Deciphering Immune Responses to Immunization via Transcriptional Analysis: A Narrative Review of the Current Evidence towards Personalized Vaccination Strategies
Abstract
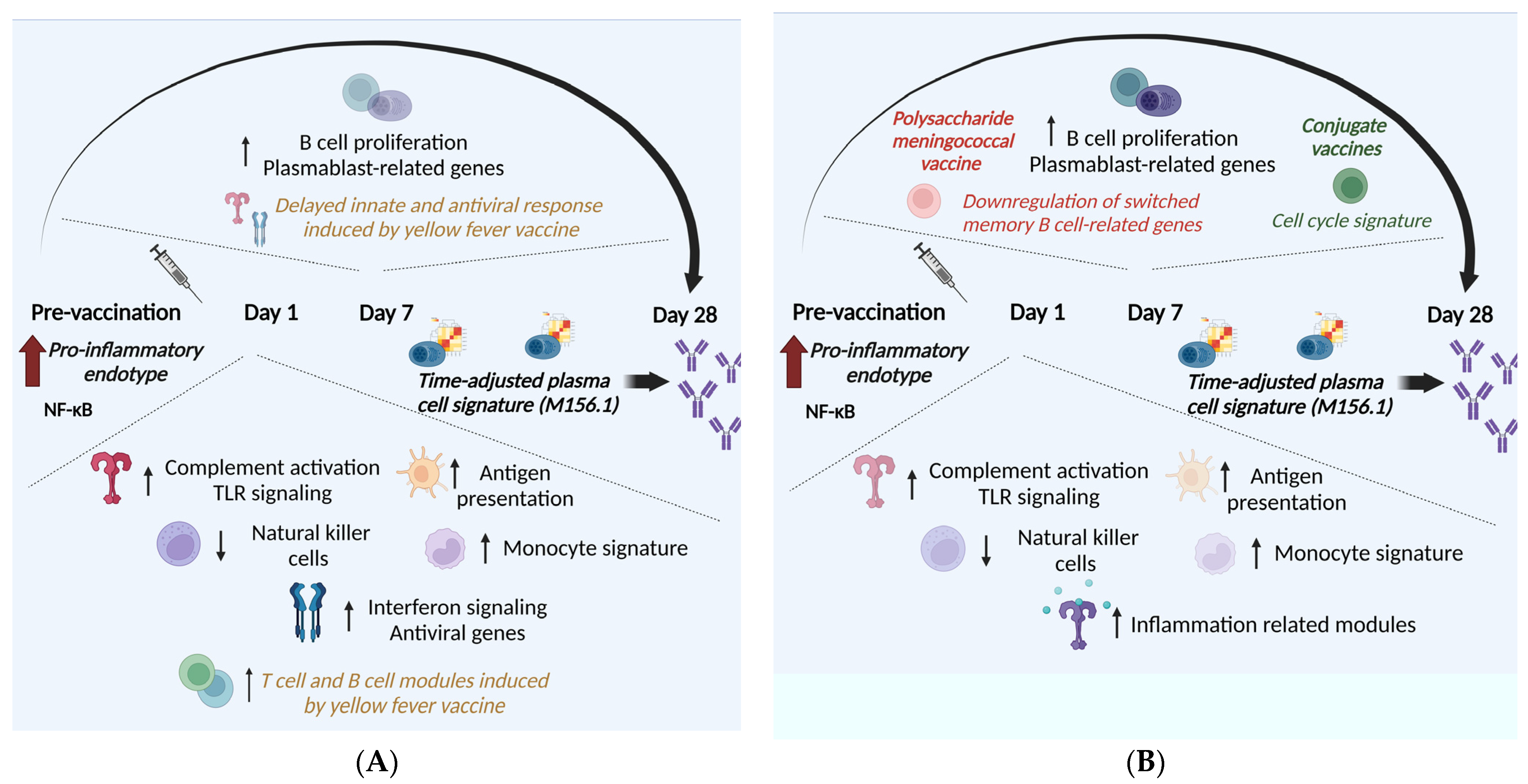
:1. Introduction
2. Vaccine-Induced Transcriptional Responses in Healthy Adults: Commonalities and Differences
2.1. Vaccines against Viral Infections
2.2. Vaccines against Bacterial and Parasitic Infections
3. Delineating Predictive Signatures of Vaccine Immunogenicity
3.1. Predictive Biomarkers of Immunogenicity for Vaccines against Viral Infections
3.2. Predictive Biomarkers of Immunogenicity for Vaccines against Bacterial and Parasitic Infections
4. The Impact of Age on Transcriptional Response to Immunization
5. Immunizations of High-Risk Populations—What Is Known So Far
6. Future Perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bugya, Z.; Prechl, J.; Szénási, T.; Nemes, É.; Bácsi, A.; Koncz, G. Multiple Levels of Immunological Memory and Their Association with Vaccination. Vaccines 2021, 9, 174. [Google Scholar] [CrossRef] [PubMed]
- Pollard, A.J.; Bijker, E.M. A Guide to Vaccinology: From Basic Principles to New Developments. Nat. Rev. Immunol. 2021, 21, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Pulendran, B.; Davis, M.M. The Science and Medicine of Human Immunology. Science 2020, 396, 6511. [Google Scholar] [CrossRef] [PubMed]
- Pulendran, B. Systems Vaccinology: Probing Humanity’s Diverse Immune Systems with Vaccines. Proc. Natl. Acad. Sci. USA 2014, 111, 12300–12306. [Google Scholar] [CrossRef] [PubMed]
- Hagan, T.; Nakaya, H.I.; Subramaniam, S.; Pulendran, B. Systems Vaccinology: Enabling Rational Vaccine Design with Systems Biological Approaches. Vaccine 2015, 33, 5294–5301. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, D.; Pollard, A.J. Characterizing Vaccine Responses Using Host Genomic and Transcriptomic Analysis. Clin. Infect. Dis. 2013, 57, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Pulendran, B.; Oh, J.Z.; Nakaya, H.I.; Ravindran, R.; Kazmin, D.A. Immunity to viruses: Learning from successful human vaccines. Immunol. Rev. 2013, 255, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Ross, T.M. Preexisting Influenza Specific Immunity and Vaccine Effectiveness. Expert Rev. Vaccines 2019, 18, 1043–1051. [Google Scholar] [CrossRef]
- Querec, T.D.; Akondy, R.S.; Lee, E.K.; Cao, W.; Nakaya, H.; Teuwen, D.; Pirani, A.; Gernert, K.; Deng, J.; Marzolf, B.; et al. Systems Biology Approach Predicts Immunogenicity of the Yellow Fever Vaccine in Humans. Nat. Immunol. 2009, 10, 116–125. [Google Scholar] [CrossRef]
- Gaucher, D.; Therrien, R.; Kettaf, N.; Angermann, B.R.; Boucher, G.; Filali-Mouhim, A.; Moser, J.M.; Mehta, R.S.; Drake, D.R.; Castro, E.; et al. Yellow Fever Vaccine Induces Integrated Multilineage and Polyfunctional Immune Responses. J. Exp. Med. 2008, 205, 3119–3131. [Google Scholar] [CrossRef]
- Tsang, J.S.; Schwartzberg, P.L.; Kotliarov, Y.; Biancotto, A.; Xie, Z.; Germain, R.N.; Wang, E.; Olnes, M.J.; Narayanan, M.; Golding, H.; et al. Global Analyses of Human Immune Variation Reveal Baseline Predictors of Post-Vaccination Responses. Cell 2014, 157, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Nakaya, H.I.; Hagan, T.; Duraisingham, S.S.; Lee, E.K.; Kwissa, M.; Rouphael, N.; Frasca, D.; Gersten, M.; Mehta, A.K.; Gaujoux, R.; et al. Systems Analysis of Immunity to Influenza Vaccination across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures. Immunity 2015, 43, 1186–1198. [Google Scholar] [CrossRef] [PubMed]
- Bucasas, K.L.; Franco, L.M.; Shaw, C.A.; Bray, M.S.; Wells, J.M.; Niño, D.; Arden, N.; Quarles, J.M.; Couch, R.B.; Belmont, J.W. Early Patterns of Gene Expression Correlate with the Humoral Immune Response to Influenza Vaccination in Humans. J. Infect. Dis. 2011, 203, 921–929. [Google Scholar] [CrossRef]
