Targeting Macrophage Polarization for Reinstating Homeostasis following Tissue Damage
Abstract
:1. Introduction
2. Macrophage Biological Roles
2.1. Macrophage Origins
2.2. Macrophage Phagocytic Ability
2.3. Macrophages as Antigen-Presenting Cells
2.4. Different Phenotypes of Macrophages
2.4.1. M1 Macrophages
| M1 (Classically Activated) | M2 (Alternatively Activated) | |||||
|---|---|---|---|---|---|---|
| Subtype | M1 [7,10,16,22,24,28,32,38,39,40,41,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,162,163,164,166,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186] | M2a [7,10,16,32,152,153,174,175,187,188,189,190,191,192,193,194] | M2b [9,10,11,16,32,154,175,195,196,197,198] | M2c [9,10,11,16,18,32,155,156,174,175,194,199,200,201,202] | M2d [10,11,16,32,157,158,159,203,204] | M2f [16,160,161,205,206,207,208] |
| Stimuli | LPS, TNF, IFNs, TLR, TLR ligands, GM-CSF, IL-17A, IL-12, ANG-1 | IL-4 (+M-CSF), IL-13 | LPS, TLRs, IL-1R ligands, immune complexes | Glucocorticoids, IL-10, TGF-β | TLR, A2AR agonists, IL-6 | Macrophage clearance of apoptotic cells |
| Markers | ↑ROS, ↑TNF-α, ↑ IL-12, ↓IL-10 | ↑IL-10, ↑MR, ↓ROS, ↓TNF-α, ↓IL-12 | ↑IL-10, ↑MR, ↓ROS, ↓IL-12 | ↑IL-10, ↑MR, ↓ROS | ↑IL-10, ↑VEGF, ↓TNF-α, ↓IL-12 | ↑TGF-β1, ↑MR |
| IL-1β, IL-6, IL-8, IL-23, iNOS, CCL2, CD14, CD16, CD32, CD80, CD86, Calprotectin, MHC-II, PKM2, MARCO, PFKFB3, ACOD1 | TGF-β, CCL17, CCL18, CCL22, CD36, CD163, CD301, IL-1Ra, Arg-1, IGF-1, MHC-II, CARKL, Ym1, Fizz-1, TREM2, IL1RN | IL-1β, IL-6, CCL1, CCL2, TNF-α, CD64, CD86, CD163, CCR8, VEGF, IGF-1, MHC-II, TNFSF14, PD-L1, SPHK-1 | TGF-β, CD163, TLR1, TLR8, SLAM, SPHK-1, THBS1, HMOX-1 | VEGF, MR, CD204, CD163, Arg-1, IDO, PGE2 | IL-10, PGE2, PAF | |
| Signaling factors | STAT1, STAT3, NF-κB (p65), IRF4, IRF5, Notch, AP-1, HIF1α | STAT6, SOCS1, PPARs, IRF4, GA TA3, KFL2, PI3K/AKT | STAT3, NF-κB (p50), IRF3, IRF4, Notch1, MAPKs, PI3K/AKT | STAT3, STAT6, NF-κB (p50), IRF4 | STAT1, NF-κB (p50), IRF3 | FcR pathway |
| Functions | Pro-inflammatory | Wound healing | Immunoregulatory | Immunosuppressive | Angiogenesis | Vessel morphogenesis |
| Boost inflammation, sterilization, apoptotic cell removal, tumor killing, Th1 response | Anti-inflammatory, cell proliferation, cell migration, growth factors production, tissue remodeling, cell debris removal, Th2 response | Cell maturation, tissue stabilization, angiogenesis, ECM synthesis, tumor progression, tissue remodeling, Th2 response | Inflammatory resolution, tissue repair, ECM synthesis, growth factors production | Anti-inflammatory, tumor progression | Anti-inflammatory, cell differentiation, vessel stabilization and maturation | |
2.4.2. M2 Macrophages
2.4.3. Transitions between M1 and M2
3. Drug Delivery for Modulating Macrophage Polarization Therapeutically
3.1. Proteins
3.2. Nucleic Acids
3.3. Other Molecules
4. Advanced Biomaterials for Macrophage Polarization Therapeutically
4.1. Biomaterials
4.1.1. Metallic Materials
4.1.2. Natural Polymers
4.1.3. Synthetic Polymers
4.2. Chemical and Physical Modification
4.2.1. Surface Topography
4.2.2. Surface Wettability and Charge
4.2.3. Substrate Stiffness and Geometry
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Martin, P.; Leibovich, S.J. Inflammatory cells during wound repair: The good, the bad and the ugly. Trends Cell Biol. 2005, 15, 599–607. [Google Scholar] [CrossRef]
- Nourshargh, S.; Alon, R. Leukocyte migration into inflamed tissues. Immunity 2014, 41, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A.; Vannella, K.M. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity 2016, 44, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Chazaud, B. Macrophages: Supportive cells for tissue repair and regeneration. Immunobiology 2014, 219, 172–178. [Google Scholar] [CrossRef]
- Oishi, Y.; Manabe, I. Macrophages in inflammation, repair and regeneration. Int. Immunol. 2018, 30, 511–528. [Google Scholar] [CrossRef]
- Hirayama, D.; Iida, T.; Nakase, H. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int. J. Mol. Sci. 2017, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Novak, M.L.; Koh, T.J. Macrophage phenotypes during tissue repair. J. Leukoc. Biol. 2013, 93, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Muntjewerff, E.M.; Meesters, L.D.; van den Bogaart, G. Antigen Cross-Presentation by Macrophages. Front. Immunol. 2020, 11, 1276. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Sun, L.; Huang, C.; Chen, B.-C.; Zhou, Z. Induction of macrophage M2b/c polarization by adipose tissue-derived mesenchymal stem cells. J. Immunol. Res. 2019, 2019, 7059680. [Google Scholar] [CrossRef]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef]
- Colin, S.; Chinetti-Gbaguidi, G.; Staels, B. Macrophage phenotypes in atherosclerosis. Immunol. Rev. 2014, 262, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Hesketh, M.; Sahin, K.B.; West, Z.E.; Murray, R.Z. Macrophage Phenotypes Regulate Scar Formation and Chronic Wound Healing. Int. J. Mol. Sci. 2017, 18, 1545. [Google Scholar] [CrossRef] [PubMed]
- Lis-López, L.; Bauset, C.; Seco-Cervera, M.; Cosín-Roger, J. Is the Macrophage Phenotype Determinant for Fibrosis Development? Biomedicines 2021, 9, 1747. [Google Scholar] [CrossRef]
- Jung, M.; Ma, Y.; Iyer, R.P.; DeLeon-Pennell, K.Y.; Yabluchanskiy, A.; Garrett, M.R.; Lindsey, M.L. IL-10 improves cardiac remodeling after myocardial infarction by stimulating M2 macrophage polarization and fibroblast activation. Basic. Res. Cardiol. 2017, 112, 33. [Google Scholar] [CrossRef] [PubMed]
- Laskin, D.L.; Sunil, V.R.; Gardner, C.R.; Laskin, J.D. Macrophages and tissue injury: Agents of defense or destruction? Annu. Rev. Pharmacol. Toxicol. 2011, 51, 267–288. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Peng, J.; Han, G.H.; Ding, X.; Wei, S.; Gao, G.; Huang, K.; Chang, F.; Wang, Y. Role of macrophages in peripheral nerve injury and repair. Neural Regen. Res. 2019, 14, 1335–1342. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.E.; Garcia, A.J. Macrophage phenotypes in tissue repair and the foreign body response: Implications for biomaterial-based regenerative medicine strategies. Acta Biomater. 2021, 133, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Biswas, S.K.; Galdiero, M.R.; Sica, A.; Locati, M. Macrophage plasticity and polarization in tissue repair and remodelling. J. Pathol. 2013, 229, 176–185. [Google Scholar] [CrossRef]
- Black, L.M.; Lever, J.M.; Agarwal, A. Renal Inflammation and Fibrosis: A Double-edged Sword. J. Histochem. Cytochem. 2019, 67, 663–681. [Google Scholar] [CrossRef]
- Yan, L.; Han, K.; Pang, B.; Jin, H.; Zhao, X.; Xu, X.; Jiang, C.; Cui, N.; Lu, T.; Shi, J. Surfactin-reinforced gelatin methacrylate hydrogel accelerates diabetic wound healing by regulating the macrophage polarization and promoting angiogenesis. Chem. Eng. J. 2021, 414, 128836. [Google Scholar] [CrossRef]
- Brady, R.V.; Thamm, D.H. Tumor-associated macrophages: Prognostic and therapeutic targets for cancer in humans and dogs. Front. Immunol. 2023, 14, 1176807. [Google Scholar] [CrossRef] [PubMed]
- Müller, E.; Christopoulos, P.F.; Halder, S.; Lunde, A.; Beraki, K.; Speth, M.; Øynebråten, I.; Corthay, A. Toll-Like Receptor Ligands and Interferon-γ Synergize for Induction of Antitumor M1 Macrophages. Front. Immunol. 2017, 8, 1383. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Guo, N.; Zhou, Y.; Chen, J.; Wei, Q.; Han, M. The role of tumor-associated macrophages (TAMs) in tumor progression and relevant advance in targeted therapy. Acta Pharm. Sin. B 2020, 10, 2156–2170. [Google Scholar] [CrossRef] [PubMed]
- Atri, C.; Guerfali, F.Z.; Laouini, D. Role of human macrophage polarization in inflammation during infectious diseases. Int. J. Mol. Sci. 2018, 19, 1801. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, D.; Harada, N.; Hayashi, T.; Tahara, Y.; Momose, F.; Sawada, S.; Mukai, S.A.; Akiyoshi, K.; Shiku, H. Nanogel-based immunologically stealth vaccine targets macrophages in the medulla of lymph node and induces potent antitumor immunity. ACS Nano 2014, 8, 9209–9218. [Google Scholar] [CrossRef]
- Kim, Y.K.; Que, R.; Wang, S.W.; Liu, W.F. Modification of biomaterials with a self-protein inhibits the macrophage response. Adv. Healthc. Mater. 2014, 3, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Talekar, M.; Raikar, A.; Amiji, M. Macrophage-targeted delivery systems for nucleic acid therapy of inflammatory diseases. J. Control. Release 2014, 190, 515–530. [Google Scholar] [CrossRef] [PubMed]
- Kashfi, K.; Kannikal, J.; Nath, N. Macrophage reprogramming and cancer therapeutics: Role of iNOS-derived NO. Cells 2021, 10, 3194. [Google Scholar] [CrossRef] [PubMed]
- Navegantes, K.C.; de Souza Gomes, R.; Pereira, P.A.T.; Czaikoski, P.G.; Azevedo, C.H.M.; Monteiro, M.C. Immune modulation of some autoimmune diseases: The critical role of macrophages and neutrophils in the innate and adaptive immunity. J. Transl. Med. 2017, 15, 36. [Google Scholar] [CrossRef]
- Vishwakarma, A.; Bhise, N.S.; Evangelista, M.B.; Rouwkema, J.; Dokmeci, M.R.; Ghaemmaghami, A.M.; Vrana, N.E.; Khademhosseini, A. Engineering Immunomodulatory Biomaterials to Tune the Inflammatory Response. Trends Biotechnol. 2016, 34, 470–482. [Google Scholar] [CrossRef]
- Wu, Y.; Hirschi, K.K. Tissue-Resident Macrophage Development and Function. Front. Cell Dev. Biol. 2020, 8, 617879. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Li, Y.; Fu, M.; Xin, H.-B. Polarizing macrophages in vitro. Macrophages Methods Protoc. 2018, 1784, 119–126. [Google Scholar]
- Sreejit, G.; Fleetwood, A.J.; Murphy, A.J.; Nagareddy, P.R. Origins and diversity of macrophages in health and disease. Clin. Transl. Immunol. 2020, 9, e1222. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, E.G.; Klapproth, K.; Schulz, C.; Busch, K.; de Bruijn, M.; Rodewald, H.R.; Geissmann, F. The Origin of Tissue-Resident Macrophages: When an Erythro-myeloid Progenitor Is an Erythro-myeloid Progenitor. Immunity 2015, 43, 1023–1024. [Google Scholar] [CrossRef] [PubMed]
- van Furth, R.; Cohn, Z.A. The origin and kinetics of mononuclear phagocytes. J. Exp. Med. 1968, 128, 415–435. [Google Scholar] [CrossRef] [PubMed]
- Epelman, S.; Lavine, K.J.; Randolph, G.J. Origin and functions of tissue macrophages. Immunity 2014, 41, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Bajgar, A.; Krejčová, G. On the origin of the functional versatility of macrophages. Front. Physiol. 2023, 14, 1128984. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Fessler, M.B.; Qu, P.; Heymann, J.; Kopp, J.B. Macrophage polarization in innate immune responses contributing to pathogenesis of chronic kidney disease. BMC Nephrol. 2020, 21, 270. [Google Scholar] [CrossRef] [PubMed]
- Yunna, C.; Mengru, H.; Lei, W.; Weidong, C. Macrophage M1/M2 polarization. Eur. J. Pharmacol. 2020, 877, 173090. [Google Scholar] [CrossRef]
- Arango Duque, G.; Descoteaux, A. Macrophage cytokines: Involvement in immunity and infectious diseases. Front. Immunol. 2014, 5, 491. [Google Scholar] [CrossRef]
- Lee, K.Y. M1 and M2 polarization of macrophages: A mini-review. Med. Biol. Sci. Eng. 2019, 2, 1–5. [Google Scholar] [CrossRef]
- Jain, S.; Tran, T.H.; Amiji, M. Macrophage repolarization with targeted alginate nanoparticles containing IL-10 plasmid DNA for the treatment of experimental arthritis. Biomaterials 2015, 61, 162–177. [Google Scholar] [CrossRef]
- Taylor, P.C.; Feldmann, M. Anti-TNF biologic agents: Still the therapy of choice for rheumatoid arthritis. Nat. Rev. Rheumatol. 2009, 5, 578–582. [Google Scholar] [CrossRef]
