Bioactivity of Eugenol: A Potential Antibiotic Adjuvant with Minimal Ecotoxicological Impact
Abstract
:1. Introduction
- (a)
- To assess the antimicrobial activity of EUG alone and its ability to enhance the bacterial inhibitory effect of commercial ABXs against clinically relevant Gram-positive and Gram-negative bacteria to find the synergistic combinations (in which the EUG would be the adjuvant) lowering MIC and thus allowing the reduction of ABXs’ doses.
- (b)
- To quantify the environmental impact of EUG by employing terrestrial and aquatic individual indicators alongside genetically sequenced microbial communities sourced from soil and river environments to obtain a more realistic environmental perspective of its potential use as an ABX adjuvant.
2. Results and Discussion
2.1. Assessment of Eugenol Antimicrobial Activity
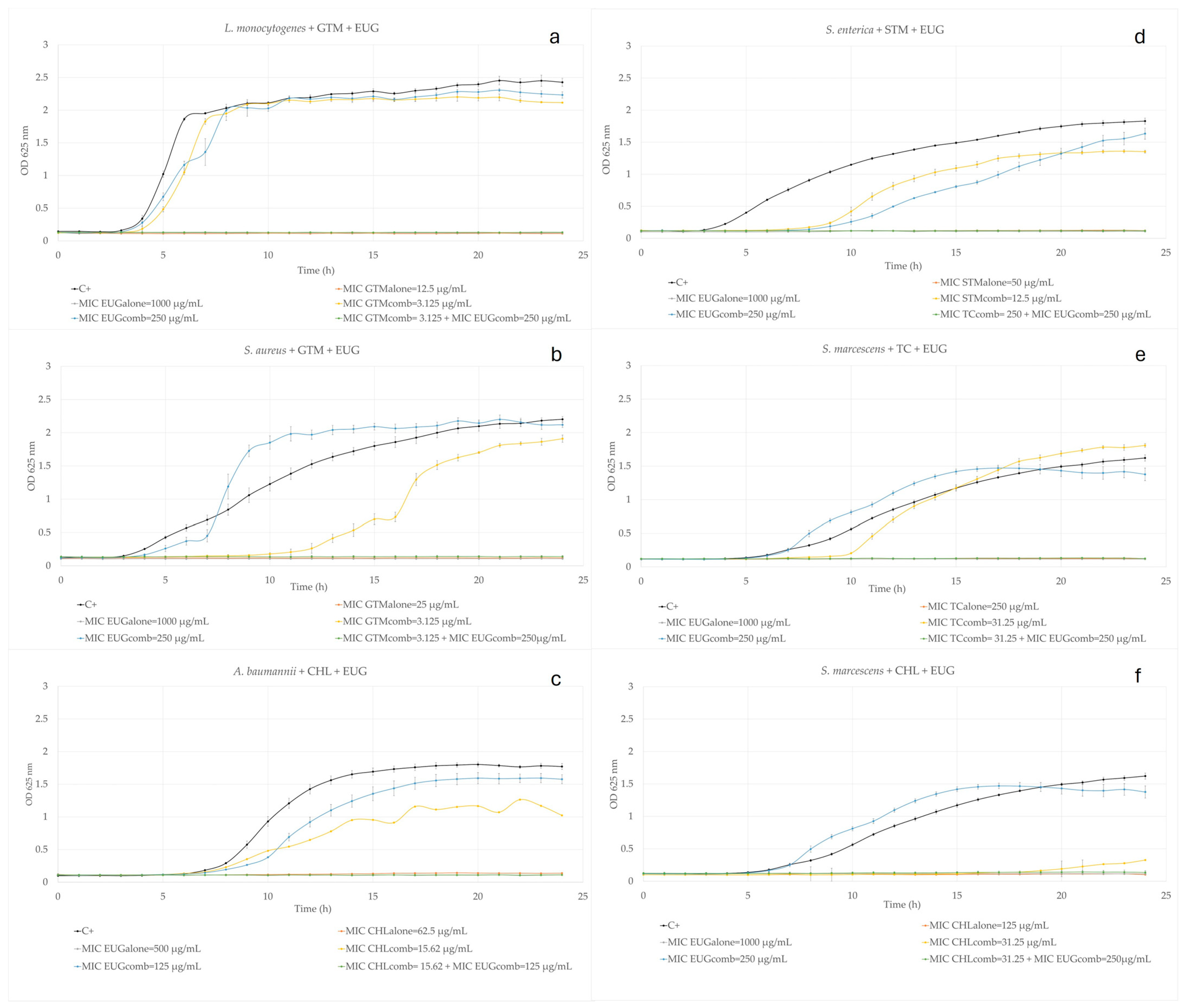
2.2. Eugenol Combined with Commercial Antibiotics
2.2.1. Aminoglycosides
2.2.2. Beta-Lactams
2.2.3. Amphenicols
2.2.4. Tetracyclines
2.2.5. Macrolides
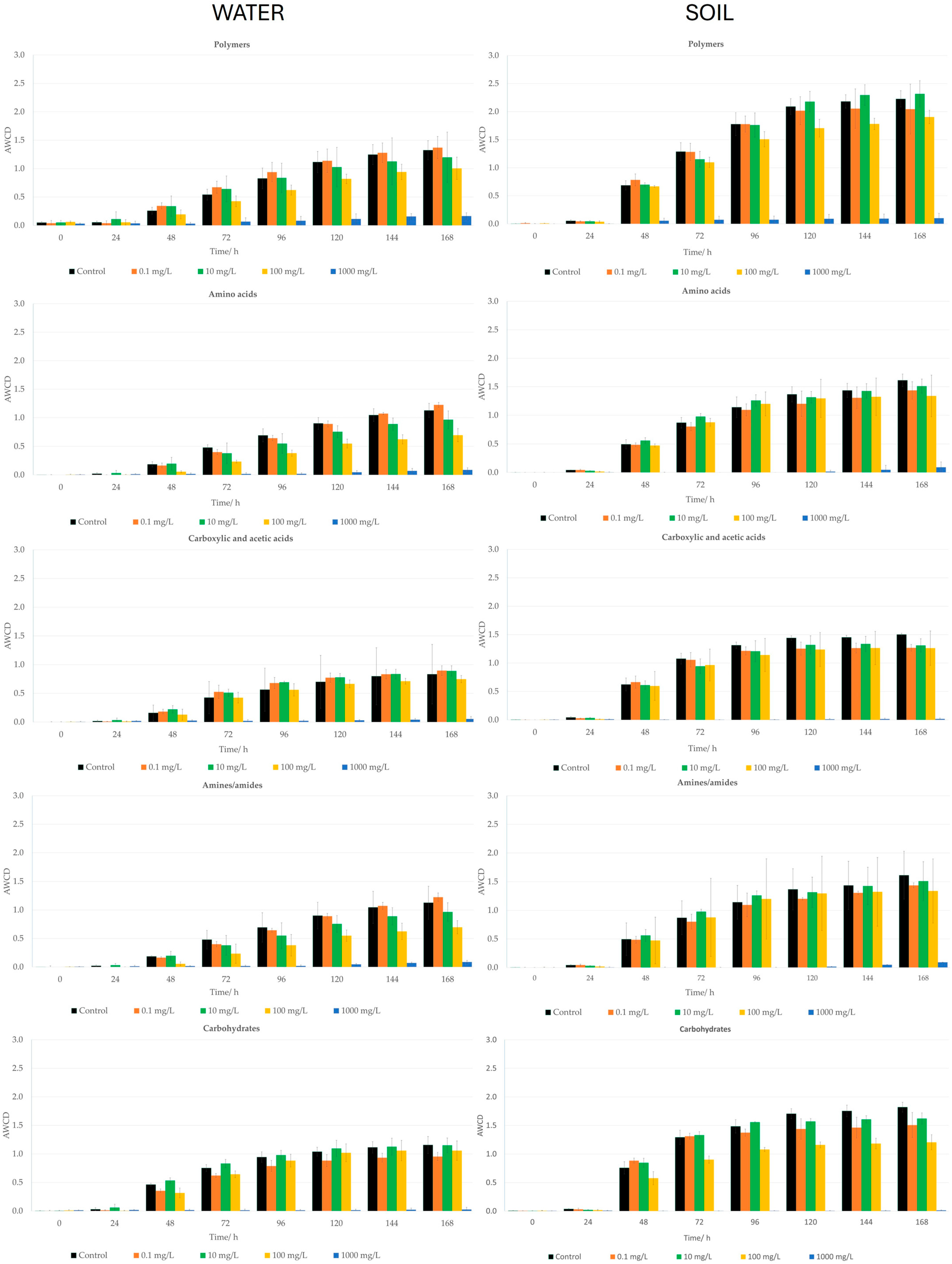
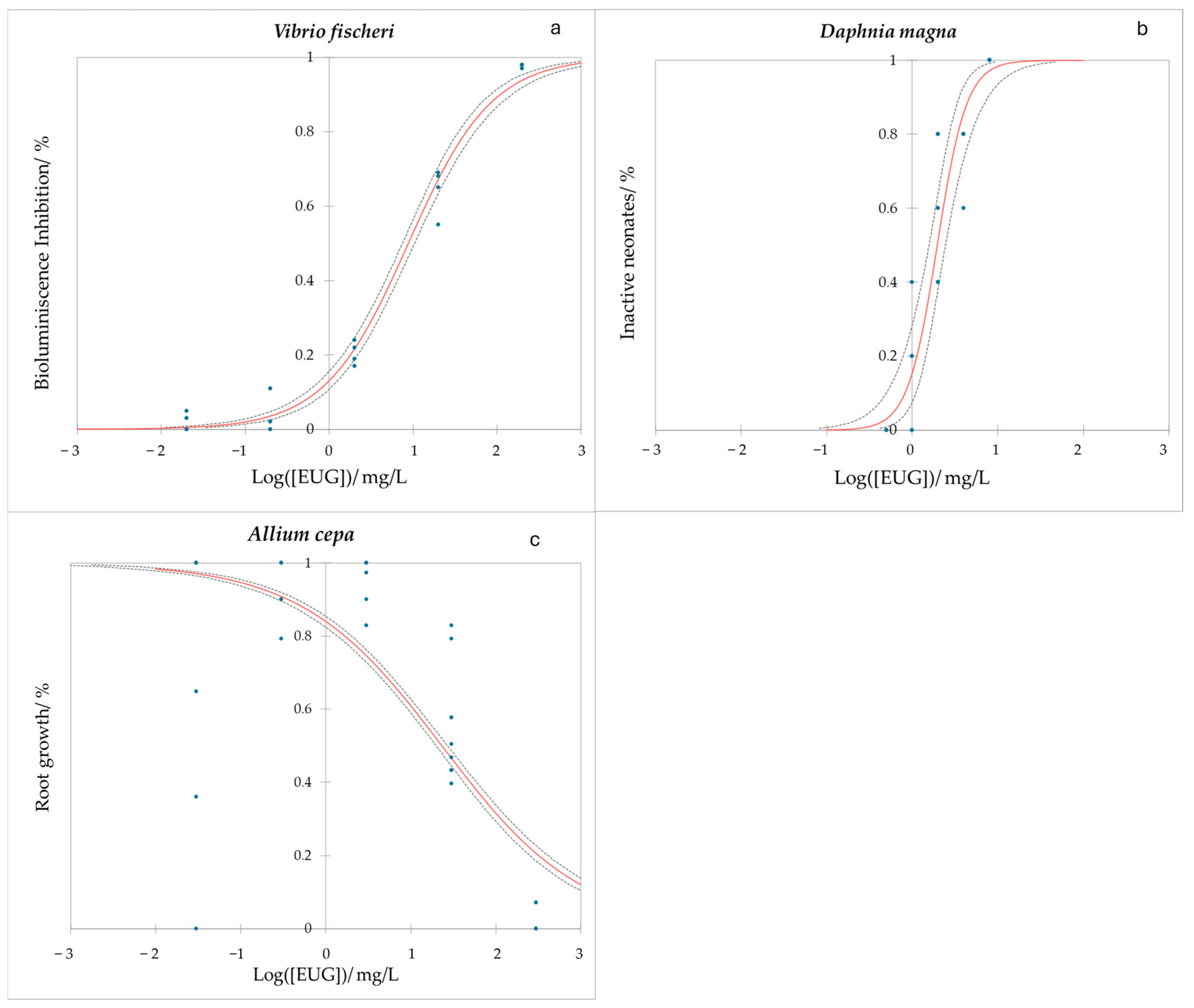
2.3. Eugenol Ecotoxicity
2.3.1. Water Ecotoxicity of Eugenol
- Average Well Color Development (AWCD) of water microbial populations and Community-Level Physiological Profiling (CLPP) after treatment with eugenol.
- Effect of EUG on water non-target organisms: Vibrio fischeri and Daphnia magna.
2.3.2. Soil Ecotoxicity of Eugenol
- Average Well Color Development (AWCD) of soil microbial populations and Community-Level Physiological Profiling (CLPP) after treatment with eugenol.
- Soil non-target organisms: Eisenia foetida and Allium cepa.
3. Materials and Methods
3.1. Reagents
