Gene Expression and Phenotypic Assessment of Egg Quality across Developmental Stages of Atlantic Cod throughout the Spawning Season
Abstract
1. Introduction
2. Results
2.1. Egg and Larvae Characteristics
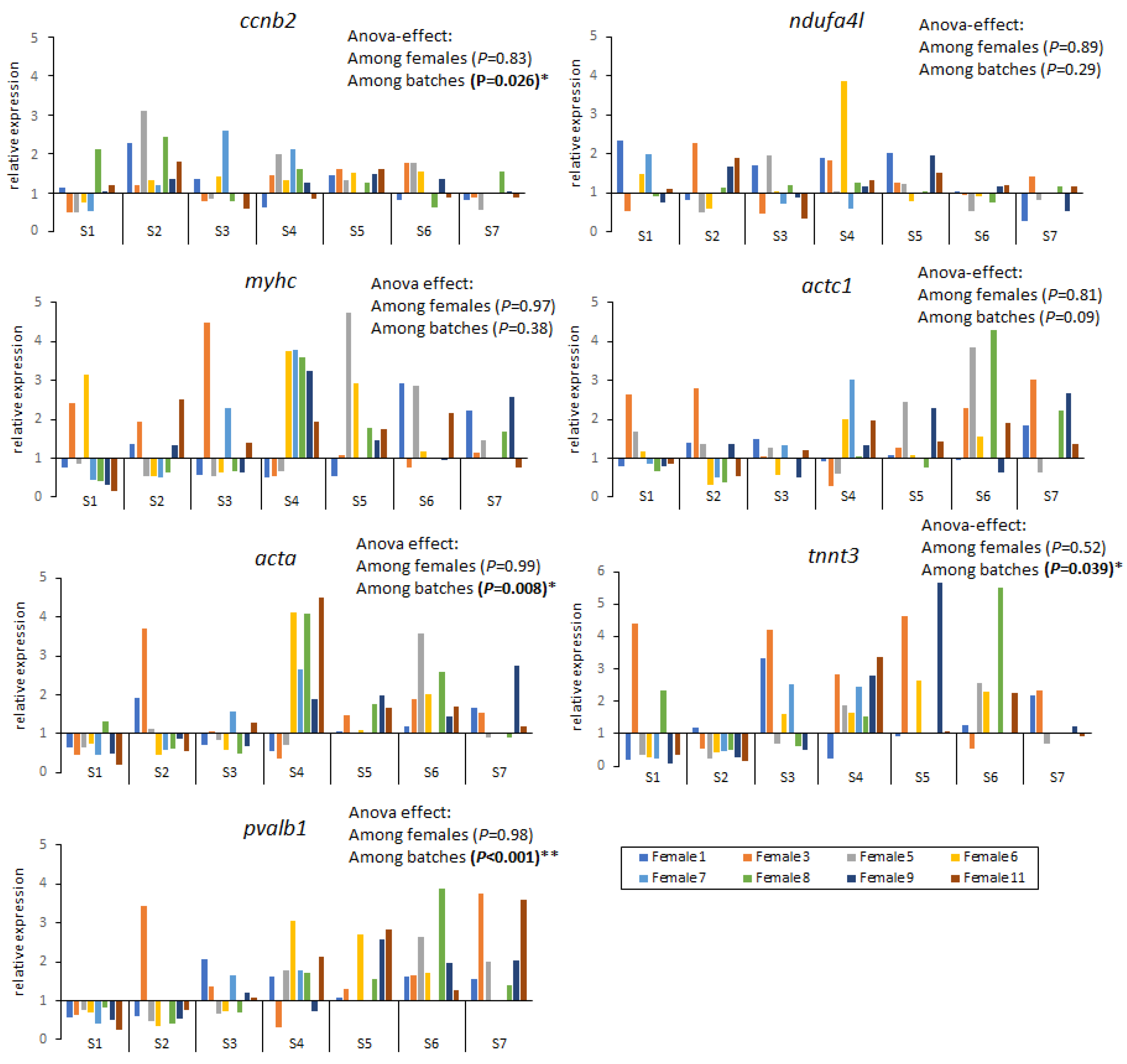
2.2. Gene Expression
2.3. Molecular and Phenotypic Data Correlation
3. Discussion
3.1. Quality Estimations Based on Morphology
3.2. Variation of Gene Expression throughout Developmental Stages
3.3. Variation of Gene Expression throughout the Spawning Season
3.4. Embryo Survival in Relation to Gene Expression
4. Materials and Methods
4.1. Broodstock Maintenance, Egg Incubation and Larval Rearing
4.1.1. Egg collection
4.1.2. Female Selection
4.1.3. Larval Rearing
4.2. Gene Expression Analysis
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petersen, P.E.; Penman, D.J.; Dahle, G.; Patursson, Ø.; Taggart, J.B. Differential Survival among Batches of Atlantic Cod (Gadus morhua L.) from Fertilisation through to post-metamorphosis. PLoS ONE 2016, 11, e0158091. [Google Scholar] [CrossRef]
- Puvanendran, V.; Mortensen, A.; Johansen, L.H.; Kettunen, A.; Hansen, Ø.J.; Henriksen, E.; Heide, M. Development of cod farming in Norway: Past and current biological and market status and future prospects and directions. Rev. Aquac. 2022, 14, 308–342. [Google Scholar] [CrossRef]
- Hansen, Ø.J.; Puvanendran, V.; Jøstensen, J.P.; Falk-Petersen, I.B. Early introduction of an inert diet and unenriched Artemia enhances growth and quality of Atlantic cod (Gadus morhua) larvae. Aquac. Nutr. 2018, 24, 102–111. [Google Scholar] [CrossRef]
- Puvanendran, V.; Swain, T.; Tveiten, H.; Hansen, Ø.J.; Mortensen, A. Optimizing intensive culture protocols for Atlantic cod (Gadus morhua) larvae. Aquac. Int. 2023, 31, 3457–3472. [Google Scholar] [CrossRef]
- Kjesbu, O.S. The spawning activity of cod. J. Fish Biol. 1989, 34, 195–206. [Google Scholar] [CrossRef]
- Salze, G.; Tocher, D.R.; Roy, W.J.; Robertson, D.A. Egg quality determinants in cod (Gadus morhua L.): Egg performance and lipids in eggs from farmed and wild broodstock. Aquac. Res. 2005, 36, 1488–1499. [Google Scholar] [CrossRef]
- Lahnsteiner, F.; Patarnello, P. Egg quality determination in the gilthead seabream, Sparus aurata, with biochemical parameters. Aquaculture 2004, 237, 443–459. [Google Scholar] [CrossRef]
