Cytological and Transcriptome Analyses Provide Insights into Persimmon Fruit Size Formation (Diospyros kaki Thunb.)
Abstract
1. Introduction
2. Results
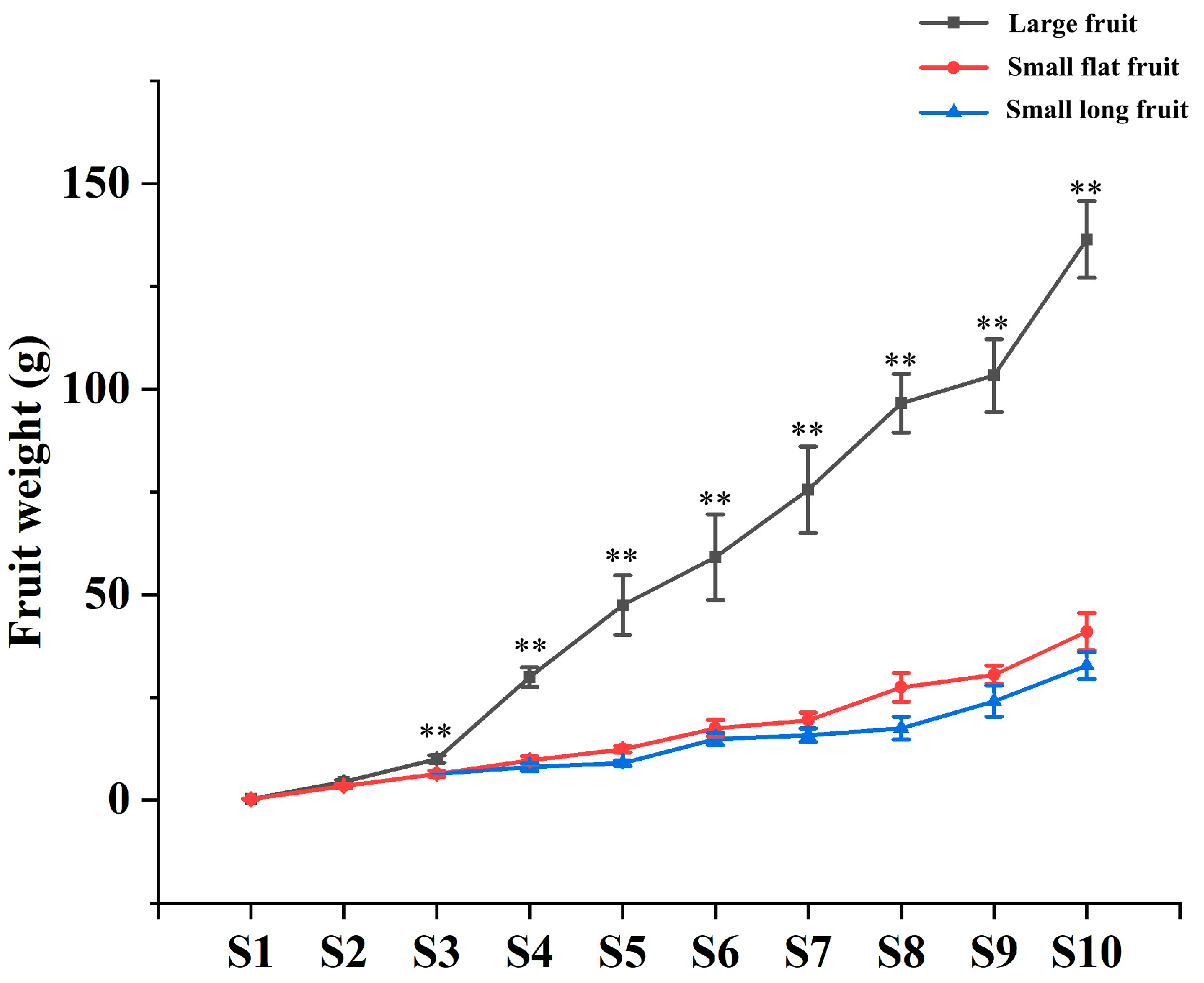
2.1. Morphological Comparison of Fruit
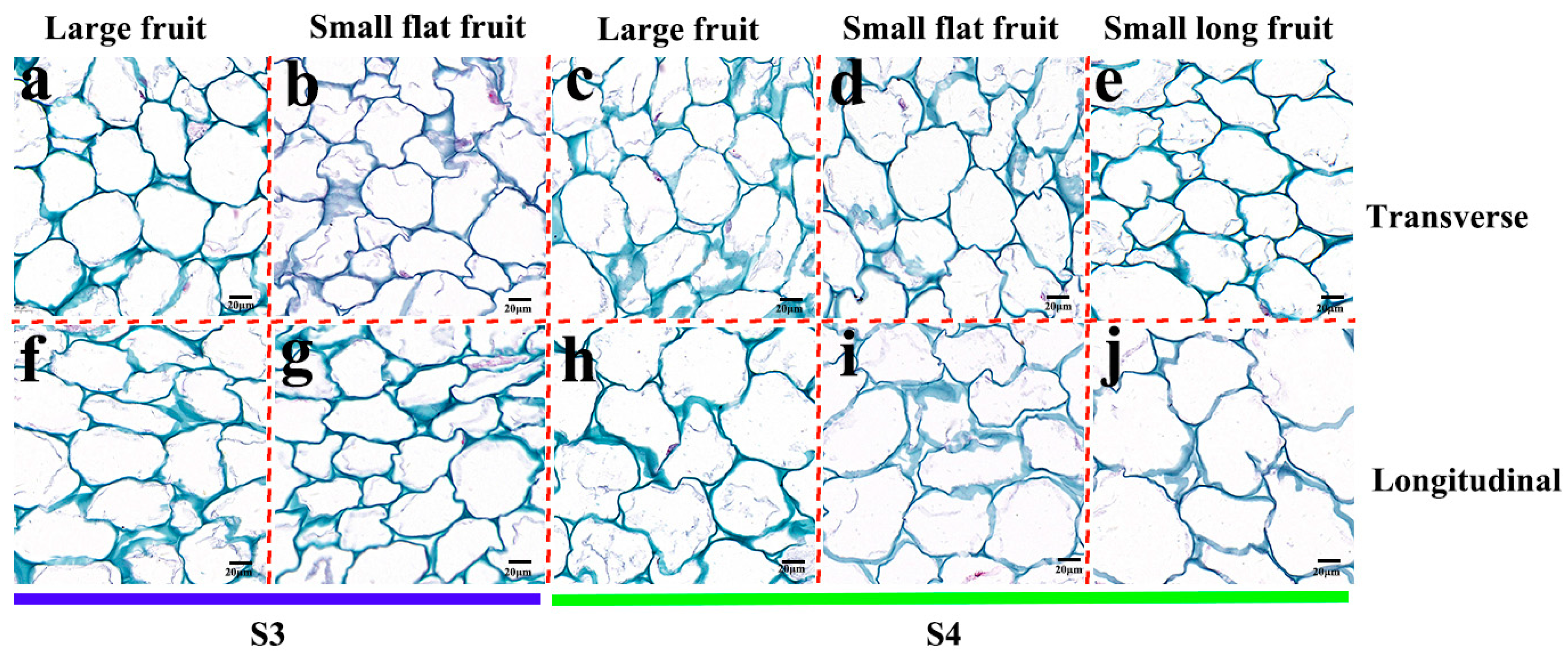
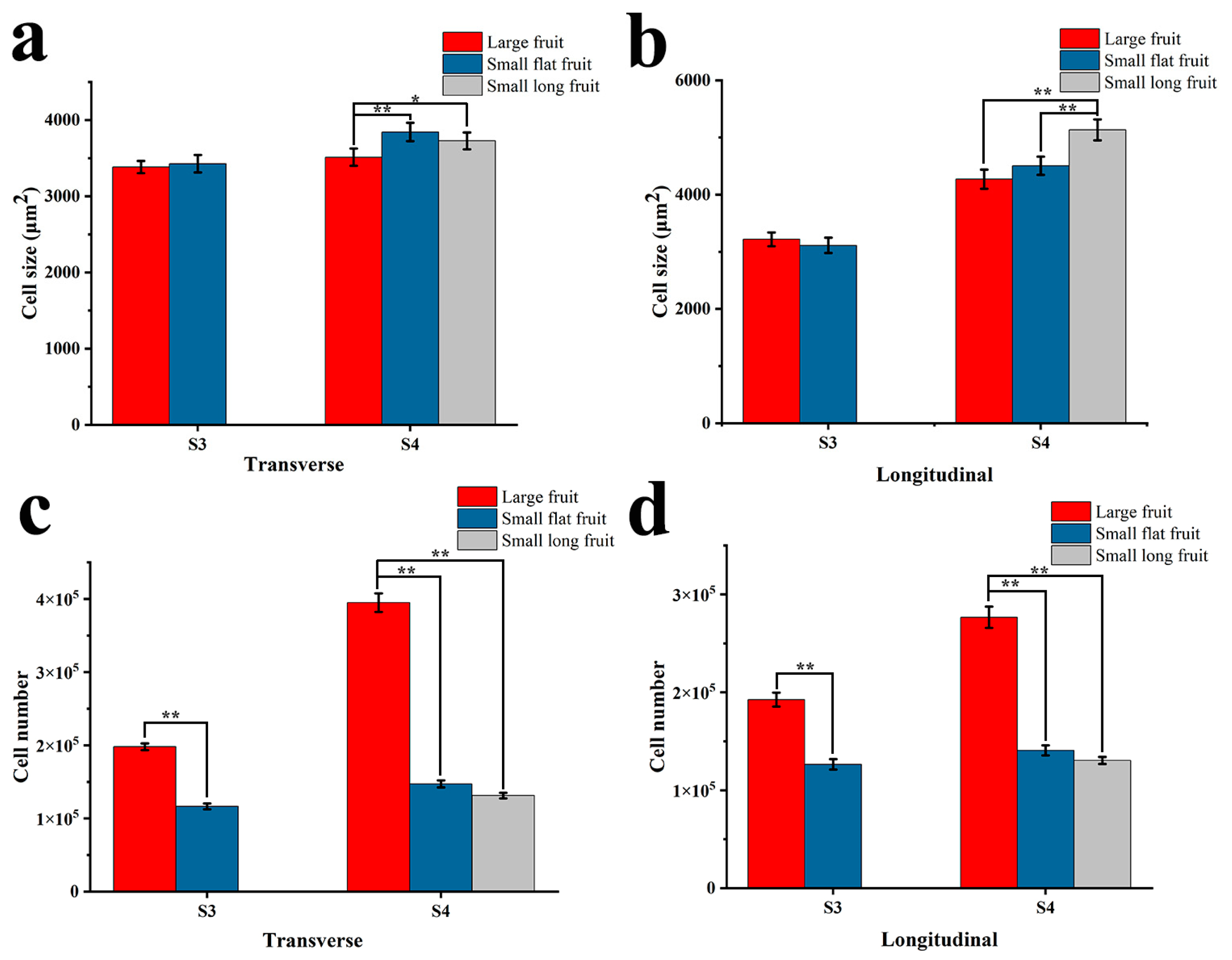
2.2. Cytological Observation
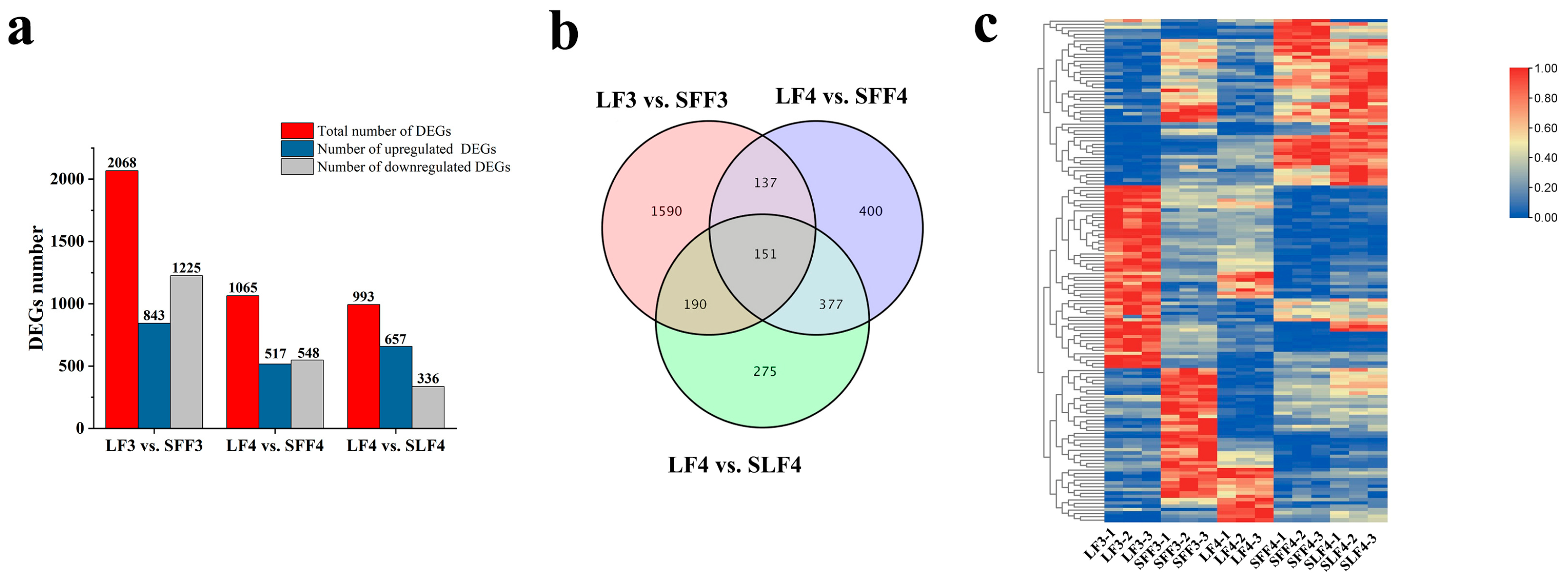
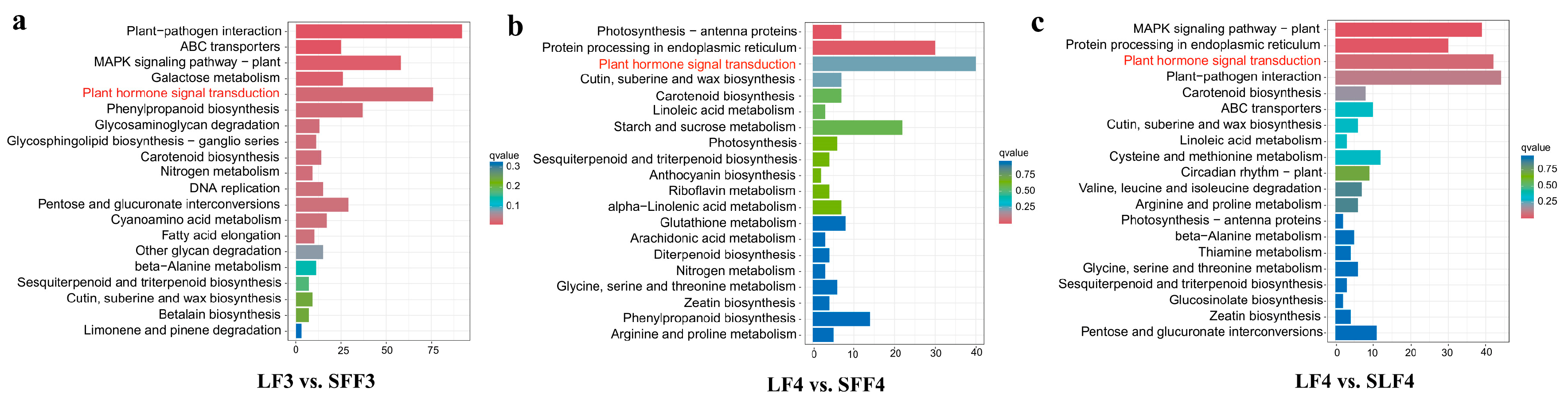
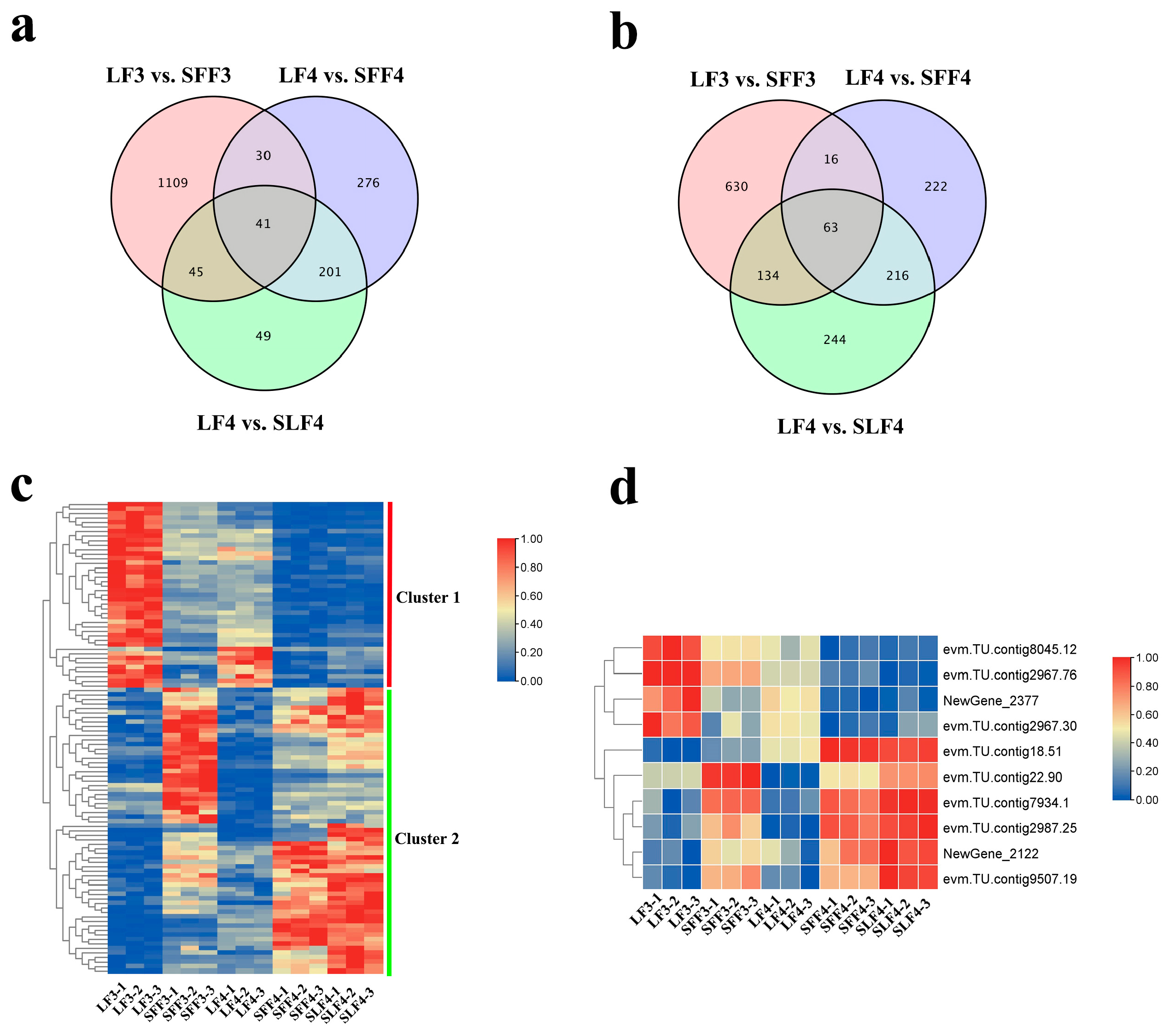
2.3. Transcriptome Sequencing
2.4. Candidate Genes Identification
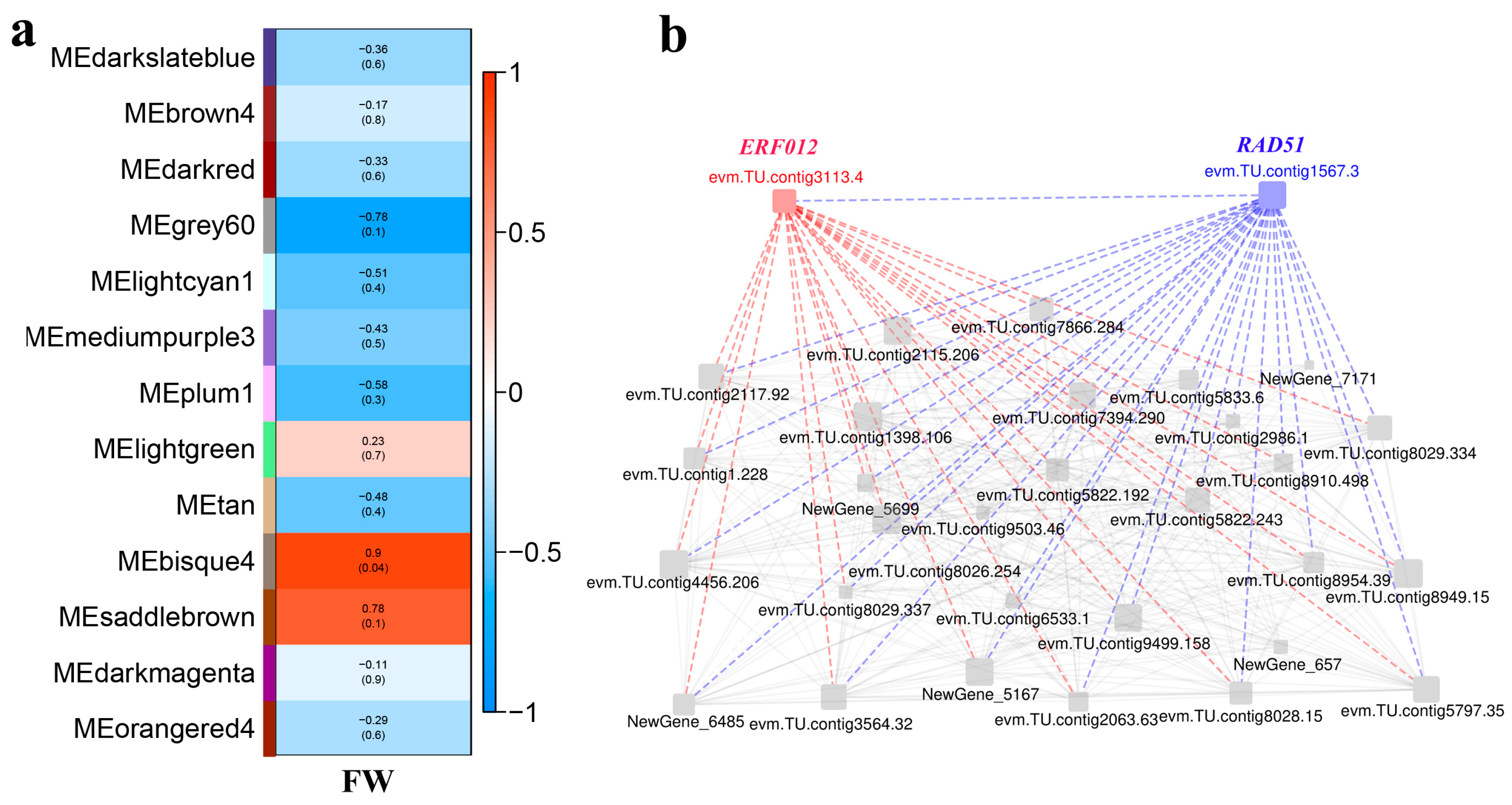
2.5. Weighted Gene Co-Expression Network (WGCNA)
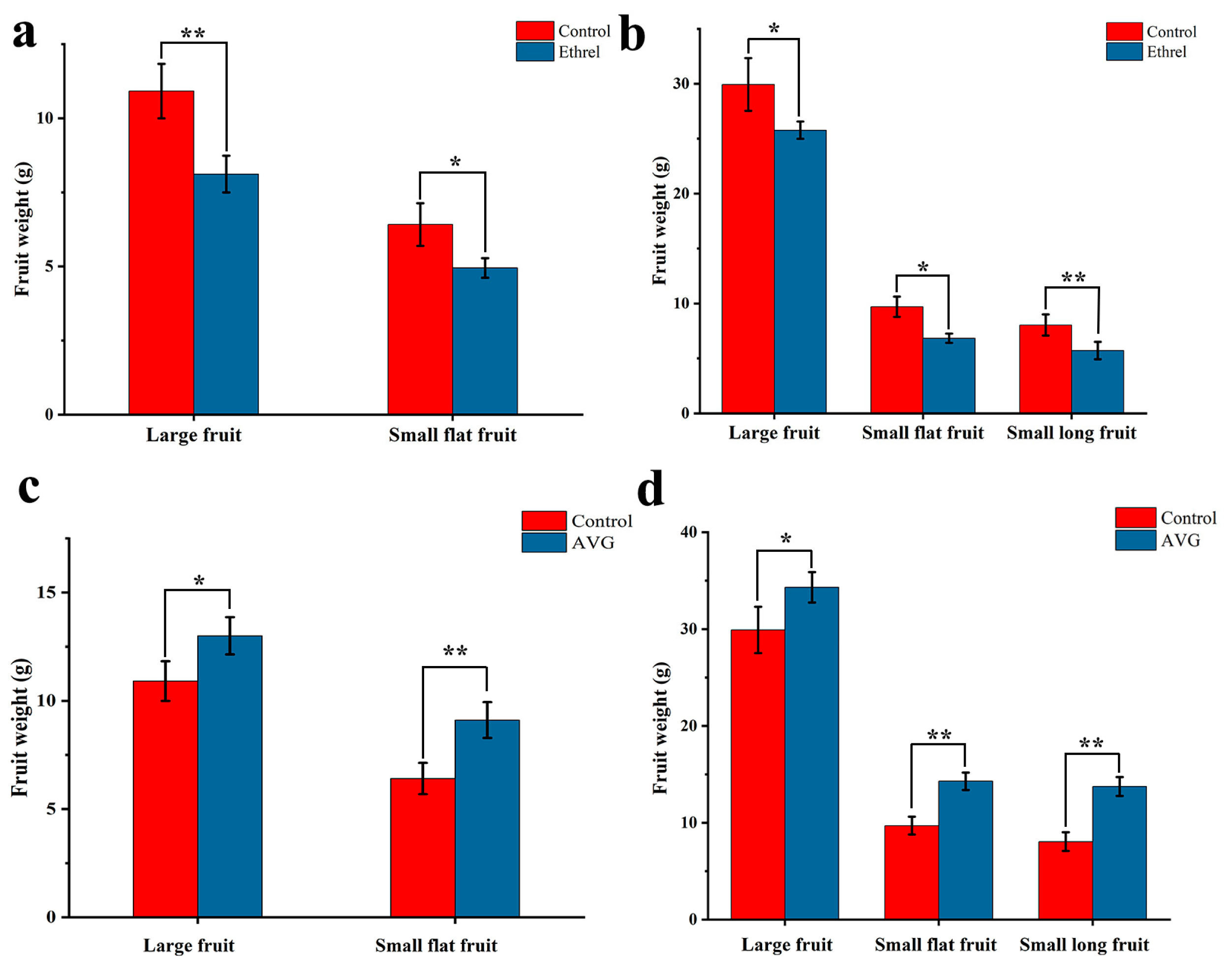
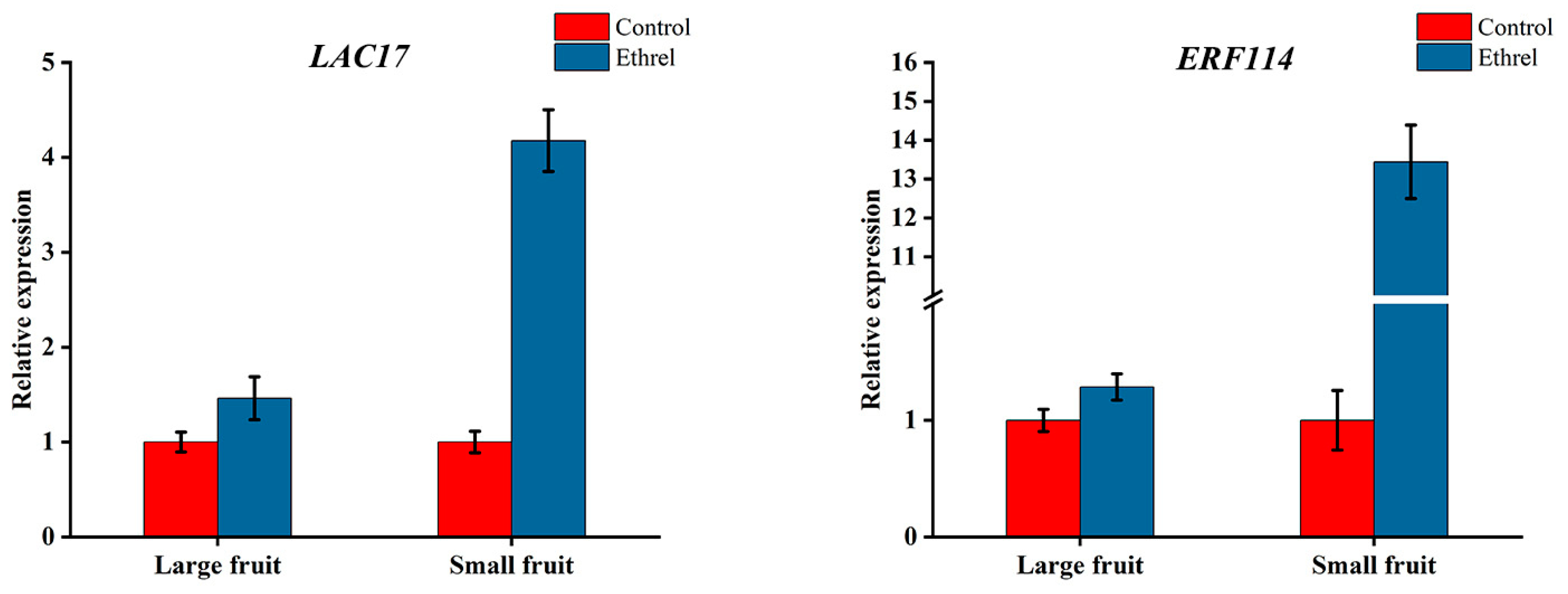
2.6. Application of Ethrel and Aminoethoxyinylglycine
2.7. qRT-qPCR
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Paraffin Section
4.3. Transcriptome
4.4. Ethrel and Aminoethoxyinylglycine Treatment
4.5. qRT-qPCR
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, Z.; Wang, R. Persimmon in China: Domestication and traditional utilization of genetic resources. Adv. Hortic. Sci. 2008, 22, 239–243. [Google Scholar]
- Seymour, G.B.; Østergaard, L.; Chapman, N.H.; Knapp, S.; Martin, C. Fruit development and ripening. Annu. Rev. Plant Biol. 2013, 64, 219–241. [Google Scholar] [CrossRef] [PubMed]
- Tanksley, S.D. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 2004, 16 (Suppl. S1), S181–S189. [Google Scholar] [CrossRef] [PubMed]