3.2. Bacterial Strains Growth
3.3. Minimum Inhibitory Concentration (MIC)
3.4. Bacterial Checkboard Assay
3.5. Bacterial Kinetic Growth Assay
3.6. Average Well Color Development (AWCD) Tests of Soil and Water Microorganisms
3.6.1. Water Samples
3.6.2. Soil Samples
3.6.3. Biolog Ecoplates Preparation
3.7. Genetic Analysis of Water and Soil Sample
3.8. Vibrio Fischeri Bioluminiscence Assay
3.9. Daphnia Magna Assay
3.10. Eisenia Foetida Assay
3.11. Allium Cepa Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural Products in Drug Discovery: Advances and Opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Ferrando, N.; Pino-Otín, M.R.; Ballestero, D.; Lorca, G.; Terrado, E.M.; Langa, E. Enhancing Commercial Antibiotics with Trans-Cinnamaldehyde in Gram-Positive and Gram-Negative Bacteria: An In Vitro Approach. Plants 2024, 13, 192. [Google Scholar] [CrossRef]
- Bergman, M.E.; Davis, B.; Phillips, M.A. Medically Useful Plant Terpenoids: Biosynthesis, Occurrence, and Mechanism of Action. Molecules 2019, 24, 3961. [Google Scholar] [CrossRef] [PubMed]
- Song, M.C.; Kim, E.J.; Kim, E.; Rathwell, K.; Nam, S.J.; Yoon, Y.J. Microbial Biosynthesis of Medicinally Important Plant Secondary Metabolites. Nat. Prod. Rep. 2014, 31, 1497–1509. [Google Scholar] [CrossRef]
- Gallucci, M.N.; Oliva, M.; Casero, C.; Dambolena, J.; Luna, A.; Zygadlo, J.; Demo, M. Antimicrobial Combined Action of Terpenes against the Food-Borne Microorganisms Escherichia coli, Staphylococcus aureus and Bacillus cereus. Flavour Fragr. J. 2009, 24, 348–354. [Google Scholar] [CrossRef]
- Eugenol|C10H12O2|CID 3314—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/3314#section=Solubility (accessed on 2 April 2024).
