To Erase or Not to Erase: Non-Canonical Catalytic Functions and Non-Catalytic Functions of Members of Histone Lysine Demethylase Families
Abstract
1. Introduction
2. Different Functions Besides Histone Demethylation Activity
2.1. Proteins Containing Jumonji Domains But Retaining No or Uncertain Histone Demethylase Activity
2.1.1. JARID2: Inactive But Essential
2.1.2. UTY: A Catalytically Inactive KDM6 Subfamily Member?
2.2. The JmjC Domain-Only Family: Enzymes with Unclear Demethylase Activity But Which Show Different Catalytic Functions
2.2.1. HSPBAP1
2.2.2. RIOX1/RIOX2
2.2.3. HIF1AN
2.2.4. JMJD4
2.2.5. KDM8 (Alias JMJD5) and JMJD7
2.2.6. JMJD6
2.2.7. JMJD8
2.2.8. TYW5
2.3. Non-Catalytic Functions of Active Histone Demethylases
2.3.1. KDM6A (Alias UTX) and KDM6B (Alias JMJD3)
2.3.2. KDM1A
2.3.3. KDM1B
2.3.4. KDM2B
2.3.5. KDM4A
2.3.6. KDM4B
2.3.7. KDM5 Demethylases
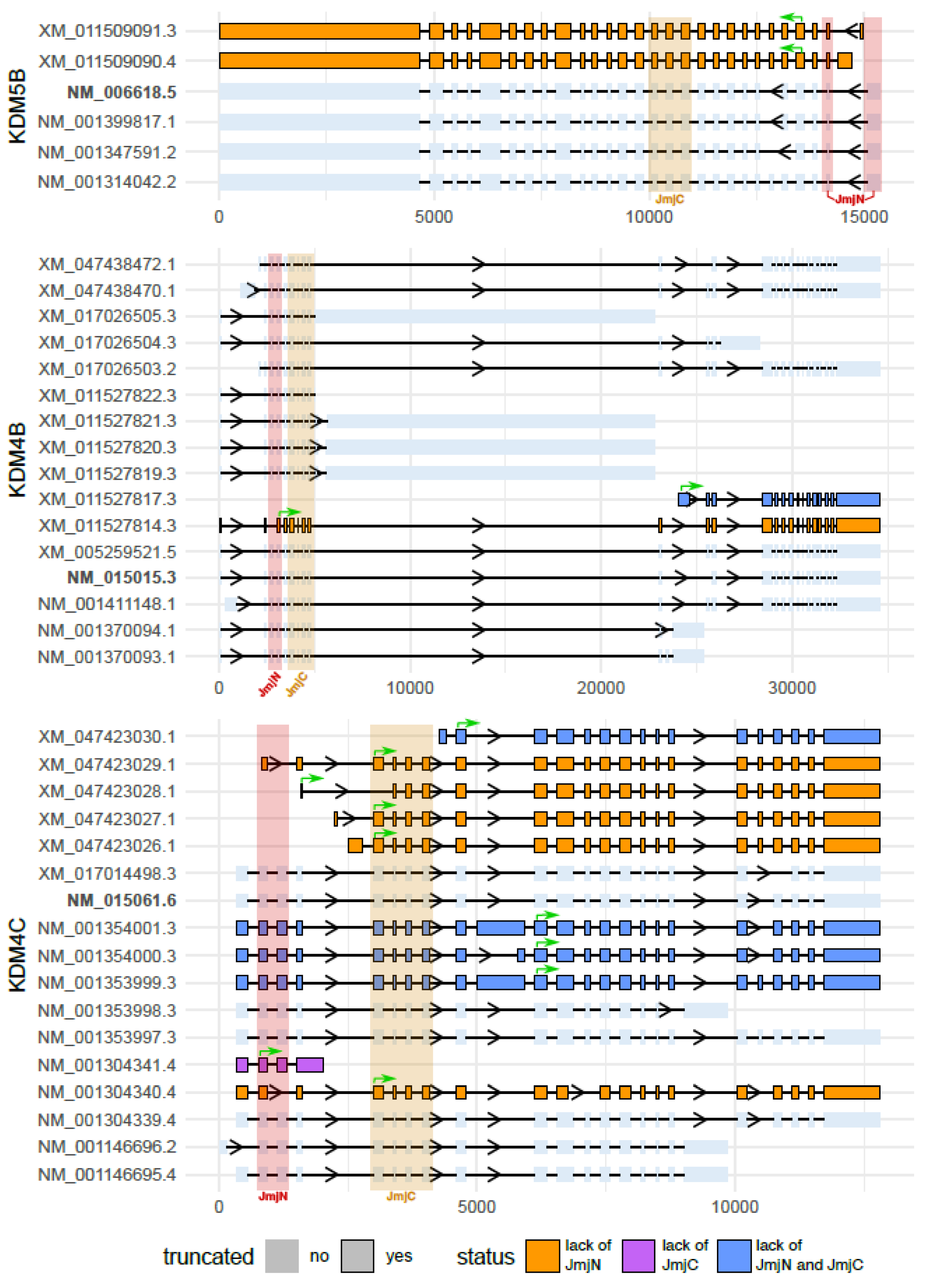
3. KDM Isoforms without Catalytic Activity
4. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Q.; Zhang, Y.; Han, J. The Role of Histone Modification in DNA Replication-Coupled Nucleosome Assembly and Cancer. Int. J. Mol. Sci. 2023, 24, 4939. [Google Scholar] [CrossRef] [PubMed]
- Frigerio, C.; Di Nisio, E.; Galli, M.; Colombo, C.V.; Negri, R.; Clerici, M. The Chromatin Landscape around DNA Double-Strand Breaks in Yeast and Its Influence on DNA Repair Pathway Choice. Int. J. Mol. Sci. 2023, 24, 3248. [Google Scholar] [CrossRef] [PubMed]
- Di Nisio, E.; Lupo, G.; Licursi, V.; Negri, R. The Role of Histone Lysine Methylation in the Response of Mammalian Cells to Ionizing Radiation. Front. Genet. 2021, 12, 639602. [Google Scholar] [CrossRef] [PubMed]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Jambhekar, A.; Dhall, A.; Shi, Y. Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 2019, 20, 625–641. [Google Scholar] [CrossRef] [PubMed]
- Klose, R.J.; Kallin, E.M.; Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006, 7, 715–727. [Google Scholar] [CrossRef]
- Pedersen, M.T.; Helin, K. Histone demethylases in development and disease. Trends Cell Biol. 2010, 20, 662–671. [Google Scholar] [CrossRef]
- Kooistra, S.M.; Helin, K. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 2012, 13, 297–311. [Google Scholar] [CrossRef]
- Hamamoto, R.; Saloura, V.; Nakamura, Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat. Rev. Cancer 2015, 15, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, R.; Sekirnik, R.; Brissett, N.C.; Krojer, T.; Ho, C.-H.; Ng, S.S.; Clifton, I.J.; Ge, W.; Kershaw, N.J.; Fox, G.C.; et al. Ribosomal oxygenases are structurally conserved from prokaryotes to humans. Nature 2014, 510, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Zang, J.; White, J.; Wang, C.; Pan, C.-H.; Zhao, R.; Murphy, R.C.; Dai, S.; Henson, P.; Kappler, J.W.; et al. Interaction of JMJD6 with single-stranded RNA. Proc. Natl. Acad. Sci. USA 2010, 107, 14568–14572. [Google Scholar] [CrossRef] [PubMed]
- Landeira, D.; Fisher, A.G. Inactive yet indispensable: The tale of Jarid2. Trends Cell Biol. 2011, 21, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Shpargel, K.B.; Sengoku, T.; Yokoyama, S.; Magnuson, T. UTX and UTY Demonstrate Histone Demethylase-Independent Function in Mouse Embryonic Development. PLoS Genet. 2012, 8, e1002964. [Google Scholar] [CrossRef]
- Walport, L.J.; Hopkinson, R.J.; Vollmar, M.; Madden, S.K.; Gileadi, C.; Oppermann, U.; Schofield, C.J.; Johansson, C. Human UTY(KDM6C) Is a Male-specific Nϵ-Methyl Lysyl Demethylase. J. Biol. Chem. 2014, 289, 18302–18313. [Google Scholar] [CrossRef] [PubMed]
- Gažová, I.; Lengeling, A.; Summers, K.M. Lysine demethylases KDM6A and UTY: The X and Y of histone demethylation. Mol. Genet. Metab. 2019, 127, 31–44. [Google Scholar] [CrossRef]
- Liu, C.; Gilmont, R.R.; Benndorf, R.; Welsh, M.J. Identification and characterization of a novel protein from Sertoli cells, PASS1, that associates with mammalian small stress protein hsp27. J. Biol. Chem. 2000, 275, 18724–18731. [Google Scholar] [CrossRef]
- Sinha, K.M.; Yasuda, H.; Coombes, M.M.; Dent, S.Y.R.; de Crombrugghe, B. Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J. 2010, 29, 68–79. [Google Scholar] [CrossRef]
- Ge, W.; Wolf, A.; Feng, T.; Ho, C.-H.; Sekirnik, R.; Zayer, A.; Granatino, N.; E Cockman, M.; Loenarz, C.; Loik, N.D.; et al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat. Chem. Biol. 2012, 8, 960–962. [Google Scholar] [CrossRef]
- Bundred, J.R.; Hendrix, E.; Coleman, M.L. The emerging roles of ribosomal histidyl hydroxylases in cell biology, physiology and disease. Cell. Mol. Life Sci. 2018, 75, 4093–4105. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Shin, S.; Janknecht, R. The small members of the JMJD protein family: Enzymatic jewels or jinxes? Biochim. Biophys. Acta (BBA)-Rev. Cancer 2019, 1871, 406–418. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Beezhold, K.; Chang, Q.; Zhang, Y.; Rojanasakul, Y.; Zhao, H.; Castranova, V.; Shi, X.; Chen, F. Lung cancer-associated JmjC domain protein mdig suppresses formation of tri-methyl lysine 9 of histone H3. Cell Cycle 2009, 8, 2101–2109. [Google Scholar] [CrossRef] [PubMed]
- Fiorini, G.; Schofield, C.J. Biochemistry of the hypoxia-inducible factor hydroxylases. Curr. Opin. Chem. Biol. 2024, 79, 102428. [Google Scholar] [CrossRef] [PubMed]
- Volkova, Y.L.; Pickel, C.; Jucht, A.E.; Wenger, R.H.; Scholz, C.C. The Asparagine Hydroxylase FIH: A Unique Oxygen Sensor. Antioxid. Redox Signal 2022, 37, 913–935. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Yamamoto, A.; Wilkins, S.E.; Sokolova, E.; Yates, L.A.; Münzel, M.; Singh, P.; Hopkinson, R.J.; Fischer, R.; Cockman, M.E.; et al. Optimal translational termination requires C4 lysyl hydroxylation of eRF1. Mol. Cell 2014, 53, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Feng, M.; Su, Y.; Ma, T.; Zhang, H.; Wu, H.; Wang, X.; Shi, S.; Zhang, Y.; Xu, Y.; et al. Jmjd4 Facilitates Pkm2 Degradation in Cardiomyocytes and Is Protective Against Dilated Cardiomyopathy. Circulation 2023, 147, 1684–1704. [Google Scholar] [CrossRef] [PubMed]
- Youn, M.-Y.; Yokoyama, A.; Fujiyama-Nakamura, S.; Ohtake, F.; Minehata, K.-I.; Yasuda, H.; Suzuki, T.; Kato, S.; Imai, Y. JMJD5, a Jumonji C (JmjC) Domain-containing Protein, Negatively Regulates Osteoclastogenesis by Facilitating NFATc1 Protein Degradation. J. Biol. Chem. 2012, 287, 12994–13004. [Google Scholar] [CrossRef] [PubMed]
