Comparative Cytological and Gene Expression Analysis Reveals That a Common Wild Rice Inbred Line Showed Stronger Drought Tolerance Compared with the Cultivar Rice
Abstract
:1. Introduction
2. Results
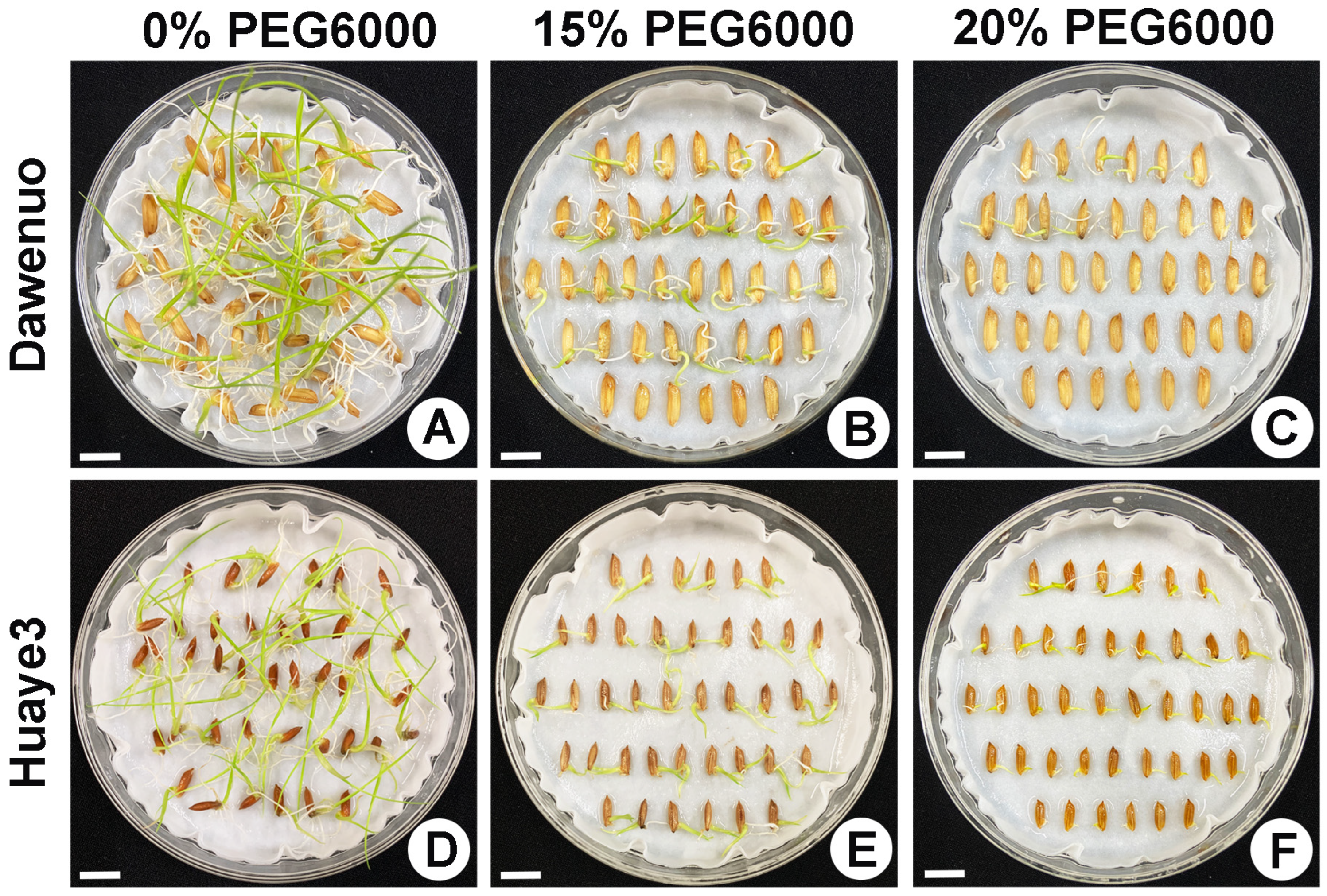
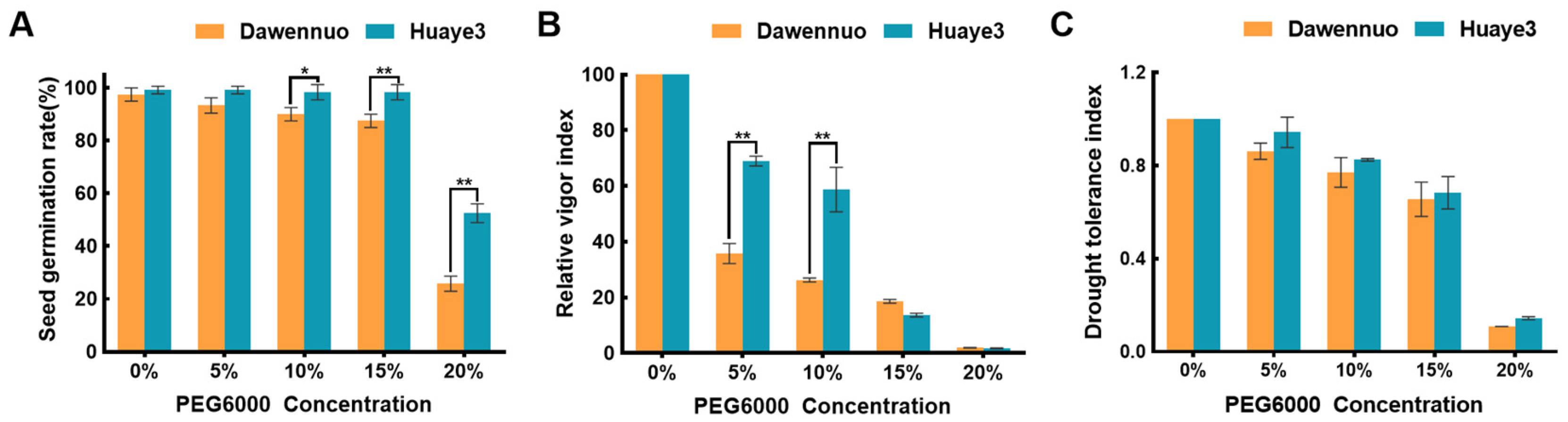
2.1. Phenotypic Variation of Seed Germination in Huaye3 and Dawennuo under Drought Stress
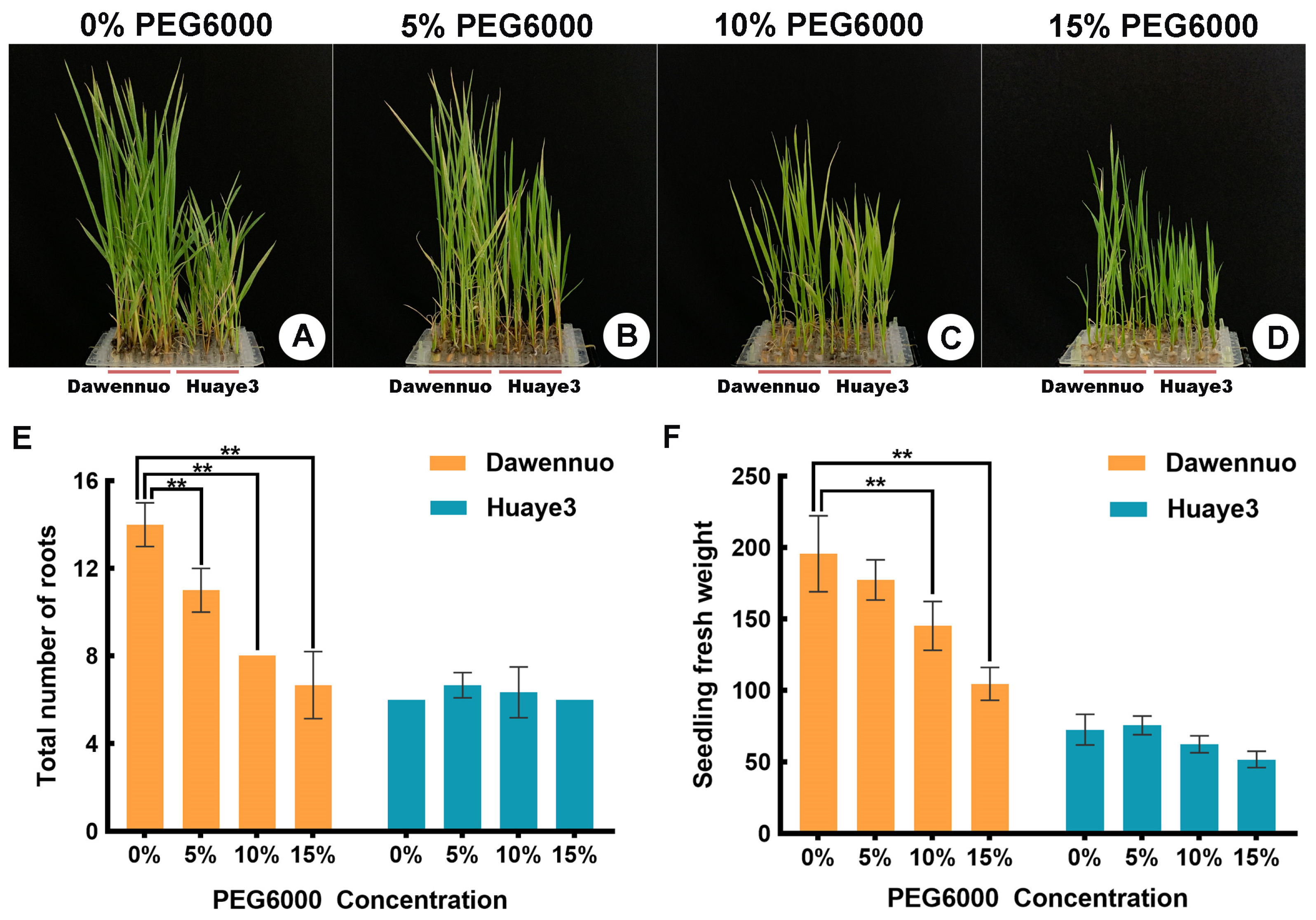
2.2. Effects of Drought Stress on Seedling Stage between the Huaye3 and Dawennuo
2.3. Effects of Drought Stress on Root/Shoot Ratio and Moisture Content between the Huaye3 and Dawennuo
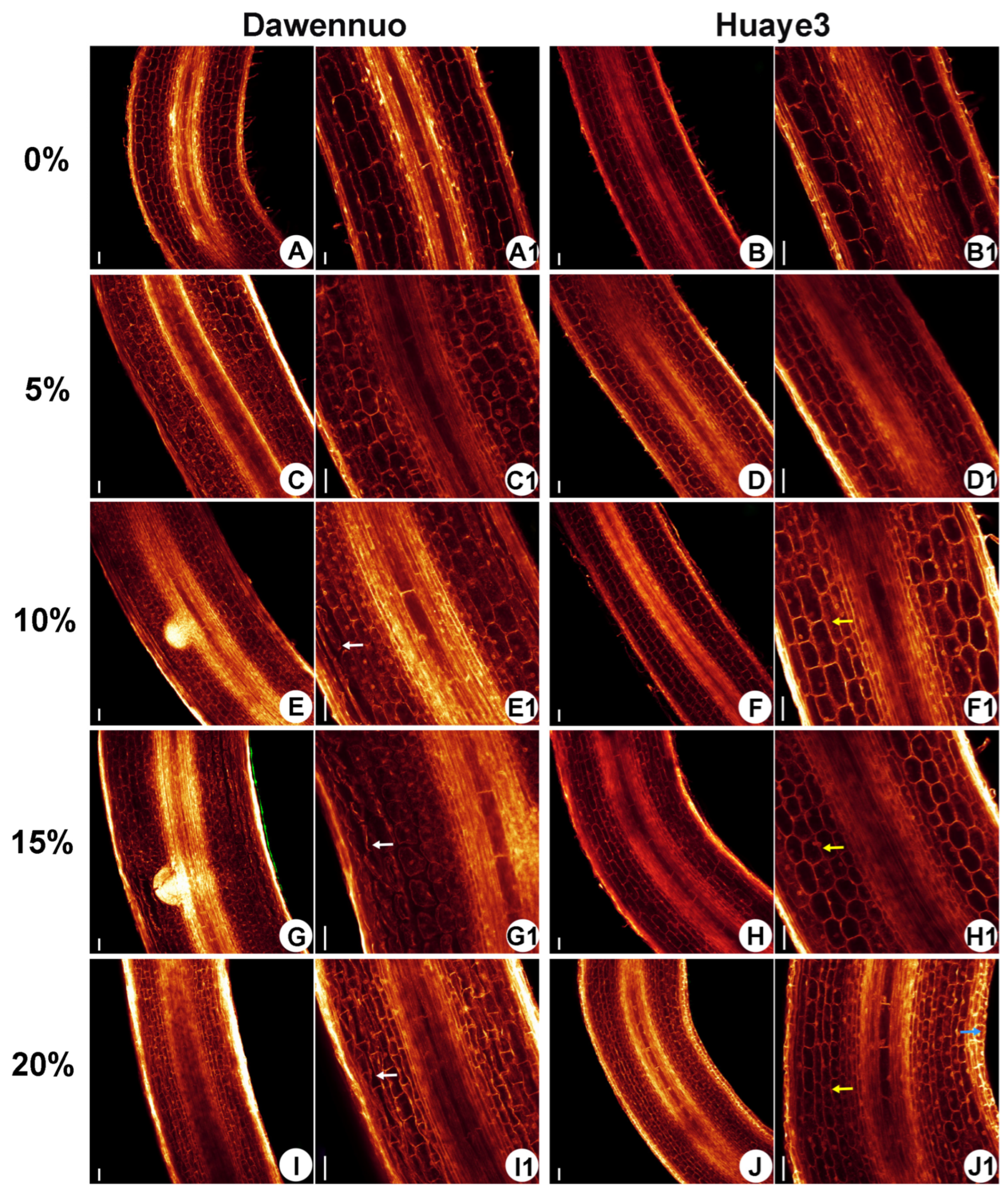
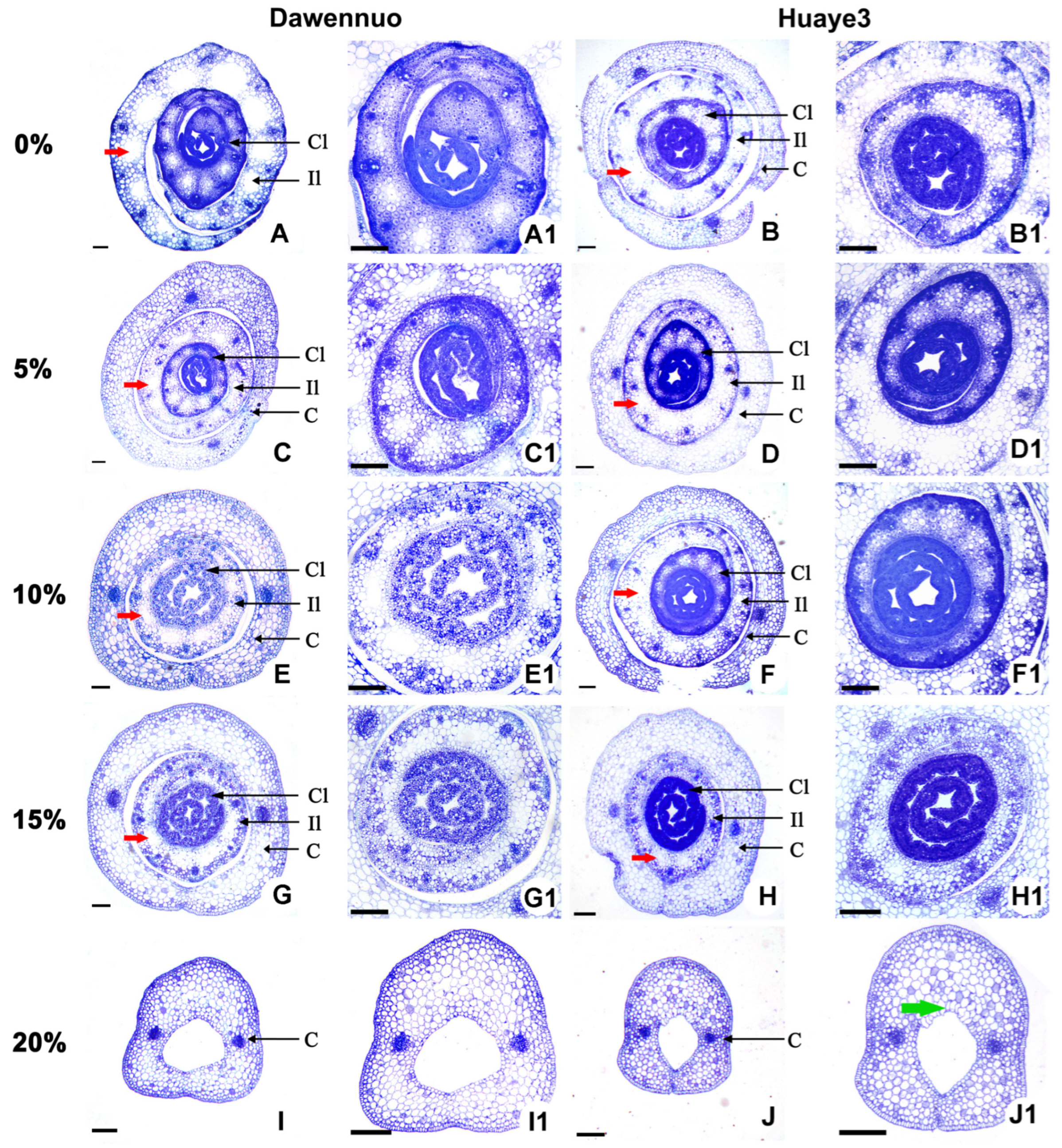
2.4. Effects of Drought Stress on Root Cell Structure between the Huaye3 and Dawennuo
2.5. Effects of Drought Stress on Rice Bud Cells between the Huaye3 and Dawennuo
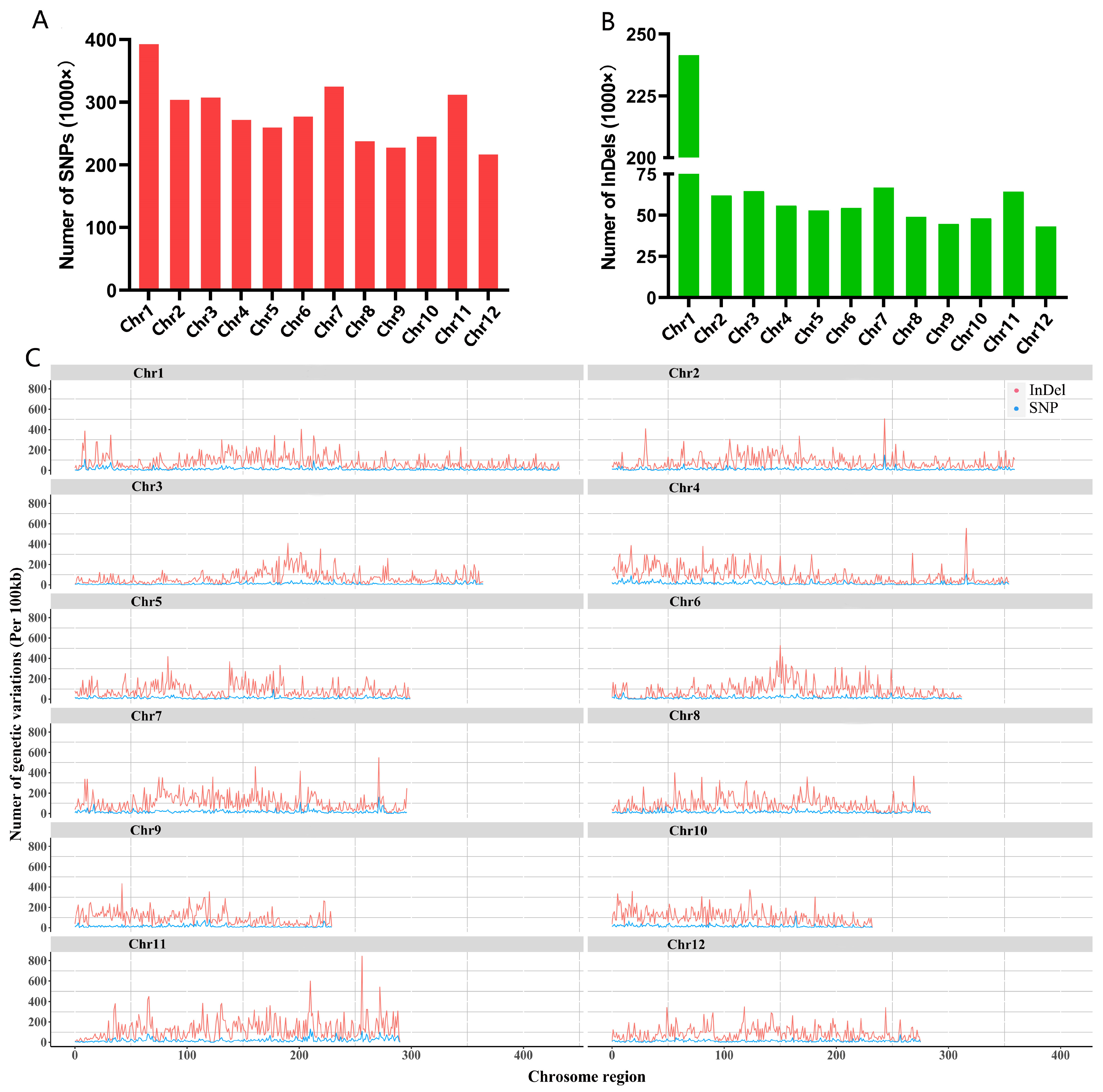
2.6. Re-Sequencing Analysis Revealed Significant Variations in Huaye3
2.7. Drought-Related Genes Were the Primary Up-Regulation in Huaye3 Compared with Cultivar Rice
3. Discussion
3.1. Huaye3 Showed Stronger Germination Characteristics Compared with the Cultivar Rice
3.2. Stable Adaptability in Root Tissue Is Probably the Primary Cause for the Drought Resistance in Huaye3
3.3. Huaye3 Existed the Significant Genetic Variation Compared with Cultivar Rice
4. Materials and Methods
4.1. Plant Materials
4.2. Measurement of Seed Germination Characteristics
4.3. Measurement of the Morphological Index in the Seedling Stage in Two Materials
4.4. Cytological Analysis of Root Tip and Bud Tissue under the Drought Stress
4.5. Re-Sequencing Analysis of Huaye3 and Dawennuo
4.6. RT-qPCR Analysis
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shanker, A.K.; Maheswari, M.; Yadav, S.K.; Desai, S.; Bhanu, D.; Attal, N.B.; Venkateswarlu, B. Drought stress responses in crops. Funct. Integr. Genom. 2014, 14, 11–22. [Google Scholar] [CrossRef]