- Dhar, N.; Sarangapani, S.; Reddy, V.A.; Kumar, N.; Panicker, D.; Jin, J.; Chua, N.H.; Sarojam, R. Characterization of a Sweet Basil Acyltransferase Involved in Eugenol Biosynthesis. J. Exp. Bot. 2020, 71, 3638–3652. [Google Scholar] [CrossRef]
- Dexter, R.; Qualley, A.; Kish, C.M.; Ma, C.J.; Koeduka, T.; Nagegowda, D.A.; Dudareva, N.; Pichersky, E.; Clark, D. Characterization of a Petunia Acetyltransferase Involved in the Biosynthesis of the Floral Volatile Isoeugenol. Plant J. 2007, 49, 265–275. [Google Scholar] [CrossRef]
- St-Pierre, B.; De Luca, V. Evolution of Acyltransferase Genes: Origin and Diversification of the BAHD Superfamily of Acyltransferases Involved in Secondary Metabolism. Recent Adv. Phytochem. 2000, 34, 285–315. [Google Scholar]
- Kamatou, G.P.; Vermaak, I.; Viljoen, A.M. Eugenol—From the Remote Maluku Islands to the International Market Place: A Review of a Remarkable and Versatile Molecule. Molecules 2012, 17, 6953–6981. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.-Q.; Kenney, P.M.; Lam, L.K.T. Sesquiterpenes from clove (Eugenia caryophyllata) as potential anticarcinogenic agents. J. Nat. Prod. 1992, 55, 999–1003. [Google Scholar] [CrossRef]
- Barceloux, D.G. Frontmatter. In Medical Toxicology of Natural Substances; Wiley: Hoboken, NJ, USA, 2008. [Google Scholar]
- Ulanowska, M.; Olas, B. Biological Properties and Prospects for the Application of Eugenol—A Review. Int. J. Mol. Sci. 2021, 22, 3671. [Google Scholar] [CrossRef]
- Marchese, A.; Barbieri, R.; Coppo, E.; Orhan, I.E.; Daglia, M.; Nabavi, S.F.; Izadi, M.; Abdollahi, M.; Nabavi, S.M.; Ajami, M. Antimicrobial Activity of Eugenol and Essential Oils Containing Eugenol: A Mechanistic Viewpoint. Crit. Rev. Microbiol. 2017, 43, 668–689. [Google Scholar] [CrossRef]
- Mohammadi Nejad, S.; Özgüneş, H.; Başaran, N. Öjenolün Farmakolojik ve Toksikolojik Özellikleri. Turk. J. Pharm. Sci. 2017, 14, 201–206. [Google Scholar] [CrossRef]
- Pramod, K.; Ansari, S.H.; Ali, J. Eugenol: A Natural Compound with Versatile Pharmacological Actions. Nat. Prod. Commun. 2010, 5, 1999–2006. [Google Scholar] [CrossRef]
- Elbestawy, M.K.M.; El-Sherbiny, G.M.; Moghannem, S.A. Antibacterial, Antibiofilm and Anti-Inflammatory Activities of Eugenol Clove Essential Oil against Resistant Helicobacter Pylori. Molecules 2023, 28, 2448. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.K.; Chae, S.W.; Im, G.J.; Chung, J.W.; Song, J.J. Eugenol: A Phyto-Compound Effective against Methicillin-Resistant and Methicillin-Sensitive Staphylococcus aureus Clinical Strain Biofilms. PLoS ONE 2015, 10, e0119564. [Google Scholar] [CrossRef] [PubMed]
- Batiha, G.E.S.; Alkazmi, L.M.; Wasef, L.G.; Beshbishy, A.M.; Nadwa, E.H.; Rashwan, E.K. Syzygium aromaticum L. (Myrtaceae): Traditional Uses, Bioactive Chemical Constituents, Pharmacological and Toxicological Activities. Biomolecules 2020, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- da Silva, F.F.M.; Monte, F.J.Q.; de Lemos, T.L.G.; do Nascimento, P.G.G.; de Medeiros Costa, A.K.; de Paiva, L.M.M. Eugenol Derivatives: Synthesis, Characterization, and Evaluation of Antibacterial and Antioxidant Activities. Chem. Cent. J. 2018, 12, 34. [Google Scholar] [CrossRef]
- Khalil, A.A.; Rahman, U.U.; Khan, M.R.; Sahar, A.; Mehmood, T.; Khan, M. Essential Oil Eugenol: Sources, Extraction Techniques and Nutraceutical Perspectives. RSC Adv. 2017, 7, 32669–32681. [Google Scholar] [CrossRef]
- Ashraf, M.V.; Pant, S.; Khan, M.A.H.; Shah, A.A.; Siddiqui, S.; Jeridi, M.; Alhamdi, H.W.S.; Ahmad, S. Phytochemicals as Antimicrobials: Prospecting Himalayan Medicinal Plants as Source of Alternate Medicine to Combat Antimicrobial Resistance. Pharmaceuticals 2023, 16, 881. [Google Scholar] [CrossRef]
- Keita, K.; Darkoh, C.; Okafor, F. Secondary Plant Metabolites as Potent Drug Candidates against Antimicrobial-Resistant Pathogens. SN Appl. Sci. 2022, 4, 209. [Google Scholar] [CrossRef]
- Javier Álvarez-Martínez, F.; Barrajón-Catalán, E.; Micol, V. Biomedicines Tackling Antibiotic Resistance with Compounds of Natural Origin: A Comprehensive Review. Biomedicines 2020, 8, 405. [Google Scholar] [CrossRef]
- Ahmed, A.; Azim, A.; Gurjar, M.; Baronia, A.K. Current Concepts in Combination Antibiotic Therapy for Critically Ill Patients. Indian J. Crit. Care Med. 2014, 18, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Lázár, V.; Snitser, O.; Barkan, D.; Kishony, R. Antibiotic Combinations Reduce Staphylococcus aureus Clearance. Nature 2022, 610, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Si, Z.; Pethe, K.; Chan-Park, M.B. Chemical Basis of Combination Therapy to Combat Antibiotic Resistance. JACS Au 2023, 3, 276–292. [Google Scholar] [CrossRef] [PubMed]
- Tängdén, T. Combination Antibiotic Therapy for Multidrug-Resistant Gram-Negative Bacteria. Ups J. Med. Sci. 2014, 119, 149–153. [Google Scholar] [CrossRef]
- Hall, M.J.; Middleton, R.F.; Westmacott, D. The Fractional Inhibitory Concentration (FIC) Index as a Measure of Synergy. J. Antimicrob. Chemother. 1983, 11, 427–433. [Google Scholar] [CrossRef]
- Yang, S.K.; Yusoff, K.; Mai, C.W.; Lim, W.M.; Yap, W.S.; Lim, S.H.E.; Lai, K.S. Additivity vs. Synergism: Investigation of the Additive Interaction of Cinnamon Bark Oil and Meropenem in Combinatory Therapy. Molecules 2017, 22, 1733. [Google Scholar] [CrossRef]
- U. S. Food and Drug Administration. CFR—Code of Federal Regulations Title 21. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?fr=184.1257 (accessed on 29 May 2024).
- World Health Organization. Evaluation of Certain Food Additives: Sixty-Fifth Report of the Joint FAO/WHO Expert Committee on Food Additives; World Health Organization: Geneva, Switzerland, 2006; ISBN 9241209348. [Google Scholar]
- Cohen, S.M.; Eisenbrand, G.; Fukushima, S.; Gooderham, N.J.; Guengerich, F.P.; Hecht, S.S.; Rietjens, I.M.C.M.; Rosol, T.J.; Harman, C.; Taylor, S.V. GRAS 29 Flavoring Substances. Food Technol. 2020, 74, 44–65. [Google Scholar]
- Banchereau, T.; Mousset, J.L.; Mousset, J. Food Additive, Useful for Dairy Ruminants to Increase the Dairy (Milk) Production, Comprises Combination of Eugenol and Thymol as Active Compounds in a Medium; Bureau Tech Animales Sa; Techna France Nutrition: Couëron, France, 2006. [Google Scholar]
- Wang, W.; Weng, Y.; Luo, T.; Wang, Q.; Yang, G.; Jin, Y. Antimicrobial and the Resistances in the Environment: Ecological and Health Risks, Influencing Factors, and Mitigation Strategies. Toxics 2023, 11, 185. [Google Scholar] [CrossRef]
- Löffler, P.; Escher, B.I.; Baduel, C.; Virta, M.P.; Lai, F.Y. Antimicrobial Transformation Products in the Aquatic Environment: Global Occurrence, Ecotoxicological Risks, and Potential of Antibiotic Resistance. Environ. Sci. Technol. 2023, 57, 9474–9494. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.-L.; Song, C.; He, L.-Y.; Wang, B.; Gao, F.-Z.; Zhang, M.; Ying, G.-G. Antibiotics in Soil and Water: Occurrence, Fate, and Risk. Curr. Opin. Environ. Sci. Health 2023, 32, 100437. [Google Scholar] [CrossRef]
- Cycoń, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment—Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, P. One Health WASH: An AMR-Smart Integrative Approach to Preventing and Controlling Infection in Farming Communities. BMJ Glob. Health 2023, 8, 11263. [Google Scholar] [CrossRef]
- Moemenbellah-Fard, M.D.; Abdollahi, A.; Ghanbariasad, A.; Osanloo, M. Antibacterial and Leishmanicidal Activities of Syzygium aromaticum Essential Oil versus Its Major Ingredient, Eugenol. Flavour Fragr. J. 2020, 35, 534–540. [Google Scholar] [CrossRef]
- Auezova, L.; Najjar, A.; Kfoury, M.; Fourmentin, S.; Greige-Gerges, H. Antibacterial Activity of Free or Encapsulated Selected Phenylpropanoids against Escherichia coli and Staphylococcus epidermidis. J. Appl. Microbiol. 2020, 128, 710–720. [Google Scholar] [CrossRef]