- Hsia, D.A.; Tepper, C.G.; Pochampalli, M.R.; Hsia, E.Y.C.; Izumiya, C.; Huerta, S.B.; Wright, M.E.; Chen, H.-W.; Kung, H.-J.; Izumiya, Y. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc. Natl. Acad. Sci. USA 2010, 107, 9671–9676. [Google Scholar] [CrossRef]
- Marcon, E.; Ni, Z.; Pu, S.; Turinsky, A.L.; Trimble, S.S.; Olsen, J.B.; Silverman-Gavrila, R.; Silverman-Gavrila, L.; Phanse, S.; Guo, H.; et al. Human-chromatin-related protein interactions identify a demethylase complex required for chromosome segregation. Cell Rep. 2014, 8, 297–310. [Google Scholar] [CrossRef]
- Liu, H.; Wei, P.; Zhang, Q.; Chen, Z.; Liu, J.; Zhang, G. The Novel Protease Activities of JMJD5-JMJD6-JMJD7 and Arginine Methylation Activities of Arginine Methyltransferases Are Likely Coupled. Biomolecules 2022, 12, 347. [Google Scholar] [CrossRef]
- Chang, B.; Chen, Y.; Zhao, Y.; Bruick, R.K. JMJD6 Is a Histone Arginine Demethylase. Science 2007, 318, 444–447. [Google Scholar] [CrossRef]
- Liu, W.; Ma, Q.; Wong, K.; Li, W.; Ohgi, K.; Zhang, J.; Aggarwal, A.K.; Rosenfeld, M.G. Brd4 and JMJD6-Associated Anti-Pause Enhancers in Regulation of Transcriptional Pause Release. Cell 2013, 155, 1581–1595. [Google Scholar] [CrossRef]
- Webby, C.J.; Wolf, A.; Gromak, N.; Dreger, M.; Kramer, H.; Kessler, B.; Nielsen, M.L.; Schmitz, C.; Butler, D.S.; Yates, J.R.; et al. Jmjd6 Catalyses Lysyl-Hydroxylation of U2AF65, a Protein Associated with RNA Splicing. Science 2009, 325, 90–93. [Google Scholar] [CrossRef]
- Hahn, P.; Wegener, I.; Burrells, A.; Böse, J.; Wolf, A.; Erck, C.; Butler, D.; Schofield, C.J.; Böttger, A.; Lengeling, A. Analysis of Jmjd6 Cellular Localization and Testing for Its Involvement in Histone Demethylation. PLoS ONE 2010, 5, e13769. [Google Scholar] [CrossRef]
- Jablonowski, C.M.; Quarni, W.; Singh, S.; Tan, H.; Bostanthirige, D.H.; Jin, H.; Fang, J.; Chang, T.-C.; Finkelstein, D.; Cho, J.-H.; et al. Metabolic reprogramming of cancer cells by JMJD6-mediated pre-mRNA splicing associated with therapeutic response to splicing inhibitor. eLife 2024, 12, RP90993. [Google Scholar] [CrossRef]
- Manfredi, J.; Wang, F.; He, L.; Huangyang, P.; Liang, J.; Si, W.; Yan, R.; Han, X.; Liu, S.; Gui, B.; et al. JMJD6 Promotes Colon Carcinogenesis through Negative Regulation of p53 by Hydroxylation. PLoS Biol. 2014, 12, e1001819. [Google Scholar] [CrossRef]
- Markolovic, S.; Zhuang, Q.; Wilkins, S.E.; Eaton, C.D.; Abboud, M.I.; Katz, M.J.; McNeil, H.E.; Leśniak, R.K.; Hall, C.; Struwe, W.B.; et al. The Jumonji-C oxygenase JMJD7 catalyzes (3S)-lysyl hydroxylation of TRAFAC GTPases. Nat. Chem. Biol. 2018, 14, 688–695. [Google Scholar] [CrossRef]
- Boeckel, J.-N.; Derlet, A.; Glaser, S.F.; Luczak, A.; Lucas, T.; Heumüller, A.W.; Krüger, M.; Zehendner, C.M.; Kaluza, D.; Doddaballapur, A.; et al. JMJD8 Regulates Angiogenic Sprouting and Cellular Metabolism by Interacting With Pyruvate Kinase M2 in Endothelial Cells. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 1425–1433. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Li, Z.; Wang, J. JMJD8 Functions as a Novel AKT1 Lysine Demethylase. Int. J. Mol. Sci. 2022, 24, 460. [Google Scholar] [CrossRef]
- Kato, M.; Araiso, Y.; Noma, A.; Nagao, A.; Suzuki, T.; Ishitani, R.; Nureki, O. Crystal structure of a novel JmjC-domain-containing protein, TYW5, involved in tRNA modification. Nucleic Acids Res. 2011, 39, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Perillo, B.; Tramontano, A.; Pezone, A.; Migliaccio, A. LSD1: More than demethylation of histone lysine residues. Exp. Mol. Med. 2020, 52, 1936–1947. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Yang, H.; Dong, Z.; Fang, J.; Wang, P.; Zhu, T.; Gong, W.; Fang, R.; Shi, Y.G.; Li, Z.; et al. Structural insight into substrate recognition by histone demethylase LSD2/KDM1b. Cell Res. 2013, 23, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Chen, F.; Dong, Z.; Hu, D.; Barbera, A.J.; Clark, E.A.; Fang, J.; Yang, Y.; Mei, P.; Rutenberg, M.; et al. LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation. Mol. Cell 2013, 49, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Marabelli, C.; Marrocco, B.; Pilotto, S.; Chittori, S.; Picaud, S.; Marchese, S.; Ciossani, G.; Forneris, F.; Filippakopoulos, P.; Schoehn, G.; et al. A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex. Cell Rep. 2019, 27, 387–399.e7. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Qian, X.; Shen, J.; Wang, Y.; Li, X.; Liu, R.; Xia, Y.; Chen, Q.; Peng, G.; Lin, S.-Y.; et al. Local generation of fumarate promotes DNA repair through inhibition of histone H3 demethylation. Nat. Cell Biol. 2015, 17, 1158–1168. [Google Scholar] [CrossRef] [PubMed]
- Frescas, D.; Guardavaccaro, D.; Bassermann, F.; Koyama-Nasu, R.; Pagano, M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 2007, 450, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, Y.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-Y.; Kim, J.; Kim, K.; Park, J.W.; Cho, H.; Hahm, J.Y.; Chae, Y.; Kim, D.; Kook, H.; Rhee, S.; et al. KDM2B is a histone H3K79 demethylase and induces transcriptional repression via sirtuin-1-mediated chromatin silencing. FASEB J. 2018, 32, 5737–5750. [Google Scholar] [CrossRef]
- Young, N.L.; Dere, R. Mechanistic insights into KDM4A driven genomic instability. Biochem. Soc. Trans. 2021, 49, 93–105. [Google Scholar] [CrossRef]
- Walport, L.J.; Hopkinson, R.J.; Chowdhury, R.; Zhang, Y.; Bonnici, J.; Schiller, R.; Kawamura, A.; Schofield, C.J. Mechanistic and structural studies of KDM-catalysed demethylation of histone 1 isotype 4 at lysine 26. FEBS Lett. 2018, 592, 3264–3273. [Google Scholar] [CrossRef] [PubMed]
- Whetstine, J.R.; Nottke, A.; Lan, F.; Huarte, M.; Smolikov, S.; Chen, Z.; Spooner, E.; Li, E.; Zhang, G.; Colaiacovo, M.; et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 2006, 125, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Tu, S.; Teng, Y.-C.; Yuan, C.; Wu, Y.-T.; Chan, M.-Y.; Cheng, A.-N.; Lin, P.-H.; Juan, L.-J.; Tsai, M.-D. The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nat. Struct. Mol. Biol. 2008, 15, 419–421. [Google Scholar] [CrossRef] [PubMed]