- Pattison, R.J.; Catalá, C. Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J. 2012, 70, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Li, M.; Liu, B.; Yan, M.; Yu, X.; Zi, H.; Liu, R.; Yamamuro, C. Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proc. Natl. Acad. Sci. USA 2018, 115, E11542–E11550. [Google Scholar] [CrossRef] [PubMed]
- Mounet, F.; Moing, A.; Kowalczyk, M.; Rohrmann, J.; Petit, J.; Garcia, V.; Maucourt, M.; Yano, K.; Deborde, C.; Aoki, K. Down-regulation of a single auxin efflux transport protein in tomato induces precocious fruit development. J. Exp. Bot. 2012, 63, 4901–4917. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhong, J.; Xu, K.; Wei, Q.; Wei, Z. Effects of exogenous GA3 on fruit development and endogenous hormones in Fujiminori grape. J. Fruit Sci. 2001, 4, 209–212. [Google Scholar]
- Pang, Y.; Zang, X.; Pang, F.; Zhou, T.; Tian, F. Changes of CTK and few nitrogen index during development of flower and fruit in Zhanhua jujube. J. North China Agric. 2017, 5, 101–104. [Google Scholar]
- Zhao, Z.; Gu, Y.; Li, B.; Guo, S.; Qi, G.; Hu, Y. Study on the development of bagging changfu 2 fruit and the changes of endogenous hormones content after bag removal. J. Hebei Agric. Univ. 2006, 5, 12–15. [Google Scholar]
- Nardozza, S.; Cooney, J.; Boldingh, H.L.; Hewitt, K.G.; Trower, T.; Jones, D.; Thrimawithana, A.H.; Allan, A.C.; Richardson, A.C. Phytohormone and transcriptomic analysis reveals endogenous cytokinins affect kiwifruit growth under restricted carbon supply. Metabolites 2020, 10, 23. [Google Scholar] [CrossRef]