- Moore, K.J.; Sheedy, F.J.; Fisher, E.A. Macrophages in atherosclerosis: A dynamic balance. Nat. Rev. Immunol. 2013, 13, 709–721. [Google Scholar] [CrossRef]
- Saqib, U.; Sarkar, S.; Suk, K.; Mohammad, O.; Baig, M.S.; Savai, R. Phytochemicals as modulators of M1-M2 macrophages in inflammation. Oncotarget 2018, 9, 17937–17950. [Google Scholar] [CrossRef] [PubMed]
- dos Santos Luz, R.B.; Nicolazzi, L.H.C.N.; Saraiva Camara, N.O.; Braga, T.T. Chapter 1—Macrophages: From Metchnikoff to 2020 and ahead. In Macrophages in the Human Body; Saraiva Camara, N.O., Braga, T.T., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 1–18. [Google Scholar] [CrossRef]
- Hoeffel, G.; Ginhoux, F. Fetal monocytes and the origins of tissue-resident macrophages. Cell Immunol. 2018, 330, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Cline, M.J.; Moore, M.A. Embryonic origin of the mouse macrophage. Blood 1972, 39, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Winkler, I.G.; Sims, N.A.; Pettit, A.R.; Barbier, V.; Nowlan, B.; Helwani, F.; Poulton, I.J.; van Rooijen, N.; Alexander, K.A.; Raggatt, L.J.; et al. Bone marrow macrophages maintain hematopoietic stem cell (HSC) niches and their depletion mobilizes HSCs. Blood 2010, 116, 4815–4828. [Google Scholar] [CrossRef] [PubMed]
- Heideveld, E.; van den Akker, E. Digesting the role of bone marrow macrophages on hematopoiesis. Immunobiology 2017, 222, 814–822. [Google Scholar] [CrossRef]
- Mass, E. Delineating the origins, developmental programs and homeostatic functions of tissue-resident macrophages. Int. Immunol. 2018, 30, 493–501. [Google Scholar] [CrossRef]
- Gentek, R.; Molawi, K.; Sieweke, M.H. Tissue macrophage identity and self-renewal. Immunol. Rev. 2014, 262, 56–73. [Google Scholar] [CrossRef] [PubMed]
- Cannon, G.J.; Swanson, J.A. The macrophage capacity for phagocytosis. J. Cell Sci. 1992, 101 Pt 4, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Rosales, C.; Uribe-Querol, E. Phagocytosis: A Fundamental Process in Immunity. BioMed Res. Int. 2017, 2017, 9042851. [Google Scholar] [CrossRef]
- Martinez-Pomares, L.; Gordon, S. Antigen presentation the macrophage way. Cell 2007, 131, 641–643. [Google Scholar] [CrossRef] [PubMed]
- Unanue, E.R. Antigen-presenting function of the macrophage. Annu. Rev. Immunol. 1984, 2, 395–428. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.; Martinez, F.O. Alternative activation of macrophages: Mechanism and functions. Immunity 2010, 32, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J. Macrophage Polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef] [PubMed]
- Teti, G.; Biondo, C.; Beninati, C. The Phagocyte, Metchnikoff, and the Foundation of Immunology. Microbiol. Spectr. 2016, 4, 17–29. [Google Scholar] [CrossRef]
- Maulitz, R.C. Rudolf Virchow, Julius Cohnheim and the program of pathology. Bull. Hist. Med. 1978, 52, 162–182. [Google Scholar]
- Silva, H.M.; Báfica, A.; Rodrigues-Luiz, G.F.; Chi, J.; Santos, P.D.A.; Reis, B.S.; Hoytema van Konijnenburg, D.P.; Crane, A.; Arifa, R.D.N.; Martin, P.; et al. Vasculature-associated fat macrophages readily adapt to inflammatory and metabolic challenges. J. Exp. Med. 2019, 216, 786–806. [Google Scholar] [CrossRef]
- Thao, N.P.; Cuong, N.X.; Luyen, B.T.; Quang, T.H.; Hanh, T.T.; Kim, S.; Koh, Y.S.; Nam, N.H.; Van Kiem, P.; Van Minh, C.; et al. Anti-inflammatory components of the starfish Astropecten polyacanthus. Mar. Drugs 2013, 11, 2917–2926. [Google Scholar] [CrossRef] [PubMed]
- Aderem, A.; Underhill, D.M. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 1999, 17, 593–623. [Google Scholar] [CrossRef] [PubMed]
- Uribe-Querol, E.; Rosales, C. Phagocytosis: Our Current Understanding of a Universal Biological Process. Front. Immunol. 2020, 11, 1066. [Google Scholar] [CrossRef] [PubMed]
- Botelho, R.J.; Grinstein, S. Phagocytosis. Curr. Biol. 2011, 21, R533–R538. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kang, R.; Coyne, C.B.; Zeh, H.J.; Lotze, M.T. PAMP s and DAMP s: Signal 0s that spur autophagy and immunity. Immunol. Rev. 2012, 249, 158–175. [Google Scholar] [CrossRef] [PubMed]
- Mueller, R.B.; Sheriff, A.; Gaipl, U.S.; Wesselborg, S.; Lauber, K. Attraction of phagocytes by apoptotic cells is mediated by lysophosphatidylcholine. Autoimmunity 2007, 40, 342–344. [Google Scholar] [CrossRef] [PubMed]
- Segawa, K.; Nagata, S. An apoptotic ‘eat me’signal: Phosphatidylserine exposure. Trends Cell Biol. 2015, 25, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.-H.; Shih, C.-H.; Feng, S.-Y.; Li, I.; Chang, S.-C.; Lin, Y.-C.; Hsu, H.-C. CX3CL1 (+) microparticles mediate the chemoattraction of alveolar macrophages toward apoptotic acute promyelocytic leukemic cells. Cell. Physiol. Biochem. 2014, 33, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Sokolowski, J.D.; Chabanon-Hicks, C.N.; Han, C.Z.; Heffron, D.S.; Mandell, J.W. Fractalkine is a “find-me” signal released by neurons undergoing ethanol-induced apoptosis. Front. Cell. Neurosci. 2014, 8, 360. [Google Scholar] [CrossRef]
- Gude, D.R.; Alvarez, S.E.; Paugh, S.W.; Mitra, P.; Yu, J.; Griffiths, R.; Barbour, S.E.; Milstien, S.; Spiegel, S. Apoptosis induces expression of sphingosine kinase 1 to release sphingosine-1-phosphate as a “come-and-get-me” signal. FASEB J. 2008, 22, 2629. [Google Scholar] [CrossRef]
- Elliott, M.R.; Chekeni, F.B.; Trampont, P.C.; Lazarowski, E.R.; Kadl, A.; Walk, S.F.; Park, D.; Woodson, R.I.; Ostankovich, M.; Sharma, P. Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature 2009, 461, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Chekeni, F.B.; Elliott, M.R.; Sandilos, J.K.; Walk, S.F.; Kinchen, J.M.; Lazarowski, E.R.; Armstrong, A.J.; Penuela, S.; Laird, D.W.; Salvesen, G.S. Pannexin 1 channels mediate ‘find-me’signal release and membrane permeability during apoptosis. Nature 2010, 467, 863–867. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, K.S. Find-me and eat-me signals in apoptotic cell clearance: Progress and conundrums. J. Exp. Med. 2010, 207, 1807–1817. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Zhang, X. Measuring opsonic phagocytosis via Fcγ receptors and complement receptors on macrophages. Curr. Protoc. Immunol. 2011, 95, 14.27.1–14.27.11. [Google Scholar] [CrossRef] [PubMed]
- Barth, N.D.; Marwick, J.A.; Vendrell, M.; Rossi, A.G.; Dransfield, I. The “phagocytic synapse” and clearance of apoptotic cells. Front. Immunol. 2017, 8, 1708. [Google Scholar] [CrossRef] [PubMed]
- Kuhlman, M.; Joiner, K.; Ezekowitz, R. The human mannose-binding protein functions as an opsonin. J. Exp. Med. 1989, 169, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Areschoug, T.; Gordon, S. Scavenger receptors: Role in innate immunity and microbial pathogenesis. Cell. Microbiol. 2009, 11, 1160–1169. [Google Scholar] [CrossRef] [PubMed]
- Palecanda, A.; Kobzik, L. Receptors for unopsonized particles: The role of alveolar macrophage scavenger receptors. Curr. Mol. Med. 2001, 1, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, R.L.; Febbraio, M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2009, 2, re3. [Google Scholar] [CrossRef]
- Canton, J.; Neculai, D.; Grinstein, S. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 2013, 13, 621–634. [Google Scholar] [CrossRef]
- Richardson, M.B.; Williams, S.J. MCL and Mincle: C-type lectin receptors that sense damaged self and pathogen-associated molecular patterns. Front. Immunol. 2014, 5, 288. [Google Scholar] [CrossRef]
- Mayer, S.; Raulf, M.-K.; Lepenies, B. C-type lectins: Their network and roles in pathogen recognition and immunity. Histochem. Cell Biol. 2017, 147, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Albacker, L.A.; Yu, S.; Bedoret, D.; Lee, W.L.; Umetsu, S.E.; Monahan, S.; Freeman, G.J.; Umetsu, D.T.; DeKruyff, R.H. TIM-4, expressed by medullary macrophages, regulates respiratory tolerance by mediating phagocytosis of antigen-specific T cells. Mucosal Immunol. 2013, 6, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Park, D.; Tosello-Trampont, A.-C.; Elliott, M.R.; Lu, M.; Haney, L.B.; Ma, Z.; Klibanov, A.L.; Mandell, J.W.; Ravichandran, K.S. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 2007, 450, 430–434. [Google Scholar] [CrossRef] [PubMed]
- Lemke, G.; Burstyn-Cohen, T. TAM receptors and the clearance of apoptotic cells. Ann. N. Y Acad. Sci. 2010, 1209, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Burstyn-Cohen, T.; Fresia, R. TAM receptors in phagocytosis: Beyond the mere internalization of particles. Immunol. Rev. 2023, 319, 7–26. [Google Scholar] [CrossRef] [PubMed]
- Poirier, M.B.; Fiorino, C.; Rajasekar, T.K.; Harrison, R.E. F-actin flashes on phagosomes mechanically deform contents for efficient digestion in macrophages. J. Cell Sci. 2020, 133, jcs239384. [Google Scholar] [CrossRef] [PubMed]
- Dingjan, I.; Linders, P.T.A.; Verboogen, D.R.J.; Revelo, N.H.; Ter Beest, M.; van den Bogaart, G. Endosomal and Phagosomal SNAREs. Physiol. Rev. 2018, 98, 1465–1492. [Google Scholar] [CrossRef] [PubMed]
- Hatsuzawa, K.; Tamura, T.; Hashimoto, H.; Hashimoto, H.; Yokoya, S.; Miura, M.; Nagaya, H.; Wada, I. Involvement of Syntaxin 18, an Endoplasmic Reticulum (ER)-localized SNARE Protein, in ER-mediated Phagocytosis. Mol. Biol. Cell 2006, 17, 3964–3977. [Google Scholar] [CrossRef]
- Tjelle, T.E.; Lovdal, T.; Berg, T. Phagosome dynamics and function. Bioessays 2000, 22, 255–263. [Google Scholar] [CrossRef]
- Canton, J. Phagosome maturation in polarized macrophages. J. Leukoc. Biol. 2014, 96, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Fountain, A.; Inpanathan, S.; Alves, P.; Verdawala, M.B.; Botelho, R.J. Phagosome maturation in macrophages: Eat, digest, adapt, and repeat. Adv. Biol. Regul. 2021, 82, 100832. [Google Scholar] [CrossRef] [PubMed]
- Stuart, L.M.; Ezekowitz, R.A. Phagocytosis: Elegant complexity. Immunity 2005, 22, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Kloc, M.; Uosef, A.; Kubiak, J.Z.; Ghobrial, R.M. Macrophage Proinflammatory Responses to Microorganisms and Transplanted Organs. Int. J. Mol. Sci. 2020, 21, 9669. [Google Scholar] [CrossRef] [PubMed]
- Banga, S.; Gao, P.; Shen, X.; Fiscus, V.; Zong, W.-X.; Chen, L.; Luo, Z.-Q. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc. Natl. Acad. Sci. USA 2007, 104, 5121–5126. [Google Scholar] [CrossRef] [PubMed]
- Lanzavecchia, A. Antigen-specific interaction between T and B cells. Nature 1985, 314, 537–539. [Google Scholar] [CrossRef]
- den Haan, J.M.; Arens, R.; van Zelm, M.C. The activation of the adaptive immune system: Cross-talk between antigen-presenting cells, T cells and B cells. Immunol. Lett. 2014, 162, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.G.; Green, J.A.; Gray, E.E.; Xu, Y.; Cyster, J.G. Immune complex relay by subcapsular sinus macrophages and noncognate B cells drives antibody affinity maturation. Nat. Immunol. 2009, 10, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, S.F.; Degn, S.E.; Pitcher, L.A.; Woodruff, M.; Heesters, B.A.; Carroll, M.C. Trafficking of B cell antigen in lymph nodes. Annu. Rev. Immunol. 2011, 29, 215–233. [Google Scholar] [CrossRef]
- Junt, T.; Moseman, E.A.; Iannacone, M.; Massberg, S.; Lang, P.A.; Boes, M.; Fink, K.; Henrickson, S.E.; Shayakhmetov, D.M.; Di Paolo, N.C.; et al. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature 2007, 450, 110–114. [Google Scholar] [CrossRef]