- Nakaya, H.I.; Wrammert, J.; Lee, E.K.; Racioppi, L.; Marie-Kunze, S.; Haining, W.N.; Means, A.R.; Kasturi, S.P.; Li, G.; Mccausland, M.; et al. Systems Biology of Seasonal Influenza Vaccination in Humans. Nat. Immunol. 2012, 12, 786–795. [Google Scholar] [CrossRef]
- Avey, S.; Mohanty, S.; Chawla, D.G.; Meng, H.; Bandaranayake, T.; Ueda, I.; Zapata, H.J.; Park, K.; Blevins, T.P.; Tsang, S.; et al. Seasonal Variability and Shared Molecular Signatures of Inactivated Influenza Vaccination in Young and Older Adults. J. Immunol. 2020, 204, 1661–1673. [Google Scholar] [CrossRef] [PubMed]
- Furman, D.; Jojic, V.; Kidd, B.; Shen-Orr, S.; Price, J.; Jarrell, J.; Tse, T.; Huang, H.; Lund, P.; Maecker, H.T.; et al. Apoptosis and Other Immune Biomarkers Predict Influenza Vaccine Responsiveness. Mol. Syst. Biol. 2013, 9, 659. [Google Scholar] [CrossRef]
- HIPC-CHI Signatures Project Team; HIPC-I Consortium. Multicohort Analysis Reveals Baseline Transcriptional Predictors of Influenza Vaccination Responses. Sci. Immunol. 2017, 2, eaal4656. [Google Scholar] [CrossRef]
- Cao, R.G.; Suarez, N.M.; Obermoser, G.; Lopez, S.M.C.; Flano, E.; Mertz, S.E.; Albrecht, R.A.; García-Sastre, A.; Mejias, A.; Xu, H.; et al. Differences in Antibody Responses between Trivalent Inactivated Influenza Vaccine and Live Attenuated Influenza Vaccine Correlate with the Kinetics and Magnitude of Interferon Signaling in Children. J. Infect. Dis. 2014, 210, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, J.; Fan, R.; Zhao, W.; Han, T.; Duan, K.; Li, X.; Zeng, P.; Deng, J.; Zhang, J.; et al. Identifying Potential Candidate Hub Genes and Functionally Enriched Pathways in the Immune Responses to Quadrivalent Inactivated Influenza Vaccines in the Elderly Through Co-Expression Network Analysis. Front. Immunol. 2020, 11, 603337. [Google Scholar] [CrossRef]
- Alcorn, J.F.; Avula, R.; Chakka, A.B.; Schwarzmann, W.E.; Nowalk, M.P.; Lin, C.J.; Ortiz, M.A.; Horne, W.T.; Chandran, U.R.; Nagg, J.P.; et al. Differential Gene Expression in Peripheral Blood Mononuclear Cells from Children Immunized with Inactivated Influenza Vaccine. Hum. Vaccines Immunother. 2020, 16, 1782–1790. [Google Scholar] [CrossRef]
- Zhu, W.; Higgs, B.W.; Morehouse, C.; Streicher, K.; Ambrose, C.S.; Woo, J.; Kemble, G.W.; Jallal, B.; Yao, Y. A Whole Genome Transcriptional Analysis of the Early Immune Response Induced by Live Attenuated and Inactivated Influenza Vaccines in Young Children. Vaccine 2010, 28, 2865–2876. [Google Scholar] [CrossRef]
- de Armas, L.R.; George, V.; Filali-Mouhim, A.; Steel, C.; Parmigiani, A.; Cunningham, C.K.; Weinberg, A.; Trautmann, L.; Sekaly, R.P.; Cameron, M.J.; et al. Transcriptional and Immunologic Correlates of Response to Pandemic Influenza Vaccine in Aviremic, HIV-Infected Children. Front. Immunol. 2021, 12, 639358. [Google Scholar] [CrossRef]
- Qiu, S.; He, P.; Fang, X.; Tong, H.; Lv, J.; Liu, J.; Zhang, L.; Zhai, X.; Wang, L.; Hu, Z.; et al. Significant Transcriptome and Cytokine Changes in Hepatitis B Vaccine Non-Responders Revealed by Genome-Wide Comparative Analysis. Hum. Vaccines Immunother. 2018, 14, 1763–1772. [Google Scholar] [CrossRef]
- Shannon, C.P.; Blimkie, T.M.; Ben-Othman, R.; Gladish, N.; Amenyogbe, N.; Drissler, S.; Edgar, R.D.; Chan, Q.; Krajden, M.; Foster, L.J.; et al. Multi-Omic Data Integration Allows Baseline Immune Signatures to Predict Hepatitis B Vaccine Response in a Small Cohort. Front. Immunol. 2020, 11, 578801. [Google Scholar] [CrossRef] [PubMed]