- Bahuguna, R.N.; Gupta, P.; Bagri, J.; Singh, D.; Dewi, A.K.; Tao, L.; Islam, M.; Sarsu, F.; Singla-Pareek, S.L.; Pareek, A. Forward and reverse genetics approaches for combined stress tolerance in rice. Indian J. Plant Physiol. 2018, 23, 630–646. [Google Scholar] [CrossRef]
- Ishimaru, K.; Shirota, K.; Higa, M.; Kawamitsu, Y. Identification of quantitative trait loci for adaxial and abaxial stomatal frequencies in Oryza sativa. Plant Physiol. Biochem. 2001, 39, 173–177. [Google Scholar] [CrossRef]
- Xia, H.; Zhang, X.; Liu, Y.; Bi, J.; Ma, X.; Zhang, A.; Liu, H.; Chen, L.; Zhou, S.; Gao, H.; et al. Blue revolution for food security under carbon neutrality: A case from the water-saving and drought-resistance rice. Mol. Plant 2022, 15, 1401–1404. [Google Scholar] [CrossRef]
- Yue, B.; Xue, W.; Xiong, L.; Yu, X.; Lou, L.; Cui, K.; Jin, D.; Xing, Y.; Zhang, Q. Genetic basis of drought resistance at reproductive stage in rice: Separation of drought tolerance from drought avoidance. Genetics 2006, 172, 1213–1228. [Google Scholar] [CrossRef] [PubMed]
- Ji, K.; Wang, Y.; Sun, W.; Lou, Q.; Mei, H.; Shen, S.; Chen, H. Drought-responsive mechanisms in rice genotypes with contrasting drought tolerance during reproductive stage. J. Plant Physiol. 2012, 169, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Barbez, E.; Dünser, K.; Gaidora, A.; Lendl, T.; Busch, W. Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, E4884–E4893. [Google Scholar] [CrossRef]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.S. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Kadam, N.N.; Tamilselvan, A.; Lawas, L.; Quinones, C.; Bahuguna, R.N.; Thomson, M.J.; Dingkuhn, M.; Muthurajan, R.; Struik, P.C.; Yin, X.; et al. Genetic control of plasticity in root morphology and anatomy of rice in response to water deficit. Plant Physiol. 2017, 174, 2302–2315. [Google Scholar] [CrossRef]
- Cantu De Leija, A.; King, S.L.; Hawkins, T.S. Seed germination responses to salinity for three rare wetland plants of spring-fed arid systems. J. Arid. Environ. 2022, 199, 104705. [Google Scholar] [CrossRef]