- Nitsch, L.; Kohlen, W.; Oplaat, C.; Charnikhova, T.; Cristescu, S.; Michieli, P.; Wolters-Arts, M.; Bouwmeester, H.; Mariani, C.; Vriezen, W. ABA-deficiency results in reduced plant and fruit size in tomato. J. Plant Physiol. 2012, 169, 878–883. [Google Scholar] [CrossRef] [PubMed]
- Xin, T.; Zhang, Z.; Li, S.; Zhang, S.; Li, Q.; Zhang, Z.-H.; Huang, S.; Yang, X. Genetic regulation of ethylene dosage for cucumber fruit elongation. Plant Cell 2019, 31, 1063–1076. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Luo, Z.; Wang, L.; Deng, W.; Wei, H.; Liu, P.; Liu, M. Morphological, cytological and nutritional changes of autotetraploid compared to its diploid counterpart in Chinese jujube (Ziziphus jujuba Mill.). Sci. Hortic. 2019, 249, 263–270. [Google Scholar] [CrossRef]
- Upadhyay, R.K.; Gupta, A.; Ranjan, S.; Singh, R.; Pathre, U.V.; Nath, P.; Sane, A.P. The EAR motif controls the early flowering and senescence phenotype mediated by over-expression of SlERF36 and is partly responsible for changes in stomatal density and photosynthesis. PLoS ONE 2014, 9, e101995. [Google Scholar] [CrossRef] [PubMed]
- Yuste-Lisbona, F.J.; Fernández-Lozano, A.; Pineda, B.; Bretones, S.; Ortíz-Atienza, A.; García-Sogo, B.; Müller, N.A.; Angosto, T.; Capel, J.; Moreno, V. ENO regulates tomato fruit size through the floral meristem development network. Proc. Natl. Acad. Sci. USA 2020, 117, 8187–8195. [Google Scholar] [CrossRef] [PubMed]