- Fourati, S.; Cristescu, R.; Loboda, A.; Talla, A.; Filali, A.; Railkar, R.; Schaeffer, A.K.; Favre, D.; Gagnon, D.; Peretz, Y.; et al. Pre-Vaccination Inflammation and B-Cell Signalling Predict Age-Related Hyporesponse to Hepatitis B Vaccination. Nat. Commun. 2016, 7, 10369. [Google Scholar] [CrossRef]
- Weinberger, B.; Haks, M.C.; de Paus, R.A.; Ottenhoff, T.H.M.; Bauer, T.; Grubeck-Loebenstein, B. Impaired Immune Response to Primary but Not to Booster Vaccination against Hepatitis B in Older Adults. Front. Immunol. 2018, 9, 1035. [Google Scholar] [CrossRef]
- Rechtien, A.; Richert, L.; Lorenzo, H.; Martrus, G.; Hejblum, B.; Dahlke, C.; Kasonta, R.; Zinser, M.; Stubbe, H.; Matschl, U.; et al. Systems Vaccinology Identifies an Early Innate Immune Signature as a Correlate of Antibody Responses to the Ebola Vaccine RVSV-ZEBOV. Cell Rep. 2017, 20, 2251–2261. [Google Scholar] [CrossRef] [PubMed]
- Vianello, E.; Gonzalez-Dias, P.; van Veen, S.; Engele, C.G.; Quinten, E.; Monath, T.P.; Medaglini, D.; Agnandij, S.T.; Ahmed, R.; Anderson, J.; et al. Transcriptomic Signatures Induced by the Ebola Virus Vaccine RVSVΔG-ZEBOV-GP in Adult Cohorts in Europe, Africa, and North America: A Molecular Biomarker Study. Lancet Microbe 2022, 3, e113–e123. [Google Scholar] [CrossRef] [PubMed]
- Santoro, F.; Donato, A.; Lucchesi, S.; Sorgi, S.; Gerlini, A.; Haks, M.C.; Ottenhoff, T.H.M.; Gonzalez-Dias, P.; Nakaya, H.I.; Huttner, A.; et al. Human Transcriptomic Response to the VSV-Vectored Ebola Vaccine. Vaccines 2021, 9, 67. [Google Scholar] [CrossRef]
- Li, S.; Sullivan, N.L.; Rouphael, N.; Yu, T.; Banton, S.; Maddur, M.S.; McCausland, M.; Chiu, C.; Canniff, J.; Dubey, S.; et al. Metabolic Phenotypes of Response to Vaccination in Humans. Cell 2017, 169, 862–877.e17. [Google Scholar] [CrossRef]
- Popper, S.J.; Strouts, F.R.; Lindow, J.C.; Cheng, H.K.; Montoya, M.; Balmaseda, A.; Durbin, A.P.; Whitehead, S.S.; Harris, E.; Kirkpatrick, B.D.; et al. Early Transcriptional Responses after Dengue Vaccination Mirror the Response to Natural Infection and Predict Neutralizing Antibody Titers. J. Infect. Dis. 2018, 218, 1911–1921. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Che, Y.; Dean, H.J.; Lorenzo-Redondo, R.; Stewart, M.; Keller, C.K.; Whorf, D.; Mills, D.; Dulin, N.N.; Kim, T.; et al. Transcriptome-Wide Changes in Gene Expression, Splicing, and LncRNAs in Response to a Live Attenuated Dengue Virus Vaccine. Cell Rep. 2022, 38, 110341. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, P.S.; Scott, M.K.D.; Hagan, T.; Li, C.; Feng, Y.; Wimmers, F.; Grigoryan, L.; Trisal, M.; Edara, V.V.; Lai, L.; et al. Systems Vaccinology of the BNT162b2 MRNA Vaccine in Humans. Nature 2021, 596, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Papadatou, I.; Geropeppa, M.; Verrou, K.-Μ.; Tzanoudaki, M.; Lagousi, T.; Liatsis, E.; Spoulou, V. SARS-CoV-2 MRNA Dual Immunization Induces Innate Transcriptional Signatures, Establishes T-Cell Memory and Coordinates the Recall Response. Vaccines 2023, 11, 103. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Knabl, L.; Moliva, J.I.; Knabl, L.; Werner, A.P.; Boyoglu-Barnum, S.; Kapferer, S.; Pateter, B.; Walter, M.; Sullivan, N.J.; et al. mRNA Vaccination in Octogenarians 15 and 20 Months after Recovery from COVID-19 Elicits Robust Immune and Antibody Responses That Include Omicron. Cell Rep. 2022, 39, 110680. [Google Scholar] [CrossRef] [PubMed]