- Klein, B.J.; Piao, L.; Xi, Y.; Rincon-Arano, H.; Rothbart, S.B.; Peng, D.; Wen, H.; Larson, C.; Zhang, X.; Zheng, X.; et al. The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep. 2014, 6, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, H.; Guo, X.; Rong, N.; Song, Y.; Xu, Y.; Lan, W.; Zhang, X.; Liu, M.; Xu, Y.; et al. The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B. Protein Cell 2014, 5, 837–850. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Bayliss, P.E.; Rinn, J.L.; Whetstine, J.R.; Wang, J.K.; Chen, S.; Iwase, S.; Alpatov, R.; Issaeva, I.; Canaani, E.; et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 2007, 449, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Agger, K.; Cloos, P.A.C.; Christensen, J.; Pasini, D.; Rose, S.; Rappsilber, J.; Issaeva, I.; Canaani, E.; Salcini, A.E.; Helin, K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007, 449, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Cho, Y.-W.; Yu, L.-R.; Yu, H.; Veenstra, T.D.; Ge, K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. USA 2007, 104, 18439–18444. [Google Scholar] [CrossRef] [PubMed]
- De Santa, F.; Totaro, M.G.; Prosperini, E.; Notarbartolo, S.; Testa, G.; Natoli, G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 2007, 130, 1083–1094. [Google Scholar] [CrossRef]
- Pasini, D.; Cloos, P.A.C.; Walfridsson, J.; Olsson, L.; Bukowski, J.-P.; Johansen, J.V.; Bak, M.; Tommerup, N.; Rappsilber, J.; Helin, K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 2010, 464, 7286. [Google Scholar] [CrossRef]
- Son, J.; Shen, S.S.; Margueron, R.; Reinberg, D. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes. Dev. 2013, 27, 2663–2677. [Google Scholar] [CrossRef] [PubMed]
- Cooper, S.; Grijzenhout, A.; Underwood, E.; Ancelin, K.; Zhang, T.; Nesterova, T.B.; Anil-Kirmizitas, B.; Bassett, A.; Kooistra, S.M.; Agger, K.; et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 2016, 7, 13661. [Google Scholar] [CrossRef] [PubMed]
- Landeira, D.; Sauer, S.; Poot, R.; Dvorkina, M.; Mazzarella, L.; Jørgensen, H.F.; Pereira, C.F.; Leleu, M.; Piccolo, F.M.; Spivakov, M.; et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat. Cell Biol. 2010, 12, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Petracovici, A.; Bonasio, R. Distinct PRC2 subunits regulate maintenance and establishment of Polycomb repression during differentiation. Mol. Cell 2021, 81, 2625–2639.e5. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Mysliwiec, M.R.; Lee, Y. Roles of JUMONJI in mouse embryonic development. Dev. Dyn. 2005, 232, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Kinkel, S.A.; Galeev, R.; Flensburg, C.; Keniry, A.; Breslin, K.; Gilan, O.; Lee, S.; Liu, J.; Chen, K.; Gearing, L.J.; et al. Jarid2 regulates hematopoietic stem cell function by acting with polycomb repressive complex 2. Blood 2015, 125, 1890–1900. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.; Mysliwiec, M.R.; Carlson, C.D.; Ansari, A.; Schwartz, R.J.; Lee, Y. Cardiac-specific developmental and epigenetic functions of Jarid2 during embryonic development. J. Biol. Chem. 2018, 293, 11659–11673. [Google Scholar] [CrossRef]
- Peng, J.C.; Valouev, A.; Swigut, T.; Zhang, J.; Zhao, Y.; Sidow, A.; Wysocka, J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 2009, 139, 1290–1302. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Kim, W.; Fujiwara, Y.; Simon, M.D.; Liu, Y.; Mysliwiec, M.R.; Yuan, G.-C.; Lee, Y.; Orkin, S.H. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 2009, 139, 1303–1314. [Google Scholar] [CrossRef]
- Li, G.; Margueron, R.; Ku, M.; Chambon, P.; Bernstein, B.E.; Reinberg, D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010, 24, 368–380. [Google Scholar] [CrossRef]
- Holoch, D.; Margueron, R. Mechanisms Regulating PRC2 Recruitment and Enzymatic Activity. Trends Biochem. Sci. 2017, 42, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Al-Raawi, D.; Jones, R.; Wijesinghe, S.; Halsall, J.; Petric, M.; Roberts, S.; A Hotchin, N.; Kanhere, A. A novel form of JARID2 is required for differentiation in lineage-committed cells. EMBO J. 2019, 38, e98449. [Google Scholar] [CrossRef] [PubMed]
- Kasinath, V.; Beck, C.; Sauer, P.; Poepsel, S.; Kosmatka, J.; Faini, M.; Toso, D.; Aebersold, R.; Nogales, E. JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. Science 2021, 371, 6527. [Google Scholar] [CrossRef] [PubMed]
- Loh, C.H.; Veenstra, G.J.C. The Role of Polycomb Proteins in Cell Lineage Commitment and Embryonic Development. Epigenomes 2022, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, S.; Bonasio, R.; Saldaña-Meyer, R.; Yoshida, T.; Son, J.; Nishino, K.; Umezawa, A.; Reinberg, D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell 2014, 53, 290–300. [Google Scholar] [CrossRef] [PubMed]
- da Rocha, S.T.; Boeva, V.; Escamilla-Del-Arenal, M.; Ancelin, K.; Granier, C.; Matias, N.R.; Sanulli, S.; Chow, J.; Schulz, E.; Picard, C.; et al. Jarid2 Is Implicated in the Initial Xist-Induced Targeting of PRC2 to the Inactive X Chromosome. Mol. Cell 2014, 53, 301–316. [Google Scholar] [CrossRef] [PubMed]
- Sanulli, S.; Justin, N.; Teissandier, A.; Ancelin, K.; Portoso, M.; Caron, M.; Michaud, A.; Lombard, B.; da Rocha, S.T.; Offer, J.; et al. Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation. Mol. Cell 2015, 57, 769–783. [Google Scholar] [CrossRef]
- Mozzetta, C.; Pontis, J.; Fritsch, L.; Robin, P.; Portoso, M.; Proux, C.; Margueron, R.; Ait-Si-Ali, S. The histone H3 lysine 9 methyltransferases G9a and GLP regulate polycomb repressive complex 2-mediated gene silencing. Mol. Cell 2014, 53, 277–289. [Google Scholar] [CrossRef]
- Shirato, H.; Ogawa, S.; Nakajima, K.; Inagawa, M.; Kojima, M.; Tachibana, M.; Shinkai, Y.; Takeuchi, T. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J. Biol. Chem. 2009, 284, 733–739. [Google Scholar] [CrossRef]
- Mysliwiec, M.R.; Carlson, C.D.; Tietjen, J.; Hung, H.; Ansari, A.Z.; Lee, Y. Jarid2 (Jumonji, AT Rich Interactive Domain 2) Regulates NOTCH1 Expression via Histone Modification in the Developing Heart. J. Biol. Chem. 2012, 287, 1235–1241. [Google Scholar] [CrossRef]