- Carrasco, Y.R.; Batista, F.D. B cells acquire particulate antigen in a macrophage-rich area at the boundary between the follicle and the subcapsular sinus of the lymph node. Immunity 2007, 27, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Moran, I.; Grootveld, A.K.; Nguyen, A.; Phan, T.G. Subcapsular Sinus Macrophages: The Seat of Innate and Adaptive Memory in Murine Lymph Nodes. Trends Immunol. 2019, 40, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Pomares, L.; Gordon, S. CD169+ macrophages at the crossroads of antigen presentation. Trends Immunol. 2012, 33, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Kuka, M.; Iannacone, M. The role of lymph node sinus macrophages in host defense. Ann. N. Y. Acad. Sci. 2014, 1319, 38–46. [Google Scholar] [CrossRef]
- Louie, D.A.P.; Liao, S. Lymph node subcapsular sinus macrophages as the frontline of lymphatic immune defense. Front. Immunol. 2019, 10, 347. [Google Scholar] [CrossRef] [PubMed]
- Unanue, E.R.; Cerottini, J.C.; Bedford, M. Persistence of Antigen on the Surface of Macrophages. Nature 1969, 222, 1193–1195. [Google Scholar] [CrossRef]
- Taylor, P.R.; Martinez-Pomares, L.; Stacey, M.; Lin, H.H.; Brown, G.D.; Gordon, S. Macrophage receptors and immune recognition. Annu. Rev. Immunol. 2005, 23, 901–944. [Google Scholar] [CrossRef]
- Phan, T.G.; Grigorova, I.; Okada, T.; Cyster, J.G. Subcapsular encounter and complement-dependent transport of immune complexes by lymph node B cells. Nat. Immunol. 2007, 8, 992–1000. [Google Scholar] [CrossRef]
- Mantegazza, A.R.; Magalhaes, J.G.; Amigorena, S.; Marks, M.S. Presentation of phagocytosed antigens by MHC class I and II. Traffic 2013, 14, 135–152. [Google Scholar] [CrossRef]
- Neefjes, J.; Jongsma, M.L.M.; Paul, P.; Bakke, O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011, 11, 823–836. [Google Scholar] [CrossRef]
- Kotsias, F.; Cebrian, I.; Alloatti, A. Antigen processing and presentation. Int. Rev. Cell Mol. Biol. 2019, 348, 69–121. [Google Scholar]
- Bevan, M.J. Antigen presentation to cytotoxic T lymphocytes in vivo. J. Exp. Med. 1995, 182, 639–641. [Google Scholar] [CrossRef]
- Brutkiewicz, R.R.; Lin, Y.; Cho, S.; Hwang, Y.K.; Sriram, V.; Roberts, T.J. CD1d-mediated antigen presentation to natural killer T (NKT) cells. Crit. Rev. Immunol. 2003, 23, 18. [Google Scholar] [CrossRef]
- Cruz-Leal, Y.; Grubaugh, D.; Nogueira, C.V.; Lopetegui-González, I.; Del Valle, A.; Escalona, F.; Laborde, R.J.; Alvarez, C.; Fernández, L.E.; Starnbach, M.N. The vacuolar pathway in macrophages plays a major role in antigen cross-presentation induced by the pore-forming protein sticholysin II encapsulated into liposomes. Front. Immunol. 2018, 9, 2473. [Google Scholar] [CrossRef] [PubMed]
- Grabowska, J.; Lopez-Venegas, M.A.; Affandi, A.J.; den Haan, J.M.M. CD169(+) Macrophages Capture and Dendritic Cells Instruct: The Interplay of the Gatekeeper and the General of the Immune System. Front. Immunol. 2018, 9, 2472. [Google Scholar] [CrossRef] [PubMed]
- Schliehe, C.; Redaelli, C.; Engelhardt, S.; Fehlings, M.; Mueller, M.; van Rooijen, N.; Thiry, M.; Hildner, K.; Weller, H.; Groettrup, M. CD8- dendritic cells and macrophages cross-present poly(D,L-lactate-co-glycolate) acid microsphere-encapsulated antigen in vivo. J. Immunol. 2011, 187, 2112–2121. [Google Scholar] [CrossRef]
- Hey, Y.Y.; Tan, J.K.; O’Neill, H.C. Redefining Myeloid Cell Subsets in Murine Spleen. Front. Immunol. 2015, 6, 652. [Google Scholar] [CrossRef]
- Enders, M.; Franken, L.; Philipp, M.S.; Kessler, N.; Baumgart, A.K.; Eichler, M.; Wiertz, E.J.H.; Garbi, N.; Kurts, C. Splenic Red Pulp Macrophages Cross-Prime Early Effector CTL That Provide Rapid Defense against Viral Infections. J. Immunol. 2020, 204, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Tang-Huau, T.-L.; Gueguen, P.; Goudot, C.; Durand, M.; Bohec, M.; Baulande, S.; Pasquier, B.; Amigorena, S.; Segura, E. Human in vivo-generated monocyte-derived dendritic cells and macrophages cross-present antigens through a vacuolar pathway. Nat. Commun. 2018, 9, 2570. [Google Scholar] [CrossRef]
- Ruedl, C.; Storni, T.; Lechner, F.; Bächi, T.; Bachmann, M.F. Cross-presentation of virus-like particles by skin-derived CD8–dendritic cells: A dispensable role for TAP. Eur. J. Immunol. 2002, 32, 818–825. [Google Scholar] [CrossRef]
- Backer, R.; Schwandt, T.; Greuter, M.; Oosting, M.; Jüngerkes, F.; Tüting, T.; Boon, L.; O’Toole, T.; Kraal, G.; Limmer, A.; et al. Effective collaboration between marginal metallophilic macrophages and CD8+ dendritic cells in the generation of cytotoxic T cells. Proc. Natl. Acad. Sci. USA 2010, 107, 216–221. [Google Scholar] [CrossRef] [PubMed]
- van Dinther, D.; Veninga, H.; Iborra, S.; Borg, E.G.F.; Hoogterp, L.; Olesek, K.; Beijer, M.R.; Schetters, S.T.T.; Kalay, H.; Garcia-Vallejo, J.J.; et al. Functional CD169 on Macrophages Mediates Interaction with Dendritic Cells for CD8(+) T Cell Cross-Priming. Cell Rep. 2018, 22, 1484–1495. [Google Scholar] [CrossRef] [PubMed]
- Blohm, U.; Roth, E.; Brommer, K.; Dumrese, T.; Rosenthal, F.M.; Pircher, H. Lack of effector cell function and altered tetramer binding of tumor-infiltrating lymphocytes. J. Immunol. 2002, 169, 5522–5530. [Google Scholar] [CrossRef] [PubMed]
- Hoeve, M.A.; Savage, N.D.L.; de Boer, T.; Langenberg, D.M.L.; de Waal Malefyt, R.; Ottenhoff, T.H.M.; Verreck, F.A.W. Divergent effects of IL-12 and IL-23 on the production of IL-17 by human T cells. Eur. J. Immunol. 2006, 36, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Riabov, V.; Gudima, A.; Wang, N.; Mickley, A.; Orekhov, A.; Kzhyshkowska, J. Role of tumor associated macrophages in tumor angiogenesis and lymphangiogenesis. Front. Physiol. 2014, 5, 75. [Google Scholar] [CrossRef] [PubMed]
- Modak, M.; Mattes, A.K.; Reiss, D.; Skronska-Wasek, W.; Langlois, R.; Sabarth, N.; Konopitzky, R.; Ramirez, F.; Lehr, K.; Mayr, T.; et al. CD206+ tumor-associated macrophages cross-present tumor antigen and drive antitumor immunity. JCI Insight 2022, 7, e155022. [Google Scholar] [CrossRef] [PubMed]
- Di Gioacchino, M.; Della Valle, L.; Allegra, A.; Pioggia, G.; Gangemi, S. AllergoOncology: Role of immune cells and immune proteins. Clin. Transl. Allergy 2022, 12, e12133. [Google Scholar] [CrossRef] [PubMed]
- Germano, G.; Frapolli, R.; Belgiovine, C.; Anselmo, A.; Pesce, S.; Liguori, M.; Erba, E.; Uboldi, S.; Zucchetti, M.; Pasqualini, F.; et al. Role of macrophage targeting in the antitumor activity of trabectedin. Cancer Cell 2013, 23, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef]
- Li, M.; Hou, Q.; Zhong, L.; Zhao, Y.; Fu, X. Macrophage Related Chronic Inflammation in Non-Healing Wounds. Front. Immunol. 2021, 12, 681710. [Google Scholar] [CrossRef]
- Lu, Y.-C.; Yeh, W.-C.; Ohashi, P.S. LPS/TLR4 signal transduction pathway. Cytokine 2008, 42, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef]
- Koo, S.-J.; Garg, N.J. Metabolic programming of macrophage functions and pathogens control. Redox Biol. 2019, 24, 101198. [Google Scholar] [CrossRef]
- Jin, J.; Xiao, Y.; Hu, H.; Zou, Q.; Li, Y.; Gao, Y.; Ge, W.; Cheng, X.; Sun, S.-C. Proinflammatory TLR signalling is regulated by a TRAF2-dependent proteolysis mechanism in macrophages. Nat. Commun. 2015, 6, 5930. [Google Scholar] [CrossRef]
- Takeuch, O.; Akira, S. Epigenetic control of macrophage polarization. Eur. J. Immunol. 2011, 41, 2490–2493. [Google Scholar] [CrossRef]
- Wajant, H.; Scheurich, P. TNFR1-induced activation of the classical NF-κB pathway. FEBS J. 2011, 278, 862–876. [Google Scholar] [CrossRef] [PubMed]
- Kroner, A.; Greenhalgh, A.D.; Zarruk, J.G.; Dos Santos, R.P.; Gaestel, M.; David, S. TNF and increased intracellular iron alter macrophage polarization to a detrimental M1 phenotype in the injured spinal cord. Neuron 2014, 83, 1098–1116. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Zhou, J.; Yang, H.; Wang, X.; Zhang, X.; Wang, Q.; Wu, X.; Han, Y.; Song, Y.; Tan, Y.; et al. IL-17A Produced by Neutrophils Protects against Pneumonic Plague through Orchestrating IFN-γ–Activated Macrophage Programming. J. Immunol. 2014, 192, 704–713. [Google Scholar] [CrossRef]
- Watkins, S.K.; Egilmez, N.K.; Suttles, J.; Stout, R.D. IL-12 rapidly alters the functional profile of tumor-associated and tumor-infiltrating macrophages in vitro and in vivo. J. Immunol. 2007, 178, 1357–1362. [Google Scholar] [CrossRef]
- Fleetwood, A.J.; Dinh, H.; Cook, A.D.; Hertzog, P.J.; Hamilton, J.A. GM-CSF-and M-CSF-dependent macrophage phenotypes display differential dependence on type I interferon signaling. J. Leukoc. Biol. 2009, 86, 411–421. [Google Scholar] [CrossRef]
- Seok, S.H.; Heo, J.-I.; Hwang, J.-H.; Na, Y.-R.; Yun, J.-H.; Lee, E.H.; Park, J.-W.; Cho, C.-H. Angiopoietin-1 elicits pro-inflammatory responses in monocytes and differentiating macrophages. Mol. Cells 2013, 35, 550–556. [Google Scholar] [CrossRef]
- Kerneur, C.; Cano, C.E.; Olive, D. Major pathways involved in macrophage polarization in cancer. Front. Immunol. 2022, 13, 1026954. [Google Scholar] [CrossRef] [PubMed]
- Watford, W.T.; Moriguchi, M.; Morinobu, A.; O’Shea, J.J. The biology of IL-12: Coordinating innate and adaptive immune responses. Cytokine Growth Factor Rev. 2003, 14, 361–368. [Google Scholar] [CrossRef]
- Wojno, E.D.T.; Hunter, C.A.; Stumhofer, J.S. The immunobiology of the interleukin-12 family: Room for discovery. Immunity 2019, 50, 851–870. [Google Scholar] [CrossRef]
- Xu, J.; Chi, F.; Tsukamoto, H. Notch signaling and M1 macrophage activation in obesity-alcohol synergism. Clin. Res. Hepatol. Gastroenterol. 2015, 39 (Suppl. 1), S24–S28. [Google Scholar] [CrossRef]
- Zhu, L.; Zhao, Q.; Yang, T.; Ding, W.; Zhao, Y. Cellular metabolism and macrophage functional polarization. Int. Rev. Immunol. 2015, 34, 82–100. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.J.; Boyle, R.H.; Astin, J.W.; Flores, M.V.; Oehlers, S.H.; Sanderson, L.E.; Ellett, F.; Lieschke, G.J.; Crosier, K.E.; Crosier, P.S. Immunoresponsive gene 1 augments bactericidal activity of macrophage-lineage cells by regulating β-oxidation-dependent mitochondrial ROS production. Cell Metab. 2013, 18, 265–278. [Google Scholar] [CrossRef]
- Wilson, H.M. SOCS Proteins in Macrophage Polarization and Function. Front. Immunol. 2014, 5, 357. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, T.; Natoli, G. Transcriptional regulation of macrophage polarization: Enabling diversity with identity. Nat. Rev. Immunol. 2011, 11, 750–761. [Google Scholar] [CrossRef]
- Chen, X.; Dou, J.; Fu, Z.; Qiu, Y.; Zou, L.; Huang, D.; Tan, X. Macrophage M1 polarization mediated via the IL-6/STAT3 pathway contributes to apical periodontitis induced by Porphyromonas gingivalis. J. Appl. Oral. Sci. 2022, 30, e20220316. [Google Scholar] [CrossRef]
- Yang, T.; Wang, R.; Liu, H.; Wang, L.; Li, J.; Wu, S.; Chen, X.; Yang, X.; Zhao, Y. Berberine regulates macrophage polarization through IL-4-STAT6 signaling pathway in Helicobacter pylori-induced chronic atrophic gastritis. Life Sci. 2021, 266, 118903. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, W.; Xiong, S. Macrophage differentiation and polarization via phosphatidylinositol 3-kinase/Akt-ERK signaling pathway conferred by serum amyloid P component. J. Immunol. 2011, 187, 1764–1777. [Google Scholar] [CrossRef]
- Wang, L.-X.; Zhang, S.-X.; Wu, H.-J.; Rong, X.-L.; Guo, J. M2b macrophage polarization and its roles in diseases. J. Leukoc. Biol. 2019, 106, 345–358. [Google Scholar] [CrossRef]
- Mohapatra, S.; Pioppini, C.; Ozpolat, B.; Calin, G.A. Non-coding RNAs regulation of macrophage polarization in cancer. Mol. Cancer 2021, 20, 24. [Google Scholar] [CrossRef]
- Sureshbabu, A.; Muhsin, S.A.; Choi, M.E. TGF-β signaling in the kidney: Profibrotic and protective effects. Am. J. Physiol. -Ren. Physiol. 2016, 310, F596–F606. [Google Scholar] [CrossRef] [PubMed]