- Ryan, F.J.; Norton, T.S.; McCafferty, C.; Blake, S.J.; Stevens, N.E.; James, J.; Eden, G.L.; Tee, Y.C.; Benson, S.C.; Masavuli, M.G.; et al. A Systems Immunology Study Comparing Innate and Adaptive Immune Responses in Adults to COVID-19 MRNA and Adenovirus Vectored Vaccines. Cell Rep. Med. 2023, 4, 100971. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Go, J.; Sung, H.; Kim, S.W.; Walter, M.; Knabl, L.; Furth, P.A.; Hennighausen, L.; Huh, J.W. Heterologous ChAdOx1-BNT162b2 Vaccination in Korean Cohort Induces Robust Immune and Antibody Responses That Includes Omicron. iScience 2022, 25, 104473. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, X.; Li, C.; Kou, Z.; Lin, L.; Yao, M.; Pang, B.; Zhang, X.; Duan, Q.; Tian, X.; et al. Transcriptome Analysis of Peripheral Blood Mononuclear Cells in SARS-CoV-2 Naïve and Recovered Individuals Vaccinated with Inactivated Vaccine. Front. Cell. Infect. Microbiol. 2022, 11, 821828. [Google Scholar] [CrossRef] [PubMed]
- Bartholomeus, E.; De Neuter, N.; Meysman, P.; Suls, A.; Keersmaekers, N.; Elias, G.; Jansens, H.; Hens, N.; Smits, E.; Van Tendeloo, V.; et al. Transcriptome Profiling in Blood before and after Hepatitis B Vaccination Shows Significant Differences in Gene Expression between Responders and Non-Responders. Vaccine 2018, 36, 6282–6289. [Google Scholar] [CrossRef]
- Ong, E.Z.; Gan, E.S.; de Alwis, R.; Wijaya, L.; Ong, X.M.; Zhang, M.; Wong, A.W.; Cheung, Y.B.; Zellweger, R.M.; Ooi, E.E.; et al. Genomic Signature of Early T-Cell Response Is Associated with Lower Antibody Titer Threshold for Sterilizing Immunity. Antiviral Res. 2019, 166, 35–41. [Google Scholar] [CrossRef]
- Noho-Konteh, F.; Adetifa, J.U.; Cox, M.; Hossin, S.; Reynolds, J.; Le, M.T.; Sanyang, L.C.; Drammeh, A.; Plebanski, M.; Forster, T.; et al. Sex-Differential Non-Vaccine Specific Immunological Effects of Diphtheria-Tetanus-Pertussis and Measles Vaccination. Clin. Infect. Dis. 2016, 63, 1213–1226. [Google Scholar]
- Gómez-Carballa, A.; Barral-Arca, R.; Cebey-López, M.; Currás-Tuala, M.J.; Pischedda, S.; Gómez-Rial, J.; Habgood-Coote, D.; Herberg, J.A.; Kaforou, M.; Martinón-Torres, F.; et al. Host Transcriptomic Response Following Administration of Rotavirus Vaccine in Infants’ Mimics Wild Type Infection. Front. Immunol. 2021, 11, 580219. [Google Scholar] [CrossRef]
- Obermoser, G.; Presnell, S.; Domico, K.; Xu, H.; Wang, Y.; Anguiano, E.; Thompson-Snipes, L.; Ranganathan, R.; Zeitner, B.; Bjork, A.; et al. Systems Scale Interactive Exploration Reveals Quantitative and Qualitative Differences in Response to Influenza and Pneumococcal Vaccines. Immunity 2013, 38, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Rouphael, N.; Duraisingham, S.; Romero-Stainer, S.; Presnell, S.; Davis, C.; Schmidt, D.S.; Johnson, S.E.; Milton, A.; Rajam, G.; et al. Molecular Signatures of Antibody Responses Derived from a Systems Biological Study of 5 Human Vaccines. Nat. Immunol. 2014, 15, 195–204. [Google Scholar] [CrossRef]
- O’Connor, D.; Clutterbuck, E.A.; Thompson, A.J.; Snape, M.D.; Ramasamy, M.N.; Kelly, D.F.; Pollard, A.J. High-Dimensional Assessment of B-Cell Responses to Quadrivalent Meningococcal Conjugate and Plain Polysaccharide Vaccine. Genome Med. 2017, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, D.; Pinto, M.V.; Sheerin, D.; Tomic, A.; Drury, R.E.; Channon-Wells, S.; Galal, U.; Dold, C.; Robinson, H.; Kerridge, S.; et al. Gene Expression Profiling Reveals Insights into Infant Immunological and Febrile Responses to Group B Meningococcal Vaccine. Mol. Syst. Biol. 2020, 16, e9888. [Google Scholar] [CrossRef]
- da Silva Antunes, R.; Soldevila, F.; Pomaznoy, M.; Babor, M.; Bennett, J.; Tian, Y.; Khalil, N.; Qian, Y.; Mandava, A.; Scheuermann, R.H.; et al. A System-View of Bordetella Pertussis Booster Vaccine Responses in Adults Primed with Whole-Cell versus Acellular Vaccine in Infancy. JCI Insight 2021, 6, 17–19. [Google Scholar] [CrossRef]
- Kazmin, D.; Nakaya, H.I.; Lee, E.K.; Johnson, M.J.; Van Der Most, R.; Van Den Berg, R.A.; Ballou, W.R.; Jongert, E.; Wille-Reece, U.; Ockenhouse, C.; et al. Systems Analysis of Protective Immune Responses to RTS,S Malaria Vaccination in Humans. Proc. Natl. Acad. Sci. USA 2017, 114, 2425–2430. [Google Scholar] [CrossRef] [PubMed]