- Rock, K.D.; Folts, L.M.; Zierden, H.C.; Marx-Rattner, R.; Leu, N.A.; Nugent, B.M.; Bale, T.L. Developmental transcriptomic patterns can be altered by transgenic overexpression of Uty. Sci. Rep. 2023, 13, 21082. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Deng, X.; Watkins, R.; Disteche, C.M. Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J. Neurosci. 2008, 28, 4521–4527. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, N.A.; Sayed, R.; Zhang, Q.; Scoggin, S.; Eliazer, S.; Rothenfluh, A.; Buszczak, M. Systematic discovery of genetic modulation by Jumonji histone demethylases in Drosophila. Sci. Rep. 2017, 7, 5240. [Google Scholar] [CrossRef] [PubMed]
- Pinzón, J.H.; Reed, A.R.; Shalaby, N.A.; Buszczak, M.; Rodan, A.R.; Rothenfluh, A. Alcohol-Induced Behaviors Require a Subset of Drosophila JmjC-Domain Histone Demethylases in the Nervous System. Alcohol. Clin. Exp. Res. 2017, 41, 2015–2024. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.-Q.; Sun, J.-J.; Wang, X.-F.; Li, M.-W.; Liu, X.-Z.; Wang, L.-Y.; Zhu, X.; Xiao, F.; Li, J.-M.; Gong, Y.; et al. HSPBAP1 is found extensively in the anterior temporal neocortex of patients with intractable epilepsy. Synapse 2007, 61, 741–747. [Google Scholar] [CrossRef]
- Türkmen, V.A.; Hintzen, J.C.J.; Tumber, A.; Moesgaard, L.; Salah, E.; Kongsted, J.; Schofield, C.J.; Mecinović, J. Substrate selectivity and inhibition of histidine JmjC hydroxylases MINA53 and NO66. RSC Chem. Biol. 2023, 4, 235–243. [Google Scholar] [CrossRef]
- Bräuer, K.E.; Brockers, K.; Moneer, J.; Feuchtinger, A.; Wollscheid-Lengeling, E.; Lengeling, A.; Wolf, A. Phylogenetic and genomic analyses of the ribosomal oxygenases Riox1 (No66) and Riox2 (Mina53) provide new insights into their evolution. BMC Evol. Biol. 2018, 18, 96. [Google Scholar] [CrossRef]
- Tsuneoka, M.; Koda, Y.; Soejima, M.; Teye, K.; Kimura, H. A novel myc target gene, mina53, that is involved in cell proliferation. J. Biol. Chem. 2002, 277, 35450–35459. [Google Scholar] [CrossRef]
- Eilbracht, J.; Reichenzeller, M.; Hergt, M.; Schnölzer, M.; Heid, H.; Stöhr, M.; Franke, W.W.; Schmidt-Zachmann, M.S. NO66, a highly conserved dual location protein in the nucleolus and in a special type of synchronously replicating chromatin. Mol. Biol. Cell 2004, 15, 1816–1832. [Google Scholar] [CrossRef]
- Eilbracht, J.; Kneissel, S.; Hofmann, A.; Schmidt-Zachmann, M.S. Protein NO52—A constitutive nucleolar component sharing high sequence homologies to protein NO66. Eur. J. Cell Biol. 2005, 84, 279–294. [Google Scholar] [CrossRef]
- Shiio, Y.; Donohoe, S.; Yi, E.C.; Goodlett, D.R.; Aebersold, R.; Eisenman, R.N. Quantitative proteomic analysis of Myc oncoprotein function. EMBO J. 2002, 21, 5088–5096. [Google Scholar] [CrossRef] [PubMed]
- Grandori, C.; Gomez-Roman, N.; Felton-Edkins, Z.A.; Ngouenet, C.; Galloway, D.A.; Eisenman, R.N.; White, R.J. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005, 7, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Yu, M.; Lu, Y.; Thakur, C.; Chen, B.; Qiu, P.; Zhao, H.; Chen, F. Carcinogenic metalloid arsenic induces expression of mdig oncogene through JNK and STAT3 activation. Cancer Lett. 2014, 346, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, Y.; Yuan, B.-Z.; Castranova, V.; Shi, X.; Stauffer, J.L.; Demers, L.M.; Chen, F. The Human mineral dust-induced gene, mdig, is a cell growth regulating gene associated with lung cancer. Oncogene 2005, 24, 4873–4882. [Google Scholar] [CrossRef] [PubMed]
- Thakur, C.; Chen, B.; Li, L.; Zhang, Q.; Yang, Z.-Q.; Chen, F. Loss of mdig expression enhances DNA and histone methylation and metastasis of aggressive breast cancer. Sig Transduct. Target. Ther. 2018, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.T.; Walport, L.J.; Hopkinson, R.J.; Madden, S.K.; Chowdhury, R.; Schofield, C.J.; Kawamura, A. Studies on the catalytic domains of multiple JmjC oxygenases using peptide substrates. Epigenetics 2014, 9, 1596–1603. [Google Scholar] [CrossRef] [PubMed]
- Mahon, P.C.; Hirota, K.; Semenza, G.L. FIH-1: A novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes. Dev. 2001, 15, 2675–2686. [Google Scholar] [CrossRef] [PubMed]
- Lando, D.; Peet, D.J.; Gorman, J.J.; Whelan, D.A.; Whitelaw, M.L.; Bruick, R.K. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes. Dev. 2002, 16, 1466–1471. [Google Scholar] [CrossRef]
- Dames, S.A.; Martinez-Yamout, M.; De Guzman, R.N.; Dyson, H.J.; Wright, P.E. Structural basis for Hif-1 alpha/CBP recognition in the cellular hypoxic response. Proc. Natl. Acad. Sci. USA 2002, 99, 5271–5276. [Google Scholar] [CrossRef]
- Jaakkola, P.; Mole, D.R.; Tian, Y.-M.; Wilson, M.I.; Gielbert, J.; Gaskell, S.J.; von Kriegsheim, A.; Hebestreit, H.F.; Mukherji, M.; Schofield, C.J.; et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001, 292, 468–472. [Google Scholar] [CrossRef]
- Lando, D.; Peet, D.J.; Whelan, D.A.; Gorman, J.J.; Whitelaw, M.L. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 2002, 295, 858–861. [Google Scholar] [CrossRef] [PubMed]
- Hewitson, K.S.; McNeill, L.A.; Riordan, M.V.; Tian, Y.-M.; Bullock, A.N.; Welford, R.W.; Elkins, J.M.; Oldham, N.J.; Bhattacharya, S.; Gleadle, J.M.; et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 2002, 277, 26351–26355. [Google Scholar] [CrossRef] [PubMed]
- Coleman, M.L.; McDonough, M.A.; Hewitson, K.S.; Coles, C.; Mecinovicí, J.; Edelmann, M.; Cook, K.M.; Cockman, M.E.; Lancaster, D.E.; Kessler, B.M.; et al. Asparaginyl Hydroxylation of the Notch Ankyrin Repeat Domain by Factor Inhibiting Hypoxia-inducible Factor. J. Biol. Chem. 2007, 282, 24027–24038. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, F.; Giaimo, B.D.; Friedrich, T.; Sugino, T.; Mertens, D.; Kugler, S.; Gahr, B.M.; Just, S.; Pan, L.; Bartkuhn, M.; et al. Hydroxylation of the NOTCH1 intracellular domain regulates Notch signaling dynamics. Cell Death Dis. 2022, 13, 600. [Google Scholar] [CrossRef]
- Cai, X.; Wang, R.; Zhu, J.; Li, X.; Liu, X.; Ouyang, G.; Wang, J.; Li, Z.; Zhu, C.; Deng, H.; et al. Factor inhibiting HIF negatively regulates antiviral innate immunity via hydroxylation of IKKϵ. Cell Rep. 2024, 43, 113606. [Google Scholar] [CrossRef]
- Wilkins, S.E.; Islam, S.; Gannon, J.M.; Markolovic, S.; Hopkinson, R.J.; Ge, W.; Schofield, C.J.; Chowdhury, R. JMJD5 is a human arginyl C-3 hydroxylase. Nat. Commun. 2018, 9, 1180. [Google Scholar] [CrossRef]