- Mancino, A.; Lawrence, T. Nuclear factor-κB and tumor-associated macrophages. Clin. Cancer Res. 2010, 16, 784–789. [Google Scholar] [CrossRef]
- Chen, L.; Wang, S.; Wang, Y.; Zhang, W.; Ma, K.; Hu, C.; Zhu, H.; Liang, S.; Liu, M.; Xu, N. IL-6 influences the polarization of macrophages and the formation and growth of colorectal tumor. Oncotarget 2018, 9, 17443. [Google Scholar] [CrossRef]
- Bai, Y.; Zhang, X.; Zhou, J.; Guo, J.; Liu, Y.; Liang, C.; Wang, W.; Xing, Y.; Wu, J.; Hu, D. A2aR on lung adenocarcinoma cells: A novel target for cancer therapy via recruiting and regulating tumor-associated macrophages. Chem.-Biol. Interact. 2023, 382, 110543. [Google Scholar] [CrossRef] [PubMed]
- Fadok, V.A.; Bratton, D.L.; Konowal, A.; Freed, P.W.; Westcott, J.Y.; Henson, P.M. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J. Clin. Investig. 1998, 101, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Graney, P.; Ben-Shaul, S.; Landau, S.; Bajpai, A.; Singh, B.; Eager, J.; Cohen, A.; Levenberg, S.; Spiller, K. Macrophages of diverse phenotypes drive vascularization of engineered tissues. Sci. Adv. 2020, 6, eaay6391. [Google Scholar] [CrossRef]
- Yang, Z.; Min, Z.; Yu, B. Reactive oxygen species and immune regulation. Int. Rev. Immunol. 2020, 39, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.D. Anatomy of a discovery: m1 and m2 macrophages. Front. Immunol. 2015, 6, 212. [Google Scholar] [CrossRef] [PubMed]
- Mills, C. M1 and M2 macrophages: Oracles of health and disease. Crit. Rev. Immunol. 2012, 32, 463–488. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; He, Y.; Sun, X.; Li, Q.; Wang, W.; Zhao, A.; Di, W. A high M1/M2 ratio of tumor-associated macrophages is associated with extended survival in ovarian cancer patients. J. Ovarian Res. 2014, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Macciò, A.; Gramignano, G.; Cherchi, M.C.; Tanca, L.; Melis, L.; Madeddu, C. Role of M1-polarized tumor-associated macrophages in the prognosis of advanced ovarian cancer patients. Sci. Rep. 2020, 10, 6096. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cao, F.; Li, M.; Li, P.; Yu, Y.; Xiang, L.; Xu, T.; Lei, J.; Tai, Y.Y.; Zhu, J.; et al. Hydroxychloroquine induced lung cancer suppression by enhancing chemo-sensitization and promoting the transition of M2-TAMs to M1-like macrophages. J. Exp. Clin. Cancer Res. 2018, 37, 259. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Yang, D.; Ma, J.; Yang, J.; Xue, J.; Song, F.; Liu, X. Modulation of Wnt/β-catenin signaling in IL-17A-mediated macrophage polarization of RAW264.7 cells. Braz. J. Med. Biol. Res. 2020, 53, e9488. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cheng, F.; Ma, T.-t.; Xiong, H.-Y.; Li, Z.-W.; Xie, C.-L.; Liu, C.-Y.; Tu, Z.-G. Interleukin-12 inhibits the hepatocellular carcinoma growth by inducing macrophage polarization to the M1-like phenotype through downregulation of Stat-3. Mol. Cell. Biochem. 2016, 415, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.A. Colony-stimulating factors in inflammation and autoimmunity. Nat. Rev. Immunol. 2008, 8, 533–544. [Google Scholar] [CrossRef]
- Jablonski, K.A.; Amici, S.A.; Webb, L.M.; Ruiz-Rosado Jde, D.; Popovich, P.G.; Partida-Sanchez, S.; Guerau-de-Arellano, M. Novel Markers to Delineate Murine M1 and M2 Macrophages. PLoS ONE 2015, 10, e0145342. [Google Scholar] [CrossRef]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.M.; Liu, J.C.; Trujillo-de Santiago, G.; Cha, B.H.; Vishwakarma, A.; Ghaemmaghami, A.M.; Khademhosseini, A. Delivery strategies to control inflammatory response: Modulating M1-M2 polarization in tissue engineering applications. J. Control. Release 2016, 240, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Cao, Q.; Zheng, D.; Sun, Y.; Wang, C.; Yu, X.; Wang, Y.; Lee, V.W.; Zheng, G.; Tan, T.K. Discrete functions of M2a and M2c macrophage subsets determine their relative efficacy in treating chronic kidney disease. Kidney Int. 2013, 84, 745–755. [Google Scholar] [CrossRef]
- Oates, T.C.; Moura, P.L.; Cross, S.; Roberts, K.; Baum, H.E.; Haydn-Smith, K.L.; Wilson, M.C.; Heesom, K.J.; Severn, C.E.; Toye, A.M. Defining the proteomic landscape of cultured macrophages and their polarization continuum. Immunol. Cell Biol. 2023, 101, 947–963. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Xu, X.-H.; Jin, L. Macrophage polarization in physiological and pathological pregnancy. Front. Immunol. 2019, 10, 792. [Google Scholar] [CrossRef] [PubMed]
- Amici, S.A.; Young, N.A.; Narvaez-Miranda, J.; Jablonski, K.A.; Arcos, J.; Rosas, L.; Papenfuss, T.L.; Torrelles, J.B.; Jarjour, W.N.; Guerau-de-Arellano, M. CD38 is robustly induced in human macrophages and monocytes in inflammatory conditions. Front. Immunol. 2018, 9, 1593. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.-M.; Tang, P.M.-K.; Li, J.; Lan, H.Y. Macrophage phenotype in kidney injury and repair. Kidney Dis. 2015, 1, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Dreschers, S.; Ohl, K.; Lehrke, M.; Möllmann, J.; Denecke, B.; Costa, I.; Vogl, T.; Viemann, D.; Roth, J.; Orlikowsky, T. Impaired cellular energy metabolism in cord blood macrophages contributes to abortive response toward inflammatory threats. Nat. Commun. 2019, 10, 1685. [Google Scholar] [CrossRef] [PubMed]
- Lateef, Z.; Stuart, G.; Jones, N.; Mercer, A.; Fleming, S.; Wise, L. The cutaneous inflammatory response to thermal burn injury in a murine model. Int. J. Mol. Sci. 2019, 20, 538. [Google Scholar] [CrossRef]
- Apostolopoulos, V.; De Courten, M.P.; Stojanovska, L.; Blatch, G.L.; Tangalakis, K.; De Courten, B. The complex immunological and inflammatory network of adipose tissue in obesity. Mol. Nutr. Food Res. 2016, 60, 43–57. [Google Scholar] [CrossRef]
- Sasaki, A. Microglia and brain macrophages: An update. Neuropathology 2017, 37, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Li, N.; Cheng, H.; Zhang, X.; Cao, X.; Qi, T.; Dai, L.; Zhang, Z.; Chen, X.; Li, C. HSPA12A is a novel player in nonalcoholic steatohepatitis via promoting nuclear PKM2-mediated M1 macrophage polarization. Diabetes 2019, 68, 361–376. [Google Scholar] [CrossRef]
- Elchaninov, A.; Lokhonina, A.; Vishnyakova, P.; Soboleva, A.; Poltavets, A.; Artemova, D.; Makarov, A.; Glinkina, V.; Goldshtein, D.; Bolshakova, G. Marco+ macrophage dynamics in regenerating liver after 70% liver resection in mice. Biomedicines 2021, 9, 1129. [Google Scholar] [CrossRef] [PubMed]
- Tawakol, A.; Singh, P.; Mojena, M.; Pimentel-Santillana, M.; Emami, H.; MacNabb, M.; Rudd, J.H.; Narula, J.; Enriquez, J.A.; Través, P.G. HIF-1α and PFKFB3 mediate a tight relationship between proinflammatory activation and anerobic metabolism in atherosclerotic macrophages. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1463–1471. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Chen, F.; Wang, N.; Tang, D.; Kang, R. ACOD1 in immunometabolism and disease. Cell. Mol. Immunol. 2020, 17, 822–833. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.-Z.; Wang, X.; Wang, Y.; Niu, A.; Wang, S.; Zou, C.; Harris, R.C. IL-4/IL-13–mediated polarization of renal macrophages/dendritic cells to an M2a phenotype is essential for recovery from acute kidney injury. Kidney Int. 2017, 91, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, T.M.; Mooney, D.J. Functional muscle recovery with nanoparticle-directed M2 macrophage polarization in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 10648–10653. [Google Scholar] [CrossRef] [PubMed]
- Nelms, K.; Keegan, A.D.; Zamorano, J.; Ryan, J.J.; Paul, W.E. The IL-4 receptor: Signaling mechanisms and biologic functions. Annu. Rev. Immunol. 1999, 17, 701–738. [Google Scholar] [CrossRef]
- Nelson, M.P.; Christmann, B.S.; Werner, J.L.; Metz, A.E.; Trevor, J.L.; Lowell, C.A.; Steele, C. IL-33 and M2a alveolar macrophages promote lung defense against the atypical fungal pathogen Pneumocystis murina. J. Immunol. 2011, 186, 2372–2381. [Google Scholar] [CrossRef]
- Mazher, M.; Moqidem, Y.A.; Zidan, M.; Sayed, A.A.; Abdellatif, A. Autophagic reprogramming of bone marrow–derived macrophages. Immunol. Res. 2023, 71, 229–246. [Google Scholar] [CrossRef]
- Little, A.C.; Pathanjeli, P.; Wu, Z.; Bao, L.; Goo, L.E.; Yates, J.A.; Oliver, C.R.; Soellner, M.B.; Merajver, S.D. IL-4/IL-13 stimulated macrophages enhance breast cancer invasion via rho-GTPase regulation of synergistic VEGF/CCL-18 signaling. Front. Oncol. 2019, 9, 456. [Google Scholar] [CrossRef] [PubMed]
- Rőszer, T. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediat. Inflamm. 2015, 2015, 816460. [Google Scholar] [CrossRef] [PubMed]
- Tseng, W.-C.; Tsai, M.-T.; Chen, N.-J.; Tarng, D.-C. Trichostatin A alleviates renal interstitial fibrosis through modulation of the M2 macrophage subpopulation. Int. J. Mol. Sci. 2020, 21, 5966. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, S.W.; Huang, L.C.; Chang, Y.P.; Hung, C.H.; Yang, Y.H. M2b macrophage subset decrement as an indicator of cognitive function in Alzheimer’s disease. Psychiatry Clin. Neurosci. 2020, 74, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-H.; Choi, E. Macrophages and Inflammation. J. Rheum. Dis. 2018, 25, 11. [Google Scholar] [CrossRef]
- Suzuki, K.; Meguro, K.; Nakagomi, D.; Nakajima, H. Roles of alternatively activated M2 macrophages in allergic contact dermatitis. Allergol. Int. 2017, 66, 392–397. [Google Scholar] [CrossRef]
- Yang, R.; Liao, Y.; Wang, L.; He, P.; Hu, Y.; Yuan, D.; Wu, Z.; Sun, X. Exosomes derived from M2b macrophages attenuate DSS-induced colitis. Front. Immunol. 2019, 10, 2346. [Google Scholar] [CrossRef] [PubMed]
- Koscsó, B.; Csóka, B.; Kókai, E.; Németh, Z.H.; Pacher, P.; Virág, L.; Leibovich, S.J.; Haskó, G. Adenosine augments IL-10-induced STAT3 signaling in M2c macrophages. J. Leukoc. Biol. 2013, 94, 1309–1315. [Google Scholar] [CrossRef]
- Lurier, E.B.; Dalton, D.; Dampier, W.; Raman, P.; Nassiri, S.; Ferraro, N.M.; Rajagopalan, R.; Sarmady, M.; Spiller, K.L. Transcriptome analysis of IL-10-stimulated (M2c) macrophages by next-generation sequencing. Immunobiology 2017, 222, 847–856. [Google Scholar]
- Hao, N.-B.; Lü, M.-H.; Fan, Y.-H.; Cao, Y.-L.; Zhang, Z.-R.; Yang, S.-M. Macrophages in Tumor Microenvironments and the Progression of Tumors. Clin. Dev. Immunol. 2012, 2012, 948098. [Google Scholar] [CrossRef]
- Stempin, C.C.; Dulgerian, L.R.; Garrido, V.V.; Cerban, F.M. Arginase in parasitic infections: Macrophage activation, immunosuppression, and intracellular signals. J. Biomed. Biotechnol. 2010, 2010, 683485. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, C.J.; Pinhal-Enfield, G.; Elson, G.; Cronstein, B.N.; Hasko, G.; Outram, S.; Leibovich, S.J. The adenosine-dependent angiogenic switch of macrophages to an M2-like phenotype is independent of interleukin-4 receptor alpha (IL-4Rα) signaling. Inflammation 2013, 36, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, D.; Cang, H.; Guo, B. Crosstalk between cancer and immune cells: Role of tumor-associated macrophages in the tumor microenvironment. Cancer Med. 2019, 8, 4709–4721. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Yang, Y.; Chu, C.; Rung, S.-A.; Wang, Z.; Man, Y.; Lin, J.; Qu, Y. Macrophage response mediated by extracellular matrix: Recent progress. Biomed. Mater. 2023, 18, 012003. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Tian, X.Y. The role of macrophages in vascular repair and regeneration after ischemic injury. Int. J. Mol. Sci. 2020, 21, 6328. [Google Scholar] [CrossRef] [PubMed]