- Meng, S.; Li, X.; Wei, X.; Liu, W.; Zhang, Y.; Bao, G. Effects of water stress on seed germination and seedling growth of Deschampsia caespitosa collected from five provenances. Pratacultural Sci. 2022, 39, 1531–1539. [Google Scholar] [CrossRef]
- Menguer, P.K.; Sperotto, R.A.; Ricachenevsky, F.K. A walk on the wild side: Oryza species as source for rice abiotic stress tolerance. Genet. Mol. Biol. 2017, 40, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Cui, F.; Zhang, L.; Wen, X.; Luo, X.; Zhou, Y.; Li, X.; Wan, Y.; Zhang, J.; Xie, J. Development and identification of a introgression line with strong drought resistance at seedling stage derived from Oryza sativa L. mating with Oryza rufipogon Griff. Euphytica 2014, 200, 1–7. [Google Scholar] [CrossRef]
- Ma, X.; Han, B.; Tang, J.; Zhang, J.; Cui, D.; Geng, L.; Zhou, H.; Li, M.; Han, L. Construction of chromosome segment substitution lines of dongxiang common wild rice (Oryza rufipogon Griff.) in the background of the japonica rice cultivar nipponbare (Oryza sativa L.). Plant Physiol. Biochem. 2019, 144, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.C.; Zhang, J.; Zhang, D.Y.; Nan, Y.H.; Ge, S.; Guo, Y.L. Identification of long noncoding natural antisense transcripts (lncNATs) correlated with drought stress response in wild rice (Oryza nivara). BMC Genom. 2021, 22, 424. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.G.; Nagaki, K.; Fujita, M.; Kawaura, K.; Uozumi, M.; Ogihara, Y. Development of an efficient maintenance and screening system for large-insert genomic DNA libraries of hexaploid wheat in a transformation-competent artificial chromosome (TAC) vector. Plant J. 2000, 23, 687–695. [Google Scholar] [CrossRef]
