tolerance primarily through modifying root morphology in soybean. Plant Sci. 2022, 315, 111148. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, X.; Ruan, H.; Zhang, J.; Xie, F.; Gai, J.; Yang, S. GmWRKY45 Enhances Tolerance to Phosphate Starvation and Salt Stress, and Changes Fertility in Transgenic Arabidopsis. Front. Plant Sci. 2020, 10, 1714. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cao, Y. GmWRKY17-mediated transcriptional regulation of GmDREB1D and GmABA2 controls drought tolerance in soybean. Plant Mol. Biol. 2023, 113, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Raineri, J.; Wang, S.; Peleg, Z.; Blumwald, E.; Chan, R.L. The rice transcription factor OsWRKY47 is a positive regulator of the response to water deficit stress. Plant Mol. Biol. 2015, 88, 401–413. [Google Scholar] [CrossRef]
- Dai, X.; Wang, Y.; Zhang, W.H. OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 2016, 67, 947–960. [Google Scholar] [CrossRef]
- Zhang, J.; Peng, Y.; Guo, Z. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 2008, 18, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Ciolkowski, I.; Wanke, D.; Birkenbihl, R.P.; Somssich, I.E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008, 68, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Maeo, K.; Hayashi, S.; Kojima-Suzuki, H.; Morikami, A.; Nakamura, K. Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci. Biotechnol. Biochem. 2001, 65, 2428–2436. [Google Scholar] [CrossRef] [PubMed]
- Hrmova, M.; Hussain, S.S. Plant Transcription Factors Involved in Drought and Associated Stresses. Int. J. Mol. Sci. 2021, 22, 5662. [Google Scholar] [CrossRef] [PubMed]
- Khoso, M.A.; Hussain, A.; Ritonga, F.N.; Ali, Q.; Channa, M.M.; Alshegaihi, R.M.; Meng, Q.; Ali, M.; Zaman, W.; Brohi, R.D.; et al. WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 2022, 13, 1039329. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cheng, X.; Yin, D.; Chen, D.; Luo, C.; Liu, H.; Huang, C. Advances in the Research on Plant WRKY Transcription Factors Responsive to External Stresses. Curr. Issues Mol. Biol. 2023, 45, 2861–2880. [Google Scholar] [CrossRef] [PubMed]
- Krupinska, K.; Desel, C.; Frank, S.; Hensel, G. WHIRLIES Are Multifunctional DNA-Binding Proteins With Impact on Plant Development and Stress Resistance. Front. Plant Sci. 2022, 13, 880423. [Google Scholar] [CrossRef]
- Qin, Y.; Yu, H.; Cheng, S.; Liu, Z.; Yu, C.; Zhang, X.; Su, X.; Huang, J.; Shi, S.; Zou, Y.; et al. Genome-Wide Analysis of the WRKY Gene Family in Malus domestica and the Role of MdWRKY70L in Response to Drought and Salt Stresses. Genes 2022, 13, 1068. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.T.; Ru, J.N.; Liu, Y.W.; Li, M.; Zhao, D.; Yang, J.F.; Fu, J.D.; Xu, Z.S. Maize WRKY Transcription Factor ZmWRKY106 Confers Drought and Heat Tolerance in Transgenic Plants. Int. J. Mol. Sci. 2018, 19, 3046. [Google Scholar] [CrossRef]
- Duan, D.; Yi, R.; Ma, Y.; Dong, Q.; Mao, K.; Ma, F. Apple WRKY transcription factor MdWRKY56 positively modulates drought stress tolerance. Environ. Exp. Bot. 2023, 212, 105400. [Google Scholar] [CrossRef]
- Zhang, P.; Chao, R.; Qiu, L.; Ge, W.; Liang, J.; Wen, P. ChaWRKY40 Enhances Drought Tolerance of ‘Dawei’ Hazelnuts by Positively Regulating Proline Synthesis. Forests 2024, 15, 407. [Google Scholar] [CrossRef]
