Genome-Wide Identification of Sorghum Paclobutrazol-Resistance Gene Family and Functional Characterization of SbPRE4 in Response to Aphid Stress
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization of the PRE Gene Family in the Sorghum Genome
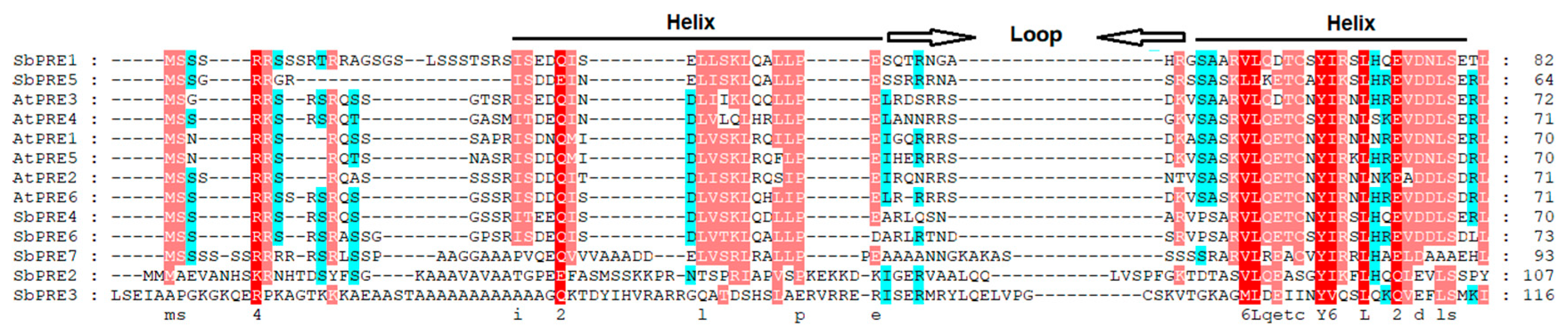
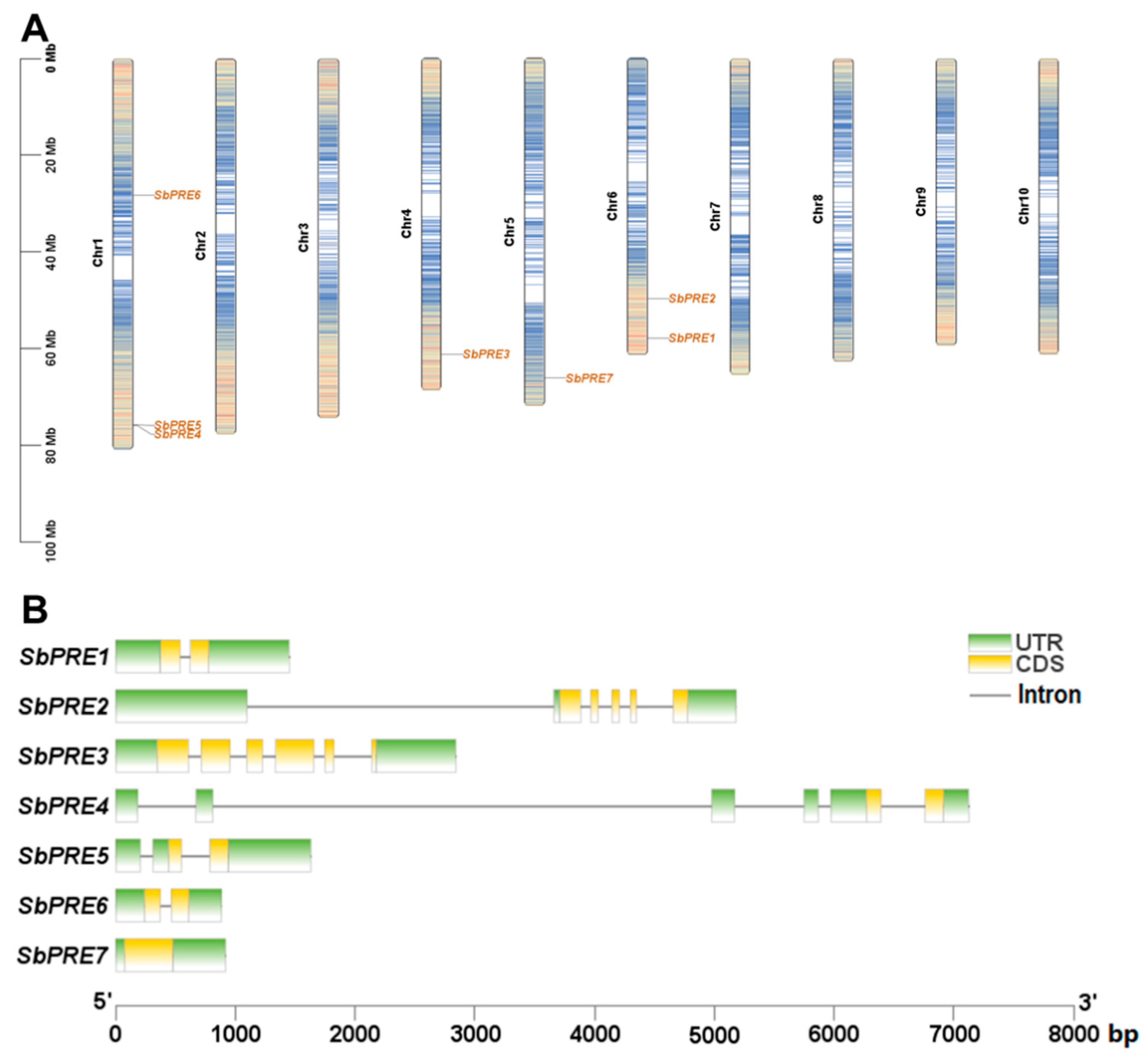
2.2. Chromosome Localization, Gene Structure, and Genetic Evolution Analysis of the SbPRE Genes
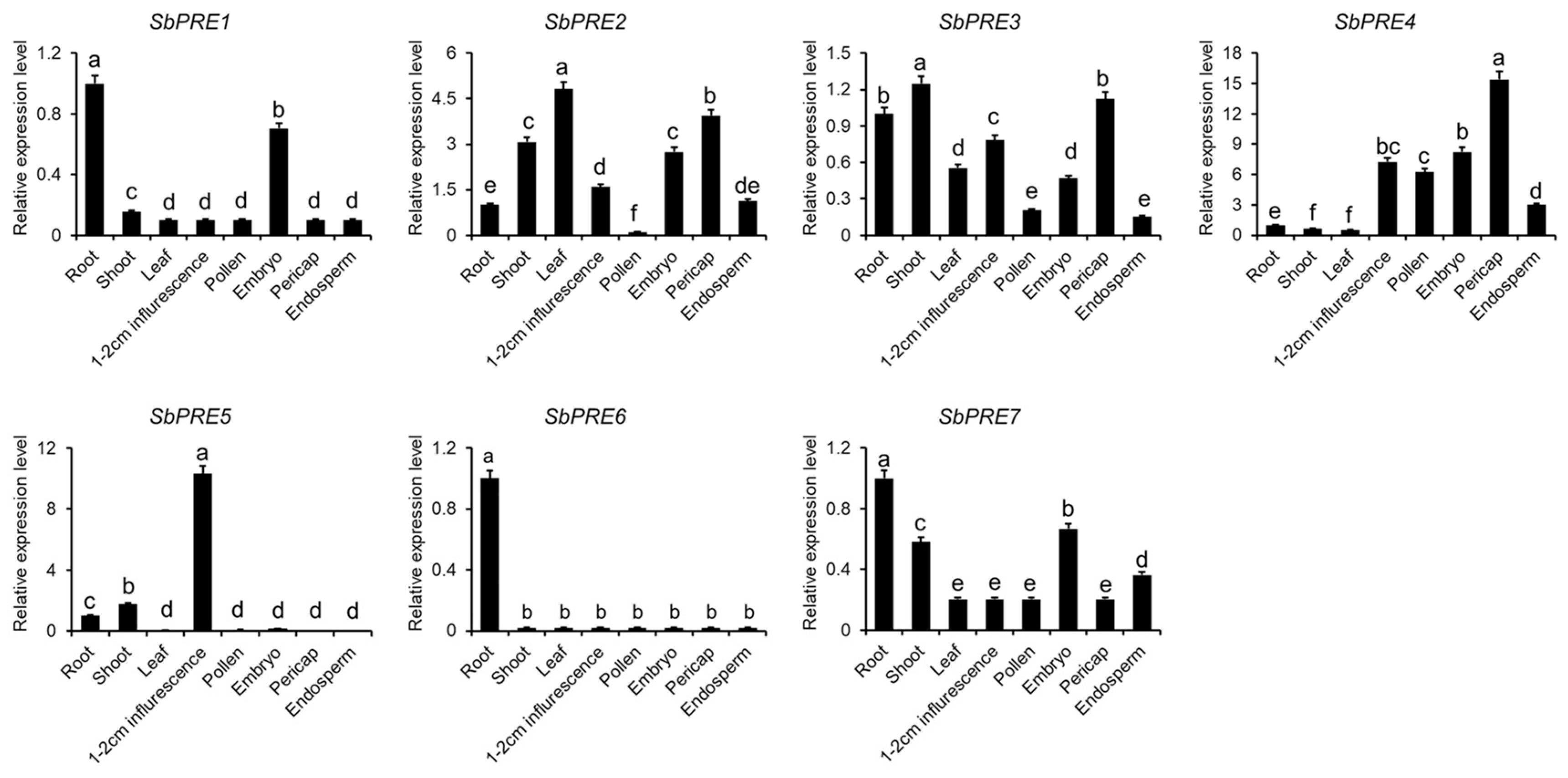
2.3. Expression Patterns of SbPREs across Sorghum Tissues
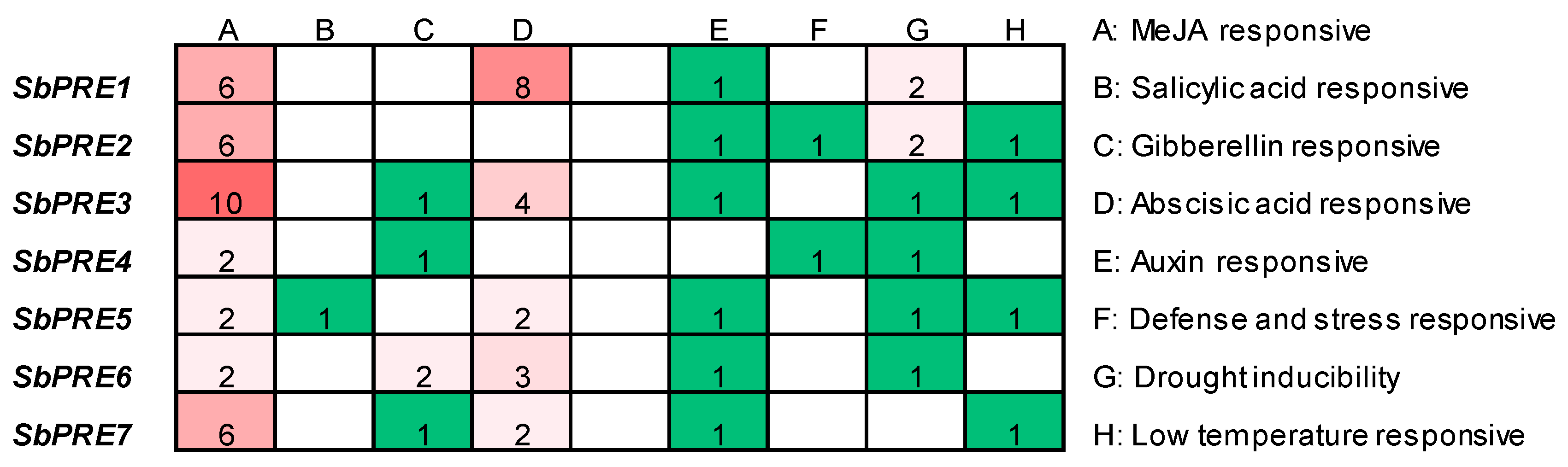
2.4. Analysis of Cis-Elements in SbPREs Promoter
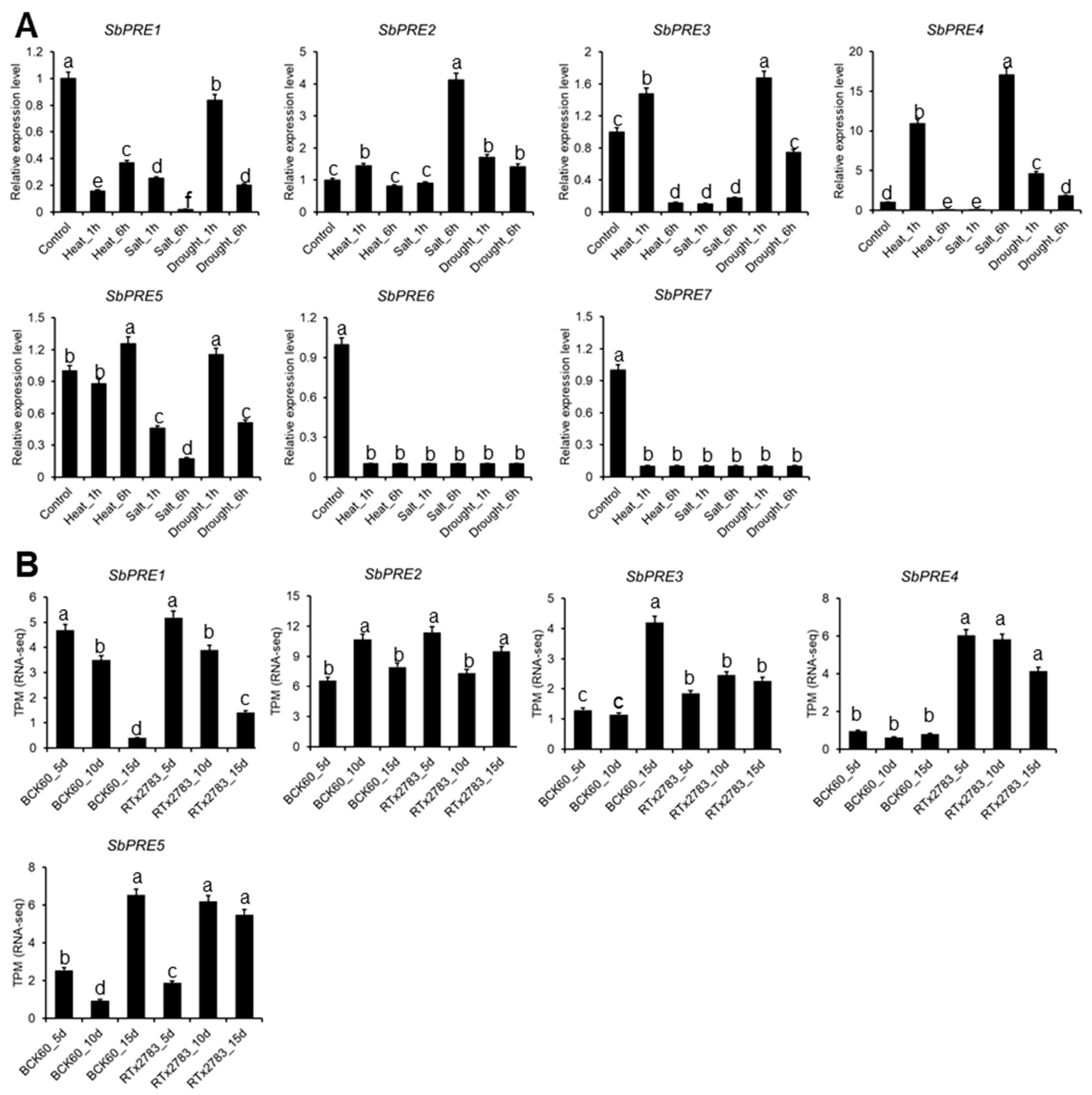
2.5. SbPRE Genes Expression under Abotic and Aphid Stress
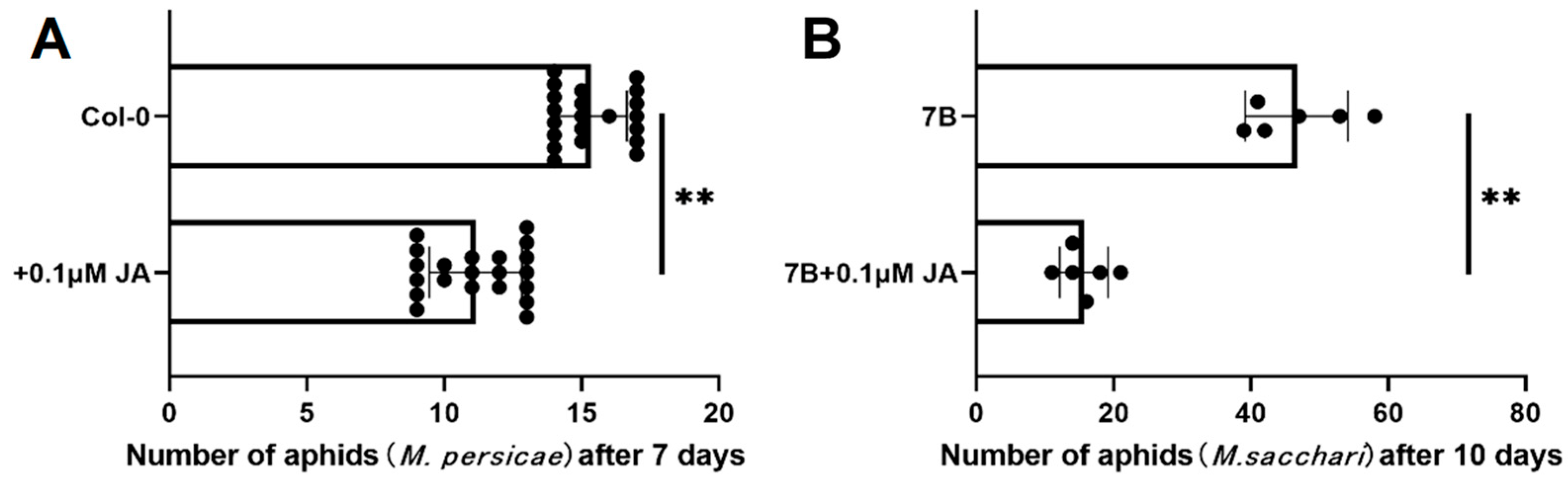
2.6. Overexpressing SbPRE4 Enhanced Aphid Resistance of Arabidopsis Thaliana by Accumulating JAs
3. Discussion
4. Materials and Methods
4.1. Identification of the PRE Genes in Sorghum
4.2. SbPRE Chromosomal Localizations and Gene Structures
4.3. Structural Domains, Phylogenetic Analysis, and Promoters Analysis of SbPREs
4.4. Expression Analysis of SbPREs by RNA-Seq Data or qRT-PCR
4.5. Plant Materials and Aphid Treatments
4.6. Construction of the SbPRE4 Expression Vector and Arabidopsis Transformation
4.7. Quantification of Phytohormones and JA Treatment