- Liu, Y.; Arai, A.; Kim, T.; Kim, S.; Park, N.-H.; Kim, R.H. Histone Demethylase Jmjd7 Negatively Regulates Differentiation of Osteoclast. Chin. J. Dent. Res. 2018, 21, 113–118. [Google Scholar] [CrossRef]
- Del Rizzo, P.A.; Krishnan, S.; Trievel, R.C. Crystal structure and functional analysis of JMJD5 indicate an alternate specificity and function. Mol. Cell Biol. 2012, 32, 4044–4052. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhou, X.; Wu, M.; Wang, C.; Zhang, X.; Tao, Y.; Chen, N.; Zang, J. Structure of the JmjC-domain-containing protein JMJD5. Acta Crystallogr. D Biol. Crystallogr. 2013, 69 Pt 10, 1911–1920. [Google Scholar] [CrossRef]
- Shen, J.; Liu, G.; Qi, H.; Xiang, X.; Shao, J. JMJD5 inhibits lung cancer progression by facilitating EGFR proteasomal degradation. Cell Death Dis. 2023, 14, 657. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, L.; Qi, H.; Shao, J.; Shen, J. Identification and functional implication of nuclear localization signals in the N-terminal domain of JMJD5. Biochimie 2013, 95, 2114–2122. [Google Scholar] [CrossRef] [PubMed]
- Vangimalla, S.S.; Ganesan, M.; Kharbanda, K.K.; Osna, N.A. Bifunctional Enzyme JMJD6 Contributes to Multiple Disease Pathogenesis: New Twist on the Old Story. Biomolecules 2017, 7, 41. [Google Scholar] [CrossRef] [PubMed]
- Böttger, A.; Islam, M.S.; Chowdhury, R.; Schofield, C.J.; Wolf, A. The oxygenase Jmjd6—A case study in conflicting assignments. Biochem. J. 2015, 468, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Fadok, V.A.; Bratton, D.L.; Rose, D.M.; Pearson, A.; Ezekewitz, R.A.B.; Henson, P.M. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature 2000, 405, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Cui, P.; Qin, B.; Liu, N.; Pan, G.; Pei, D. Nuclear localization of the phosphatidylserine receptor protein via multiple nuclear localization signals. Exp. Cell Res. 2004, 293, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.; Schmitz, C.; Böttger, A. Changing story of the receptor for phosphatidylserine-dependent clearance of apoptotic cells. EMBO Rep. 2007, 8, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Neumann, B.; oakley, S.; Giordano-Santini, R.; Linton, C.; Lee, E.S.; Nakagawa, A.; Xue, D.; Hilliard, M.A. EFF-1-mediated regenerative axonal fusion requires components of the apoptotic pathway. Nature 2015, 517, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Chen, Y.-Z.; Zhang, Y.; Wang, X.; Zhao, X.; Godfroy, J.I.; Liang, Q.; Zhang, M.; Zhang, T.; Yuan, Q.; et al. A lysine-rich motif in the phosphatidylserine receptor PSR-1 mediates recognition and removal of apoptotic cells. Nat. Commun. 2015, 6, 5717. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tsai, W.-C.; Reineke, L.C.; Jain, A.; Jung, S.Y.; Lloyd, R.E. Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule-nucleating protein G3BP1. J. Biol. Chem. 2017, 292, 18886–18896. [Google Scholar] [CrossRef]
- Han, G.; Li, J.; Wang, Y.; Li, X.; Mao, H.; Liu, Y.; Chen, C.D. The hydroxylation activity of Jmjd6 is required for its homo-oligomerization. J. Cell. Biochem. 2011, 113, 1663–1670. [Google Scholar] [CrossRef]
- Mantri, M.; Krojer, T.; Bagg, E.A.; Webby, C.J.; Butler, D.S.; Kochan, G.; Kavanagh, K.L.; Oppermann, U.; McDonough, M.A.; Schofield, C.J. Crystal Structure of the 2-Oxoglutarate- and Fe(II)-Dependent Lysyl Hydroxylase JMJD6. J. Mol. Biol. 2010, 401, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Unoki, M.; Masuda, A.; Dohmae, N.; Arita, K.; Yoshimatsu, M.; Iwai, Y.; Fukui, Y.; Ueda, K.; Hamamoto, R.; Shirakawa, M.; et al. Lysyl 5-Hydroxylation, a Novel Histone Modification, by Jumonji Domain Containing 6 (JMJD6). J. Biol. Chem. 2013, 288, 6053–6062. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; McDonough, M.A.; Chowdhury, R.; Gault, J.; Khan, A.; Pires, E.; Schofield, C.J. Biochemical and structural investigations clarify the substrate selectivity of the 2-oxoglutarate oxygenase JMJD6. J. Biol. Chem. 2019, 294, 11637–11652. [Google Scholar] [CrossRef] [PubMed]
- Cockman, M.E.; Sugimoto, Y.; Pegg, H.B.; Masson, N.; Salah, E.; Tumber, A.; Flynn, H.R.; Kirkpatrick, J.M.; Schofield, C.J.; Ratcliffe, P.J. Widespread hydroxylation of unstructured lysine-rich protein domains by JMJD6. Proc. Natl. Acad. Sci. USA 2022, 119, e2201483119. [Google Scholar] [CrossRef] [PubMed]
- Boeckel, J.-N.; Guarani, V.; Koyanagi, M.; Roexe, T.; Lengeling, A.; Schermuly, R.T.; Gellert, P.; Braun, T.; Zeiher, A.; Dimmeler, S. Jumonji domain-containing protein 6 (Jmjd6) is required for angiogenic sprouting and regulates splicing of VEGF-receptor 1. Proc. Natl. Acad. Sci. USA 2011, 108, 3276–3281. [Google Scholar] [CrossRef] [PubMed]
- Heim, A.; Grimm, C.; Müller, U.; Häußler, S.; Mackeen, M.M.; Merl, J.; Hauck, S.M.; Kessler, B.M.; Schofield, C.J.; Wolf, A.; et al. Jumonji domain containing protein 6 (Jmjd6) modulates splicing and specifically interacts with arginine-serine-rich (RS) domains of SR- and SR-like proteins. Nucleic Acids Res. 2014, 42, 7833–7850. [Google Scholar] [CrossRef] [PubMed]
- Markolovic, S.; Leissing, T.M.; Chowdhury, R.; Wilkins, S.E.; Lu, X.; Schofield, C.J. Structure–function relationships of human JmjC oxygenases—Demethylases versus hydroxylases. Curr. Opin. Struct. Biol. 2016, 41, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Long, Y.-H.; Wang, S.-Q.; Zhang, Y.-Y.; Li, Y.-F.; Mi, J.-S.; Yu, C.-H.; Li, D.-Y.; Zhang, J.-H.; Zhang, X.-J. JMJD6 regulates histone H2A.X phosphorylation and promotes autophagy in triple-negative breast cancer cells via a novel tyrosine kinase activity. Oncogene 2019, 38, 980–997. [Google Scholar] [CrossRef] [PubMed]
- Huo, D.; Chen, H.; Cheng, Y.; Song, X.; Zhang, K.; Li, M.J.; Xuan, C. JMJD6 modulates DNA damage response through downregulating H4K16ac independently of its enzymatic activity. Cell Death Differ. 2020, 27, 1052–1106. [Google Scholar] [CrossRef]
- Fages, J.; Chailleux, C.; Humbert, J.; Jang, S.-M.; Loehr, J.; Lambert, J.-P.; Côté, J.; Trouche, D.; Canitrot, Y. JMJD6 participates in the maintenance of ribosomal DNA integrity in response to DNA damage. PLoS Genet. 2020, 16, e1008511. [Google Scholar] [CrossRef]
- Yeo, K.S.; Tan, M.C.; Lim, Y.-Y.; Ea, C.-K. JMJD8 is a novel endoplasmic reticulum protein with a JmjC domain. Sci. Rep. 2017, 7, 15407. [Google Scholar] [CrossRef] [PubMed]