- Wang, D.; Chen, Q.; Chen, W.; Liu, X.; Xia, Y.; Guo, Q.; Jing, D.; Liang, G. A WRKY Transcription Factor, EjWRKY17, from Eriobotrya japonica Enhances Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2021, 22, 5593. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wang, J.; Li, Y.; Song, L.; Chen, D.; Liu, L.; Jiang, C.Z. A WRKY Protein, MfWRKY40, of Resurrection Plant Myrothamnus flabellifolia Plays a Positive Role in Regulating Tolerance to Drought and Salinity Stresses of Arabidopsis. Int. J. Mol. Sci. 2022, 23, 8145. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Noman, A.; Khan, M.I.; Zaynab, M.; Aqeel, M.; Anwar, M.; Ashraf, M.F.; Liu, Z.; Raza, A.; Mahpara, S.; et al. Molecular regulation of pepper innate immunity and stress tolerance: An overview of WRKY TFs. Microb. Pathog. 2019, 135, 103610. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Luan, Y.; Meng, J.; Sun, J.; Tao, J.; Zhao, D. WRKY Transcription Factor Response to High-Temperature Stress. Plants 2021, 10, 2211. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Seo, D.H.; Shin, H.; Kim, H.J.; Kim, C.M.; Jang, G. The Role of Stress-Responsive Transcription Factors in Modulating Abiotic Stress Tolerance in Plants. Agronomy 2020, 10, 788. [Google Scholar] [CrossRef]
- Li, S.; Khoso, M.A.; Wu, J.; Yu, B.; Wagan, S.; Liu, L. Exploring the Mechanisms of WRKY transcription factors and Regulated Pathways in Response to Abiotic Stress. Plant Stress. 2024, 12, 100429. [Google Scholar] [CrossRef]
- Manna, M.; Thakur, T.; Chirom, O.; Mandlik, R.; Deshmukh, R.; Salvi, P. Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Physiol. Plant 2021, 172, 847–868. [Google Scholar] [CrossRef]
- He, G.H.; Xu, J.Y.; Wang, Y.X.; Liu, J.M.; Li, P.S.; Chen, M.; Ma, Y.Z.; Xu, Z.S. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016, 16, 116. [Google Scholar] [CrossRef]
- Wang, Y.; Gai, W.; Yuan, L.; Shang, L.; Li, F.; Gong, Z.; Ge, P.F.; Wang, Y.R.; Tao, J.B.; Zhang, X.Y.; et al. Heat-inducible SlWRKY3 confers thermotolerance by activating the SlGRXS1 gene cluster in tomato. Hortic. Plant J. 2024, 10, 515–531. [Google Scholar] [CrossRef]
- Wu, Z.; Li, T.; Cao, X.; Zhang, D.; Teng, N. Lily WRKY factor LlWRKY22 promotes thermotolerance through autoactivation and activation of LlDREB2B. Hortic. Res. 2022, 9, 186. [Google Scholar] [CrossRef] [PubMed]
- Balfagón, D.; Pascual, L.S.; Sengupta, S.; Halliday, K.J.; Gómez-Cadenas, A.; Peláez-Vico, M.Á.; Sinha, R.; Mittler, R.; Zandalinas, S.I. WRKY48 negatively regulates plant acclimation to a combination of high light and heat stress. Plant J. 2024, 117, 1642–1655. [Google Scholar] [CrossRef] [PubMed]
- Ritonga, F.N.; Ngatia, J.N.; Wang, Y.; Khoso, M.A.; Farooq, U.; Chen, S. AP2/ERF, an important cold stress-related transcription factor family in plants: A review. Physiol. Mol. Biol. Plants 2021, 27, 1953–1968. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Li, L.; Zhou, J.; Lyu, D.; Zhao, D.; Qin, S. Comparison of transcriptome and metabolome analysis revealed differences in cold resistant metabolic pathways in different apple cultivars under low temperature stress. Hortic. Plant J. 2023, 9, 183–198. [Google Scholar] [CrossRef]
- Mi, X.; Tang, M.; Zhu, J.; Shu, M.; Wen, H.; Zhu, J.; Wei, C. Alternative splicing of CsWRKY21 positively regulates cold response in tea plant. Plant Physiol. Biochem. 2024, 208, 108473. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Li, J.; Chang, X.; Dong, N.; Chen, B.; Wang, J.; Zha, L.; Gui, S. Genome-wide identification and expression profiling of the WRKY gene family reveals abiotic stress response mechanisms in Platycodon grandiflorus. Int. J. Biol. Macromol. 2024, 257, 128617. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dong, B.; Wang, N.; Zheng, Z.; Yang, L.; Zhong, S.; Fang, Q.; Xiao, Z.; Zhao, H. A WRKY Transcription Factor PmWRKY57 from Prunus mume Improves Cold Tolerance in Arabidopsis thaliana. Mol. Biotechnol. 2023, 65, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liang, X.; Cai, W.; Wang, H.; Liu, X.; Cheng, L.; Song, P.; Luo, G.; Han, D. Isolation and Functional Analysis of VvWRKY28, a Vitis vinifera WRKY Transcription Factor Gene, with Functions in Tolerance to Cold and Salt Stress in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 13418. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, H.; Chen, G.; Luo, G.; Shen, X.; Ouyang, B.; Bie, Z. Transcription factor SlWRKY50 enhances cold tolerance in tomato by activating the jasmonic acid signaling. Plant Physiol. 2024, 194, 1075–1090. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Mishra, R.; Rai, S.; Bano, A.; Pathak, N.; Fujita, M.; Kumar, M.; Hasanuzzaman, M. Mechanistic Insights of Plant Growth Promoting Bacteria Mediated Drought and Salt Stress Tolerance in Plants for Sustainable Agriculture. Int. J. Mol. Sci. 2022, 23, 3741. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Hu, L.; Zhang, S.; Zhang, M.; Jiang, W.; Wu, T.; Du, X. Rice OsWRKY50 Mediates ABA-Dependent Seed Germination and Seedling Growth, and ABA-Independent Salt Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 8625. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Liu, F.; Chao, D.; Xin, B.; Liu, K.; Cao, S.; Chen, X.; Peng, L.; Zhang, B.; Fu, S.; et al. The WRKY Transcription Factor OsWRKY54 Is Involved in Salt Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 11999. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Li, W.; Yuan, H.; Chen, H.; Bo, C.; Ma, Q.; Cai, R. Mutation of ZmWRKY86 confers enhanced salt stress tolerance in maize. Plant Physiol. Biochem. 2021, 167, 840–850. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wu, Y.; He, L. A wheat WRKY transcription factor TaWRKY17 enhances tolerance to salt stress in transgenic Arabidopsis and wheat plant. Plant Mol. Biol. 2023, 113, 171–191. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Wang, Q.; Guan, Q.; Ma, Y.; Ni, F.; Yang, E.; Zhang, J. Heavy metal pollution levels, source apportionment and risk assessment in dust storms in key cities in Northwest China. J. Hazard. Mater. 2022, 422, 126878. [Google Scholar] [CrossRef] [PubMed]
- Mirza, Z.; Haque, M.M.; Gupta, M. WRKY transcription factors: A promising way to deal with arsenic stress in rice. Mol. Biol. Rep. 2022, 49, 10895–10904. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Asaf, S.; Jan, R.; Bilal, S.; Lubna; Khan, A.L.; Kim, K.M.; Al-Harrasi, A. Genome-wide annotation and expression analysis of WRKY and bHLH transcriptional factor families reveal their involvement under cadmium stress in tomato (Solanum lycopersicum L.). Front. Plant Sci. 2023, 14, 1100895. [Google Scholar]
- Abdullah; Wani, K.I.; Naeem, M.; Jha, P.K.; Jha, U.C.; Aftab, T.; Prasad, P.V.V. Systems biology of chromium-plant interaction: Insights from omics approaches. Front. Plant Sci. 2024, 14, 1305179. [Google Scholar] [CrossRef]
- Shen, C.; Yang, Y.M.; Sun, Y.F.; Zhang, M.; Chen, X.J.; Huang, Y.Y. The regulatory role of abscisic acid on cadmium uptake, accumulation and translocation in plants. Front. Plant Sci. 2022, 13, 953717. [Google Scholar] [CrossRef]
- Gu, L.; Hou, Y.; Sun, Y.; Chen, X.; Wang, G.; Wang, H.; Zhu, B.; Du, X. The maize WRKY transcription factor ZmWRKY64 confers cadmium tolerance in Arabidopsis and maize (Zea mays L.). Plant Cell Rep. 2024, 43, 44. [Google Scholar] [CrossRef]
- Jia, Z.; Li, M.; Wang, H.; Zhu, B.; Gu, L.; Du, X.; Ren, M. TaWRKY70 Positively Regulates TaCAT5 Enhanced Cd Tolerance in Transgenic Arabidopsis. Environ. Exp. Bot. 2021, 190, 104591. [Google Scholar] [CrossRef]
- Xian, P.; Yang, Y.; Xiong, C.; Guo, Z.; Alam, I.; He, Z.; Zhang, Y.; Cai, Z.; Nian, H. Overexpression of GmWRKY172 enhances cadmium tolerance in plants and reduces cadmium accumulation in soybean seeds. Front. Plant Sci. 2023, 14, 1133892. [Google Scholar] [CrossRef]