- Chawla, A. Control of macrophage activation and function by PPARs. Circ. Res. 2010, 106, 1559–1569. [Google Scholar] [CrossRef] [PubMed]
- Colegio, O.R.; Chu, N.Q.; Szabo, A.L.; Chu, T.; Rhebergen, A.M.; Jairam, V.; Cyrus, N.; Brokowski, C.E.; Eisenbarth, S.C.; Phillips, G.M.; et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 2014, 513, 559–563. [Google Scholar] [CrossRef]
- Mantovani, A.; Sica, A.; Locati, M. Macrophage polarization comes of age. Immunity 2005, 23, 344–346. [Google Scholar] [CrossRef]
- Tu, G.-w.; Shi, Y.; Zheng, Y.-j.; Ju, M.-j.; He, H.-y.; Ma, G.-g.; Hao, G.-w.; Luo, Z. Glucocorticoid attenuates acute lung injury through induction of type 2 macrophage. J. Transl. Med. 2017, 15, 181. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, L.; Ma, K.; Zhao, Y.; Liu, X.; Wang, Y.; Liu, M.; Liang, S.; Zhu, H.; Xu, N. Polarization of macrophages in the tumor microenvironment is influenced by EGFR signaling within colon cancer cells. Oncotarget 2016, 7, 75366. [Google Scholar] [CrossRef]
- Liu, F.; Qiu, H.; Xue, M.; Zhang, S.; Zhang, X.; Xu, J.; Chen, J.; Yang, Y.; Xie, J. MSC-secreted TGF-β regulates lipopolysaccharide-stimulated macrophage M2-like polarization via the Akt/FoxO1 pathway. Stem Cell Res. Ther. 2019, 10, 345. [Google Scholar] [CrossRef] [PubMed]
- Braune, J.; Weyer, U.; Hobusch, C.; Mauer, J.; Brüning, J.C.; Bechmann, I.; Gericke, M. IL-6 regulates M2 polarization and local proliferation of adipose tissue macrophages in obesity. J. Immunol. 2017, 198, 2927–2934. [Google Scholar] [CrossRef] [PubMed]
- Coffelt, S.B.; Tal, A.O.; Scholz, A.; De Palma, M.; Patel, S.; Urbich, C.; Biswas, S.K.; Murdoch, C.; Plate, K.H.; Reiss, Y. Angiopoietin-2 regulates gene expression in TIE2-expressing monocytes and augments their inherent proangiogenic functions. Cancer Res. 2010, 70, 5270–5280. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Chen, L.; Cai, Z.; Qian, S.; Liu, Z.; Zhao, B.; Zhang, Y.; Sun, X.; Cui, W. Advanced Biomaterials for Regulating Polarization of Macrophages in Wound Healing. Adv. Funct. Mater. 2022, 32, 2111003. [Google Scholar] [CrossRef]
- Sridharan, R.; Cameron, A.R.; Kelly, D.J.; Kearney, C.J.; O’Brien, F.J. Biomaterial based modulation of macrophage polarization: A review and suggested design principles. Mater. Today 2015, 18, 313–325. [Google Scholar] [CrossRef]
- Liu, Y.; Segura, T. Biomaterials-Mediated Regulation of Macrophage Cell Fate. Front. Bioeng. Biotechnol. 2020, 8, 609297. [Google Scholar] [CrossRef] [PubMed]
- Boersema, G.S.; Grotenhuis, N.; Bayon, Y.; Lange, J.F.; Bastiaansen-Jenniskens, Y.M. The Effect of Biomaterials Used for Tissue Regeneration Purposes on Polarization of Macrophages. Biores Open Access 2016, 5, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Rostam, H.M.; Singh, S.; Salazar, F.; Magennis, P.; Hook, A.; Singh, T.; Vrana, N.E.; Alexander, M.R.; Ghaemmaghami, A.M. The impact of surface chemistry modification on macrophage polarisation. Immunobiology 2016, 221, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Bratlie, K.M. The Influence of Polysaccharides-Based Material on Macrophage Phenotypes. Macromol. Biosci. 2021, 21, 2100031. [Google Scholar] [CrossRef]
- Brown, B.N.; Ratner, B.D.; Goodman, S.B.; Amar, S.; Badylak, S.F. Macrophage polarization: An opportunity for improved outcomes in biomaterials and regenerative medicine. Biomaterials 2012, 33, 3792–3802. [Google Scholar] [CrossRef]
- Saclier, M.; Yacoub-Youssef, H.; Mackey, A.L.; Arnold, L.; Ardjoune, H.; Magnan, M.; Sailhan, F.; Chelly, J.; Pavlath, G.K.; Mounier, R.; et al. Differentially activated macrophages orchestrate myogenic precursor cell fate during human skeletal muscle regeneration. Stem Cells 2013, 31, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, H.; Wang, X.; Jiang, G.; Liu, H.; Zhang, G.; Wang, H.; Fang, R.; Bu, X.; Cai, S. TGF-β induces M2-like macrophage polarization via SNAIL-mediated suppression of a pro-inflammatory phenotype. Oncotarget 2016, 7, 52294. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Lei, Z.; Yuan, M.; Zhu, H.; Yuan, X.; Liu, W.; Pu, H.; Jiang, J.; Zhang, Y.; Jiang, X.; et al. Cartilage repair mediated by thermosensitive photocrosslinkable TGFβ1-loaded GM-HPCH via immunomodulating macrophages, recruiting MSCs and promoting chondrogenesis. Theranostics 2020, 10, 2872–2887. [Google Scholar] [CrossRef]
- Wang, Q.; Li, H.; Xiao, Y.; Li, S.; Li, B.; Zhao, X.; Ye, L.; Guo, B.; Chen, X.; Ding, Y.; et al. Locally controlled delivery of TNFα antibody from a novel glucose-sensitive scaffold enhances alveolar bone healing in diabetic conditions. J. Control. Release 2015, 206, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, J.; Li, Z.; Yang, Y.; Li, L.; Zhao, Y.; Zhao, J. Enzyme-Loaded Catalytic Macrophage Vesicles with Cascade Amplification of Tumor-Targeting for Oxygenated Photodynamic Therapy. Int. J. Nanomed. 2021, 16, 7801–7812. [Google Scholar] [CrossRef] [PubMed]
- Guiducci, C.; Vicari, A.P.; Sangaletti, S.; Trinchieri, G.; Colombo, M.P. Redirecting in vivo elicited tumor infiltrating macrophages and dendritic cells towards tumor rejection. Cancer Res. 2005, 65, 3437–3446. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Kapate, N.; Shields, C.W.; Mitragotri, S. Drug delivery to macrophages: A review of targeting drugs and drug carriers to macrophages for inflammatory diseases. Adv. Drug Deliv. Rev. 2020, 165–166, 15–40. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Chen, G.; Chen, Q.; Li, P.Y.; Cheng, H.; Gu, Z. Engineering protein delivery depots for cancer immunotherapy. Bioconjugate Chem. 2019, 30, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Nair, L.S.; Laurencin, C.T. Biodegradable polymers as biomaterials. Progress. Polym. Sci. 2007, 32, 762–798. [Google Scholar] [CrossRef]
- Ezike, T.C.; Okpala, U.S.; Onoja, U.L.; Nwike, C.P.; Ezeako, E.C.; Okpara, O.J.; Okoroafor, C.C.; Eze, S.C.; Kalu, O.L.; Odoh, E.C.; et al. Advances in drug delivery systems, challenges and future directions. Heliyon 2023, 9, e17488. [Google Scholar] [CrossRef]
- Tabata, K.; Kurosaka, S.; Watanabe, M.; Edamura, K.; Satoh, T.; Yang, G.; Abdelfattah, E.; Wang, J.; Goltsov, A.; Floryk, D.; et al. Tumor growth and metastasis suppression by Glipr1 gene-modified macrophages in a metastatic prostate cancer model. Gene Ther. 2011, 18, 969–978. [Google Scholar] [CrossRef]
- Boehler, R.M.; Kuo, R.; Shin, S.; Goodman, A.G.; Pilecki, M.A.; Gower, R.M.; Leonard, J.N.; Shea, L.D. Lentivirus delivery of IL-10 to promote and sustain macrophage polarization towards an anti-inflammatory phenotype. Biotechnol. Bioeng. 2014, 111, 1210–1221. [Google Scholar] [CrossRef] [PubMed]
- Nikitina, E.; Larionova, I.; Choinzonov, E.; Kzhyshkowska, J. Monocytes and Macrophages as Viral Targets and Reservoirs. Int. J. Mol. Sci. 2018, 19, 2821. [Google Scholar] [CrossRef] [PubMed]
- Reid, T.; Warren, R.; Kirn, D. Intravascular adenoviral agents in cancer patients: Lessons from clinical trials. Cancer Gene Ther. 2002, 9, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Raper, S.E.; Chirmule, N.; Lee, F.S.; Wivel, N.A.; Bagg, A.; Gao, G.P.; Wilson, J.M.; Batshaw, M.L. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol. Genet. Metab. 2003, 80, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Marshall, E. Gene therapy death prompts review of adenovirus vector. Science 1999, 286, 2244–2245. [Google Scholar] [CrossRef] [PubMed]
- Hirai, H.; Satoh, E.; Osawa, M.; Inaba, T.; Shimazaki, C.; Kinoshita, S.; Nakagawa, M.; Mazda, O.; Imanishi, J. Use of EBV-based Vector/HVJ-liposome complex vector for targeted gene therapy of EBV-associated neoplasms. Biochem. Biophys. Res. Commun. 1997, 241, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Caffery, B.; Lee, J.S.; Alexander-Bryant, A.A. Vectors for Glioblastoma Gene Therapy: Viral & Non-Viral Delivery Strategies. Nanomaterials 2019, 9, 105. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Conley, S.M.; Makkia, R.; Guo, J.; Cooper, M.J.; Naash, M.I. Comparative Analysis of DNA Nanoparticles and AAVs for Ocular Gene Delivery. PLoS ONE 2012, 7, e52189. [Google Scholar] [CrossRef]
- Nayerossadat, N.; Maedeh, T.; Ali, P.A. Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 2012, 1, 27. [Google Scholar] [CrossRef]
- Al-Halifa, S.; Gauthier, L.; Arpin, D.; Bourgault, S.; Archambault, D. Nanoparticle-Based Vaccines Against Respiratory Viruses. Front. Immunol. 2019, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Leng, X.; Zhang, Q. The Current State of Nanoparticle-Induced Macrophage Polarization and Reprogramming Research. Int. J. Mol. Sci. 2017, 18, 336. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, Y.; Xu, H.; Fu, Q. Nanoparticle-Delivered IRF5 siRNA Facilitates M1 to M2 Transition, Reduces Demyelination and Neurofilament Loss, and Promotes Functional Recovery After Spinal Cord Injury in Mice. Inflammation 2016, 39, 1704–1717. [Google Scholar] [CrossRef] [PubMed]
- Shobaki, N.; Sato, Y.; Suzuki, Y.; Okabe, N.; Harashima, H. Manipulating the function of tumor-associated macrophages by siRNA-loaded lipid nanoparticles for cancer immunotherapy. J. Control. Release 2020, 325, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Cecchin, R.; Troyer, Z.; Witwer, K.; Morris, K.V. Extracellular vesicles: The next generation in gene therapy delivery. Mol. Ther. 2023, 31, 1225–1230. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Huang, L.; Mao, M.; Ding, J.; Wu, G.; Fan, W.; Yang, T.; Zhang, M.; Huang, Y.; Xie, H.-Y. Viral Protein-Pseudotyped and siRNA-Electroporated Extracellular Vesicles for Cancer Immunotherapy. Adv. Funct. Mater. 2020, 30, 2006515. [Google Scholar] [CrossRef]
- Getting, S.J. Melanocortin peptides and their receptors: New targets for anti-inflammatory therapy. Trends Pharmacol. Sci. 2002, 23, 447–449. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.W. The immunomodulating neuropeptide alpha-melanocyte-stimulating hormone (alpha-MSH) suppresses LPS-stimulated TLR4 with IRAK-M in macrophages. J. Neuroimmunol. 2005, 162, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Ju, N.; Hayashi, H.; Shimamura, M.; Baba, S.; Yoshida, S.; Morishita, R.; Rakugi, H.; Nakagami, H. Prevention of bleomycin-induced pulmonary fibrosis by a RANKL peptide in mice. Sci. Rep. 2022, 12, 12474. [Google Scholar] [CrossRef]
- Jha, A.; Larkin, J., 3rd; Moore, E. SOCS1-KIR Peptide in PEGDA Hydrogels Reduces Pro-Inflammatory Macrophage Activation. Macromol. Biosci. 2023, 23, e2300237. [Google Scholar] [CrossRef]
- Zhou, W.; Kang, S.; Wang, F.; Qin, Y.; Liu, J.; Xiao, X.; Chen, X.; Zhang, D. Chromofungin, a chromogranin A-derived peptide, protects against sepsis-induced acute lung injury by inhibiting LBP/TLR4-dependent inflammatory signaling. Eur. J. Pharmacol. 2023, 958, 176043. [Google Scholar] [CrossRef] [PubMed]
- Gunassekaran, G.R.; Poongkavithai Vadevoo, S.M.; Baek, M.C.; Lee, B. M1 macrophage exosomes engineered to foster M1 polarization and target the IL-4 receptor inhibit tumor growth by reprogramming tumor-associated macrophages into M1-like macrophages. Biomaterials 2021, 278, 121137. [Google Scholar] [CrossRef] [PubMed]
- Cha, B.H.; Shin, S.R.; Leijten, J.; Li, Y.C.; Singh, S.; Liu, J.C.; Annabi, N.; Abdi, R.; Dokmeci, M.R.; Vrana, N.E.; et al. Integrin-Mediated Interactions Control Macrophage Polarization in 3D Hydrogels. Adv. Healthc. Mater. 2017, 6, 1700289. [Google Scholar] [CrossRef]