- Mantegazza, O.; Gregis, V.; Mendes, M.A.; Morandini, P.; Alves-Ferreira, M.; Patreze, C.M.; Nardeli, S.M.; Kater, M.M.; Colombo, L. Analysis of the Arabidopsis REM gene family predicts functions during flower development. Ann. Bot. 2014, 114, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J.; Baumann, E.; Wisman, E.; Yanofsky, M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Suo, Y.; Li, H.; Sun, P.; Han, W.; Fu, J. Cytological, Phytohormone, and transcriptome analyses provide insights into persimmon fruit shape formation (Diospyros kaki Thunb.). Int. J. Mol. Sci. 2024, 25, 4812. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sun, P.; Wang, Y.; Zhang, Z.; Yang, J.; Suo, Y.; Han, W.; Diao, S.; Li, F.; Fu, J. Allele-aware chromosome-level genome assembly of the autohexaploid Diospyros kaki Thunb. Sci. Data 2023, 10, 270. [Google Scholar] [CrossRef]
- Yamane, H.; Ichiki, M.; Tao, R.; Esumi, T.; Yonemori, K.; Niikawa, T.; Motosugi, H. Growth characteristics of a small-fruit dwarf mutant arising from bud sport mutation in Japanese persimmon (Diospyros kaki Thunb.). HortScience 2008, 43, 1726–1730. [Google Scholar] [CrossRef]
- Zhao, J.; Jiang, L.; Che, G.; Pan, Y.; Li, Y.; Hou, Y.; Zhao, W.; Zhong, Y.; Ding, L.; Yan, S. A functional allele of CsFUL1 regulates fruit length through repressing CsSUP and inhibiting auxin transport in cucumber. Plant Cell 2019, 31, 1289–1307. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cao, C.; Zheng, S.; Zhang, H.; Liu, P.; Ge, Q.; Li, J.; Ren, Z. Transcriptomic analysis of short-fruit 1 (sf1) reveals new insights into the variation of fruit-related traits in Cucumis sativus. Sci. Rep. 2017, 7, 2950. [Google Scholar] [CrossRef] [PubMed]
- Harada, T.; Kurahashi, W.; Yanai, M.; Wakasa, Y.; Satoh, T. Involvement of cell proliferation and cell enlargement in increasing the fruit size of Malus species. Sci. Hortic. 2005, 105, 447–456. [Google Scholar] [CrossRef]
- Li, Y.-H.; Zhang, Z.; Sun, G.-M. Changes in cell number and cell size during pineapple (Ananas comosus L.) fruit development and their relationship with fruit size. Aust. J. Bot. 2010, 58, 673–678. [Google Scholar] [CrossRef]
- Higashi, K.; Hosoya, K.; Ezura, H. Histological analysis of fruit development between two melon (Cucumis melo L. reticulatus) genotypes setting a different size of fruit. J. Exp. Bot. 1999, 50, 1593–1597. [Google Scholar]
- Olmstead, J.W.; Iezzoni, A.F.; Whiting, M.D. Genotypic differences in sweet cherry fruit size are primarily a function of cell number. J. Am. Soc. Hortic. Sci. 2007, 132, 697–703. [Google Scholar] [CrossRef]
- Scorzal, R.; May, L.G.; Purnell, B.; Upchurch, B. Differences in number and area of mesocarp cells between small-and large-fruited peach cultivars. J. Am. Soc. Hortic. Sci. 1991, 116, 861–864. [Google Scholar] [CrossRef]