- Moncunill, G.; Carnes, J.; Young, W.C.; Carpp, L.; De Rosa, S.; Campo, J.J.; Nhabomba, A.; Mpina, M.; Jairoce, C.; Finak, G.; et al. Transcriptional Correlates of Malaria in RTS,S/AS01-Vaccinated African Children: A Matched Case-Control Study. eLife 2022, 11, e70393. [Google Scholar] [CrossRef] [PubMed]
- Vahey, M.T.; Wang, Z.; Kester, K.E.; Cummings, J.; Heppner, D.G.; Nau, M.E.; Ofori-Anyinam, O.; Cohen, J.; Coche, T.; Ripley Ballou, W.; et al. Expression of Genes Associated with Immunoproteasome Processing of Major Histocompatibility Complex Peptides Is Indicative of Protection with Adjuvanted RTS,S Malaria Vaccine. J. Infect. Dis. 2010, 201, 580–589. [Google Scholar] [CrossRef]
- van den Berg, R.A.; Coccia, M.; Ballou, W.R.; Kester, K.E.; Ockenhouse, C.F.; Vekemans, J.; Jongert, E.; Didierlaurent, A.M.; van der Most, R.G. Predicting RTS,S Vaccine-Mediated Protection from Transcriptomes in a Malaria-Challenge Clinical Trial. Front. Immunol. 2017, 8, 557. [Google Scholar] [CrossRef] [PubMed]
- Hagan, T.; Gerritsen, B.; Tomalin, L.E.; Fourati, S.; Mulè, M.P.; Chawla, D.G.; Rychkov, D.; Henrich, E.; Miller, H.E.R.; Diray-Arce, J.; et al. Transcriptional Atlas of the Human Immune Response to 13 Vaccines Reveals a Common Predictor of Vaccine-Induced Antibody Responses. Nat. Immunol. 2022, 23, 1788–1798. [Google Scholar] [CrossRef] [PubMed]
- Klotman, M.E.; Chang, T.L. Defensins in Innate Antiviral Immunity. Nat. Rev. Immunol. 2006, 6, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Dejonckheere, E.; Vandenbroucke, R.E.; Libert, C. Matrix Metalloproteinase8 Has a Central Role in Inflammatory Disorders and Cancer Progression. Cytokine Growth Factor Rev. 2011, 22, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Grimm, M.; Schmitt, S.; Teriete, P.; Biegner, T.; Stenzl, A.; Hennenlotter, J.; Muhs, H.J.; Munz, A.; Nadtotschi, T.; König, K.; et al. A Biomarker Based Detection and Characterization of Carcinomas Exploiting Two Fundamental Biophysical Mechanisms in Mammalian Cells. BMC Cancer 2013, 13, 569. [Google Scholar] [CrossRef] [PubMed]
- Atreya, I.; Schimanski, C.C.; Becker, C.; Wirtz, S.; Dornhoff, H.; Schnürer, E.; Berger, M.R.; Galle, P.R.; Herr, W.; Neurath, M.F. The T-Box Transcription Factor Eomesodermin Controls CD8 T Cell Activity and Lymph Node Metastasis in Human Colorectal Cancer. Gut 2007, 56, 1572–1578. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.; Wolfraim, L.A.; Drake, C.G.; Horton, M.R.; Powell, J.D. Cutting Edge: TCR-Induced NAB2 Enhances T Cell Function by Coactivating IL-2 Transcription. J. Immunol. 2006, 177, 8301–8305. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W.; Rice, C.M. Interferon-Stimulated Genes and Their Antiviral Effector Functions. Curr. Opin. Virol. 2011, 1, 519–525. [Google Scholar] [CrossRef]
- Huang, I.C.; Bailey, C.C.; Weyer, J.L.; Radoshitzky, S.R.; Becker, M.M.; Chiang, J.J.; Brass, A.L.; Ahmed, A.A.; Chi, X.; Dong, L.; et al. Distinct Patterns of IFITM-Mediated Restriction of Filoviruses, SARS Coronavirus, and Influenza A Virus. PLoS Pathog. 2011, 7, e1001258. [Google Scholar] [CrossRef]
- Pilla-Moffett, D.; Barber, M.F.; Taylor, G.A.; Coers, J. Interferon-Inducible GTPases in Host Resistance, Inflammation and Disease Graphical Abstract HHS Public Access. J Mol Biol. 2016, 428, 3495–3513. [Google Scholar] [CrossRef]