- Lee, D.; Nah, H.; Ko, W.-K.; Jun Kim, S.; Han, G.H.; Jeong, D.; Lee, D.; Han, I.; Sheen, S.H.; Heo, D.N.; et al. Thiolate poly(lactic-co-glycolic acid) nanofibers loaded with dexamethasone and ropivacaine show enhanced sustained release in the treatment of neuropathic pain through a local therapy technique. Chem. Eng. J. 2022, 431, 133356. [Google Scholar] [CrossRef]
- Beeraka, N.M.; Doreswamy, S.H.; Sadhu, S.P.; Srinivasan, A.; Pragada, R.R.; Madhunapantula, S.V.; Aliev, G. The Role of Exosomes in Stemness and Neurodegenerative Diseases-Chemoresistant-Cancer Therapeutics and Phytochemicals. Int. J. Mol. Sci. 2020, 21, 6818. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.B.; Águas, A.P.; Barbosa, M.A.; Barbosa, J.N. Immunomodulatory biomaterial-based wound dressings advance the healing of chronic wounds via regulating macrophage behavior. Regen. Biomater. 2022, 9, rbac065. [Google Scholar] [CrossRef]
- Dukhinova, M.S.; Prilepskii, A.Y.; Shtil, A.A.; Vinogradov, V.V. Metal Oxide Nanoparticles in Therapeutic Regulation of Macrophage Functions. Nanomaterials 2019, 9, 1631. [Google Scholar] [CrossRef]
- Dervan, A.; Franchi, A.; Almeida-Gonzalez, F.R.; Dowling, J.K.; Kwakyi, O.B.; McCoy, C.E.; O’Brien, F.J.; Hibbitts, A. Biomaterial and Therapeutic Approaches for the Manipulation of Macrophage Phenotype in Peripheral and Central Nerve Repair. Pharmaceutics 2021, 13, 2161. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, N.; Schirmer, L.; Atallah, P.; Wandel, E.; Ferrer, R.A.; Werner, C.; Simon, J.C.; Franz, S.; Freudenberg, U. Glycosaminoglycan-based hydrogels capture inflammatory chemokines and rescue defective wound healing in mice. Sci. Transl. Med. 2017, 9, eaai9044. [Google Scholar] [CrossRef]
- Rezvani Ghomi, E.; Nourbakhsh, N.; Akbari Kenari, M.; Zare, M.; Ramakrishna, S. Collagen-based biomaterials for biomedical applications. J. Biomed. Mater. Res. Part B Appl. Biomater. 2021, 109, 1986–1999. [Google Scholar] [CrossRef]
- Fakhri, E.; Eslami, H.; Maroufi, P.; Pakdel, F.; Taghizadeh, S.; Ganbarov, K.; Yousefi, M.; Tanomand, A.; Yousefi, B.; Mahmoudi, S.; et al. Chitosan biomaterials application in dentistry. Int. J. Biol. Macromol. 2020, 162, 956–974. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Zhou, C.; Fan, H.; Fan, Y.; Jiang, Q.; Song, P.; Fan, H.; Chen, Y.; Zhang, X. Bio-Functional Design, Application and Trends in Metallic Biomaterials. Int. J. Mol. Sci. 2018, 19, 24. [Google Scholar] [CrossRef] [PubMed]
- Eliaz, N. Corrosion of Metallic Biomaterials: A Review. Materials 2019, 12, 407. [Google Scholar] [CrossRef]
- Choi, S.-r.; Kwon, J.-w.; Suk, K.-s.; Kim, H.-s.; Moon, S.-h.; Park, S.-y.; Lee, B.H. The Clinical Use of Osteobiologic and Metallic Biomaterials in Orthopedic Surgery: The Present and the Future. Materials 2023, 16, 3633. [Google Scholar] [CrossRef] [PubMed]
- Lima, F.d.S.; Fock, R.A. A Review of the Action of Magnesium on Several Processes Involved in the Modulation of Hematopoiesis. Int. J. Mol. Sci. 2020, 21, 7084. [Google Scholar] [CrossRef]
- Kazakova, G.; Safronova, T.; Golubchikov, D.; Shevtsova, O.; Rau, J.V. Resorbable Mg2+-containing phosphates for bone tissue repair. Materials 2021, 14, 4857. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo Borgognoni, C.; Kim, J.H.; Zucolotto, V.; Fuchs, H.; Riehemann, K. Human macrophage responses to metal-oxide nanoparticles: A review. Artif. Cells Nanomed. Biotechnol. 2018, 46, 694–703. [Google Scholar] [CrossRef] [PubMed]
- Nagajyothi, P.C.; Cha, S.J.; Yang, I.J.; Sreekanth, T.V.; Kim, K.J.; Shin, H.M. Antioxidant and anti-inflammatory activities of zinc oxide nanoparticles synthesized using Polygala tenuifolia root extract. J. Photochem. Photobiol. B 2015, 146, 10–17. [Google Scholar] [CrossRef]
- Antonoglou, O.; Lafazanis, K.; Mourdikoudis, S.; Vourlias, G.; Lialiaris, T.; Pantazaki, A.; Dendrinou-Samara, C. Biological relevance of CuFeO(2) nanoparticles: Antibacterial and anti-inflammatory activity, genotoxicity, DNA and protein interactions. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 99, 264–274. [Google Scholar] [CrossRef]
- Ali, S.S.; Morsy, R.; El-Zawawy, N.A.; Fareed, M.F.; Bedaiwy, M.Y. Synthesized zinc peroxide nanoparticles (ZnO(2)-NPs): A novel antimicrobial, anti-elastase, anti-keratinase, and anti-inflammatory approach toward polymicrobial burn wounds. Int. J. Nanomed. 2017, 12, 6059–6073. [Google Scholar] [CrossRef]
- Seisenbaeva, G.A.; Fromell, K.; Vinogradov, V.V.; Terekhov, A.N.; Pakhomov, A.V.; Nilsson, B.; Ekdahl, K.N.; Vinogradov, V.V.; Kessler, V.G. Dispersion of TiO(2) nanoparticles improves burn wound healing and tissue regeneration through specific interaction with blood serum proteins. Sci. Rep. 2017, 7, 15448. [Google Scholar] [CrossRef] [PubMed]
- Serebrovska, Z.; Swanson, R.J.; Portnichenko, V.; Shysh, A.; Pavlovich, S.; Tumanovska, L.; Dorovskych, A.; Lysenko, V.; Tertykh, V.; Bolbukh, Y.; et al. Anti-inflammatory and antioxidant effect of cerium dioxide nanoparticles immobilized on the surface of silica nanoparticles in rat experimental pneumonia. Biomed. Pharmacother. 2017, 92, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Heckman, K.L.; DeCoteau, W.; Estevez, A.; Reed, K.J.; Costanzo, W.; Sanford, D.; Leiter, J.C.; Clauss, J.; Knapp, K.; Gomez, C.; et al. Custom cerium oxide nanoparticles protect against a free radical mediated autoimmune degenerative disease in the brain. ACS Nano 2013, 7, 10582–10596. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; You, Q.; Chen, X. Transition Metal-Based Therapies for Inflammatory Diseases. Adv. Mater. 2023, 35, e2212102. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fan, Y.; Liu, H. Macrophage Polarization in Response to Biomaterials for Vascularization. Ann. Biomed. Eng. 2021, 49, 1992–2005. [Google Scholar] [CrossRef] [PubMed]
- Torregrossa, M.; Kakpenova, A.; Simon, J.C.; Franz, S. Modulation of macrophage functions by ECM-inspired wound dressings—A promising therapeutic approach for chronic wounds. Biol. Chem. 2021, 402, 1289–1307. [Google Scholar] [CrossRef] [PubMed]
- Huleihel, L.; Dziki, J.L.; Bartolacci, J.G.; Rausch, T.; Scarritt, M.E.; Cramer, M.C.; Vorobyov, T.; LoPresti, S.T.; Swineheart, I.T.; White, L.J.; et al. Macrophage phenotype in response to ECM bioscaffolds. Semin. Immunol. 2017, 29, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Elhadad, A.A.; Alcudia, A.; Begines, B.; Pérez-Soriano, E.M.; Torres, Y. A multidisciplinary perspective on the latest trends in artificial cartilage fabrication to mimic real tissue. Appl. Mater. Today 2022, 29, 101603. [Google Scholar] [CrossRef]
- Shoulders, M.D.; Raines, R.T. Collagen Structure and Stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef]
- Belda Marín, C.; Fitzpatrick, V.; Kaplan, D.L.; Landoulsi, J.; Guénin, E.; Egles, C. Silk Polymers and Nanoparticles: A Powerful Combination for the Design of Versatile Biomaterials. Front. Chem. 2020, 8, 604398. [Google Scholar] [CrossRef]
- Ghatak, S.; Maytin, E.V.; Mack, J.A.; Hascall, V.C.; Atanelishvili, I.; Moreno Rodriguez, R.; Markwald, R.R.; Misra, S. Roles of Proteoglycans and Glycosaminoglycans in Wound Healing and Fibrosis. Int. J. Cell Biol. 2015, 2015, 834893. [Google Scholar] [CrossRef]
- Berdiaki, A.; Neagu, M.; Giatagana, E.-M.; Kuskov, A.; Tsatsakis, A.M.; Tzanakakis, G.N.; Nikitovic, D. Glycosaminoglycans: Carriers and Targets for Tailored Anti-Cancer Therapy. Biomolecules 2021, 11, 395. [Google Scholar] [CrossRef]
- Litwiniuk, M.; Krejner, A.; Speyrer, M.S.; Gauto, A.R.; Grzela, T. Hyaluronic acid in inflammation and tissue regeneration. Wounds 2016, 28, 78–88. [Google Scholar]
- Snetkov, P.; Zakharova, K.; Morozkina, S.; Olekhnovich, R.; Uspenskaya, M. Hyaluronic Acid: The Influence of Molecular Weight on Structural, Physical, Physico-Chemical, and Degradable Properties of Biopolymer. Polymers 2020, 12, 1800. [Google Scholar] [CrossRef]
- Stern, R.; Asari, A.A.; Sugahara, K.N. Hyaluronan fragments: An information-rich system. Eur. J. Cell Biol. 2006, 85, 699–715. [Google Scholar] [CrossRef]
- Hintze, V.; Schnabelrauch, M.; Rother, S. Chemical modification of hyaluronan and their biomedical applications. Front. Chem. 2022, 10, 830671. [Google Scholar] [CrossRef]
- Zhang, M.; James, S. Synthesis and properties of melt-processable hyaluronan esters. J. Mater. Sci. 2005, 40, 2937–2943. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Matsukawa, K.; Ishigami, Y. The relation between the adsorption behavior at the interface and the conformational changes in hyaluronates partially modified with various acyl chains. Carbohydr. Polym. 1995, 26, 149–154. [Google Scholar] [CrossRef]
- Zhang, R.; Huang, Z.; Xue, M.; Yang, J.; Tan, T. Detailed characterization of an injectable hyaluronic acid-polyaspartylhydrazide hydrogel for protein delivery. Carbohydr. Polym. 2011, 85, 717–725. [Google Scholar] [CrossRef]
- Dahl, L.B.; Laurent, T.C.; Smedsrød, B. Preparation of biologically intact radioiodinated hyaluronan of high specific radioactivity: Coupling of 125I-tyramine-cellobiose to amino groups after partial N-deacetylation. Anal. Biochem. 1988, 175, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Balazs, E.; Högberg, B.; Laurent, T. The biological activity of hyaluron sulfuric acid. Acta Physiol. Scand. 1951, 23, 168–178. [Google Scholar] [CrossRef]
- Magnani, A.; Albanese, A.; Lamponi, S.; Barbucci, R. Blood-interaction performance of differently sulphated hyaluronic acids. Thromb. Res. 1996, 81, 383–395. [Google Scholar] [CrossRef]
- Christgen, S.; Place, D.E.; Kanneganti, T.D. Toward targeting inflammasomes: Insights into their regulation and activation. Cell Res. 2020, 30, 315–327. [Google Scholar] [CrossRef]
- Jouy, F.; Lohmann, N.; Wandel, E.; Ruiz-Gómez, G.; Pisabarro, M.T.; Beck-Sickinger, A.G.; Schnabelrauch, M.; Möller, S.; Simon, J.C.; Kalkhof, S.; et al. Sulfated hyaluronan attenuates inflammatory signaling pathways in macrophages involving induction of antioxidants. Proteomics 2017, 17, e1700082. [Google Scholar] [CrossRef]
- Hauck, S.; Zager, P.; Halfter, N.; Wandel, E.; Torregrossa, M.; Kakpenova, A.; Rother, S.; Ordieres, M.; Räthel, S.; Berg, A.; et al. Collagen/hyaluronan based hydrogels releasing sulfated hyaluronan improve dermal wound healing in diabetic mice via reducing inflammatory macrophage activity. Bioact. Mater. 2021, 6, 4342–4359. [Google Scholar] [CrossRef]
- Hatano, S.; Watanabe, H. Regulation of Macrophage and Dendritic Cell Function by Chondroitin Sulfate in Innate to Antigen-Specific Adaptive Immunity. Front. Immunol. 2020, 11, 232. [Google Scholar] [CrossRef]
- Couchman, J.R.; Pataki, C.A. An introduction to proteoglycans and their localization. J. Histochem. Cytochem. 2012, 60, 885–897. [Google Scholar] [CrossRef]
- Taylor, K.R.; Gallo, R.L. Glycosaminoglycans and their proteoglycans: Host-associated molecular patterns for initiation and modulation of inflammation. FASEB J 2006, 20, 9–22. [Google Scholar] [CrossRef]
- Hascall, V.C.; Majors, A.K.; De La Motte, C.A.; Evanko, S.P.; Wang, A.; Drazba, J.A.; Strong, S.A.; Wight, T.N. Intracellular hyaluronan: A new frontier for inflammation? Biochim. Biophys. Acta 2004, 1673, 3–12. [Google Scholar] [CrossRef]
- Tremmel, M.; Matzke, A.; Albrecht, I.; Laib, A.M.; Olaku, V.; Ballmer-Hofer, K.; Christofori, G.; Héroult, M.; Augustin, H.G.; Ponta, H.; et al. A CD44v6 peptide reveals a role of CD44 in VEGFR-2 signaling and angiogenesis. Blood 2009, 114, 5236–5244. [Google Scholar] [CrossRef]
- Rayahin, J.E.; Buhrman, J.S.; Zhang, Y.; Koh, T.J.; Gemeinhart, R.A. High and low molecular weight hyaluronic acid differentially influence macrophage activation. ACS Biomater. Sci. Eng. 2015, 1, 481–493. [Google Scholar] [CrossRef]