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tao, Y.; Luo, H.; Xu, J.; Cruickshank, A.; Zhao, X.; Teng, F.; Hathorn, A.; Wu, X.; Liu, Y.; Shatte, T.; et al. Extensive variation within the pan-genome of cultivated and wild sorghum. Nat. Plants 2021, 7, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Dahlberg, J. The Role of Sorghum in Renewables and Biofuels. Methods Mol. Biol. 2019, 1931, 269–277. [Google Scholar] [PubMed]
- Chadalavada, K.; Kumari, B.D.R.; Kumar, T.S. Sorghum mitigates climate variability and change on crop yield and quality. Planta 2021, 253, 113. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.U.; Padmaja, P.G.; Seetharama, N. Biology and management of the sugarcane aphid, Melanaphis sacchari (Zehntner) (Homoptera: Aphididae), in sorghum: A review. Crop Prot. 2004, 23, 739–755. [Google Scholar] [CrossRef]
- Berg, J.V.D. Status of resistance of sorghum hybrids to the aphid, Melanaphis sacchari (Zehntner) (Homoptera: Aphididae). S. Afr. J. Plant Soil 2002, 19, 151–155. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martinez-Garcia, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Yi, P.; Li, X.; Olson, E.N. Hand, an evolutionarily conserved bHLH transcription factor required for Drosophila cardiogenesis and hematopoiesis. Development 2006, 133, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Hyun, Y.; Lee, I. KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 2006, 61, 283–296. [Google Scholar] [CrossRef]