- Kong, J.; Wang, Y.; Yao, Z.; Lin, Y.; Zhang, Y.; Han, Y.; Zhou, T.; Ye, J.; Cao, J. Eugenol Works Synergistically with Colistin against Colistin-Resistant Pseudomonas aeruginosa and Klebsiella pneumoniae Isolates by Enhancing Membrane Permeability. Microbiol. Spectr. 2023, 11, e0366622. [Google Scholar] [CrossRef]
- Nazer, A.I.; Kobilinsky, A.; Tholozan, J.L.; Dubois-Brissonnet, F. Combinations of Food Antimicrobials at Low Levels to Inhibit the Growth of Salmonella sv. Typhimurium: A Synergistic Effect? Food Microbiol. 2005, 22, 391–398. [Google Scholar] [CrossRef]
- Fekrirad, Z.; Gattali, B.; Kashef, N. Quorum Sensing-Regulated Functions of Serratia Marcescens Are Reduced by Eugenol. Iran. J. Microbiol. 2020, 12, 451. [Google Scholar] [CrossRef] [PubMed]
- Walsh, S.E.; Maillard, J.Y.; Russell, A.D.; Catrenich, C.E.; Charbonneau, D.L.; Bartolo, R.G. Activity and Mechanisms of Action of Selected Biocidal Agents on Gram-Positive and -Negative Bacteria. J. Appl. Microbiol. 2003, 94, 240–247. [Google Scholar] [CrossRef]
- Hammer, K.A.; Heel, K.A. Use of Multiparameter Flow Cytometry to Determine the Effects of Monoterpenoids and Phenylpropanoids on Membrane Polarity and Permeability in Staphylococci and Enterococci. Int. J. Antimicrob. Agents 2012, 40, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Perugini Biasi-Garbin, R.; Saori Otaguiri, E.; Morey, A.T.; Fernandes Da Silva, M.; Belotto Morguette, A.E.; Armando Contreras Lancheros, C.; Kian, D.; Perugini, M.R.E.; Nakazato, G.; Durán, N.; et al. Effect of Eugenol against Streptococcus agalactiae and Synergistic Interaction with Biologically Produced Silver Nanoparticles. Evid.-Based Complement. Altern. Med. 2015, 2015, 861497. [Google Scholar] [CrossRef] [PubMed]
- Kawacka, I.; Olejnik-Schmidt, A.; Schmidt, M.; Sip, A. Natural Plant-Derived Chemical Compounds as Listeria Monocytogenes Inhibitors In Vitro and in Food Model Systems. Pathogens 2021, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Niu, H.; Zhang, W.; Mu, H.; Sun, C.; Duan, J. Synergy among Thymol, Eugenol, Berberine, Cinnamaldehyde and Streptomycin against Planktonic and Biofilm-Associated Food-Borne Pathogens. Lett. Appl. Microbiol. 2015, 60, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Alves, F.C.B.; Barbosa, L.N.; Andrade, B.F.M.T.; Albano, M.; Furtado, F.B.; Marques Pereira, A.F.; Rall, V.L.M.; Júnior, A.F. Short Communication: Inhibitory Activities of the Lantibiotic Nisin Combined with Phenolic Compounds against Staphylococcus aureus and Listeria monocytogenes in Cow Milk. J. Dairy Sci. 2016, 99, 1831–1836. [Google Scholar] [CrossRef] [PubMed]
- Mith, H.; Duré, R.; Delcenserie, V.; Zhiri, A.; Daube, G.; Clinquart, A. Antimicrobial Activities of Commercial Essential Oils and Their Components against Food-borne Pathogens and Food Spoilage Bacteria. Food Sci. Nutr. 2014, 2, 403–416. [Google Scholar] [CrossRef]
- Purkait, S.; Bhattacharya, A.; Bag, A.; Chattopadhyay, R.R. Evaluation of Antibiofilm Efficacy of Essential Oil Components β-Caryophyllene, Cinnamaldehyde and Eugenol Alone and in Combination against Biofilm Formation and Preformed Biofilms of Listeria monocytogenes and Salmonella typhimurium. Lett. Appl. Microbiol. 2020, 71, 195–202. [Google Scholar] [CrossRef]
- Apolónio, J.; Faleiro, M.L.; Miguel, M.G.; Neto, L. No Induction of Antimicrobial Resistance in Staphylococcus aureus and Listeria monocytogenes during Continuous Exposure to Eugenol and Citral. FEMS Microbiol. Lett. 2014, 354, 92–101. [Google Scholar] [CrossRef]
- Gill, A.O.; Holley, R.A. Disruption of Escherichia coli, Listeria monocytogenes and Lactobacillus sakei Cellular Membranes by Plant Oil Aromatics. Int. J. Food Microbiol. 2006, 108, 1–9. [Google Scholar] [CrossRef]
- Devi, K.P.; Nisha, S.A.; Sakthivel, R.; Pandian, S.K. Eugenol (an Essential Oil of Clove) Acts as an Antibacterial Agent against Salmonella typhi by Disrupting the Cellular Membrane. J. Ethnopharmacol. 2010, 130, 107–115. [Google Scholar] [CrossRef]
- Yadav, M.K.; Park, S.; Chae, S.; Song, J.; Kim, H.C. Antimicrobial Activities of Eugenia caryophyllata Extract and Its Major Chemical Constituent Eugenol against Streptococcus pneumoniae. APMIS 2013, 121, 1198–1206. [Google Scholar] [CrossRef] [PubMed]
- Gill, A.O.; Holley, R.A. Mechanisms of Bactericidal Action of Cinnamaldehyde against Listeria monocytogenes and of Eugenol against L. monocytogenes and Lactobacillus sakei. Appl. Environ. Microbiol. 2004, 70, 5750–5755. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.; Johny, A.K.; Amalaradjou, M.A.R.; Ananda Baskaran, S.; Kim, K.S.; Venkitanarayanan, K. Plant-Derived Antimicrobials Reduce Listeria Monocytogenes Virulence Factors in Vitro, and down-Regulate Expression of Virulence Genes. Int. J. Food Microbiol. 2012, 157, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Braschi, G.; Serrazanetti, D.I.; Siroli, L.; Patrignani, F.; De Angelis, M.; Lanciotti, R. Gene Expression Responses of Listeria Monocytogenes Scott a Exposed to Sub-Lethal Concentrations of Natural Antimicrobials. Int. J. Food Microbiol. 2018, 286, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Liu, H.; Zheng, Y.; Guo, D.; Shi, C.; Xu, Y.; Xia, X. Inhibitory Effect of Thymoquinone on Listeria monocytogenes ATCC 19115 Biofilm Formation and Virulence Attributes Critical for Human Infection. Front. Cell. Infect. Microbiol. 2019, 9, 304. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.; Upadhyaya, I.; Kollanoor-Johny, A.; Venkitanarayanan, K. Antibiofilm Effect of Plant Derived Antimicrobials on Listeria monocytogenes. Food Microbiol. 2013, 36, 79–89. [Google Scholar] [CrossRef]
- Upadhyay, A.; Upadhyaya, I.; Mooyottu, S.; Venkitanarayanan, K. Eugenol in Combination with Lactic Acid Bacteria Attenuates Listeria monocytogenes Virulence in Vitro and in Invertebrate Model Galleria mellonella. J. Med. Microbiol. 2016, 65, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Aleksic Sabo, V.; Nikolic, I.; Mimica-Dukic, N.; Knezevic, P. Anti-Acinetobacter Baumannii Activity of Selected Phytochemicals Alone, in Binary Combinations and in Combinations with Conventional Antibiotics. Nat. Prod. Res. 2021, 35, 5964–5967. [Google Scholar] [CrossRef] [PubMed]
- Karumathil, D.P.; Nair, M.S.; Gaffney, J.; Kollanoor-Johny, A.; Venkitanarayanan, K. Trans-Cinnamaldehyde and Eugenol Increase Acinetobacter Baumannii Sensitivity to Beta-Lactam Antibiotics. Front. Microbiol. 2018, 9, 1011. [Google Scholar] [CrossRef]
- Liu, W.; Chen, G.; Dou, K.; Yi, B.; Wang, D.; Zhou, Q.; Sun, Y. Eugenol Eliminates Carbapenem-Resistant Klebsiella Pneumoniae via Reactive Oxygen Species Mechanism. Front. Microbiol. 2023, 14, 1090787. [Google Scholar] [CrossRef]
- Ezraty, B.; Gennaris, A.; Barras, F.; Collet, J.-F. Oxidative Stress, Protein Damage and Repair in Bacteria. Nat. Rev. Microbiol. 2017, 15, 385–396. [Google Scholar] [CrossRef]
- Qi, X.; Zhang, Y.; Guo, H.; Hai, Y.; Luo, Y.; Yue, T. Mechanism and Intervention Measures of Iron Side Effects on the Intestine. Crit. Rev. Food Sci. Nutr. 2020, 60, 2113–2125. [Google Scholar] [CrossRef]
- Morgaan, H.A.; Omar, H.M.G.; Zakaria, A.S.; Mohamed, N.M. Repurposing Carvacrol, Cinnamaldehyde, and Eugenol as Potential Anti-Quorum Sensing Agents against Uropathogenic Escherichia coli Isolates in Alexandria, Egypt. BMC Microbiol. 2023, 23, 300. [Google Scholar] [CrossRef] [PubMed]
- Gill, A.O.; Holley, R.A. Inhibition of Membrane Bound ATPases of Escherichia coli and Listeria monocytogenes by Plant Oil Aromatics. Int. J. Food Microbiol. 2006, 111, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Cesar Silva, J.; Luiz Silva Pereira, R.; Esmeraldo Rocha, J.; Silva Macedo, N.; Leite Linhares, M.; Assis Bezerra da Cunha, F.; Douglas Melo Coutinho, H.; Nascimento Pereira Junior, F.; Fernandes Galvão Rodrigues, F.; Joaquim Garcia Santos, G. Evaluation of Antibacterial and Toxicological Activities of Essential Oil of Ocimum gratissimum L. and Its Major Constituent Eugenol. Food Biosci. 2022, 50, 102128. [Google Scholar] [CrossRef]