- He, G.; Saleem, M.; Deng, T.; Zhong, Z.; He, T.; Wu, J. Unraveling the Mechanism of StWRKY6 in Potato (Solanum tuberosum)’s Cadmium Tolerance for Ensuring Food Safety. Foods 2023, 12, 2303. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Xian, P.; Wang, H.; Lin, R.; Lian, T.; Cheng, Y.; Ma, Q.; Nian, H. Transcription Factor GmWRKY142 Confers Cadmium Resistance by Up-Regulating the Cadmium Tolerance 1-Like Genes. Front. Plant Sci. 2020, 11, 724. [Google Scholar] [CrossRef]
- Chen, Y.; Kong, X.; Yang, L.; Fu, M.; Zhang, S. Genome-Wide Identification of WRKY Family Genes and the Expression Profiles in Response to Nitrogen Deficiency in Poplar. Genes 2022, 13, 2324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, C.; Zhang, X.; Liang, Z.; Xia, P. Multi-algorithm cooperation research of WRKY genes under nitrogen stress in Panax notoginseng. Protoplasma 2023, 260, 1081–1096. [Google Scholar] [CrossRef] [PubMed]
- Poll, A.A.; Lee, J.; Sanderson, R.A.; Byrne, E.; Gatehouse, J.A.; Sadanandom, A.; Gatehouse, A.M.R.; Edwards, M.G. Septoria Leaf Blotch and Reduced Nitrogen Availability Alter WRKY Transcription Factor Expression in a Codependent Manner. Int. J. Mol. Sci. 2020, 21, 4165. [Google Scholar] [CrossRef]
- Javed, T.; Zhou, J.R.; Li, J.; Hu, Z.T.; Wang, Q.N.; Gao, S.J. Identification and Expression Profiling of WRKY Family Genes in Sugarcane in Response to Bacterial Pathogen Infection and Nitrogen Implantation Dosage. Front. Plant Sci. 2022, 13, 917953. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, T.; Xu, X.; Yang, Y.; Tong, H.; Mur, L.A.J.; Yuan, H. Transcriptomic Characterization of Nitrate-Enhanced Stevioside Glycoside Synthesis in Stevia (Stevia rebaudiana) Bertoni. Int. J. Mol. Sci. 2021, 22, 8549. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, C.; Xie, F.; Hua, Q.; Zhang, Z.; Zhang, R.; Chen, J.; Zhao, J.; Hu, G.; Qin, Y. A Novel WRKY Transcription Factor HmoWRKY40 Associated with Betalain Biosynthesis in Pitaya (Hylocereus monacanthus) through Regulating HmoCYP76AD1. Int. J. Mol. Sci. 2021, 22, 2171. [Google Scholar] [CrossRef]
- Tang, W.; Wang, F.; Chu, H.; You, M.; Lv, Q.; Ji, W.; Deng, X.; Zhou, B.; Peng, D. WRKY transcription factors regulate phosphate uptake in plants. Environ. Exp. Bot. 2023, 208, 105241. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, H.; Liang, Y.; Lu, T.; Liu, Z.; Jin, X.; Hou, L.; Xu, J.; Zhao, H.; Shi, Y.; et al. Comparative transcriptomic and metabolomic analyses reveal the protective effects of silicon against low phosphorus stress in tomato plants. Plant Physiol. Biochem. 2021, 166, 78–87. [Google Scholar] [CrossRef]
- Nilsson, L.; Müller, R.; Nielsen, T.H. Dissecting the plant transcriptome and the regulatory responses to phosphate deprivation. Physiol. Plant 2010, 139, 129–143. [Google Scholar] [CrossRef]
- Shukla, D.; Waigel, S.; Rouchka, E.C.; Sandhu, G.; Trivedi, P.K.; Sahi, S.V. Genome-wide expression analysis reveals contrasting regulation of phosphate starvation response (PSR) in root and shoot of Arabidopsis and its association with biotic stress. Environ. Exp. Bot. 2021, 188, 104483. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, J.; Gu, M.; Xu, G. OsWRKY108 is an integrative regulator of phosphorus homeostasis and leaf inclination in rice. Plant Signal Behav. 2021, 16, 1976545. [Google Scholar] [CrossRef]
- Wang, S.; Lv, B.; Wang, A.; Hu, J.; Wu, Q.; Li, C. Cloning and functional characterization of FtWRKY29, a low phosphorus stress-inducible transcription factor in Tartary buckwheat. Plant Physiol. Biochem. 2023, 203, 107997. [Google Scholar] [CrossRef]
- Zhang, J.; Gu, M.; Liang, R.; Shi, X.; Chen, L.; Hu, X.; Wang, S.; Dai, X.; Qu, H.; Li, H.; et al. OsWRKY21 and OsWRKY108 function redundantly to promote phosphate accumulation through maintaining the constitutive expression of OsPHT1;1 under phosphate-replete conditions. New Phytol. 2021, 229, 1598–1614. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xu, T.; Chen, M.; Geng, L.; Huang, Z.; Dai, X.; Qu, H.; Zhang, J.; Li, H.; Gu, M.; et al. The transcription factor OsWRKY10 inhibits phosphate uptake via suppressing OsPHT1;2 expression under phosphate-replete conditions in rice. J. Exp. Bot. 2023, 74, 1074–1089. [Google Scholar] [CrossRef] [PubMed]