- Mara, C.D.; Huang, T.; Irish, V.F. The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell 2010, 22, 690–702. [Google Scholar] [CrossRef]
- Bai, M.Y.; Fan, M.; Oh, E.; Wang, Z.Y. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 2012, 24, 4917–4929. [Google Scholar] [CrossRef] [PubMed]
- Castelain, M.; Le Hir, R.; Bellini, C. The non-DNA-binding bHLH transcription factor PRE3/bHLH135/ATBS1/TMO7 is involved in the regulation of light signaling pathway in Arabidopsis. Physiol. Plant 2012, 145, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhu, Y.; Fujioka, S.; Asami, T.; Li, J.; Li, J. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 2009, 21, 3781–3791. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Y.; Bai, M.Y.; Wu, J.; Zhu, J.Y.; Wang, H.; Zhang, Z.; Wang, W.; Sun, Y.; Zhao, J.; Sun, X.; et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Oh, E.; Choi, G.; Liang, Z.; Wang, Z.Y. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 2012, 5, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.Y.; Wang, Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Wang, Y.; Wang, S. The non-DNA binding bHLH transcription factor Paclobutrazol Resistances are involved in the regulation of ABA and salt responses in Arabidopsis. Plant Physiol. Biochem. 2019, 139, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Lai, D.; Yang, H.; Xue, G.; He, A.; Chen, L.; Feng, L.; Ruan, J.; Xiang, D.; Yan, J.; et al. Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in foxtail millet (Setaria italica L.). BMC Genom. 2021, 22, 778. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Shi, Y.; Zhu, B.; Zhang, T.; Feng, Z.; Wang, X.; Li, X.; You, C. Genome-Wide Identification of Apple Atypical bHLH Subfamily PRE Members and Functional Characterization of MdPRE4.3 in Response to Abiotic Stress. Front. Genet. 2022, 13, 846559. [Google Scholar] [CrossRef]
- Wang, B.; Regulski, M.; Tseng, E.; Olson, A.; Goodwin, S.; McCombie, W.R.; Ware, D. A comparative transcriptional landscape of maize and sorghum obtained by single-molecule sequencing. Genome. Res. 2018, 28, 921–932. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, M.; de Lorenzo, L.; Abdel-Ghany, S.E.; Reddy, A.S.N.; Hunt, A.G. Wide-ranging transcriptome remodelling mediated by alternative polyadenylation in response to abiotic stresses in Sorghum. Plant J. 2020, 102, 916–930. [Google Scholar] [CrossRef] [PubMed]
- Tetreault, H.M.; Grover, S.; Scully, E.D.; Gries, T.; Palmer, N.A.; Sarath, G.; Louis, J.; Sattler, S.E. Global Responses of Resistant and Susceptible Sorghum (Sorghum bicolor) to Sugarcane Aphid (Melanaphis sacchari). Front. Plant Sci. 2019, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.M.; Marchi-Werle, L.; Hunt, T.E.; Heng-Moss, T.M.; Louis, J. Abscisic and Jasmonic Acids Contribute to Soybean Tolerance to the Soybean Aphid (Aphis glycines Matsumura). Sci. Rep. 2018, 8, 15148. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Cao, J.F.; Hu, G.J.; Chen, Z.W.; Wang, L.Y.; Shangguan, X.X.; Wang, L.J.; Mao, Y.B.; Zhang, T.Z.; Wendel, J.F.; et al. Core cis-element variation confers subgenome-biased expression of a transcription factor that functions in cotton fiber elongation. New Phytol. 2018, 218, 1061–1075. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, S.; Yang, K.Y.; Kim, Y.M.; Park, S.Y.; Kim, S.Y.; Soh, M.S. Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Heang, D.; Sassa, H. An atypical bHLH protein encoded by positive regulator of grain length 2 is involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG. Breed Sci. 2012, 62, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Medina-Puche, L.; Martinez-Rivas, F.J.; Molina-Hidalgo, F.J.; Mercado, J.A.; Moyano, E.; Rodriguez-Franco, A.; Caballero, J.L.; Munoz-Blanco, J.; Blanco-Portales, R. An atypical HLH transcriptional regulator plays a novel and important role in strawberry ripened receptacle. BMC Plant Biol. 2019, 19, 586. [Google Scholar] [CrossRef]
- Zhu, Z.; Chen, G.; Guo, X.; Yin, W.; Yu, X.; Hu, J.; Hu, Z. Overexpression of SlPRE2, an atypical bHLH transcription factor, affects plant morphology and fruit pigment accumulation in tomato. Sci. Rep. 2017, 7, 5786. [Google Scholar] [CrossRef]