- Song, J.; Dong, F.; Lilly, J.W.; Stupar, R.M.; Jiang, J. Instability of bacterial artificial chromosome (BAC) clones containing tandemly repeated DNA sequences. Genome 2001, 44, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, S.; Fu, Y.; Su, Z.; Wang, X.; Sun, C. Identification of a drought tolerant introgression line derived from dongxiang common wild rice (O. rufipogon Griff.). Plant Mol. Biol. 2006, 62, 247–259. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Huang, D.; Li, R.; Qiu, Y.; Song, J.; Yang, H.; Zheng, J.; Huang, Y.; Li, X.; Liu, C.; et al. Identification of a resistance gene bls1 to bacterial leaf streak in wild rice Oryza rufipogon Griff. J. Integr. Agric. 2012, 11, 962–969. [Google Scholar] [CrossRef]
- Huang, P.; Molina, J.; Flowers, J.M.; Rubinstein, S.; Jackson, S.A.; Purugganan, M.D.; Schaal, B.A. Phylogeography of asian wild rice, Oryza rufipogon: A genome-wide view. Mol. Ecol. 2012, 21, 4593–4604. [Google Scholar] [CrossRef]
- Xiao, N.; Huang, W.; Li, A.; Gao, Y.; Li, Y.; Pan, C.; Ji, H.; Zhang, X.; Dai, Y.; Dai, Z.; et al. Fine mapping of the qLOP2 and qPSR2-1 loci associated with chilling stress tolerance of wild rice seedlings. Theor. Appl. Genet. 2015, 128, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009, 27, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Lu, T.; Han, B. Resequencing rice genomes: An emerging new era of rice genomics. Trends Genet. 2013, 29, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Han, B. Natural variations and genome-wide association studies in crop plants. Annu. Rev. Plant Biol. 2014, 65, 531–551. [Google Scholar] [CrossRef]
- Bin Rahman, A.N.M.R.; Zhang, J. Flood and drought tolerance in rice: Opposite but may coexist. Food Energy Secur. 2016, 5, 76–88. [Google Scholar] [CrossRef]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Chung, Y.S.; Lee, E.; Tripathi, P.; Heo, S.; Kim, K.H. Root response to drought stress in rice (Oryza sativa L.). Int. J. Mol. Sci. 2020, 21, 1513. [Google Scholar] [CrossRef]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 attenuates drought resistance by regulating JA and ABA signaling in Rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef]