- Aljuwayd, M.; Malli, I.A.; Kwon, Y.M. Application of Eugenol in Poultry to Control Salmonella Colonization and Spread. Vet. Sci. 2023, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zheng, S.M.; Wei, S.M.; Tian, Q.M.; Tao, Y.; Bo, R.N.; Liu, M.J.; Li, J.G. The Protective Effect and Potential Mechanisms of Eugenol against Salmonella In Vivo and In Vitro. Poult. Sci. 2022, 101, 101801. [Google Scholar] [CrossRef]
- Garneau-Tsodikova, S.; Labby, K.J. Mechanisms of Resistance to Aminoglycoside Antibiotics: Overview and Perspectives. Medchemcomm 2016, 7, 11–27. [Google Scholar] [CrossRef]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [Google Scholar] [CrossRef] [PubMed]
- Albano, M.; Alves, F.C.B.; Andrade, B.F.M.T.; Barbosa, L.N.; Pereira, A.F.M.; de Lourdes Ribeiro de Souza da Cunha, M.; Rall, V.L.M.; Júnior, A.F. Antibacterial and Anti-Staphylococcal Enterotoxin Activities of Phenolic Compounds. Innov. Food Sci. Emerg. Technol. 2016, 38, 83–90. [Google Scholar] [CrossRef]
- Özel, Y.; Yilmaz, U.; Ünlü, M.; Vardar Ünlü, G. Antibacterial Activity and Synergistic Interaction of Various Essential Oil Components and Antibiotics. Mikrobiyoloji Bul. 2022, 56, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Masi, M.; Réfregiers, M.; Pos, K.M.; Pagès, J.M. Mechanisms of Envelope Permeability and Antibiotic Influx and Efflux in Gram-Negative Bacteria. Nat. Microbiol. 2017, 2, 17001. [Google Scholar] [CrossRef] [PubMed]
- Calvo, J.; Martínez-Martínez, L. Mecanismos de Acción de Los Antimicrobianos. Enferm. Infecc. Microbiol. Clin. 2009, 27, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, R.; Amador, P.; Prudêncio, C. β-Lactams: Chemical Structure, Mode of Action and Mechanisms of Resistance. Rev. Res. Med. Microbiol. 2013, 24, 7–17. [Google Scholar] [CrossRef]
- Cristani, M.; D’Arrigo, M.; Mandalari, G.; Castelli, F.; Sarpietro, M.G.; Micieli, D.; Venuti, V.; Bisignano, G.; Saija, A.; Trombetta, D. Interaction of Four Monoterpenes Contained in Essential Oils with Model Membranes: Implications for Their Antibacterial Activity. J. Agric. Food Chem. 2007, 55, 6300–6308. [Google Scholar] [CrossRef] [PubMed]
- Zunino, M.P.; Turina, A.V.; Zygadlo, J.A.; Perillo, M.A. Stereoselective Effects of Monoterpenes on the Microviscosity and Curvature of Model Membranes Assessed by DPH Steady-state Fluorescence Anisotropy and Light Scattering Analysis. Chirality 2011, 23, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Hemaiswarya, S.; Doble, M. Synergistic Interaction of Eugenol with Antibiotics against Gram Negative Bacteria. Phytomedicine 2009, 16, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, K.; Holley, R.A. Use of Natural Antimicrobials to Increase Antibiotic Susceptibility of Drug Resistant Bacteria. Int. J. Food Microbiol. 2010, 140, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Gowda, J.; Tavarageri, A.; Kulkarni, R.; Anegundi, R.T.; Janardhan, A.; Bhat, M.A. Comparative Assessment of the Antimicrobial Efficacy of Triclosan, Amoxicillin and Eugenol against Enterococcus Faecalis. Int. J. Clin. Pediatr. Dent. 2021, 14, 59–62. [Google Scholar] [CrossRef]
- Subhadra, B.; Kim, D.H.; Woo, K.; Surendran, S.; Choi, C.H. Control of Biofilm Formation in Healthcare: Recent Advances Exploiting Quorum-Sensing Interference Strategies and Multidrug Efflux Pump Inhibitors. Materials 2018, 11, 1676. [Google Scholar] [CrossRef]
- Gaynor, M.; Mankin, A.S. Macrolide Antibiotics: Binding Site, Mechanism of Action, Resistance. Curr. Top. Med. Chem. 2003, 3, 949–960. [Google Scholar] [CrossRef]
- Prajapati, J.D.; Kleinekathöfer, U.; Winterhalter, M. How to Enter a Bacterium: Bacterial Porins and the Permeation of Antibiotics. Chem. Rev. 2021, 121, 5158–5192. [Google Scholar] [CrossRef] [PubMed]
- Hemaiswarya, S.; Kruthiventi, A.K.; Doble, M. Synergism between Natural Products and Antibiotics against Infectious Diseases. Phytomedicine 2008, 15, 639–652. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.P.; Grant, J.A. Eugenol Profile. In Publications (NYS Integrated Pest Management Program); New York State IPM Program: Geneva, NY, USA, 2018; pp. 1–15. [Google Scholar]
- Adhikari, K.; Khanikor, B.; Sarma, R. Persistent Susceptibility of Aedes aegypti to Eugenol. Sci. Rep. 2022, 12, 2277. [Google Scholar] [CrossRef]
- Battistuzzi, F.U.; Hedges, S.B. A Major Clade of Prokaryotes with Ancient Adaptations to Life on Land. Mol. Biol. Evol. 2009, 26, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Pino-Otín, M.R.; Lorca, G.; Langa, E.; Roig, F.; Terrado, E.M.; Ballestero, D. Assessing the Ecotoxicity of Eight Widely Used Antibiotics on River Microbial Communities. Int. J. Mol. Sci. 2023, 24, 16960. [Google Scholar] [CrossRef]
- Weber, K.P.; Legge, R.L. One-Dimensional Metric for Tracking Bacterial Community Divergence Using Sole Carbon Source Utilization Patterns. J. Microbiol. Methods 2009, 79, 55–61. [Google Scholar] [CrossRef]
- Pathak, A.; Angst, D.C.; León-Sampedro, R.; Hall, A.R. Antibiotic-Degrading Resistance Changes Bacterial Community Structure via Species-Specific Responses. ISME J. 2023, 17, 1495–1503. [Google Scholar] [CrossRef]
- Marin Municipal Water District Vegetation Management Plan Marin Municipal Water District Vegetation Management Plan. Herbicide Risk Assessment; Draft 1/1/2010. Chapter 6-Clove Oil (Eugenol); Pesticide Research Institute: Berkley, CA, USA, 2010.
- Zheng, X.; Bossier, P. Toxicity Assessment and Anti-Vibrio Activity of Essential Oils: Potential for Application in Shrimp Aquaculture. Rev. Aquac. 2023, 15, 1554–1573. [Google Scholar] [CrossRef]
- Lal, H.; Misra, V.; Viswanathan, P.N.; Murti, C.R.K. The Water Flea (Daphnia magna) as a Sensitive Indicator for the Assessment of Toxicity of Synthetic Detergents. Ecotoxicol. Environ. Saf. 1984, 8, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Gueretz, J.S.; Somensi, C.A.; Martins, M.L.; Souza, A.P.D. Avaliação da toxicidade do eugenol em bioensaios com organismos-teste. Cienc. Rural 2017, 47, e20170194. [Google Scholar] [CrossRef]
- Ioele, G.; De Luca, M.; Ragno, G. Acute Toxicity of Antibiotics in Surface Waters by Bioluminescence Test. Curr. Pharm. Anal. 2015, 12, 220–226. [Google Scholar] [CrossRef]
- Ji, J.Y.; Xing, Y.J.; Ma, Z.T.; Zhang, M.; Zheng, P. Acute Toxicity of Pharmaceutical Wastewaters Containing Antibiotics to Anaerobic Digestion Treatment. Chemosphere 2013, 91, 1094–1098. [Google Scholar] [CrossRef]
- Park, S.; Choi, K. Hazard Assessment of Commonly Used Agricultural Antibiotics on Aquatic Ecosystems. Ecotoxicology 2008, 17, 526–538. [Google Scholar] [CrossRef]
- Havelkova, B.; Beklova, M.; Kovacova, V.; Hlavkova, D.; Pikula, J. Ecotoxicity of Selected Antibiotics for Organisms of Aquatic and Terrestrial Ecosystems. Neuroendocrinol. Lett. 2016, 37, 38–44. [Google Scholar]
- Neghi, N.; Kumar, M. Photocatalytic and Adsorptive Performance of Polyvinyl Alcohol/Chitosan/TiO2 Composite for Antibiotics Removal: Single- And Multi-Pollutant Conditions. Water Sci. Technol. 2022, 86, 800–813. [Google Scholar] [CrossRef]
- Czech, B.; Jośko, I.; Oleszczuk, P. Ecotoxicological Evaluation of Selected Pharmaceuticals to Vibrio Fischeri and Daphnia Magna before and after Photooxidation Process. Ecotoxicol. Environ. Saf. 2014, 104, 247–253. [Google Scholar] [CrossRef]
- Calleja, M.C.; Persoone, G.; Geladi, P. Comparative Acute Toxicity of the First 50 Multicentre Evaluation of In Vitro Cytotoxicity Chemicals to Aquatic Non-Vertebrates. Arch. Environ. Contam. Toxicol. 1994, 26, 69–78. [Google Scholar] [CrossRef]
- Müller, J.B.; Melegari, S.P.; Perreault, F.; Matias, W.G. Comparative Assessment of Acute and Chronic Ecotoxicity of Water Soluble Fractions of Diesel and Biodiesel on Daphnia magna and Aliivibrio fischeri. Chemosphere 2019, 221, 640–646. [Google Scholar] [CrossRef]