- Papadatou, I.; Spoulou, V. Pneumococcal Vaccination in High-Risk Individuals: Are We Doing It Right? Clin. Vaccine Immunol. 2016, 23, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Fourati, S.; Tomalin, L.E.; Mulè, M.P.; Chawla, D.G.; Gerritsen, B.; Rychkov, D.; Henrich, E.; Miller, H.E.R.; Hagan, T.; Diray-Arce, J.; et al. Pan-Vaccine Analysis Reveals Innate Immune Endotypes Predictive of Antibody Responses to Vaccination. Nat. Immunol. 2022, 23, 1777–1787. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, J.A.; Radtke, D.; Maurberger, A.; Winkler, T.H.; Nitschke, L. Grb2 Regulates B-Cell Maturation, B-Cell Memory Responses and Inhibits B-Cell Ca2+ Signalling. EMBO J. 2011, 30, 1621–1633. [Google Scholar] [CrossRef] [PubMed]
- Packard, T.A.; Cambier, J.C. B Lymphocyte Antigen Receptor Signaling: Initiation, Amplification, and Regulation. F1000Prime Rep. 2013, 5, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Pei, G.; Bronietzki, M.; Gutierrez, M.G. Immune Regulation of Rab Proteins Expression and Intracellular Transport. J. Leukoc. Biol. 2012, 92, 41–50. [Google Scholar] [CrossRef]
- Meliopoulos, V.A.; Andersen, L.E.; Brooks, P.; Yan, X.; Bakre, A.; Coleman, J.K.; Tompkins, S.M.; Tripp, R.A. MicroRNA Regulation of Human Protease Genes Essential for Influenza Virus Replication. PLoS ONE 2012, 7, e37169. [Google Scholar] [CrossRef]
- Parruti, G.; Peracchia, F.; Sallese, M.; Ambrosini, G.; Masini, M.; Rotilio, D.; De Blasi, A. Molecular Analysis of Human β-Arrestin-1: Cloning, Tissue Distribution, and Regulation of Expression. Identification of Two Isoforms Generated by Alternative Splicing. J. Biol. Chem. 1993, 268, 9753–9761. [Google Scholar] [CrossRef]
- Nakaya, H.I.; Clutterbuck, E.; Kazmin, D.; Wang, L.; Cortese, M.; Bosinger, S.E.; Patel, N.B.; Zak, D.E.; Aderem, A.; Dong, T.; et al. Systems Biology of Immunity to MF59-Adjuvanted versus Nonadjuvanted Trivalent Seasonal Influenza Vaccines in Early Childhood. Proc. Natl. Acad. Sci. USA 2016, 113, 1853–1858. [Google Scholar] [CrossRef]
- Shaw, A.C.; Goldstein, D.R.; Montgomery, R.R. Age-Dependent Dysregulation of Innate Immunity. Nat. Rev. Immunol. 2013, 13, 875–887. [Google Scholar] [CrossRef]
- Ciabattini, A.; Nardini, C.; Santoro, F.; Garagnani, P.; Franceschi, C.; Medaglini, D. Vaccination in the Elderly: The Challenge of Immune Changes with Aging. Semin. Immunol. 2018, 40, 83–94. [Google Scholar] [CrossRef]
- Kotliarov, Y.; Sparks, R.; Martins, A.J.; Mulè, M.P.; Lu, Y.; Goswami, M.; Kardava, L.; Banchereau, R.; Pascual, V.; Biancotto, A.; et al. Broad Immune Activation Underlies Shared Set Point Signatures for Vaccine Responsiveness in Healthy Individuals and Disease Activity in Patients with Lupus. Nat. Med. 2020, 26, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Kedersha, N. Visibly Stressed: The Role of EIF2, TIA-1, and Stress Granules in Protein Translation. Cell Stress Chaperones 2002, 7, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Krebs, J.; Wilson, A.; Kisielow, P. Calmodulin-Dependent Protein Kinase IV during T-Cell Development. Biochem. Biophys. Res. Commun. 1997, 241, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Illario, M.; Giardino-Torchia, M.L.; Sankar, U.; Ribar, T.J.; Galgani, M.; Vitiello, L.; Masci, A.M.; Bertani, F.R.; Ciaglia, E.; Astone, D.; et al. Calmodulin-Dependent Kinase IV Links Toll-like Receptor 4 Signaling with Survival Pathway of Activated Dendritic Cells. Blood 2008, 111, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Hagan, T.; Pulendran, B. Will Systems Biology Deliver Its Promise and Contribute to the Development of New or Improved Vaccines?: From Data to Understanding through Systems Biology. Cold Spring Harb. Perspect. Biol. 2018, 10, a033308. [Google Scholar] [CrossRef] [PubMed]
- de Mot, L.; Bechtold, V.; Bol, V.; Callegaro, A.; Coccia, M.; Essaghir, A.; Hasdemir, D.; Ulloa-Montoya, F.; Siena, E.; Smilde, A.; et al. Transcriptional Profiles of Adjuvanted Hepatitis B Vaccines Display Variable Interindividual Homogeneity but a Shared Core Signature. Sci. Transl. Med. 2020, 12, eaay8618. [Google Scholar] [CrossRef] [PubMed]