- Long, Q.; Qiu, S.; Man, J.; Ren, D.; Xu, N.; Luo, R. OsAAI1 increases rice yield and drought tolerance dependent on ABA-mediated regulatory and ROS scavenging pathway. Rice 2023, 16, 35. [Google Scholar] [CrossRef]
- Yang, C.; Wang, H.; Ouyang, Q.; Chen, G.; Fu, X.; Hou, D.; Xu, H. Deficiency of Auxin efflux carrier OsPIN1b impairs chilling and drought tolerance in rice. Plants 2023, 12, 4058. [Google Scholar] [CrossRef]
- Yu, H.; Shahid, M.Q.; Li, R.; Li, W.; Liu, W.; Ghouri, F.; Liu, X. Genome-wide analysis of genetic variations and the detection of rich variants of NBS-LRR encoding genes in common wild rice lines. Plant Mol. Biol. Rep. 2018, 36, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.S.; Jeong, H.I.; Bang, S.W.; Jung, S.E.; Kim, G.; Kim, Y.S.; Redillas, M.; Oh, S.J.; Seo, J.S.; Kim, J.K. DROUGHT-INDUCED BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE enhances drought tolerance in rice. Plant Physiol. 2023, 191, 1435–1447. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Huang, S.; Meynard, D.; Chaine, C.; Michel, R.; Roelfsema, M.; Guiderdoni, E.; Sentenac, H.; Véry, A.A. A dual role for the OsK5.2 Ion channel in stomatal movements and K+ loading into Xylem sap. Plant Physiol. 2017, 174, 2409–2418. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Liu, Z.; Liu, Y.; Kong, D.; Li, T.; Yu, S.; Mei, H.; Xu, X.; Liu, H.; Chen, L.; et al. A novel gene OsAHL1 improves both drought avoidance and drought tolerance in rice. Sci. Rep. 2016, 6, 30264. [Google Scholar] [CrossRef]
- Fatimah, S.; Amzeri, A.; Syafii, M.; Purwaningsih, Y. Screening of red rice (Oryza sativa L.) landraces for drought tolerance at early stages using PEG 6000. Agrivita 2023, 45, 199–208. [Google Scholar] [CrossRef]
- Rahim, D.; Kalousek, P.; Tahir, N.; Vyhnánek, T.; Tarkowski, P.; Trojan, V.; Abdulkhaleq, D.; Ameen, A.H.; Havel, L. In vitro assessment of kurdish rice genotypes in response to PEG-induced drought stress. Appl. Sci. 2020, 10, 4471. [Google Scholar] [CrossRef]