- Zheng, K.; Wang, Y.; Zhang, N.; Jia, Q.; Wang, X.; Hou, C.; Chen, J.G.; Wang, S. Involvement of paclobutrazol resistance6/kidari, an Atypical bHLH Transcription Factor, in Auxin Responses in Arabidopsis. Front. Plant Sci. 2017, 8, 1813. [Google Scholar] [CrossRef]
- McCormick, R.F.; Truong, S.K.; Sreedasyam, A.; Jenkins, J.; Shu, S.; Sims, D.; Kennedy, M.; Amirebrahimi, M.; Weers, B.D.; McKinley, B.; et al. The Sorghum bicolor reference genome: Improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 2018, 93, 338–354. [Google Scholar] [CrossRef] [PubMed]
- Siefers, N.; Dang, K.K.; Kumimoto, R.W.; Bynum, W.E.t.; Tayrose, G.; Holt, B.F., 3rd. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009, 149, 625–641. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt, B.F., 3rd; Mantovani, R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.F.; Zhao, B.; Huang, C.C.; Chen, Z.W.; Zhao, T.; Liu, H.R.; Hu, G.J.; Shangguan, X.X.; Shan, C.M.; Wang, L.J.; et al. The miR319-Targeted GhTCP4 Promotes the Transition from Cell Elongation to Wall Thickening in Cotton Fiber. Mol. Plant 2020, 13, 1063–1077. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, S.; Wang, C.; Ding, G.; Cai, H.; Shi, L.; Xu, F. Induction of jasmonic acid biosynthetic genes inhibits Arabidopsis growth in response to low boron. J. Integr. Plant Biol. 2021, 63, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Moran, P.J.; Thompson, G.A. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 2001, 125, 1074–1085. [Google Scholar] [CrossRef]