- Zhang, C.; Tanabe, K.; Tani, H.; Nakajima, H.; Mori, M.; Sakuno, E. Biologically active gibberellins and abscisic acid in fruit of two late-maturing Japanese pear cultivars with contrasting fruit size. J. Am. Soc. Hortic. Sci. 2007, 132, 452–458. [Google Scholar] [CrossRef]
- Frary, A.; Nesbitt, T.C.; Frary, A.; Grandillo, S.; Knaap, E.V.D.; Cong, B.; Liu, J.; Meller, J.; Elber, R.; Alpert, K.B. fw2. 2: A quantitative trait locus key to the evolution of tomato fruit size. Science 2000, 289, 85–88. [Google Scholar] [CrossRef]
- Cong, B.; Liu, J.; Tanksley, S.D. Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc. Natl. Acad. Sci. USA 2002, 99, 13606–13611. [Google Scholar] [CrossRef]
- Guo, M.; Rupe, M.A.; Dieter, J.A.; Zou, J.; Spielbauer, D.; Duncan, K.E.; Howard, R.J.; Hou, Z.; Simmons, C.R. Cell Number Regulator1 affects plant and organ size in maize: Implications for crop yield enhancement and heterosis. Plant Cell 2010, 22, 1057–1073. [Google Scholar] [CrossRef] [PubMed]
- Klucher, K.M.; Chow, H.; Reiser, L.; Fischer, R.L. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 1996, 8, 137–153. [Google Scholar]
- Mizukami, Y.; Fischer, R.L. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 942–947. [Google Scholar] [CrossRef] [PubMed]
- Kohoutová, L.; Kourová, H.; Nagy, S.K.; Volc, J.; Halada, P.; Mészáros, T.; Meskiene, I.; Bögre, L.; Binarová, P. The Arabidopsis mitogen—Activated protein kinase 6 is associated with γ-tubulin on microtubules, phosphorylates EB 1c and maintains spindle orientation under nitrosative stress. New Phytol. 