- Lilius, H.; Isomaa, B.; Holmstr6m, T. A Comparison of the Toxicity of 50 Reference Chemicals to Freshly Isolated Rainbow Trout Hepatocytes and Daphnia magna; Elsevier: Amsterdam, The Netherlands, 1994; Volume 30. [Google Scholar]
- Choi, K.; Kim, Y.; Jung, J.; Kim, M.-H.; Kim, C.-S.; Kim, N.-H.; Park, J. Hazard/Risk Assessment Occurrences and Ecological Risks of Roxithromycin, Trimethoprim, and Chloramphenicol in the Han River, Korea. Environ. Toxicol. Chem. Int. J. 2008, 27, 711–719. [Google Scholar] [CrossRef]
- Lomba, L.; Lapeña, D.; Ros, N.; Aso, E.; Cannavò, M.; Errazquin, D.; Giner, B. Ecotoxicological Study of Six Drugs in Aliivibrio fischeri, Daphnia magna and Raphidocelis subcapitata. Environ. Sci. Pollut. Res. 2020, 27, 9891–9900. [Google Scholar] [CrossRef] [PubMed]
- Iannacone, J.; Alvariño, L. Evaluación del riesgo acuático de siete productos farmacéuticos sobre Daphnia magna. Ecol. Apl. 2009, 8, 71. [Google Scholar] [CrossRef]
- Wollenberger, L.; Halling-Sùrensen, B.; Kusk, K.O. Acute and Chronic Toxicity of Veterinary Antibiotics to Daphnia magna. Chemosphere 2000, 40, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Dojmi Di Delupis, G.; Macrib, A.; Civitarealeb, C.; Migliorec, L. Antibiotics of Zootechnical Use: Effects of Acute and Low Ose Contamination on Aphnia magna Traus. Aquat. Toxicol. 1992, 22, 53–59. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, P.; Wang, M.; Wu, Y.; Sun, Y.; Su, H.; Deng, J. Mixture Toxicity Effects of Chloramphenicol, Thiamphenicol, Florfenicol in Daphnia Magna under Different Temperatures. Ecotoxicology 2021, 30, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Yuxuan, Z.; Peiyong, G.; Yanmei, W.; Xiaoyan, Z.; Meixian, W.; Simin, Y.; Yinshi, S.; Jun, D.; Haitao, S. Evaluation of the Subtle Effects and Oxidative Stress Response of Chloramphenicol, Thiamphenicol, and Florfenicol in Daphnia Magna. Environ. Toxicol. Chem. 2019, 38, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Bownik, A. Protective Effects of Ectoine on Physiological Parameters of Daphnia magna Subjected to Clove Oil-Induced Anaesthesia. Turk. J. Fish. Aquat. Sci. 2016, 16, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Bekker, J.M.; Krijgsman, B.J. Physiological Investigations into the Heart Function of Daphnia. J. Physiol. 1951, 115, 249–257. [Google Scholar] [CrossRef]
- Dyson, D.; Maxie, M.; Schnurr, D. Morbidity and Mortality Associated with Anesthetic Management in Small Animal Veterinary Practice in Ontario. J. Am. Anim. Hosp. Assoc. 1998, 34, 325–335. [Google Scholar] [CrossRef]
- Chung, G.; Rhee, J.N.; Jung, S.J.; Kim, J.S.; Oh, S.B. Modulation of CaV2.3 Calcium Channel Currents by Eugenol. J. Dent. Res. 2008, 87, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, B.; Zhang, Z.; Zhu, C.; Tian, Y.; Ye, J. Evolution of Microbial Communities during Electrokinetic Treatment of Antibiotic-Polluted Soil. Ecotoxicol. Environ. Saf. 2018, 148, 842–850. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Jia, S.; He, X.; Zhang, X.; Ye, L. Different Impacts of Manure and Chemical Fertilizers on Bacterial Community Structure and Antibiotic Resistance Genes in Arable Soils. Chemosphere 2017, 188, 455–464. [Google Scholar] [CrossRef]
- Pino-Otín, M.R.; Ferrando, N.; Ballestero, D.; Langa, E.; Roig, F.J.; Terrado, E.M. Impact of Eight Widely Consumed Antibiotics on the Growth and Physiological Profile of Natural Soil Microbial Communities. Chemosphere 2022, 305, 135473. [Google Scholar] [CrossRef] [PubMed]
- Pérès, G.; Vandenbulcke, F.; Guernion, M.; Hedde, M.; Beguiristain, T.; Douay, F.; Houot, S.; Piron, D.; Richard, A.; Bispo, A.; et al. Earthworm Indicators as Tools for Soil Monitoring, Characterization and Risk Assessment. An Example from the National Bioindicator Programme (France). Pedobiologia 2011, 54, S77–S87. [Google Scholar] [CrossRef]
- El-Hassanin, A.S.; Samak, M.R.; Ahmed, S.M.; Afifi, M.M.I.; Abd El-Satar, A.M. Bioaccumulation of Heavy Metals during Composting and Vermicomposting Processes of Sewage Sludge. Egypt J. Chem. 2022, 65, 1155–1162. [Google Scholar] [CrossRef]
- Garg, P.; Satya, S.; Sharma, S. Effect of Heavy Metal Supplementation on Local (Allolobophora Parva) and Exotic (Eisenia Fetida) Earthworm Species: A Comparative Study. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 2009, 44, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xu, Z.; Wu, J.; Tian, G. Bioaccumulation of Heavy Metals in the Earthworm Eisenia Fetida in Relation to Bioavailable Metal Concentrations in Pig Manure. Bioresour. Technol. 2010, 101, 3430–3436. [Google Scholar] [CrossRef] [PubMed]
- Almadiy, A.; Nenaah, G. Chemical Profile, Bioactivity, and Biosafety Evaluations of Essential Oils and Main Terpenes of Two Plant Species against Trogoderma granarium. Agronomy 2022, 12, 3112. [Google Scholar] [CrossRef]
- Alemu, D.; Kitila, C.; Garedew, W.; Jule, L.T.; Badassa, B.; Nagaprasad, N.; Seenivasan, V.; Saka, A.; Ramaswamy, K. Growth, Yield, and Yield Variables of Onion (Allium cepa L.) Varieties as Influenced by Plantspacing at DambiDollo, Western Ethiopia. Sci. Rep. 2022, 12, 20563. [Google Scholar] [CrossRef]
- Nwajuaku, I.I.; Nweke, C.N. Comparative Analysis of Allium cepa L. and White Onion as Bioindicators of Heavy Metals InGround water. IRE J. 2019, 3, 96–101. [Google Scholar]
- Przygocka-Cyna, K.; Barlóg, P.; Grzebisz, W.; Spizewski, T. Onion (Allium cepa L.) Yield and Growth Dynamics Response to in-Season Patterns of Nitrogen and Sulfur Uptake. Agronomy 2020, 10, 1146. [Google Scholar] [CrossRef]
- Gogoi, R.; Sarma, N.; Loying, R.; Pandey, S.K.; Begum, T.; Lal, M. A Comparative Analysis of Bark and Leaf Essential Oil and Their Chemical Composition, Antioxidant, Anti-Inflammatory, Antimicrobial Activities and Genotoxicity of North East Indian Cinnamomum Zeylanicum Blume. Nat. Prod. J. 2021, 11, 74–84. [Google Scholar] [CrossRef]
- Tworkoski, T. Herbicide Effects of Essential Oils. Weed Sci. 2002, 50, 425–431. [Google Scholar] [CrossRef]
- Wilson, G.B. Cytological effects of some antibiotics. J. Hered. 1950, 41, 227–231. [Google Scholar] [CrossRef]
- Druhy, R.E.; Khan, A.A. The Inhibition of Seedling Growth by Streptomycin Sulfate. Physiol. Plant 1969, 22, 751–758. [Google Scholar] [CrossRef]
- Mann, S.K. Interaction of Tetracycline (TC) with Chromosomes in Allium cepa. Environ. Exp. Bot. 1978, 18, 201–205. [Google Scholar] [CrossRef]
- Bluggs, C.W.; Pilling, M.A. The Absorption, Distribution, and Excretion of Streptomycin in Man. J. Clin. Investig. 1946, 25, 94–102. [Google Scholar] [CrossRef]
- Kunin, C.M.; Dornbush, A.C.; Finland, M. Distribution and excretion of four tetracycline analogues in normal young men. J. Clin. Investig. 1959, 38, 1950–1963. [Google Scholar] [CrossRef]
- Wood, M.J.; Farrell, W. Comparison of Urinary Excretion of Tobramycin and Gentamicin in Adults. J. Infect. Dis. 1937, 134, S133–S138. [Google Scholar] [CrossRef]
- Fischer, I.U.; Von Unruh, G.E.; Dengler, H.J. The Metabolism of Eugenol in Man. Xenobiotica 1990, 20, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Graham, G.G.; Pile, K.D. Compendium of Inflammatory Diseases; Springer: Basel, Switzerland, 2016. [Google Scholar]
- Aranda, F.L.; Rivas, B.B. Removal of amoxicillin through different methods, emphasizing removal by biopolymers and its derivatives. An overview. J. Chil. Chem. Soc. 2022, 67, 5643–5655. [Google Scholar] [CrossRef]
- Farzam, K.; Nessel, T.A.; Quick, J. Erythromycin; StatPearls Publ. LLC: Treasure Island, FL, USA, 2023. [Google Scholar]
- Finnah, A.; Piñeiro, S.; Reuss, R.; Sanders, P. Ampicillin. In Proceedings of the Residue Monograph Prepared by the Meeting of the Joint FAO/WHO Expert Committee on Food Additives (JECFA), 85th Meeting 2017, Geneva, Switzerland, 17–26 October 2017; FAO/WHO: Rome, Italy, 2007. [Google Scholar]
- ATCC: The Global Bioresource Center|ATCC. Available online: https://www.atcc.org/ (accessed on 29 May 2024).