- Harandi, A.M. Systems Analysis of Human Vaccine Adjuvants. Semin. Immunol. 2018, 39, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Victora, G.D.; Nussenzweig, M.C. Germinal Centers. Annu. Rev. Immunol. 2012, 30, 429–457. [Google Scholar] [CrossRef]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human Nutrition, the Gut Microbiome and the Immune System. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef]
- Duraisingham, S.S.; Rouphael, N.; Cavanagh, M.M.; Nakaya, H.I.; Goronzy, J.J.; Pulendran, B. Systems Biology of Vaccination in the Elderly. Curr. Top. Microbiol. Immunol. 2013, 363, 117–142. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Vastrik, I.; D’Eustachio, P.; Schmidt, E.; Joshi-Tope, G.; Gopinath, G.; Croft, D.; de Bono, B.; Gillespie, M.; Jassal, B.; Lewis, S.; et al. Reactome: A Knowledge Base of Biologic Pathways and Processes. Genome Biol. 2007, 8, R39. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From Genomics to Chemical Genomics: New Developments in KEGG. Nucleic Acids Res. 2006, 34, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular Signatures Database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
| Reference | Vaccine (Doses) | Population | Vaccination Schedule (Days) | Bleeding Points (Days) |
|---|---|---|---|---|
| Studies on vaccines against viral infections | ||||
| Querec et al., 2009, Nat. Immunol [9] | YF-17D (1) | 15 adults (18–45 y.o.) | 0 | 0, 3, 7 |
| Gaucher et al., 2008, J. Exp. Med [10] | YF-17D (1) | 15 adults | 0 | 0, 3, 7, 10, 14, 28, 60 |
| Tsang et al., 2014, Cell [11] | TIV (1) | 63 adults (>18 y.o.) | 0 | 0, 1, 7, 70 |
| Nakaya et al., Immunity, 2015 [12] | TIV (1) | 430 adults (>18 y.o.) | 0 | 0, 1, 3, 7 |
| Bucasas et al., 2011, J. Infect. Dis. [13] | TIV (1) | 119 adults (18–40 y.o.) | 0 | 0, 1, 3, 14 |
| Nakaya et al., 2011, Nat. Immunol. [14] | TIV or LAIV (1) | 56 adults (18–50 y.o.) | 0 | 0, 3, 7 |
| Avey et al., 2020, J. Immunol. [15] | TIV (1) | 317 adults (21–30 y.o., >65 y.o.) | 0 | 0, 2–4, 7, 28 |
| Furman et al., 2013, Mol. Syst. Biol. [16] | TIV (1) | 89 adults (20–89 y.o.) | 0 | 0, 28 |
| HIPC-CHI Signatures Project Team, 2017, Sci. Immunol. [17] | TIV (1) | 516 adults (<35 y.o., >60 y.o.) | 0 | 0 |
| Cao et al., 2014, J. Infect. Dis. [18] | LAIV or TIV (1) | 40 infants, children (6 m.o.–14 y.o.) | 0 | 0, 1, 7, 30 |
| Yang et al., 2020, Front. Immunol. [19] | QIV (1) | 16 adults (>60 y.o.) | 0 | 0, 3, 28, 180 |
| Alcorn et al., 2020, Hum. Vaccines Immunother. [20] | TIV (1) | 16 children (3–17 y.o.) | 0 | 0, 3, 7 |
| Zhu et al., 2010, Vaccine [21] | LAIV or TIV (1) | 85 infants, children (1–3 y.o.) | 0 | 0, 7–10 |
| de Armas et al., 2021, Front. Immunol. [22] | TIV (2) | 40 children, adults (4–24 y.o.) | 0, 21–28 | 0, 21–28 |
| Qiu et al., 2018, Hum. Vaccines Immunother. [23] | HBV (3) | 14 adults (18–50 y.o.) | 0, 28, 56 | 0 3, 7, 28, 35, 56 |
| Shannon et al., 2020, Front. Immunol. [24] | HBV (3) | 15 adults (44–73 y.o.) | 0, 28, 180 | 0, 1, 3, 7, 14 |
| Fourati et al., 2016, Nat. Commun. [25] | HBV (2) | 174 adults (25–40 y.o., >65 y.o.) | 0, 30 | 0, 7 |
| Weinberger et al., 2018, Front. Immunol. [26] | HBV (3 primary or 1 booster) | 41 adults (20–40 y.o., >60 y.o.) | 0, 30, 180 | 0, 1 |
| Rechtien et al., 2017, Cell Rep. [27] | rVSV-ZEBOV (1) | 20 adults (18–55 y.o.) | 0 | 0, 1, 3, 7 |
| Vianello et al., 2022, The Lancet Microbe [28] | rVSV-ZEBOV (1) | 354 adults (18–55 y.o.) | 0 | 0, 1, 2, 3, 7, 14, 28 |
| Santoro et al., 2021, Vaccines [29] | rVSV-ZEBOV (1) | 115 adults (18–65 y.o.) | 0 | 0, 1, 3, 7, 14, 28, 35, 168 |