- León, J.; Gayubas, B.; Castillo, M.C. Valine-Glutamine Proteins in Plant Responses to Oxygen and Nitric Oxide. Front. Plant Sci. 2021, 11, 632678. [Google Scholar] [CrossRef] [PubMed]
- Tolosa, L.N.; Zhang, Z. The Role of Major Transcription Factors in Solanaceous Food Crops under Different Stress Conditions: Current and Future Perspectives. Plants 2020, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Rai, G.K.; Mishra, S.; Chouhan, R.; Mushtaq, M.; Chowdhary, A.A.; Rai, P.K.; Kumar, R.R.; Kumar, P.; Perez-Alfocea, F.; Colla, G.; et al. Plant salinity stress, sensing, and its mitigation through WRKY. Front. Plant Sci. 2023, 14, 1238507. [Google Scholar] [CrossRef]
- Khedia, J.; Agarwal, P.; Agarwal, P.K. Deciphering hydrogen peroxide-induced signalling towards stress tolerance in plants. 3 Biotech. 2019, 9, 395. [Google Scholar] [CrossRef]
- Jia, H.; Wang, C.; Wang, F.; Liu, S.; Li, G.; Guo, X. GhWRKY68 reduces resistance to salt and drought in transgenic Nicotiana benthamiana. PLoS ONE 2015, 10, e0120646. [Google Scholar] [CrossRef]
- Sun, Y.; Yu, D. Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep. 2015, 34, 1295–1306. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Cao, Y.; Zhao, L.; Zhang, J.; Li, S. Review: WRKY transcription factors: Understanding the functional divergence. Plant Sci. 2023, 334, 111770. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Sunarti, S.; Kissoudis, C.; Visser, R.G.F.; van der Linden, C.G. The Role of Tomato WRKY Genes in Plant Responses to Combined Abiotic and Biotic Stresses. Front. Plant Sci. 2018, 9, 801. [Google Scholar] [CrossRef] [PubMed]
- Schluttenhofer, C.; Yuan, L. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 2015, 167, 295–306. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Samad, A.F.A.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.M.A.; Zainal, Z.; Ismail, I. MicroRNA and Transcription Factor: Key Players in Plant Regulatory Network. Front. Plant Sci. 2017, 8, 565. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Finatto, T.; Viana, V.E.; Woyann, L.G.; Busanello, C.; Maia, L.C.D.; Oliveira, A.C. Can WRKY transcription factors help plants to overcome environmental challenges? Genet. Mol. Biol. 2018, 41, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Cui, R.; Huang, Y.; Shi, L.; Wang, S.; Xu, F. Repression of transcription factor AtWRKY47 confers tolerance to boron toxicity in Arabidopsis thaliana. Ecotoxicol. Environ. Saf. 2021, 220, 112406. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Yan, X.; Huang, Y.; Han, Y.; Zhang, C.; Ren, Y.; Fan, T.; Xiao, F.; Liu, Y.; Cao, S. The WRKY transcription factor, WRKY13, activates PDR8 expression to positively regulate cadmium tolerance in Arabidopsis. Plant Cell Environ. 2019, 42, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Babitha, K.C.; Ramu, S.V.; Pruthvi, V.; Mahesh, P.; Nataraja, K.N.; Udayakumar, M. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 2013, 22, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, P.; Vishal, B.; Ho, W.J.; Lok, F.C.J.; Lee, F.S.M.; Kumar, P.P. Regulation of a Cytochrome P450 Gene CYP94B1 by WRKY33 Transcription Factor Controls Apoplastic Barrier Formation in Roots to Confer Salt Tolerance. Plant Physiol. 2020, 184, 2199–2215. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Li, C.X.; Li, G.X.; Wu, Y.R.; Zheng, S.J. Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 2015, 84, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.Y.; Du, Y.T.; Ma, J.; Min, D.H.; Jin, L.G.; Chen, J.; Chen, M.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S.; et al. The WRKY Transcription Factor GmWRKY12 Confers Drought and Salt Tolerance in Soybean. Int. J. Mol. Sci. 2018, 19, 4087. [Google Scholar] [CrossRef]