- Pant, S.; Huang, Y. Genome-wide studies of PAL genes in sorghum and their responses to aphid infestation. Sci. Rep. 2022, 12, 22537. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.D.; Hua, Y.P.; Huang, J.Y.; Yu, S.T.; Wu, T.B.; Zhang, Y.; Chen, H.L.; Yue, C.P. Multiomic Analysis Reveals Core Regulatory Mechanisms underlying Steroidal Glycoalkaloid Metabolism in Potato Tubers. J. Agric. Food Chem. 2022, 70, 415–426. [Google Scholar] [CrossRef] [PubMed]


| Gene | Gene ID | Chromosome Location | CDS Length (bp) | Protein Length (aa) | bHLH Domain | Molecular Weight (Da) | pI |
|---|---|---|---|---|---|---|---|
| SbPRE1 | Sobic.006G236600 | Chr06:57805968–57807417 | 315 | 105 | 2–104 | 11,180.38 | 8 |
| SbPRE2 | Sobic.006G131900 | Chr06:49616617–49621803 | 483 | 161 | 39–109 | 17,385.78 | 9.48 |
| SbPRE3 | Sobic.004G267400 | Chr04:61174563–61177405 | 1080 | 360 | 153–237 | 37,275.27 | 6.35 |
| SbPRE4 | Sobic.001G488600 | Chr01:75864770–75871892 | 279 | 93 | 1–92 | 10,304.59 | 6.57 |
| SbPRE5 | Sobic.001G488400 | Chr01:75826747–75828379 | 261 | 87 | 1–86 | 9690.97 | 9.03 |
| SbPRE6 | Sobic.001G254100 | Chr01:28215024–28215913 | 288 | 96 | 1–95 | 10,442.79 | 7.98 |