- CLSI M100-S24; Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement an Informational Supplement for Global Application Developed through the Clinical and Laboratory Standards Institute Consensus Process. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2014.
- EUCAST. Clinical Breakpoints—Bacteria (v 13.1). European Society of Clinical Microbiology and Infectious Disease. 2023. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_13.1_Breakpoint_Tables.pdf (accessed on 29 May 2024).
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and Broth Dilution Methods to Determine the Minimal Inhibitory Concentration (MIC) of Antimicrobial Substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Odds, F.C. Synergy, Antagonism, and What the Chequerboard Puts between Them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef] [PubMed]
- Rand, K.H.; Houck, H.J.; Brown, P.; Bennett, D. Reproducibility of the Microdilution Checkerboard Method for Antibiotic Synergy. Antimicrob. Agents Chemother. 1993, 37, 613–615. [Google Scholar] [CrossRef]
- Bonapace, C.R.; White, R.L.; Friedrich, L.V.; Bosso, J.A. Evaluation of Antibiotic Synergy against Acinetobacter baumannii: A Comparison with Etest, Time-Kill, and Checkerboard Methods. Diagn. Microbiol. Infect. Dis. 2000, 38, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.H.; Yu, C.M.; Yu, V.L.; Chow, J.W. Synergy Assessed by Checkerboard a Critical Analysis. Diagn. Microbiol. Infect. Dis. 1993, 16, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Caesar, L.K.; Cech, N.B. Synergy and Antagonism in Natural Product Extracts: When 1 + 1 Does Not Equal 2. Nat. Prod. Rep. 2019, 36, 869–888. [Google Scholar] [CrossRef] [PubMed]
- CLSI M26-A; Methods for Determining Bactericidal Activity of Antimicrobial Agents; Approved Guideline This Document Provides Procedures for Determining the Lethal Activity of Antimicrobial Agents. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 1999.
- Chadha, J.; Ravi; Singh, J.; Chhibber, S.; Harjai, K. Gentamicin Augments the Quorum Quenching Potential of Cinnamaldehyde In Vitro and Protects Caenorhabditis elegans from Pseudomonas aeruginosa Infection. Front. Cell. Infect. Microbiol. 2022, 12, 899566. [Google Scholar] [CrossRef]
- Siriwong, S.; Teethaisong, Y.; Thumanu, K.; Dunkhunthod, B.; Eumkeb, G. The Synergy and Mode of Action of Quercetin plus Amoxicillin against Amoxicillin-Resistant Staphylococcus epidermidis. BMC Pharmacol. Toxicol. 2016, 17, 39. [Google Scholar] [CrossRef]
- Gan, C.; Langa, E.; Valenzuela, A.; Ballestero, D.; Pino-Otín, M.R. Synergistic Activity of Thymol with Commercial Antibiotics against Critical and High WHO Priority Pathogenic Bacteria. Plants 2023, 12, 1868. [Google Scholar] [CrossRef] [PubMed]
- ISO 19458:2006; Norma Española Grupo 18. ISO: Geneva, Switzerland, 2007.
- Pino-Otín, M.R.; Langa, E.; Val, J.; Mainar, A.M.; Ballestero, D. Impact of Citronellol on River and Soil Environments Using Non-Target Model Organisms and Natural Populations. J. Environ. Manag. 2021, 287, 112303. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Zhang, Z.; Wang, W.; Tang, Q.; Song, S.; Zhu, J.; Xie, Y.; Zhang, L. The Effects of Radiation Pollution on the Population Diversities and Metabolic Characteristics of Soil Microorganisms. Water Air Soil Pollut. 2014, 225, 2133. [Google Scholar] [CrossRef]
- Kelly, J.J.; Iii, R.L.T. Use of BIOLOG for the Analysis of Microbial Communities from Zinc-Contaminated Soils; American Society of Agronomy, Crop Science Society of America, and Soil Science Society of America: Madison, WI, USA, 1998. [Google Scholar]
- Tiquia, S.M. Metabolic Diversity of the Heterotrophic Microorganisms and Potential Link to Pollution of the Rouge River. Environ. Pollut. 2010, 158, 1435–1443. [Google Scholar] [CrossRef] [PubMed]
- Stefanowicz, A.M.; Szafer, W. The Biolog Plates Technique as a Tool in Ecological Studies of Microbial Communities. Pol. J. Environ. Stud. 2006, 15, 669–676. [Google Scholar]
- Pino-Otín, M.R.; Lorca, G.; Val, J.; Ferrando, N.; Ballestero, D.; Langa, E. Ecotoxicological Study of Tannic Acid on Soil and Water Non-Target Indicators and Its Impact on Fluvial and Edaphic Communities. Plants 2023, 12, 4041. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acid Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- ISO 11348; Water Quality-Determination of the Inhibitory Effect of Water Samples on the Light Emission of Vibrio Fischeri (Luminescent Bacteria Test). International Organization for Standardization: Geneva, Switzerland, 2007.
- OECD. OECD Test No. 202: Daphnia sp. Acute Immobilisation Test, OECD Guidelines for the Testing of Chemicals, Section 2; OECD: Paris, France, 2004. [Google Scholar]
- AENOR UNE-EN ISO 6341; Water Quality—Determination of the Inhibition of the Mobility of Daphnia Magna Straus (Cladocera, Crustacea)—Acute Toxicity Test. Asociación Española de Normalización: Madrid, Spain, 2012.