- Zhao, J.; Feng, Y.; Liu, X.; Li, H.; Guo, H.; Ke, J.; Long, X. The relationship of ALPK1, hyaluronic acid and M1 macrophage polarization in the temporomandibular joint synovitis. J. Cell. Mol. Med. 2024, 28, e18172. [Google Scholar] [CrossRef]
- Gao, Y.; Sun, Y.; Yang, H.; Qiu, P.; Cong, Z.; Zou, Y.; Song, L.; Guo, J.; Anastassiades, T.P. A Low Molecular Weight Hyaluronic Acid Derivative Accelerates Excisional Wound Healing by Modulating Pro-Inflammation, Promoting Epithelialization and Neovascularization, and Remodeling Collagen. Int. J. Mol. Sci. 2019, 20, 3722. [Google Scholar] [CrossRef]
- Jiang, D.; Liang, J.; Fan, J.; Yu, S.; Chen, S.; Luo, Y.; Prestwich, G.D.; Mascarenhas, M.M.; Garg, H.G.; Quinn, D.A. Regulation of lung injury and repair by Toll-like receptors and hyaluronan. Nat. Med. 2005, 11, 1173–1179. [Google Scholar] [CrossRef]
- Petrey, A.C.; de la Motte, C.A. Hyaluronan, a crucial regulator of inflammation. Front. Immunol. 2014, 5, 101. [Google Scholar] [CrossRef]
- Kobayashi, T.; Chanmee, T.; Itano, N. Hyaluronan: Metabolism and Function. Biomolecules 2020, 10, 1525. [Google Scholar] [CrossRef]
- Shi, Q.; Zhao, L.; Xu, C.; Zhang, L.; Zhao, H. High Molecular Weight Hyaluronan Suppresses Macrophage M1 Polarization and Enhances IL-10 Production in PM2.5-Induced Lung Inflammation. Molecules 2019, 24, 1766. [Google Scholar] [CrossRef]
- Lee, B.M.; Park, S.J.; Noh, I.; Kim, C.-H. The effects of the molecular weights of hyaluronic acid on the immune responses. Biomater. Res. 2021, 25, 27. [Google Scholar] [CrossRef]
- Lee, C.-H.; Chiang, C.-F.; Kuo, F.-C.; Su, S.-C.; Huang, C.-L.; Liu, J.-S.; Lu, C.-H.; Hsieh, C.-H.; Wang, C.-C.; Lee, C.-H.; et al. High-Molecular-Weight Hyaluronic Acid Inhibits IL-1β-Induced Synovial Inflammation and Macrophage Polarization through the GRP78-NF-κB Signaling Pathway. Int. J. Mol. Sci. 2021, 22, 11917. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.X.; Jin, H.; Chung, Y.S.; Shin, J.Y.; Woo, M.A.; Lee, K.H.; Palmos, G.N.; Choi, B.D.; Cho, M.H. Chondroitin sulfate extracted from the Styela clava tunic suppresses TNF-alpha-induced expression of inflammatory factors, VCAM-1 and iNOS by blocking Akt/NF-kappaB signal in JB6 cells. Cancer Lett. 2008, 264, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Jomphe, C.; Gabriac, M.; Hale, T.M.; Héroux, L.; Trudeau, L.E.; Deblois, D.; Montell, E.; Vergés, J.; du Souich, P. Chondroitin sulfate inhibits the nuclear translocation of nuclear factor-kappaB in interleukin-1beta-stimulated chondrocytes. Basic. Clin. Pharmacol. Toxicol. 2008, 102, 59–65. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, F.; Niu, H.; Wang, Q.; Duan, J. Mechanistic insights into cellular immunity of chondroitin sulfate A and its zwitterionic N-deacetylated derivatives. Carbohydr. Polym. 2015, 123, 331–338. [Google Scholar] [CrossRef]
- Campo, G.M.; Avenoso, A.; Campo, S.; Traina, P.; D’Ascola, A.; Calatroni, A. Glycosaminoglycans reduced inflammatory response by modulating toll-like receptor-4 in LPS-stimulated chondrocytes. Arch. Biochem. Biophys. 2009, 491, 7–15. [Google Scholar] [CrossRef]
- Kastana, P.; Choleva, E.; Poimenidi, E.; Karamanos, N.; Sugahara, K.; Papadimitriou, E. Insight into the role of chondroitin sulfate E in angiogenesis. FEBS J. 2019, 286, 2921–2936. [Google Scholar] [CrossRef]
- Taraballi, F.; Corradetti, B.; Minardi, S.; Powel, S.; Cabrera, F.; Van Eps, J.L.; Weiner, B.K.; Tasciotti, E. Biomimetic collagenous scaffold to tune inflammation by targeting macrophages. J. Tissue Eng. 2016, 7, 2041731415624667. [Google Scholar] [CrossRef]
- Tan, G.K.; Tabata, Y. Chondroitin-6-sulfate attenuates inflammatory responses in murine macrophages via suppression of NF-κB nuclear translocation. Acta Biomater. 2014, 10, 2684–2692. [Google Scholar] [CrossRef]
- Pudełko, A.; Wisowski, G.; Olczyk, K.; Koźma, E.M. The dual role of the glycosaminoglycan chondroitin-6-sulfate in the development, progression and metastasis of cancer. FEBS J 2019, 286, 1815–1837. [Google Scholar] [CrossRef]
- Coombe, D.R. Biological implications of glycosaminoglycan interactions with haemopoietic cytokines. Immunol. Cell Biol. 2008, 86, 598–607. [Google Scholar] [CrossRef]
- Hasan, M.; Najjam, S.; Gordon, M.Y.; Gibbs, R.V.; Rider, C.C. IL-12 is a heparin-binding cytokine. J. Immunol. 1999, 162, 1064–1070. [Google Scholar] [CrossRef] [PubMed]
- Esko, J.D.; Lindahl, U. Molecular diversity of heparan sulfate. J. Clin. Investig. 2001, 108, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Nelson, S.M.; Greer, I.A. The potential role of heparin in assisted conception. Hum. Reprod. Update 2008, 14, 623–645. [Google Scholar] [CrossRef]
- Page, C. Heparin and related drugs: Beyond anticoagulant activity. ISRN Pharmacol. 2013, 2013, 910743. [Google Scholar] [CrossRef]
- Vivès, R.R.; Sadir, R.; Imberty, A.; Rencurosi, A.; Lortat-Jacob, H. A kinetics and modeling study of RANTES(9-68) binding to heparin reveals a mechanism of cooperative oligomerization. Biochemistry 2002, 41, 14779–14789. [Google Scholar] [CrossRef]
- Li, J.P.; Vlodavsky, I. Heparin, heparan sulfate and heparanase in inflammatory reactions. Thromb. Haemost. 2009, 102, 823–828. [Google Scholar] [CrossRef]
- Stringer, S.E.; Nelson, M.S.; Gupta, P. Identification of an MIP-1alpha -binding heparan sulfate oligosaccharide that supports long-term in vitro maintenance of human LTC-ICs. Blood 2003, 101, 2243–2245. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Wu, X.; Sun, J.; Zhou, Z.; Kang, M.; Hu, Y.; Teng, L. N-desulfated and reacetylated modification of heparin modulates macrophage polarization. Int. J. Biol. Macromol. 2023, 229, 354–362. [Google Scholar] [CrossRef]
- Gordts, P.; Foley, E.M.; Lawrence, R.; Sinha, R.; Lameda-Diaz, C.; Deng, L.; Nock, R.; Glass, C.K.; Erbilgin, A.; Lusis, A.J.; et al. Reducing macrophage proteoglycan sulfation increases atherosclerosis and obesity through enhanced type I interferon signaling. Cell Metab. 2014, 20, 813–826. [Google Scholar] [CrossRef]
- Funderburgh, J.L. MINI REVIEW Keratan sulfate: Structure, biosynthesis, and function. Glycobiology 2000, 10, 951–958. [Google Scholar] [CrossRef]
- Funderburgh, J.L. Keratan sulfate biosynthesis. IUBMB Life 2002, 54, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Caterson, B.; Melrose, J. Keratan sulfate, a complex glycosaminoglycan with unique functional capability. Glycobiology 2018, 28, 182–206. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Kadomatsu, K.; Kojima, T.; Ishiguro, N. Keratan sulfate and related murine glycosylation can suppress murine cartilage damage in vitro and in vivo. Biochem. Biophys. Res. Commun. 2011, 409, 732–737. [Google Scholar] [CrossRef]
- Xu, H.; Kurihara, H.; Ito, T.; Kikuchi, H.; Yoshida, K.; Yamanokuchi, H.; Asari, A. The keratan sulfate disaccharide Gal(6S03) beta1,4-GlcNAc(6S03) modulates interleukin 12 production by macrophages in murine Thy-1 type autoimmune disease. J. Biol. Chem. 2005, 280, 20879–20886. [Google Scholar] [CrossRef]
- Sharma, R.; Kuche, K.; Thakor, P.; Bhavana, V.; Srivastava, S.; Mehra, N.K.; Jain, S. Chondroitin Sulfate: Emerging biomaterial for biopharmaceutical purpose and tissue engineering. Carbohydr. Polym. 2022, 286, 119305. [Google Scholar] [CrossRef]
- Kwon, H.; Han, Y. Chondroitin sulfate-based biomaterials for tissue engineering. Turk. J. Biol. 2016, 40, 290–299. [Google Scholar] [CrossRef]
- Li, C.; Wang, K.; Zhou, X.; Li, T.; Xu, Y.; Qiang, L.; Peng, M.; Xu, Y.; Xie, L.; He, C.; et al. Controllable fabrication of hydroxybutyl chitosan/oxidized chondroitin sulfate hydrogels by 3D bioprinting technique for cartilage tissue engineering. Biomed. Mater. 2019, 14, 025006. [Google Scholar] [CrossRef]
- Ashikari-Hada, S.; Habuchi, H.; Kariya, Y.; Kimata, K. Heparin Regulates Vascular Endothelial Growth Factor165-dependent Mitogenic Activity, Tube Formation, and Its Receptor Phosphorylation of Human Endothelial Cells: Comparison of the Effects of Heparin and Modified Heparins*. J. Biol. Chem. 2005, 280, 31508–31515. [Google Scholar] [CrossRef]
- Rabenstein, D.L. Heparin and heparan sulfate: Structure and function. Nat. Prod. Rep. 2002, 19, 312–331. [Google Scholar] [CrossRef]
- Liang, Y.; Kiick, K.L. Heparin-functionalized polymeric biomaterials in tissue engineering and drug delivery applications. Acta Biomater. 2014, 10, 1588–1600. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Li, Q.; Wu, D.; Niu, Y.; Yang, C.; Dong, L.; Wang, C. A macrophage-activating, injectable hydrogel to sequester endogenous growth factors for in situ angiogenesis. Biomaterials 2017, 134, 128–142. [Google Scholar] [CrossRef]
- Belvedere, R.; Novizio, N.; Palazzo, M.; Pessolano, E.; Petrella, A. The pro-healing effects of heparan sulfate and growth factors are enhanced by the heparinase enzyme: New association for skin wound healing treatment. Eur. J. Pharmacol. 2023, 960, 176138. [Google Scholar] [CrossRef]
- Devernois, E.; Coradin, T. Synthesis, Characterization and Biological Properties of Type I Collagen–Chitosan Mixed Hydrogels: A Review. Gels 2023, 9, 518. [Google Scholar] [CrossRef]
- Negm, N.A.; Hefni, H.H.H.; Abd-Elaal, A.A.A.; Badr, E.A.; Abou Kana, M.T.H. Advancement on modification of chitosan biopolymer and its potential applications. Int. J. Biol. Macromol. 2020, 152, 681–702. [Google Scholar] [CrossRef]
- von Boxberg, Y.; Soares, S.; Giraudon, C.; David, L.; Viallon, M.; Montembault, A.; Nothias, F. Macrophage polarization in vitro and in vivo modified by contact with fragmented chitosan hydrogel. J. Biomed. Mater. Res. Part A 2022, 110, 773–787. [Google Scholar] [CrossRef]
- Oliveira, M.I.; Santos, S.G.; Oliveira, M.J.; Torres, A.L.; Barbosa, M.A. Chitosan drives anti-inflammatory macrophage polarisation and pro-inflammatory dendritic cell stimulation. Eur. Cell Mater. 2012, 24, 133–136. [Google Scholar] [CrossRef]
- Shen, T.; Dai, K.; Yu, Y.; Wang, J.; Liu, C. Sulfated chitosan rescues dysfunctional macrophages and accelerates wound healing in diabetic mice. Acta Biomater. 2020, 117, 192–203. [Google Scholar] [CrossRef]
- You, C.; Zhu, Z.; Wang, S.; Wang, X.; Han, C.; Shao, H. Nanosilver alleviates foreign body reaction and facilitates wound repair by regulating macrophage polarization. J. Zhejiang Univ. Sci. B 2023, 24, 510–523. [Google Scholar] [CrossRef]
- You, C.; Li, Q.; Wang, X.; Wu, P.; Ho, J.K.; Jin, R.; Zhang, L.; Shao, H.; Han, C. Silver nanoparticle loaded collagen/chitosan scaffolds promote wound healing via regulating fibroblast migration and macrophage activation. Sci. Rep. 2017, 7, 10489. [Google Scholar] [CrossRef]
- You, C.; Han, C.; Wang, X.; Zheng, Y.; Li, Q.; Hu, X.; Sun, H. The progress of silver nanoparticles in the antibacterial mechanism, clinical application and cytotoxicity. Mol. Biol. Rep. 2012, 39, 9193–9201. [Google Scholar] [CrossRef]
- Bhol, K.; Schechter, P. Topical nanocrystalline silver cream suppresses inflammatory cytokines and induces apoptosis of inflammatory cells in a murine model of allergic contact dermatitis. Br. J. Dermatol. 2005, 152, 1235–1242. [Google Scholar] [CrossRef]
- Gil, E.S.; Panilaitis, B.; Bellas, E.; Kaplan, D.L. Functionalized silk biomaterials for wound healing. Adv. Healthc. Mater. 2013, 2, 206–217. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Zhang, J.; You, R.; Qu, J.; Li, M. Functionalized silk fibroin dressing with topical bioactive insulin release for accelerated chronic wound healing. Mater. Sci. Eng. C 2017, 72, 394–404. [Google Scholar] [CrossRef]