- Yeo, K.S.; Tan, M.C.; Wong, W.Y.; Loh, S.W.; Lam, Y.L.; Tan, C.L.; Lim, Y.-Y.; Ea, C.-K. JMJD8 is a positive regulator of TNF-induced NF-κB signaling. Sci. Rep. 2016, 6, 34125. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Wang, L.; Du, J.; Wang, M.; Shen, H.; Liu, Z.; Qin, Y.; Liu, J.; Hu, G.; Xiao, R.; et al. ER-localized JmjC domain-containing protein JMJD8 targets STING to promote immune evasion and tumor growth in breast cancer. Dev. Cell 2023, 58, 760–778.e6. [Google Scholar] [CrossRef] [PubMed]
- Noma, A.; Ishitani, R.; Kato, M.; Nagao, A.; Nureki, O.; Suzuki, T. Expanding role of the jumonji C domain as an RNA hydroxylase. J. Biol. Chem. 2010, 285, 34503–34507. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, X.; Zhao, L.; Liang, R.; Deng, W.; Guo, W.; Wang, Q.; Hu, X.; Du, X.; Sham, P.C.; et al. Comprehensive and integrative analyses identify TYW5 as a schizophrenia risk gene. BMC Med. 2022, 20, 169. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.G.; Villa, R.; Trojer, P.; Norman, J.; Yan, K.-P.; Reinberg, D.; Di Croce, L.; Shiekhattar, R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 2007, 318, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, A. The UTX gene escapes X inactivation in mice and humans. Human. Mol. Genet. 1998, 7, 737–742. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; Mohn, S.E.; Weinmann, A.S. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell 2010, 40, 594–605. [Google Scholar] [CrossRef]
- Wang, C.; Lee, J.-E.; Cho, Y.-W.; Xiao, Y.; Jin, Q.; Liu, C.; Ge, K. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc. Natl. Acad. Sci. USA 2012, 109, 15324–15329. [Google Scholar] [CrossRef]
- Tran, N.; Broun, A.; Ge, K. Lysine Demethylase KDM6A in Differentiation, Development, and Cancer. Mol. Cell Biol. 2020, 40, e00341-20. [Google Scholar] [CrossRef]
- Vandamme, J.; Lettier, G.; Sidoli, S.; Di Schiavi, E.; Jensen, O.N.; Salcini, A.E. The C. elegans H3K27 Demethylase UTX-1 Is Essential for Normal Development, Independent of Its Enzymatic Activity. PLoS Genet. 2012, 8, e1002647. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, H.C.; Sabari, B.R. Disordered and dead, but in good company: How a catalytically inactive UTX retains its function. Mol. Cell 2021, 81, 4577–4578. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Shi, B.; Li, W.; Li, W.; Song, Y.; Song, Y.; Wang, Z.; Wang, Z.; Ju, R.; Ju, R.; et al. UTX condensation underlies its tumour-suppressive activity. Nature 2021, 597, 726–731. [Google Scholar] [CrossRef]
- Wang, S.-P.; Tang, Z.; Chen, C.-W.; Shimada, M.; Koche, R.P.; Wang, L.-H.; Nakadai, T.; Chramiec, A.; Krivtsov, A.V.; Armstrong, S.A.; et al. A UTX-MLL4-p300 Transcriptional Regulatory Network Coordinately Shapes Active Enhancer Landscapes for Eliciting Transcription. Mol. Cell 2017, 67, 308–321.e6. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Li, Q.; Ayers, S.; Gu, Y.; Shi, Z.; Zhu, Q.; Chen, Y.; Wang, H.Y.; Wang, R.-F. Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination. Cell 2013, 152, 1037–1050. [Google Scholar] [CrossRef] [PubMed]
- Lan, H.; Tan, M.; Zhang, Q.; Yang, F.; Wang, S.; Li, H.; Xiong, X.; Sun, Y. LSD1 destabilizes FBXW7 and abrogates FBXW7 functions independent of its demethylase activity. Proc. Natl. Acad. Sci. USA 2019, 116, 12311–12320. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Lin, Y.; Wang, Z.; Wu, X.; Ye, Z.; Wang, Y.; Lan, H. Biological roles of LSD1 beyond its demethylase activity. Cell Mol. Life Sci. 2020, 77, 3341–3350. [Google Scholar] [CrossRef]
- Welcker, M.; Clurman, B. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 2008, 8, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Chao, A.; Lin, C.-Y.; Chao, A.-N.; Tsai, C.-L.; Chen, M.-Y.; Lee, L.-Y.; Chang, T.-C.; Wang, T.-H.; Lai, C.-H.; Wang, H.-S. Lysine-specific demethylase 1 (LSD1) destabilizes p62 and inhibits autophagy in gynecologic malignancies. Oncotarget 2017, 8, 74434–74450. [Google Scholar] [CrossRef]
- Carnesecchi, J.; Forcet, C.; Zhang, L.; Tribollet, V.; Barenton, B.; Boudra, R.; Cerutti, C.; Billas, I.M.L.; Sérandour, A.A.; Carroll, J.S.; et al. ERRα induces H3K9 demethylation by LSD1 to promote cell invasion. Proc. Natl. Acad. Sci. USA 2017, 114, 3909–3914. [Google Scholar] [CrossRef]
- Zeng, C.; Chen, J.; Cooke, E.W.; Subuddhi, A.; Roodman, E.T.; Chen, F.X.; Cao, K. Demethylase-independent roles of LSD1 in regulating enhancers and cell fate transition. Nat. Commun. 2023, 14, 4944. [Google Scholar] [CrossRef] [PubMed]
- Kumaraswamy, A.; Duan, Z.; Flores, D.; Zhang, C.; Sehrawat, A.; Hu, Y.-M.; Swaim, O.A.; Rodansky, E.; Storck, W.K.; Kuleape, J.A.; et al. LSD1 promotes prostate cancer reprogramming by repressing TP53 signaling independently of its demethylase function. JCI Insight 2023, 8, e167440. [Google Scholar] [CrossRef]
- Marayati, B.F.; Tucker, J.F.; De La Cerda, D.A.; Hou, T.-C.; Chen, R.; Sugiyama, T.; Pease, J.B.; Zhang, K. The Catalytic-Dependent and -Independent Roles of Lsd1 and Lsd2 Lysine Demethylases in Heterochromatin Formation in Schizosaccharomyces pombe. Cells 2020, 9, 955. [Google Scholar] [CrossRef]
- Yang, Z.; Jiang, J.; Stewart, D.M.; Qi, S.; Yamane, K.; Li, J.; Zhang, Y.; Wong, J. AOF1 is a histone H3K4 demethylase possessing demethylase activity-independent repression function. Cell Res. 2010, 20, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Farcas, A.M.; Blackledge, N.P.; Sudbery, I.; Long, H.K.; McGouran, J.F.; Rose, N.R.; Lee, S.; Sims, D.; Cerase, A.; Sheahan, T.W.; et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife 2012, 1, e00205. [Google Scholar] [CrossRef]
- He, J.; Shen, L.; Wan, M.; Taranova, O.; Wu, H.; Zhang, Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat. Cell Biol. 2013, 15, 373–384. [Google Scholar] [CrossRef]
- Taylor-Papadimitriou, J.; Burchell, J.M. Histone Methylases and Demethylases Regulating Antagonistic Methyl Marks: Changes Occurring in Cancer. Cells 2022, 11, 1113. [Google Scholar] [CrossRef] [PubMed]
- Couture, J.-F.; Collazo, E.; Ortiz-Tello, P.A.; Brunzelle, J.S.; Trievel, R.C. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 2007, 14, 689–695. [Google Scholar] [CrossRef]
- Huang, Y.; Fang, J.; Bedford, M.T.; Zhang, Y.; Xu, R.-M. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 2006, 312, 748–751. [Google Scholar] [CrossRef]