- OECD. OECD Test No. 207: Earthworm, Acute Toxicity Tests, OECD Guidelines for the Testing of Chemicals, Section 2; OECD Publication: Paris, France, 1984; Volume 1, pp. 1–9. [Google Scholar] [CrossRef]
- Pino, M.R.; Val, J.; Mainar, A.M.; Zuriaga, E.; Español, C.; Langa, E. Acute Toxicological Effects on the Earthworm Eisenia Fetida of 18 Common Pharmaceuticals in Artificial Soil. Sci. Total Environ. 2015, 518–519, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Fiskesjö, G. The Allium Test in Wastewater Monitoring. Environ. Toxicol. Water Qual. 1993, 8, 291–298. [Google Scholar] [CrossRef]

| MICalone (μg/mL) | |||
|---|---|---|---|
| Gram | Bacteria | EUG (Exp) | EUG (Lit) |
| + | Bacillus subtilis (ATCC 6633) | 2000 | - |
| Listeria monocytogenes (ATCC 7644) | 1000 | - | |
| Streptococcus agalactiae (ATCC 12386) | 1000 | - | |
| Staphylococcus aureus (ATCC 9144) | 1000 | - | |
| − | Acinetobacter baumannii (ATCC 19606) | 500 | - |
| Enterococcus faecalis (ATCC 19433) | 2000 | - | |
| Escherichia coli (ATCC 25922) | 1000 | >2000 a 197.4 b | |
| Klebsiella aerogenes (ATCC 13048) | 500 | - | |
| Klebsiella pneumoniae (C6) | 500 | - | |
| Pasteurella aerogenes (ATCC 27883) | 500 | - | |
| Proteus mirabilis (ATCC 35659) | - | - | |
| Pseudomonas aeruginosa (ATCC 27853) | 2000 | >2000 a >1000 c | |
| Salmonella enterica (ATCC 13311) | 1000 | 525 d | |
| Serratia marcescens (ATCC 13880) | 1000 | 460 e | |
| Gram+/− | Bacteria | Compounds | MICcomb (μg/mL) | Ratio [EUG]/[ABX] * | FIC | ΣFIC | Conclusion | MIC Reduction (%) |
|---|---|---|---|---|---|---|---|---|
| GRAM+ | L. monocytogenes | EUG | 250 | 714.3 | 0.25 | 0.50 | Synergy | 75 |
| GTM | 3.1 a | 0.25 | 75 | |||||
| EUG | 500 | 270.3 | 0.50 | 0.53 | Addition | 50 | ||
| AMP | 3.9 a | 0.03 | 97 | |||||
| EUG | 500 | 1111.1 | 0.50 | 0.51 | 50 | |||
| AMO | 1.0 a | 0.01 | 98 | |||||
| EUG | 500 | 44.4 | 0.50 | 1.00 | 50 | |||
| STM | 100 a | 0.50 | 50 | |||||
| EUG | 500 | 35.7 | 0.50 | 1.00 | 50 | |||
| ERY | 62.5 a | 0.50 | 50 | |||||
| EUG | 500 | 909.1 | 0.50 | 0.62 | 50 | |||
| TC | 1.6 a | 0.12 | 88 | |||||
| EUG | 500 | 196.1 | 0.50 | 1.00 | 50 | |||
| CHL | 5 a | 0.50 | 50 | |||||
| S. agalactiae | EUG | 500 | 34.1 | 0.50 | 1.00 | Addition | 50 | |
| AMP | 31.2 a | 0.50 | 50 | |||||
| EUG | 500 | 35.7 | 0.50 | 1.00 | 50 | |||
| AMO | 31.2 a | 0.50 | 50 | |||||
| EUG | 500 | 1428.6 | 0.50 | 0.62 | 50 | |||
| GTM | 3.3 a | 0.12 | 88 | |||||
| EUG | 500 | 178.6 | 0.50 | 0.75 | 50 | |||
| STM | 25 a | 0.25 | 75 | |||||
| EUG | 500 | 35.7 | 0.50 | 1.00 | 50 | |||
| ERY | 62.5 a | 0.50 | 50 | |||||
| EUG | 500 | 158.7 | 0.50 | 0.62 | 50 | |||
| CHL | 6.2 a | 0.12 | 88 | |||||
| S. aureus | EUG | 250 | 714.3 | 0.25 | 0.37 | Synergy | 75 | |
| GTM | 3.1 a | 0.12 | 88 | |||||
| EUG | 500 | 357.1 | 0.50 | 0.62 | Addition | 50 | ||
| STM | 12.5 a | 0.12 | 88 | |||||
| EUG | 125 | 29.3 | 0.12 | 1.12 | 88 | |||
| TC | 12.5 a | 1.00 | 0 | |||||
| EUG | 500 | 65.8 | 0.50 | 1.00 | 50 | |||
| CHL | 15 a | 0.50 | 50 | |||||
| GRAM− | A. baumannii | EUG | 125 | 15.8 | 0.25 | 0.50 | Synergy | 75 |
| CHL | 15.6 a | 0.25 | 75 | |||||
| EUG | 250 | 8.5 | 0.50 | 1.00 | Addition | 50 | ||
| AMP | 62.5 a | 0.50 | 50 | |||||
| EUG | 250 | 8.9 | 0.50 | 1.50 | 50 | |||
| AMO | 62.5 a | 1.00 | 0 | |||||
| EUG | 250 | 181.8 | 0.50 | 1.00 | 50 | |||
| GTM | 12.5 a | 0.50 | 50 | |||||
| EUG | 250 | 112.4 | 0.50 | 1.50 | 50 | |||
| ERY | 10 a | 1.00 | 0 | |||||
| EUG | 250 | 7.4 | 0.50 | 4.50 | Antagonism | 50 | ||
| STM | 300 a | 4.00 | −300 | |||||
| E. coli | EUG | 500 | 294.1 | 0.50 | 1.50 | Addition | 50 | |
| GTM | 15.6 a | 1.00 | 0 | |||||
| EUG | 500 | 588.2 | 0.50 | 1.00 | 50 | |||
| STM | 7.8 a | 0.50 | 50 | |||||
| EUG | 250 | 17.9 | 0.50 | 1.50 | 50 | |||
| ERY | 62.5 a | 1.00 | 0 | |||||
| K. aerogenes | EUG | 250 | 0.7 | 0.50 | 1.00 | Addition | 50 | |
| AMP | 750 a | 0.50 | 50 | |||||
| EUG | 250 | 0.6 | 0.50 | 1.50 | 50 | |||
| AMO | 1000 a | 1.00 | 0 | |||||
| EUG | 500 | 454.5 | 0.50 | 1.00 | 50 | |||
| ERY | 5 a | 0.50 | −300 | |||||
| EUG | 250 | 3.9 | 0.50 | 4.50 | Antagonism | 50 | ||
| CHL | 125 a | 4.00 | 50 | |||||
| K. pneumoniae | EUG | 250 | 2.1 | 0.50 | 1.00 | Addition | 50 | |
| AMP | 250 a | 0.50 | 50 | |||||
| EUG | 250 | 17.8 | 0.50 | 0.75 | 50 | |||
| AMO | 31.2 a | 0.25 | 75 | |||||
| EUG | 250 | 909.1 | 0.50 | 0.75 | 50 | |||
| STM | 2.5 a | 0.25 | 75 | |||||
| EUG | 250 | 71.4 | 0.50 | 0.75 | 50 | |||
| ERY | 15.6 a | 0.25 | 75 | |||||
| EUG | 250 | 256.4 | 0.50 | 0.62 | 50 | |||
| CHL | 1.9 a | 0.12 | 88 | |||||
| P. aerogenes | EUG | 250 | 4.3 | 0.50 | 1.50 | Addition | 50 | |
| AMP | 125 a | 1.00 | 0 | |||||
| EUG | 250 | 1.1 | 0.5 | 1.50 | 50 | |||
| AMO | 500 a | 1.00 | 0 | |||||
| EUG | 250 | 142.9 | 0.50 | 1.50 | 50 | |||
| STM | 15.6 a | 1.00 | 0 | |||||
| EUG | 250 | 35.8 | 0.50 | 1.00 | 50 | |||
| ERY | 31.2 a | 0.50 | 50 | |||||
| EUG | 250 | 147.1 | 0.50 | 1.00 | 50 | |||
| TC | 5 a | 0.50 | 50 | |||||
| S. enterica | EUG | 250 | 178.6 | 0.25 | 0.50 | Synergy | 75 | |
| STM | 12.5 a | 0.25 | 75 | |||||
| EUG | 500 | 71.4 | 0.50 | 1.50 | Addition | 50 | ||
| ERY | 31.2 a | 1.00 | 0 | |||||
| S. marcescens | EUG | 250 | 23.5 | 0.25 | 0.37 | Synergy | 75 | |
| TC | 31.2 a | 0.12 | 88 | |||||
| EUG | 250 | 15.8 | 0.25 | 0.50 | 75 | |||
| CHL | 31.2 a | 0.25 | 75 | |||||
| EUG | 250 | 8.5 | 0.25 | 0.75 | Addition | 75 | ||
| AMP | 62.5 a | 0.50 | 50 | |||||
| EUG | 250 | 8.9 | 0.25 | 0.75 | 75 | |||
| AMO | 62.5 a | 0.50 | 50 | |||||
| EUG | 250 | 8.9 | 0.25 | 0.75 | 75 | |||
| ERY | 125 a | 0.50 | 50 |
| Bacterial Strains | ||||
|---|---|---|---|---|
| Gram | Name | T/°C | Broth | Provider |
| + | Bacillus subtilis (ATCC 6633) | 30 | BHI | Thermo Scientific |
| Enterococcus faecalis (ATCC 19433) | 37 | |||
| Streptococcus agalactiae (ATCC 12386) | ||||
| Staphylococcus aureus (ATCC 9144) | TSB | |||
| − | Acinetobacter baumannii (ATCC 19606) | |||
| Escherichia coli (ATCC 25922) | ||||
| Klebsiella aerogenes (ATCC 13048) | 30 | NB | ||
| Klebsiella pneumoniae (C6) | 37 | |||
| Listeria monocytogenes (ATCC 7644) | BHI | |||
| Pasteurella aerogenes (ATCC 27883) | ||||
| Proteus mirabilis (ATCC 35659) | TSB | |||
| Pseudomonas aeruginosa (ATCC 27853) | ||||
| Salmonella enterica (ATCC 13311) | NB | |||
| Serratia marcescens (ATCC 13880) | 26 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrando, N.; Pino-Otín, M.R.; Terrado, E.; Ballestero, D.; Langa, E. Bioactivity of Eugenol: A Potential Antibiotic Adjuvant with Minimal Ecotoxicological Impact. Int. J. Mol. Sci. 2024, 25, 7069. https://doi.org/10.3390/ijms25137069
Ferrando N, Pino-Otín MR, Terrado E, Ballestero D, Langa E. Bioactivity of Eugenol: A Potential Antibiotic Adjuvant with Minimal Ecotoxicological Impact. International Journal of Molecular Sciences. 2024; 25(13):7069. https://doi.org/10.3390/ijms25137069
Chicago/Turabian StyleFerrando, Natalia, María Rosa Pino-Otín, Eva Terrado, Diego Ballestero, and Elisa Langa. 2024. "Bioactivity of Eugenol: A Potential Antibiotic Adjuvant with Minimal Ecotoxicological Impact" International Journal of Molecular Sciences 25, no. 13: 7069. https://doi.org/10.3390/ijms25137069