2015, 207, 1061–1074. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Ding, H.; Fan, Y.; Wu, X.; Liu, X.; Niu, J.; Cao, F.; Li, M. Overexpression of a laccase gene, DiLAC17, from Davidia involucrata causes severe seed abortion in Arabidopsis. Plant Physiol. Biochem. 2023, 202, 107956. [Google Scholar] [CrossRef] [PubMed]
- Vandenbussche, F.; Vaseva, I.; Vissenberg, K.; Van Der Straeten, D. Ethylene in vegetative development: A tale with a riddle. New Phytol. 2012, 194, 895–909. [Google Scholar] [CrossRef] [PubMed]
- Christians, M.J.; Robles, L.M.; Zeller, S.M.; Larsen, P.B. The eer5 mutation, which affects a novel proteasome—Related subunit, indicates a prominent role for the COP9 signalosome in resetting the ethylene—Signaling pathway in Arabidopsis. Plant J. 2008, 55, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Byers, R.E.; Carbaugh, D.H.; Combs, L.D. Ethylene inhibitors delay fruit drop, maturity, and increase fruit size of ‘Arlet’ apples. HortScience 2005, 40, 2061–2065. [Google Scholar] [CrossRef]
- Zhang, Y.; Park, C.; Bennett, C.; Thornton, M.; Kim, D. Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res. 2021, 31, 1290–1295. [Google Scholar] [CrossRef]
- Shumate, A.; Wong, B.; Pertea, G.; Pertea, M. Improved transcriptome assembly using a hybrid of long and short reads with StringTie. PLoS Comput. Biol. 2022, 18, e1009730. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Du, G.; Wang, L.; Li, H.; Sun, P.; Fu, J.; Suo, Y.; Han, W.; Diao, S.; Mai, Y.; Li, F. Selection and validation of reference genes for quantitative gene expression analyses in persimmon (Diospyros kaki Thunb.) using real-time quantitative PCR. Biol. Futur. 2019, 70, 261–267. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Suo, Y.; Li, H.; Sun, P.; Li, S.; Yuan, D.; Han, W.; Fu, J. Cytological and Transcriptome Analyses Provide Insights into Persimmon Fruit Size Formation (Diospyros kaki Thunb.). Int. J. Mol. Sci. 2024, 25, 7238. https://doi.org/10.3390/ijms25137238
Li H, Suo Y, Li H, Sun P, Li S, Yuan D, Han W, Fu J. Cytological and Transcriptome Analyses Provide Insights into Persimmon Fruit Size Formation (Diospyros kaki Thunb.). International Journal of Molecular Sciences. 2024; 25(13):7238. https://doi.org/10.3390/ijms25137238
Chicago/Turabian StyleLi, Huawei, Yujing Suo, Hui Li, Peng Sun, Shuzhan Li, Deyi Yuan, Weijuan Han, and Jianmin Fu. 2024. "Cytological and Transcriptome Analyses Provide Insights into Persimmon Fruit Size Formation (Diospyros kaki Thunb.)" International Journal of Molecular Sciences 25, no. 13: 7238. https://doi.org/10.3390/ijms25137238
APA StyleLi, H., Suo, Y., Li, H., Sun, P., Li, S., Yuan, D., Han, W., & Fu, J. (2024). Cytological and Transcriptome Analyses Provide Insights into Persimmon Fruit Size Formation (Diospyros kaki Thunb.). International Journal of Molecular Sciences, 25(13), 7238. https://doi.org/10.3390/ijms25137238