- Johmura, Y.; Sun, J.; Kitagawa, K.; Nakanishi, K.; Kuno, T.; Naiki-Ito, A.; Sawada, Y.; Miyamoto, T.; Okabe, A.; Aburatani, H.; et al. SCF(Fbxo22)-KDM4A targets methylated p53 for degradation and regulates senescence. Nat. Commun. 2016, 7, 10574. [Google Scholar] [CrossRef]
- Shi, L.; Sun, L.; Li, Q.; Liang, J.; Yu, W.; Yi, X.; Yang, X.; Li, Y.; Han, X.; Zhang, Y.; et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 7541–7546. [Google Scholar] [CrossRef] [PubMed]
- Kawazu, M.; Saso, K.; Tong, K.I.; McQuire, T.; Goto, K.; Son, D.-O.; Wakeham, A.; Miyagishi, M.; Mak, T.W.; Okada, H. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS ONE 2011, 6, e17830. [Google Scholar] [CrossRef]
- Gaughan, L.; Stockley, J.; Coffey, K.; O’neill, D.; Jones, D.L.; Wade, M.; Wright, J.; Moore, M.; Tse, S.; Rogerson, L.; et al. KDM4B is a master regulator of the estrogen receptor signalling cascade. Nucleic Acids Res. 2013, 41, 6892–6904. [Google Scholar] [CrossRef] [PubMed]
- Johmura, Y.; Maeda, I.; Suzuki, N.; Wu, W.; Goda, A.; Morita, M.; Yamaguchi, K.; Yamamoto, M.; Nagasawa, S.; Kojima, Y.; et al. Fbxo22-mediated KDM4B degradation determines selective estrogen receptor modulator activity in breast cancer. J. Clin. Investig. 2018, 128, 5603–5619. [Google Scholar] [CrossRef] [PubMed]
- Johmura, Y.; Harris, A.S.; Ohta, T.; Nakanishi, M. FBXO22, an epigenetic multiplayer coordinating senescence, hormone signaling, and metastasis. Cancer Sci. 2020, 111, 2718–2725. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, S.; Catchpole, S.; Chen, J.; So, C.W.E.; Burchell, J.; Rahman, K.M.; Taylor-Papadimitriou, J. KDM5B protein expressed in viable and fertile ΔARID mice exhibit no demethylase activity. Int. J. Oncol. 2021, 59, 96. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Kim, A.; Huang, Y.-T.; Pi, W.-C.; Gong, W.; Yu, X.; Qi, J.; Jin, J.; Cai, L.; Roeder, R.G.; et al. A PRC2-Kdm5b axis sustains tumorigenicity of acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2022, 119, e2122940119. [Google Scholar] [CrossRef]
- Zhang, S.-M.; Cai, W.L.; Liu, X.; Thakral, D.; Luo, J.; Chan, L.H.; McGeary, M.K.; Song, E.; Blenman, K.R.M.; Micevic, G.; et al. KDM5B promotes immune evasion by recruiting SETDB1 to silence retroelements. Nature 2021, 598, 682–687. [Google Scholar] [CrossRef]
- DiTacchio, L.; Le, H.D.; Vollmers, C.; Hatori, M.; Witcher, M.; Secombe, J.; Panda, S. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science 2011, 333, 1881–1885. [Google Scholar] [CrossRef]
- Gaillard, S.; Charasson, V.; Ribeyre, C.; Salifou, K.; Pillaire, M.-J.; Hoffmann, J.-S.; Constantinou, A.; Trouche, D.; Vandromme, M. KDM5A and KDM5B histone-demethylases contribute to HU-induced replication stress response and tolerance. Biol. Open 2021, 10, bio057729. [Google Scholar] [CrossRef]
- Ohguchi, Y.; Ohguchi, H. Diverse Functions of KDM5 in Cancer: Transcriptional Repressor or Activator? Cancers 2022, 14, 3270. [Google Scholar] [CrossRef] [PubMed]
- Pippa, S.; Mannironi, C.; Licursi, V.; Bombardi, L.; Colotti, G.; Cundari, E.; Mollica, A.; Coluccia, A.; Naccarato, V.; La Regina, G.; et al. Small Molecule Inhibitors of KDM5 Histone Demethylases Increase the Radiosensitivity of Breast Cancer Cells Overexpressing JARID1B. Molecules 2019, 24, 1739. [Google Scholar] [CrossRef] [PubMed]
- Pavlenko, E.; Ruengeler, T.; Engel, P.; Poepsel, S. Functions and Interactions of Mammalian KDM5 Demethylases. Front. Genet. 2022, 13, 906662. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Lam, Y.M.; He, Z.; Zheng, Y.; Li, L.; Zhang, Y.; Li, C.; Mbadhi, M.N.; Zheng, L.; Cheng, Z.; et al. Histone lysine demethylase KDM5B maintains chronic myeloid leukemia via multiple epigenetic actions. Exp. Hematol. 2020, 82, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Secombe, J. The Histone Demethylase KDM5 Activates Gene Expression by Recognizing Chromatin Context through Its PHD Reader Motif. Cell Rep. 2015, 13, 2219–2231. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.F.; Marshall, O.J.; Secombe, J. KDM5-mediated activation of genes required for mitochondrial biology is necessary for viability in Drosophila. Development 2023, 150, dev202024. [Google Scholar] [CrossRef]
- Deogharia, M.; Venegas-Zamora, L.; Agrawal, A.; Shi, M.; Jain, A.K.; McHugh, K.J.; Altamirano, F.; Marian, A.J.; Gurha, P. Histone demethylase KDM5 regulates cardiomyocyte maturation by promoting fatty acid oxidation, oxidative phosphorylation, and myofibrillar organization. Cardiovasc. Res. 2024, 120, 630–643. [Google Scholar] [CrossRef] [PubMed]
- Yheskel, M.; Sidoli, S.; Secombe, J. Proximity labeling reveals a new in vivo network of interactors for the histone demethylase KDM5. Epigenetics Chromatin 2023, 16, 8. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.R.; Engstrom, A.; Zoeller, E.L.; Liu, X.; Shanks, J.R.; Zhang, X.; Johns, M.A.; Vertino, P.M.; Fu, H.; Cheng, X.l. Characterization of a Linked Jumonji Domain of the KDM5/JARID1 Family of Histone H3 Lysine 4 Demethylases. J. Biol. Chem. 2016, 291, 2631–2646. [Google Scholar] [CrossRef]
- Johansson, C.; Velupillai, S.; Tumber, A.; Szykowska, A.; Hookway, E.S.; Nowak, R.P.; Strain-Damerell, C.; Gileadi, C.; Philpott, M.; Burgess-Brown, N.; et al. Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat. Chem. Biol. 2016, 12, 539–545. [Google Scholar] [CrossRef]
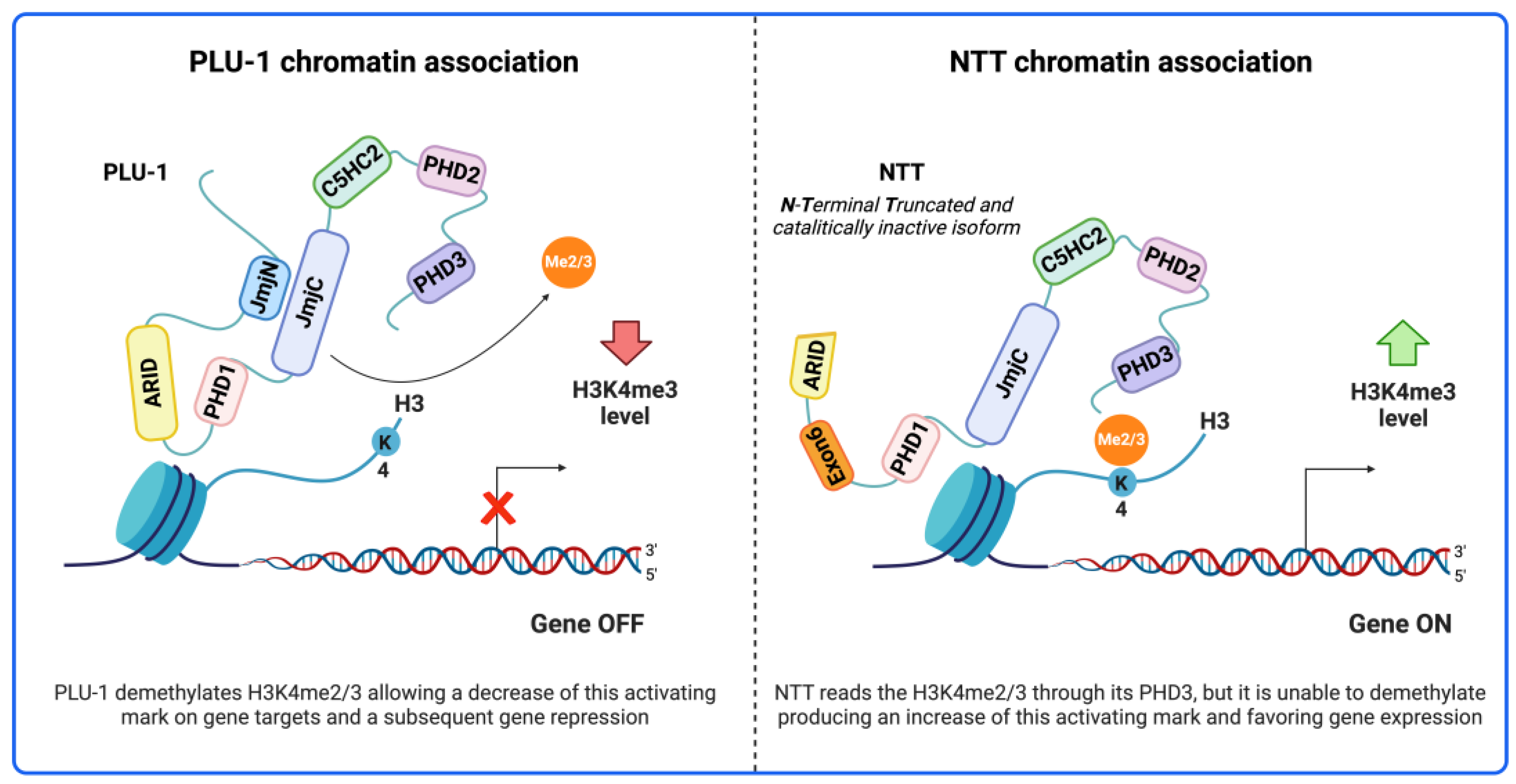
- Di Nisio, E.; Licursi, V.; Mannironi, C.; Buglioni, V.; Paiardini, A.; Robusti, G.; Noberini, R.; Bonaldi, T.; Negri, R. A truncated and catalytically inactive isoform of KDM5B histone demethylase accumulates in breast cancer cells and regulates H3K4 tri-methylation and gene expression. Cancer Gene Ther. 2023, 30, 822–832. [Google Scholar] [CrossRef] [PubMed]
- Roesch, A.; Becker, B.; Meyer, S.; Wild, P.; Hafner, C.; Landthaler, M.; Vogt, T. Retinoblastoma-binding protein 2-homolog 1: A retinoblastoma-binding protein downregulated in malignant melanomas. Mod. Pathol. 2005, 18, 1249–1257. [Google Scholar] [CrossRef] [PubMed]
- National Library of Medicine (US), National Center for Biotechnology Information. NCBI Gene RefSeq Database; National Library of Medicine (US), National Center for Biotechnology Information: Bethesda, MD, USA, 1988. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 6 April 2017).