- Ma, Q.; Xia, Z.; Cai, Z.; Li, L.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 Enhances Drought and Salt Tolerance Through an ABA-Mediated Pathway in Arabidopsis thaliana. Front. Plant Sci. 2019, 9, 1979. [Google Scholar] [CrossRef]
- Luo, X.; Bai, X.; Sun, X.; Zhu, D.; Liu, B.; Ji, W.; Cai, H.; Cao, L.; Wu, J.; Hu, M.; et al. Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 2013, 64, 2155–2169. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, H.W.; Li, Q.T.; Wei, W.; Li, W.; Zhang, W.K.; Ma, B.; Bi, Y.D.; Lai, Y.C.; Liu, X.L.; et al. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 2015, 83, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Liang, D.W.; Bian, X.H.; Shen, M.; Xiao, J.H.; Zhang, W.K.; Ma, B.; Lin, Q.; Lv, J.; Chen, X.; et al. GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+ signaling pathways in transgenic soybean. Plant J. 2019, 100, 384–398. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, P.; Hou, L.; Zhao, S.; Zhao, C.; Xia, H.; Li, P.; Zhang, Y.; Bian, X.; Wang, X. Global Analysis of WRKY Genes and Their Response to Dehydration and Salt Stress in Soybean. Front. Plant Sci. 2016, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Dai, W.; Zhang, C.; Wang, Y.; Wu, M.; Zhao, Y.; Ma, Q.; Xiang, Y.; Cheng, B. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 2017, 246, 1215–1231. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yan, J.; Xiang, Y.; Sun, Y.; Zhang, A. ZmWRKY104 Transcription Factor Phosphorylated by ZmMPK6 Functioning in ABA-Induced Antioxidant Defense and Enhance Drought Tolerance in Maize. Biology 2021, 10, 893. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Sato, Y.; Tanabe, S.; Chujo, T.; Shimizu, T.; Okada, K.; Yamane, H.; Shimono, M.; Sugano, S.; Takatsuji, H.; et al. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 2013, 64, 5085–5097. [Google Scholar] [CrossRef] [PubMed]
- Pillai, S.E.; Kumar, C.; Patel, H.K.; Sonti, R.V. Overexpression of a cell wall damage induced transcription factor, OsWRKY42, leads to enhanced callose deposition and tolerance to salt stress but does not enhance tolerance to bacterial infection. BMC Plant Biol. 2018, 18, 177. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Wu, T.; Ma, Z.; Li, Z.; Chen, H.; Zhang, M.; Bian, M.; Bai, H.; Jiang, W.; Du, X. Rice Transcription Factor OsWRKY55 Is Involved in the Drought Response and Regulation of Plant Growth. Int. J. Mol. Sci. 2021, 22, 4337. [Google Scholar] [CrossRef]
- Lim, C.; Kang, K.; Shim, Y.; Yoo, S.C.; Paek, N.C. Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 2022, 188, 1900–1916. [Google Scholar] [CrossRef]
- Lv, M.; Hou, D.; Wan, J.; Ye, T.; Zhang, L.; Fan, J.; Li, C.; Dong, Y.; Chen, W.; Rong, S.; et al. OsWRKY97, an Abiotic Stress-Induced Gene of Rice, Plays a Key Role in Drought Tolerance. Plants 2023, 12, 3338. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Z.; Wang, Z.Q.; Yokosho, K.; Ding, B.; Fan, W.; Gong, Q.Q.; Li, G.X.; Wu, Y.R.; Yang, J.L.; Ma, J.F.; et al. Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4 expression and enhancement of citrate secretion in rice (Oryza sativa). New Phytol. 2018, 219, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Baoxiang, W.; Jingfang, L.; Zhiguang, S.; Ming, C.; Yungao, X.; Bo, X.; Bo, Y.; Jian, L.; Jinbo, L.; et al. A novel SAPK10-WRKY87-ABF1 biological pathway synergistically enhance abiotic stress tolerance in transgenic rice (Oryza sativa). Plant Physiol. Biochem. 2021, 168, 252–262. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).