- Islam, M.M.; Kayesh, E.; Zaman, E.; Urmi, T.A.; Haque, M.A. Evaluation of rice (Oryza sativa L.) genotypes for drought tolerance at germination and early seedling stage. Agriculturists 2018, 16, 34. [Google Scholar] [CrossRef]
- Sobahan, M.A.; Akter, N.; Rana, M.M. Polyethylene glycol mediated drought stress impacts on germination, growth and accumulation of proline in rice (Oryza sativa L.). SAARC J. Agric. 2022, 20, 107–119. [Google Scholar] [CrossRef]
- Swapna, S.; Shylaraj, K.S. Screening for osmotic stress responses in rice varieties under drought condition. Rice Sci. 2017, 24, 253–263. [Google Scholar] [CrossRef]
- Hu, C.; Elias, E.; Nawrocki, W.J.; Croce, R. Drought affects both photosystems in Arabidopsis thaliana. New Phytol. 2023, 240, 663–675. [Google Scholar] [CrossRef]
- Khan, M.K.; Pandey, A.; Hamurcu, M.; Vyhnánek, T.; Zargar, S.M.; Kahraman, A.; Topal, A.; Gezgin, S. Exploring strigolactones for inducing abiotic stress tolerance in plants. Czech J. Genet. Plant Breed. 2024, 60, 55–69. [Google Scholar] [CrossRef]
- Henry, A.; Cal, A.J.; Batoto, T.C.; Torres, R.O.; Serraj, R. Root attributes affecting water uptake of rice (Oryza sativa) under drought. J. Exp. Bot. 2012, 63, 4751–4763. [Google Scholar] [CrossRef] [PubMed]
- Panda, D.; Mishra, S.S.; Behera, P.K. Drought tolerance in rice: Focus on recent mechanisms and approaches. Rice Sci. 2021, 28, 119–132. [Google Scholar] [CrossRef]
- Hassan, M.A.; Ni, D.H.; Tu, H.N.; Zhu, Q.; Yi, Y.M.; Li, Y.R.; Wang, S.M. Drought stress in rice: Morpho-physiological and molecular responses and marker-assisted breeding. Front. Plant Sci. 2023, 14, 1215371. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ma, X.; Yan, L.; Li, Y.; Wei, S.; Teng, Z.; Zhang, H.; Tang, W.; Peng, S.; Li, Y. Soil-root interface hydraulic conductance determines responses of photosynthesis to drought in rice and wheat. Plant Physiol. 2023, 194, 376–390. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Ge, X.; Ma, H. The rice OsDIL gene plays a role in drought tolerance at vegetative and reproductive stages. Plant Mol. Biol. 2013, 82, 239–253. [Google Scholar] [CrossRef]