| Jumonji Histone Demethylases with No or Uncertain Histone Demethylase Activity | ||||||
| Official Symbol (Synonyms) | Protein Name | Uniprot Entry | Protein Domains | Substrates | Catalytic Activity | Refs |
| JARID2 (JMJ, DIDDF7) | Protein Jumonji | Q92833 |  | - | None | [8,14] |
| UTY (KDM6C, KDM6AL) | Ubiquitously transcribed tetratricopeptide repeat containing, Y-linked | O14607 |  | H3K27me2/3 | Demethylase (uncertain) | [15,16,17] |
| JmjC-Only Domain Proteins with Different Catalytic Functions | ||||||
| Official Symbol (Synonyms) | Protein Name | Uniprot Entry | Protein Domains | Substrates | Catalytic Activity | Refs |
| HSPBAP1 (PASS1) | HSPB1-associated protein 1 | Q96EW2 |  | HSP27 | Not investigated | [18] |
| RIOX1 (MAPJD, NO66, C14orf169, JMJD9) | Ribosomal oxygenase 1 | Q9H6W3 |  | RPL8, H3K4me1/2/3, H3K36me2/3 | Hydroxylase, Demethylase (uncertain) | [12,19,20,21,22] |
| RIOX2 (MINA53, NO52, JMJD10, MINA, MDIG) | Ribosomal oxygenase 2 | Q8IUF8 |  | RPL27A, H3K9me3 | Hydroxylase, Demethylase (uncertain) | [12,20,21,22,23] |
| HIF1AN (FIH) | Hypoxia-inducible factor 1 subunit alpha inhibitor | Q9NWT6 |  | HIF1-2α, self hydroxylation at Trp-296, ankyrin repeat domain (ARD)-containing proteins | Hydroxylase | [24,25] |
| JMJD4 | Jumonji domain-containing 4 | Q9H9V9 |  | eRF1, PKM2 | Hydroxylase | [26,27] |
| KDM8 (JMJD5) | Lysine demethylase 8 | Q8N371 |  | H3K9me1, HEK36me2, NFATc1 | Hydroxylase, Demethylase? Protease? | [28,29,30,31] |
| JMJD6 (KIAA0585, PTDSR1, PTDSR, PSR) | Jumonji domain-containing 6 | Q6NYC1 |  | Arginines, lysines H3R2me, H4R3me, 7SK, MePCE, p53, GLS RNA | Hydroxylase, Demethylase (uncertain), Protease? | [31,32,33,34,35,36,37] |
| JMJD7 | Jumonji domain-containing 7 | P0C870 |  | H3R2me2, H4R3me2, DRG1/2 | Hydroxylase, Protease? | [31,38] |
| JMJD8 (C16orf20, PP14397) | Jumonji domain-containing 8 | Q96S16 |  | AKT1 | Demethylase | [39,40] |
| TYW5 (C2orf60, HTYW5) | tRNA wybutosine-synthesizing protein 5 | A2RUC4 |  | yW-72 in tRNA(Phe) | Hydroxylase | [41] |
| Catalytically Active KDMs with Catalytic-Independent Functions | ||||||
| Official Symbol (Synonyms) | Protein Name | Uniprot Entry | Protein Domains | Substrates | Catalytic Activity | Refs |
| KDM1A (LSD1, AOF2, BHC110, CPRF, KDM1, KIAA0601) | Lysine-specific histone demethylase 1A | O60341 |  | H3K4me1/2 H3K9me1/2 | Demethylase | [8,42] |
| KDM1B (LSD2, AOF1, C6orf193) | Lysine-specific histone demethylase 2 | Q8NB78 |  | H3K4me1/2 | Demethylase | [43,44,45] |
| KDM2B (CXXC2, FBL10, FBXL10, JHDM1B, NDY1, PCCX2) | Lysine-specific demethylase 2B | Q8NHM5 | 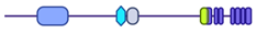 | H3K4me3 H3K36me2 H3K79me2/3 | Demethylase | [46,47,48,49] |
| KDM4A (JHDM3A, JMJD2, JMJD2A, KIAA0677, TDRD14A) | Lysine-specific demethylase 4A | O75164 |  | H3K9me3 H3K36me3 H1.4K26 | Demethylase | [8,50,51] |
| KDM4B (JHDM3B, JMJD2B, KIAA0876, MRD65, TDRD14B) | Lysine-specific demethylase 4B | O94953 |  | H3K9me3 | Demethylase | [52] |
| KDM5A (RBP2, RBBP2, JARID1A) | Lysine-specific demethylase 5A | P29375 |  | H3K4me2/3 | Demethylase | [53] |
| KDM5B (PLU-1, CT31, RBBP2H1A, PPP1R98, JARID1B) | Lysine-specific demethylase 5B | Q9UGL1 | 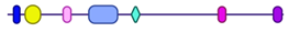 | H3K4me2/3 | Demethylase | [54,55] |
| KDM6A (UTX) | Lysine-specific demethylase 6A | O15550 |  | H3K27me2/3 | Demethylase | [56,57,58] |
| KDM6B (JMJD3, KIAA0346) | Lysine-specific demethylase 6B | O15054 |  | H3K27me2/3 | Demethylase | [56,57,58,59] |
 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Nisio, E.; Manzini, V.; Licursi, V.; Negri, R. To Erase or Not to Erase: Non-Canonical Catalytic Functions and Non-Catalytic Functions of Members of Histone Lysine Demethylase Families. Int. J. Mol. Sci. 2024, 25, 6900. https://doi.org/10.3390/ijms25136900
Di Nisio E, Manzini V, Licursi V, Negri R. To Erase or Not to Erase: Non-Canonical Catalytic Functions and Non-Catalytic Functions of Members of Histone Lysine Demethylase Families. International Journal of Molecular Sciences. 2024; 25(13):6900. https://doi.org/10.3390/ijms25136900
Chicago/Turabian StyleDi Nisio, Elena, Valeria Manzini, Valerio Licursi, and Rodolfo Negri. 2024. "To Erase or Not to Erase: Non-Canonical Catalytic Functions and Non-Catalytic Functions of Members of Histone Lysine Demethylase Families" International Journal of Molecular Sciences 25, no. 13: 6900. https://doi.org/10.3390/ijms25136900
APA StyleDi Nisio, E., Manzini, V., Licursi, V., & Negri, R. (2024). To Erase or Not to Erase: Non-Canonical Catalytic Functions and Non-Catalytic Functions of Members of Histone Lysine Demethylase Families. International Journal of Molecular Sciences, 25(13), 6900. https://doi.org/10.3390/ijms25136900