| SbPRE7 | Sobic.005G178400 | Chr05:66069136–66070056 | 405 | 135 | 4–114 | 13,862.51 | 6.73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Wang, Z.; Jiao, Z.; Yuan, G.; Cui, L.; Duan, P.; Niu, J.; Lv, P.; Wang, J.; Shi, Y. Genome-Wide Identification of Sorghum Paclobutrazol-Resistance Gene Family and Functional Characterization of SbPRE4 in Response to Aphid Stress. Int. J. Mol. Sci. 2024, 25, 7257. https://doi.org/10.3390/ijms25137257
Guo Y, Wang Z, Jiao Z, Yuan G, Cui L, Duan P, Niu J, Lv P, Wang J, Shi Y. Genome-Wide Identification of Sorghum Paclobutrazol-Resistance Gene Family and Functional Characterization of SbPRE4 in Response to Aphid Stress. International Journal of Molecular Sciences. 2024; 25(13):7257. https://doi.org/10.3390/ijms25137257
Chicago/Turabian StyleGuo, Yongchao, Zhifang Wang, Zhiyin Jiao, Guang Yuan, Li Cui, Pengwei Duan, Jingtian Niu, Peng Lv, Jinping Wang, and Yannan Shi. 2024. "Genome-Wide Identification of Sorghum Paclobutrazol-Resistance Gene Family and Functional Characterization of SbPRE4 in Response to Aphid Stress" International Journal of Molecular Sciences 25, no. 13: 7257. https://doi.org/10.3390/ijms25137257
APA StyleGuo, Y., Wang, Z., Jiao, Z., Yuan, G., Cui, L., Duan, P., Niu, J., Lv, P., Wang, J., & Shi, Y. (2024). Genome-Wide Identification of Sorghum Paclobutrazol-Resistance Gene Family and Functional Characterization of SbPRE4 in Response to Aphid Stress. International Journal of Molecular Sciences, 25(13), 7257. https://doi.org/10.3390/ijms25137257