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N.; et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, K.; Hwang, H.; Bhatnagar, N.; Kim, D.Y.; Yoon, I.S.; Byun, M.O.; Kim, S.T.; Jung, K.H.; Kim, B.G. Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J. Exp. Bot. 2014, 65, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.J.; Long, Y.; Wang, J.; Zhang, J.W.; Wang, Y.Y.; Li, W.M.; Peng, Y.F.; Yuan, Q.H.; Pei, X.W. De novo transcriptome assembly of common wild rice (Oryza rufipogon Griff.) and discovery of drought-response genes in root tissue based on transcriptomic data. PLoS ONE 2015, 10, e0131455. [Google Scholar] [CrossRef]
- Liu, J.; Hasanuzzaman, M.; Wen, H.; Zhang, J.; Peng, T.; Sun, H.; Zhao, Q. High temperature and drought stress cause abscisic acid and reactive oxygen species accumulation and suppress seed germination growth in rice. Protoplasma 2019, 256, 1217–1227. [Google Scholar] [CrossRef] [PubMed]
- Bouslama, M.; Schapaugh, W.T. Stress tolerance in soybeans. I. evaluation of three screening techniques for heat and drought tolerance. Crop Sci. 1984, 24, 933–937. [Google Scholar] [CrossRef]
- Mu, Y.; Li, Y.; Zhang, Y.; Guo, X.; Song, S.; Huang, Z.; Li, L.; Ma, Q.; Khan, M.N.; Nie, L. A comparative study on the role of conventional, chemical and nanopriming for better salt tolerance during seed germination of direct seeding rice. J. Integr. Agric. 2023, in press. [CrossRef]
- Jira-anunkul, W.; Pattanagul, W. Seed priming with hydrogen peroxide alleviates the effects of drought stress in rice (Oryza sativa L.) seedlings. Not. Bot. Horti Agrobot. 2020, 48, 273–283. [Google Scholar] [CrossRef]