- Wang, Y.; Yao, D.; Li, L.; Qian, Z.; He, W.; Ding, R.; Liu, H.; Fan, Y. Effect of electrospun silk fibroin–silk sericin films on macrophage polarization and vascularization. ACS Biomater. Sci. Eng. 2020, 6, 3502–3512. [Google Scholar] [CrossRef]
- Roy, S.; Sharma, A.; Ghosh, S. Macrophage polarization profiling on native and regenerated silk biomaterials. ACS Biomater. Sci. Eng. 2022, 8, 659–671. [Google Scholar] [CrossRef]
- Reeves, A.R.; Spiller, K.L.; Freytes, D.O.; Vunjak-Novakovic, G.; Kaplan, D.L. Controlled release of cytokines using silk-biomaterials for macrophage polarization. Biomaterials 2015, 73, 272–283. [Google Scholar] [CrossRef]
- Reddy, M.S.B.; Ponnamma, D.; Choudhary, R.; Sadasivuni, K.K. A Comparative Review of Natural and Synthetic Biopolymer Composite Scaffolds. Polymers 2021, 13, 1105. [Google Scholar] [CrossRef]
- Abbasian, M.; Massoumi, B.; Mohammad-Rezaei, R.; Samadian, H.; Jaymand, M. Scaffolding polymeric biomaterials: Are naturally occurring biological macromolecules more appropriate for tissue engineering? Int. J. Biol. Macromol. 2019, 134, 673–694. [Google Scholar] [CrossRef]
- Kyriakides, T.R.; Kim, H.J.; Zheng, C.; Harkins, L.; Tao, W.; Deschenes, E. Foreign body response to synthetic polymer biomaterials and the role of adaptive immunity. Biomed. Mater. 2022, 17, 022007. [Google Scholar] [CrossRef]
- Mir, M.; Ali, M.N.; Barakullah, A.; Gulzar, A.; Arshad, M.; Fatima, S.; Asad, M. Synthetic polymeric biomaterials for wound healing: A review. Progress. Biomater. 2018, 7, 1–21. [Google Scholar] [CrossRef]
- Liu, X.; Chen, M.; Luo, J.; Zhao, H.; Zhou, X.; Gu, Q.; Yang, H.; Zhu, X.; Cui, W.; Shi, Q. Immunopolarization-regulated 3D printed-electrospun fibrous scaffolds for bone regeneration. Biomaterials 2021, 276, 121037. [Google Scholar] [CrossRef]
- Nakkala, J.R.; Duan, Y.; Ding, J.; Muhammad, W.; Zhang, D.; Mao, Z.; Ouyang, H.; Gao, C. Macrophage membrane-functionalized nanofibrous mats and their immunomodulatory effects on macrophage polarization. Acta Biomater. 2022, 141, 24–38. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, J.; Lin, J.; Liang, R.; He, M.; Wang, Y.; Tan, H. Porous Three-Dimensional Polyurethane Scaffolds Promote Scar-Free Endogenous Regeneration After Acute Brain Hemorrhage. Transl. Stroke Res. 2023; ahead of print. [Google Scholar] [CrossRef]
- Wolf, M.T.; Dearth, C.L.; Ranallo, C.A.; LoPresti, S.T.; Carey, L.E.; Daly, K.A.; Brown, B.N.; Badylak, S.F. Macrophage polarization in response to ECM coated polypropylene mesh. Biomaterials 2014, 35, 6838–6849. [Google Scholar] [CrossRef]
- Li, Y.-M.; Wu, J.-Y.; Jiang, J.; Dong, S.-K.; Chen, Y.-S.; He, H.-Y.; Liu, C.-S.; Zhao, J.-Z. Chondroitin sulfate-polydopamine modified polyethylene terephthalate with extracellular matrix-mimetic immunoregulatory functions for osseointegration. J. Mater. Chem. B 2019, 7, 7756–7770. [Google Scholar] [CrossRef]
- Hook, A.L.; Anderson, D.G.; Langer, R.; Williams, P.; Davies, M.C.; Alexander, M.R. High throughput methods applied in biomaterial development and discovery. Biomaterials 2010, 31, 187–198. [Google Scholar] [CrossRef]
- Crawford, L.A.; Cuzzucoli Crucitti, V.; Stimpson, A.; Morgan, C.; Blake, J.; Wildman, R.D.; Hook, A.L.; Alexander, M.R.; Irvine, D.J.; Avery, S.V. A potential alternative to fungicides using actives-free (meth)acrylate polymers for protection of wheat crops from fungal attachment and infection. Green. Chem. 2023, 25, 8558–8569. [Google Scholar] [CrossRef]
- Wong, S.Y.; Hook, A.L.; Gardner, W.; Chang, C.-Y.; Mei, Y.; Davies, M.C.; Williams, P.; Alexander, M.R.; Ballabio, D.; Muir, B.W.; et al. Exploring the Relationship between Polymer Surface Chemistry and Bacterial Attachment Using ToF-SIMS and Self-Organizing maps. Adv. Mater. Interfaces 2023, 10, 2202334. [Google Scholar] [CrossRef]
- Vallieres, C.; Hook, A.L.; He, Y.; Crucitti, V.C.; Figueredo, G.; Davies, C.R.; Burroughs, L.; Winkler, D.A.; Wildman, R.D.; Irvine, D.J.; et al. Discovery of (meth)acrylate polymers that resist colonization by fungi associated with pathogenesis and biodeterioration. Sci. Adv. 2020, 6, eaba6574. [Google Scholar] [CrossRef]
- Rostam, H.M.; Fisher, L.E.; Hook, A.L.; Burroughs, L.; Luckett, J.C.; Figueredo, G.P.; Mbadugha, C.; Teo, A.C.; Latif, A.; Kämmerling, L. Immune-instructive polymers control macrophage phenotype and modulate the foreign body response in vivo. Matter 2020, 2, 1564–1581. [Google Scholar] [CrossRef]
- Latif, A.; Fisher, L.E.; Dundas, A.A.; Cuzzucoli Crucitti, V.; Imir, Z.; Lawler, K.; Pappalardo, F.; Muir, B.W.; Wildman, R.; Irvine, D.J.; et al. Microparticles Decorated with Cell-Instructive Surface Chemistries Actively Promote Wound Healing. Adv. Mater. 2022, e2208364. [Google Scholar] [CrossRef]
- Luu, T.U.; Gott, S.C.; Woo, B.W.; Rao, M.P.; Liu, W.F. Micro-and nanopatterned topographical cues for regulating macrophage cell shape and phenotype. ACS Appl. Mater. Interfaces 2015, 7, 28665–28672. [Google Scholar] [CrossRef]
- Vassey, M.J.; Figueredo, G.P.; Scurr, D.J.; Vasilevich, A.S.; Vermeulen, S.; Carlier, A.; Luckett, J.; Beijer, N.R.; Williams, P.; Winkler, D.A. Immune modulation by design: Using topography to control human monocyte attachment and macrophage differentiation. Adv. Sci. 2020, 7, 1903392. [Google Scholar] [CrossRef]
- Monteiro, N.; Casanova, M.; Quinteira, R.; Fangueiro, J.; Reis, R.; Neves, N. Biomimetic surface topography as a potential modulator of macrophages inflammatory response to biomaterials. Biomater. Adv. 2022, 141, 213128. [Google Scholar] [CrossRef]
- Singh, S.; Awuah, D.; Rostam, H.M.; Emes, R.D.; Kandola, N.K.; Onion, D.; Htwe, S.S.; Rajchagool, B.; Cha, B.-H.; Kim, D. Unbiased analysis of the impact of micropatterned biomaterials on macrophage behavior provides insights beyond predefined polarization states. ACS Biomater. Sci. Eng. 2017, 3, 969–978. [Google Scholar] [CrossRef]
- Jia, Y.; Yang, W.; Zhang, K.; Qiu, S.; Xu, J.; Wang, C.; Chai, Y. Nanofiber arrangement regulates peripheral nerve regeneration through differential modulation of macrophage phenotypes. Acta Biomater. 2019, 83, 291–301. [Google Scholar] [CrossRef]
- Han, D.; Gouma, P.-I. Electrospun bioscaffolds that mimic the topology of extracellular matrix. Nanomed. Nanotechnol. Biol. Med. 2006, 2, 37–41. [Google Scholar] [CrossRef]
- Hotchkiss, K.M.; Reddy, G.B.; Hyzy, S.L.; Schwartz, Z.; Boyan, B.D.; Olivares-Navarrete, R. Titanium surface characteristics, including topography and wettability, alter macrophage activation. Acta Biomater. 2016, 31, 425–434. [Google Scholar] [CrossRef]
- Duan, Y.; Zheng, H.; Li, Z.; Yao, Y.; Ding, J.; Wang, X.; Nakkala, J.R.; Zhang, D.; Wang, Z.; Zuo, X. Unsaturated polyurethane films grafted with enantiomeric polylysine promotes macrophage polarization to a M2 phenotype through PI3K/Akt1/mTOR axis. Biomaterials 2020, 246, 120012. [Google Scholar] [CrossRef]
- Huang, Y.-J.; Hung, K.-C.; Hung, H.-S.; Hsu, S.-h. Modulation of macrophage phenotype by biodegradable polyurethane nanoparticles: Possible relation between macrophage polarization and immune response of nanoparticles. ACS Appl. Mater. Interfaces 2018, 10, 19436–19448. [Google Scholar] [CrossRef]
- Bartneck, M.; Keul, H.A.; Singh, S.; Czaja, K.; Bornemann, J.; Bockstaller, M.; Moeller, M.; Zwadlo-Klarwasser, G.; Groll, J. Rapid uptake of gold nanorods by primary human blood phagocytes and immunomodulatory effects of surface chemistry. ACS Nano 2010, 4, 3073–3086. [Google Scholar] [CrossRef]
- Jones, J.A.; Chang, D.T.; Meyerson, H.; Colton, E.; Kwon, I.K.; Matsuda, T.; Anderson, J.M. Proteomic analysis and quantification of cytokines and chemokines from biomaterial surface-adherent macrophages and foreign body giant cells. J. Biomed. Mater. Res. Part A 2007, 83, 585–596. [Google Scholar] [CrossRef]
- Scott, R.A.; Kiick, K.L.; Akins, R.E. Substrate stiffness directs the phenotype and polarization state of cord blood derived macrophages. Acta Biomater. 2021, 122, 220–235. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, Y.; Zhou, P.; Liu, X.; Zhao, H.; Zhou, X.; Gu, Q.; Li, B.; Zhu, X.; Shi, Q. Substrate stiffness modulates bone marrow-derived macrophage polarization through NF-κB signaling pathway. Bioact. Mater. 2020, 5, 880–890. [Google Scholar] [CrossRef]
- Camarero-Espinosa, S.; Carlos-Oliveira, M.; Liu, H.; Mano, J.F.; Bouvy, N.; Moroni, L. 3D printed dual-porosity scaffolds: The combined effect of stiffness and porosity in the modulation of macrophage polarization. Adv. Healthc. Mater. 2022, 11, 2101415. [Google Scholar] [CrossRef]
- Roach, P.; Eglin, D.; Rohde, K.; Perry, C.C. Modern biomaterials: A review—Bulk properties and implications of surface modifications. J. Mater. Sci. Mater. Med. 2007, 18, 1263–1277. [Google Scholar] [CrossRef]


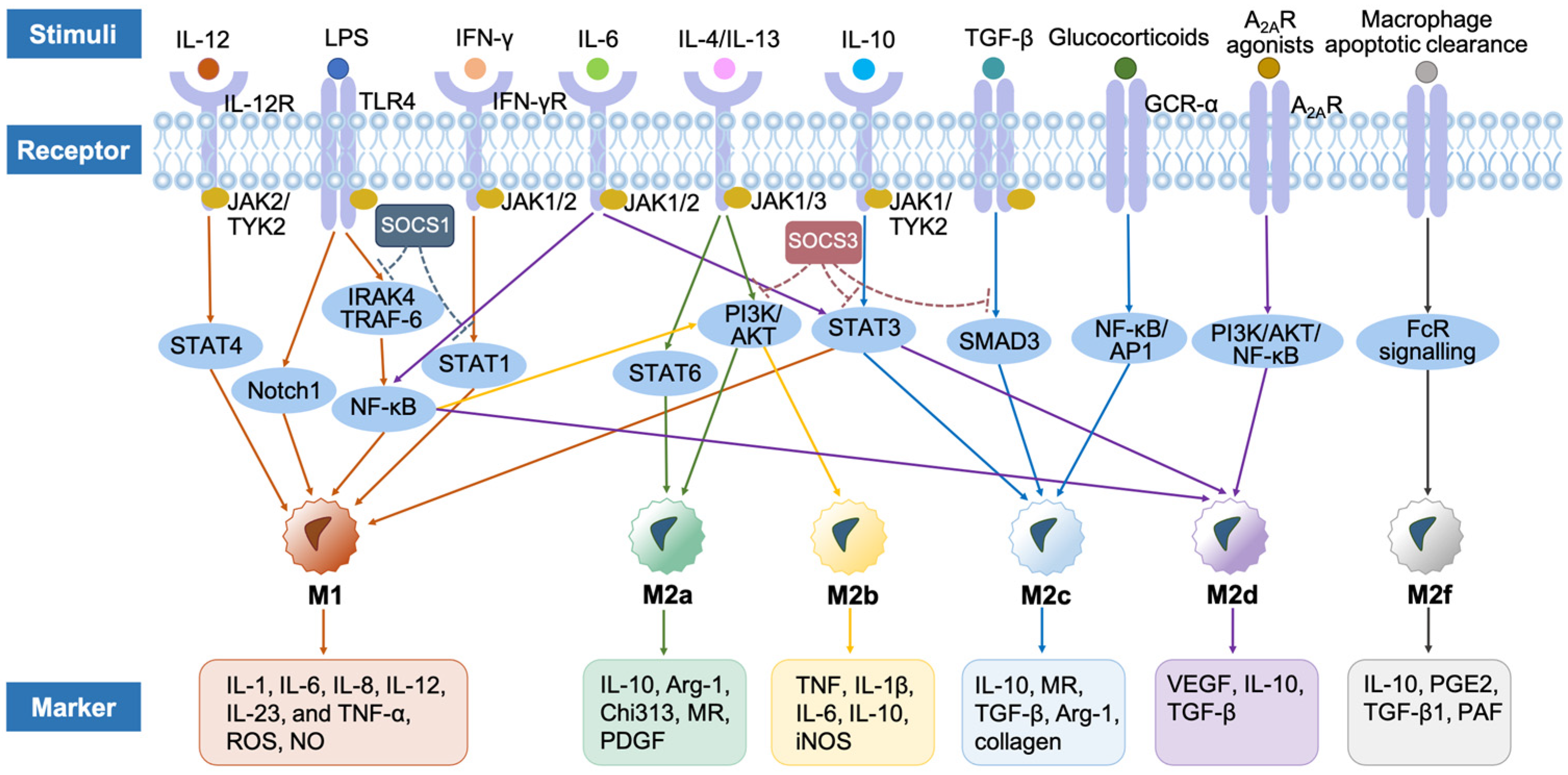
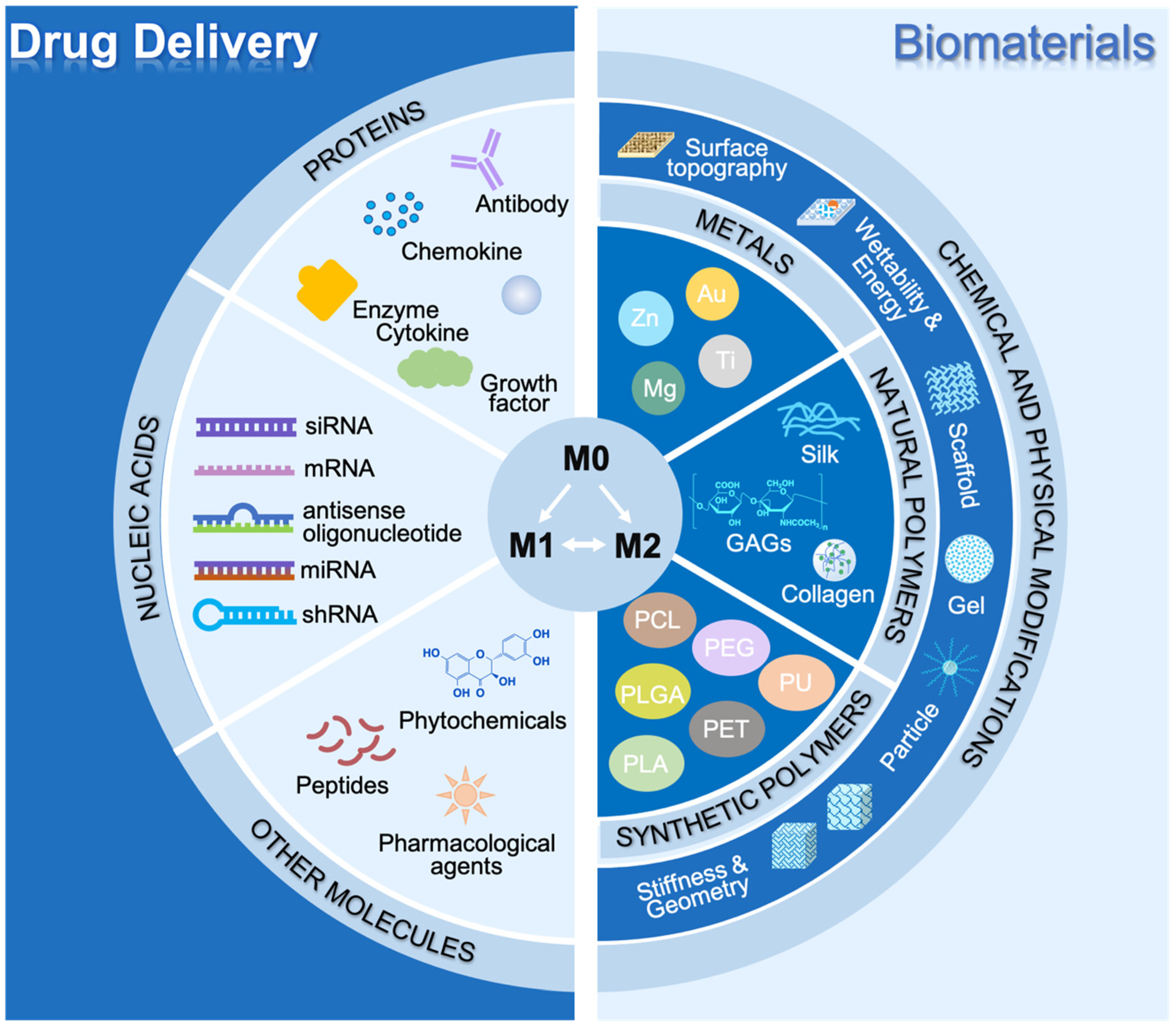
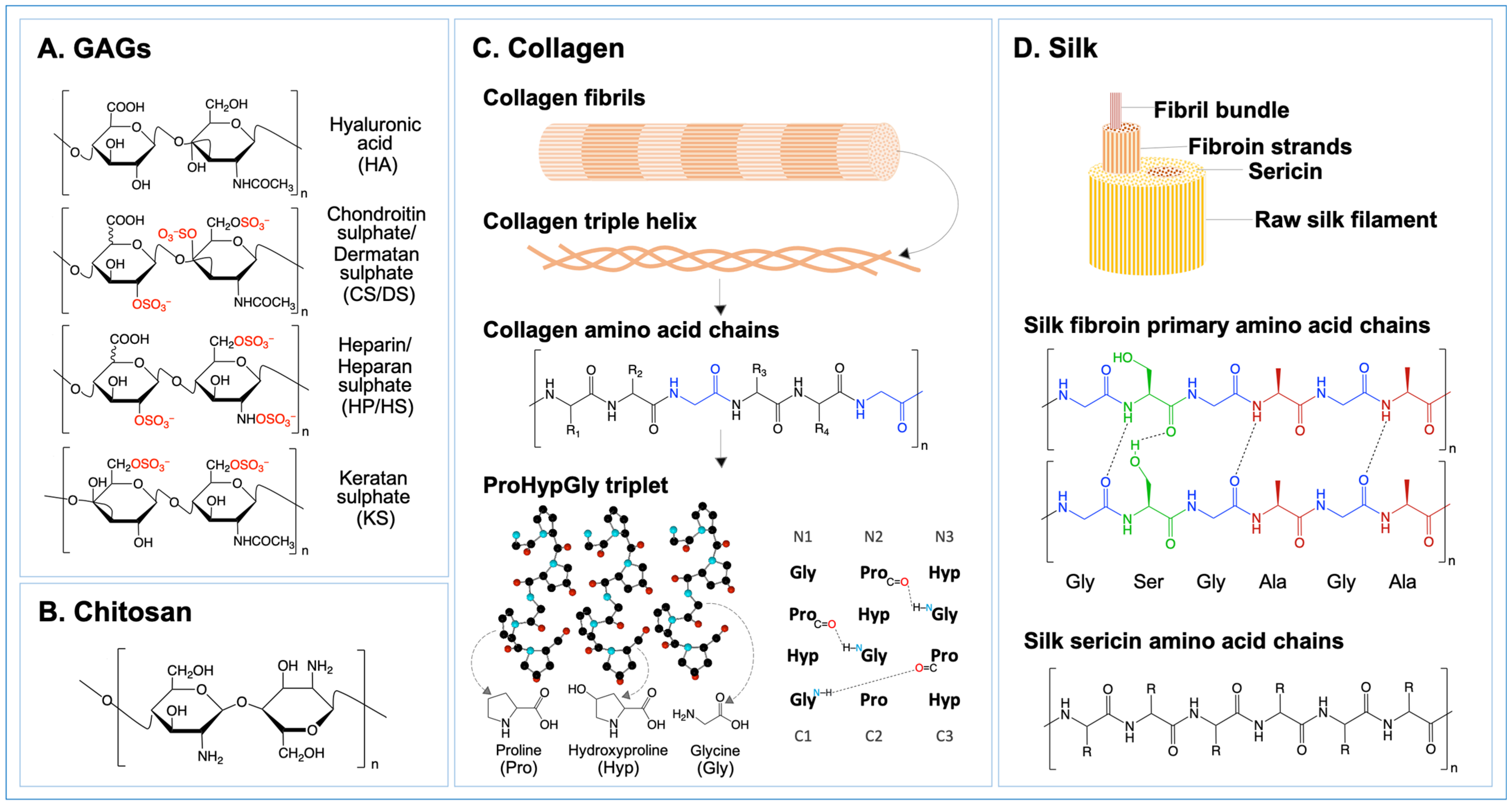

| GAGs | Functions in Inflammation | Macrophage Polarization | Signaling Factors | Markers | |
|---|---|---|---|---|---|
| HA | LMW-HA | LMW-HA induces the expression of inflammatory cytokines and participates in inflammation, immune stimulation, cell migration, and induction of angiogenesis [300,301]. | ↑M1 | TLR2, TLR4, NF-κB and MyD88 | ↑INOS, IL-2b, IL-6, TNF-α, IL-1β and CD80 [302,303,304,305] |
| HMW-HA | HMW-HA has anti-angiogenesis, anti-inflammatory, and immunosuppressive effects [306]. In malignant cells, HMW-HA is involved maintaining the homeostasis of tumors and supporting their migration [307]. | ↑M2 ↓M1 | JNK and p38 pathways | ↑Arg-1, MRC1, TGF-β1, IL-10, IL-11, CD68 and CD163 ↓IL-6, PGE2, TNF-α, CCL2 and IL-1β [302,308,309,310] | |
| CS/DS | CS mainly regulates inflammatory responses by inhibiting the release of LPS-induced pro-inflammatory factors and related enzymes [311,312]. According to the different sulphation patterns of CS, CS shows different activities in inflammation: CSA and CSE have both pro-inflammatory and anti-inflammatory activities, while CSC and CSD have anti-inflammatory activities [297,313,314,315]. | ↑M2 ↓M1 | TLR, LPS, CD44 and NF-kB | ↑TGF-β, IL-10, Arg-1 and MRC1 ↓IL-6 and TNF-α [316,317,318] | |
| HP/HS | HP/HS coordinates different levels of inflammation by interacting with numerous molecules expressed on the cell surface, such as selectins and integrins, activating cytokines and chemokines, such as the interleukin family, and inhibiting the pro-inflammatory enzymes and cytotoxic mediators [319,320,321,322,323,324,325,326]. | ↑M1 | STAT1 | ↑ IL-6, TNF-α, IFN, NO [327,328] | |
| KS | KS regulates inflammatory responses and can be used as a new type of therapeutic compound for the treatment of inflammation damage [329,330,331,332]. | ↓M1 | -- | ↓IL-12 [333] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, Q.; Dickinson, A.; Nakuleswaran, P.; Maghami, S.; Alagoda, S.; Hook, A.L.; Ghaemmaghami, A.M. Targeting Macrophage Polarization for Reinstating Homeostasis following Tissue Damage. Int. J. Mol. Sci. 2024, 25, 7278. https://doi.org/10.3390/ijms25137278
Du Q, Dickinson A, Nakuleswaran P, Maghami S, Alagoda S, Hook AL, Ghaemmaghami AM. Targeting Macrophage Polarization for Reinstating Homeostasis following Tissue Damage. International Journal of Molecular Sciences. 2024; 25(13):7278. https://doi.org/10.3390/ijms25137278
Chicago/Turabian StyleDu, Qiran, Anna Dickinson, Pruthvi Nakuleswaran, Susan Maghami, Savindu Alagoda, Andrew L. Hook, and Amir M. Ghaemmaghami. 2024. "Targeting Macrophage Polarization for Reinstating Homeostasis following Tissue Damage" International Journal of Molecular Sciences 25, no. 13: 7278. https://doi.org/10.3390/ijms25137278