- Kjesbu, O.S.; Kryvi, H.; Sundby, S.; Solemdal, P. Buoyancy variations in eggs of Atlantic cod (Gadus morhua L.) in relation to chorion thickness and egg size: Theory and observations. J. Fish Biol. 1992, 41, 581–599. [Google Scholar] [CrossRef]
- Marteinsdóttir, G.; Steinarsson, A. Maternal influence on the size and viability of Iceland cod Gadus morhua eggs and larvae. J. Fish Biol. 1998, 52, 1241–1258. [Google Scholar] [CrossRef]
- Bonnet, E.; Fostier, A.; Bobe, J. Microarray-based analysis of fish egg quality after natural or controlled ovulation. BMC Genom. 2007, 8, 55. [Google Scholar] [CrossRef]
- Coban, D.; Kamacı, H.O.; Süzer, C.; Yıldırım, Ş.; Arda, G.; Korkut, A.Y.; Saka, S.; Fırat, K. Effect of Some Morphometric Characteristics on Egg Quality in Common Dentex, Dentex dentex (Linnaeus, 1758). Turk. J. Fish. Aquat. Sci. 2011, 11, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Bromage, N.; Jones, J.; Randall, C.; Thrush, M.; Davies, B.; Springate, J.; Duston, J.; Barker, G. Broodstock management, fecundity, egg quality and the timing of egg production in the rainbow trout (Oncorhynchus mykiss). Aquaculture 1992, 100, 141–166. [Google Scholar] [CrossRef]
- Penney, R.W.; Lush, P.L.; Wade, J.; Brown, J.A.; Parrish, C.C.; Burton, M.P. Comparative utility of egg blastomere morphology and lipid biochemistry for prediction of hatching success in Atlantic cod, Gadus morhua L. Aquac. Res. 2006, 37, 272–283. [Google Scholar] [CrossRef]
- Avery, T.S.; Killen, S.S.; Hollinger, T.R. The relationship of embryonic development, mortality, hatching success, and larval quality to normal or abnormal early embryonic cleavage in Atlantic cod, Gadus morhua. Aquaculture 2009, 289, 265–273. [Google Scholar] [CrossRef]
- Brooks, S.; Tyler, C.R.; Sumpter, J.P. Egg quality in fish: What makes a good egg? Rev. Fish Biol. Fish. 1997, 7, 387–416. [Google Scholar] [CrossRef]
- McCormick, M.I. Experimental test of the effect of maternal hormones on larval quality of a coral reef fish. Oecologia 1999, 118, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Lall, S.P.; Nanton, D. Nutrition of Atlantic cod. Bull. Aquac. Assoc. Can. 2002, 102, 23–26. [Google Scholar]
- Dworkin, M.B.; Dworkin-Rastl, E. Functions of maternal mRNA in early development. Mol. Reprod. Dev. 1990, 26, 261–297. [Google Scholar] [CrossRef]