- Li, X.; Yu, H.; Jiao, Y.; Shahid, M.Q.; Wu, J.; Liu, X. Genome-wide analysis of DNA polymorphisms, the methylome and transcriptome revealed that multiple factors are associated with low pollen fertility in autotetraploid rice. PLoS ONE 2018, 13, e0201854. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.X.; Hu, C.Y.; Lu, Y.G.; Li, J.Q.; Liu, X.D. Diversity of abnormal embryo sacs in indica/japonica hybrids in rice demonstrated by confocal microscopy of ovaries. Plant Breed. 2007, 126, 574–580. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. AgriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, 122–129. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Shahid, M.Q.; Guo, H.; Yin, W.; Chen, Z.; Wang, L.; Liu, X.; Lu, Y. Comparative cytological and transcriptomic analysis of pollen development in autotetraploid and diploid rice. Plant Reprod. 2014, 27, 181–196. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.; Huang, P.; Chen, S.; Hu, M.; Yu, H.; Guo, H.; Shahid, M.Q.; Liu, X.; Wu, J. Comparative Cytological and Gene Expression Analysis Reveals That a Common Wild Rice Inbred Line Showed Stronger Drought Tolerance Compared with the Cultivar Rice. Int. J. Mol. Sci. 2024, 25, 7134. https://doi.org/10.3390/ijms25137134
Huang Z, Huang P, Chen S, Hu M, Yu H, Guo H, Shahid MQ, Liu X, Wu J. Comparative Cytological and Gene Expression Analysis Reveals That a Common Wild Rice Inbred Line Showed Stronger Drought Tolerance Compared with the Cultivar Rice. International Journal of Molecular Sciences. 2024; 25(13):7134. https://doi.org/10.3390/ijms25137134
Chicago/Turabian StyleHuang, Zijuan, Peishan Huang, Shihui Chen, Mengzhu Hu, Hang Yu, Haibin Guo, Muhammad Qasim Shahid, Xiangdong Liu, and Jinwen Wu. 2024. "Comparative Cytological and Gene Expression Analysis Reveals That a Common Wild Rice Inbred Line Showed Stronger Drought Tolerance Compared with the Cultivar Rice" International Journal of Molecular Sciences 25, no. 13: 7134. https://doi.org/10.3390/ijms25137134