| Li et al., 2017, Cell [30] | Zostavax (1) | 77 adults (25–40 y.o., 60–79 y.o.) | 0 | 0, 1, 3, 7 |
| Popper et al., 2018, J. Infect. Dis. [31] | Dengvaxia (1) | 10 adults (<65 y.o.) | 0 | 0, 2, 5, 6, 8, 9, 12, 14, 20, 29, 42, 180 |
| Kim et al., 2022, Cell Reports [32] | Qdenga (2) | 20 adults | 0, 90 | 0, 2, 4, 7, 92 |
| Arunachalam et al., 2021, Nature [33] | BNT162b2 (2) | 56 adults (19–79 y.o.) | 0, 21 | 0, 1, 7, 21, 22, 23, 28 |
| Papadatou et al., 2023, Vaccines [34] | BNT162b2 (3) | 18 adults (28–65 y.o.) | 0, 21, 180 | 21, 24 |
| Lee et al., 2022, Cell Rep. [35] | BNT162b2 (2) | 30 adults (I: median 58 y.o., II: >80 y.o.) | I: 0, 35, II: 0, 150 | 0, 1, 7 |
| Ryan et al., 2023, Cell Reports Med. [36] | BNT162b2 or ChAd/mRNA-1273 or BNT162b2 (3) | 102 adults (39 ± 11 y.o.) | 0, 28, 164 | 0, 6, 29, 165 |
| Lee et al., 2022, iScience [37] | ChAd/BNT162b2 (2) | 46 adults (median 35 y.o.) | 0, 70 | 0, 3, 7, 70, 73, 77 |
| Zhang et al., 2022, Front. Cell. Infect. Microbiol. [38] | Sinovac CoronaVac (3) | 20 adults (24–61 y.o.) | 0, 28, 194 | 0, 14, 42, 208 |
| Bartholomeus et al., 2018, Vaccine [39] | MMR (1) | 40 adults (20–30 y.o.) | 0 | 0, 3, 7 |
| Ong et al., 2019, Antiviral Res. [40] | MMR (1) | 98 adults (<65 y.o.) | 0 | 0, 1, 3 |
| Noho-Konteh et al. Clin. Infect. Dis. [41] | MV (1) | 183 infants (9 m.o.) | 0 | 0, 28 |
| Gomez-Carballa et al., 2021, Front. Immunol. [42] | RV5 (3) | 14 infants, children (2–34 m.o.) | 0, 30, 60 | 0, 90 |
| Studies on vaccines against bacterial and parasitic infections | ||||
| Obermoser et al., 2013, Immunity [43] | PPV23 or TIV (1) | 12 adults (18–64 y.o.) | 0 | 0, 1, 3, 7, 10, 14, 21, 28 |
| Li et al., 2014, Nat. Immuol [44] | MPSV4, MCV4 (1) | 30 adults (18–45 y.o.) | 0 | 0, 3, 7 |
| O’Connor et al., 2017, Genome Med. [45] | MPSV4 or MCV4/MCV4 (2) | 20 adults (30–70 y.o.) | 0, 28 | 0, 7, 28, 35, 36 |
| O’Connor et al., 2020, Mol. Syst. Biol. [46] | 4CMenB (1) | 181 infants (4 m.o.) | 0 | 0 (0 h, 4 h), 1 (24 h), 3, 7 |
| da Silva Antunes et al. 2021, JCI Insight [47] | Tdap (1) | 18 adults | 0 | 0, 1, 3, 7, 14 |
| Kazmin et al., 2017, Proc. Natl. Acad. Sci. [48] | RTS,S (3) | 46 adults (mean 30 y.o.) | 0, 28, 56 | 0, 1, 2, 6, 14, 28, 29, 34, 56, 57, 62 |
| Moncunill et al., 2022, Elife [49] | RTS,S (3) | 350 infants, children (6–12 w.o., 5–17 m.o.) | 0, 30, 60 | 0, 90 |
| Vahey et al., 2010, J. Infect. Dis. [50] | RTS,S (3) | 39 adults (18–45 y.o.) | 0, 30, 60 | 60, 61, 63, 74 |
| van den Berg et al., 2017, Front. Immunol. [51] | RTS,S (3) | 117 adults (18–45 y.o.) | 0, 30, 60 | 0, 1, 30, 31, 60, 61, 63, 74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadatou, I.; Geropeppa, M.; Piperi, C.; Spoulou, V.; Adamopoulos, C.; Papavassiliou, A.G. Deciphering Immune Responses to Immunization via Transcriptional Analysis: A Narrative Review of the Current Evidence towards Personalized Vaccination Strategies. Int. J. Mol. Sci. 2024, 25, 7095. https://doi.org/10.3390/ijms25137095
Papadatou I, Geropeppa M, Piperi C, Spoulou V, Adamopoulos C, Papavassiliou AG. Deciphering Immune Responses to Immunization via Transcriptional Analysis: A Narrative Review of the Current Evidence towards Personalized Vaccination Strategies. International Journal of Molecular Sciences. 2024; 25(13):7095. https://doi.org/10.3390/ijms25137095
Chicago/Turabian StylePapadatou, Ioanna, Maria Geropeppa, Christina Piperi, Vana Spoulou, Christos Adamopoulos, and Athanasios G. Papavassiliou. 2024. "Deciphering Immune Responses to Immunization via Transcriptional Analysis: A Narrative Review of the Current Evidence towards Personalized Vaccination Strategies" International Journal of Molecular Sciences 25, no. 13: 7095. https://doi.org/10.3390/ijms25137095