- Lubzens, E.; Bobe, J.; Young, G.; Sullivan, C.V. Maternal investment in fish oocytes and eggs: The molecular cargo and its contributions to fertility and early development. Aquaculture 2017, 472, 107–143. [Google Scholar] [CrossRef]
- Škugor, A.; Krasnov, A.; Andersen, Ø. Genome-wide microarray analysis of Atlantic cod (Gadus morhua) oocyte and embryo. BMC Genom. 2014, 15, 594. [Google Scholar] [CrossRef][Green Version]
- Jukam, D.; Shariati, S.A.M.; Skotheim, J.M. Zygotic genome activation in vertebrates. Dev. Cell 2017, 42, 316–332. [Google Scholar] [CrossRef] [PubMed]
- Skjærven, K.H.; Olsvik, P.A.; Finn, R.N.; Holen, E.; Hamre, K. Ontogenetic expression of maternal and zygotic genes in Atlantic cod embryos under ambient and thermally stressed conditions. Comp. Biochem. Physiol. Part A Mol. Integra. Physiol. 2011, 159, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Kleppe, L.; Edvardsen, R.B.; Kuhl, H.; Malde, K.; Furmanek, T.; Drivenes, Ø.; Reinhardt, R.; Taranger, G.L.; Wargelius, A. Maternal 3’UTRs: From egg to onset of zygotic transcription in Atlantic cod. BMC Genom. 2012, 13, 443. [Google Scholar] [CrossRef] [PubMed]
- Drivenes, Ø.; Taranger, G.L.; Edvardsen, R.B. Gene expression profiling of Atlantic cod (Gadus morhua) embryogenesis using microarray. J. Mar. Biotechnol. 2012, 14, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Lanes, C.F.C.; Bizuayehu, T.T.; de Oliveira Fernandes, J.M.; Kiron, V.; Babiak, I. Transcriptome of Atlantic cod (Gadus morhua L.) early embryos from farmed and wild broodstocks. Mar. Biotechnol. 2013, 15, 677–694. [Google Scholar] [CrossRef]
- Kleppe, L.; Edvardsen, R.B.; Furmanek, T.; Taranger, G.L.; Wargelius, A. Global transcriptome analysis identifies regulated transcripts and pathways activated during oogenesis and early embryogenesis in Atlantic cod. Mol. Reprod. Develop. 2014, 81, 619–635. [Google Scholar] [CrossRef] [PubMed]
- Pickova, J.; Dutta, P.C.; Larsson, P.O.; Kiessling, A. Early embryonic cleavage pattern, hatching success, and egg-lipid fatty acid composition: Comparison between two cod (Gadus morhua) stocks. Can. J. Fish. Aquat. Sci. 1997, 54, 2410–2416. [Google Scholar] [CrossRef]
- Hansen, Ø.J.; Puvanendran, V. Fertilisation success and blastomere morphology as predictors of egg and juvenile quality for domesticated Atlantic cod, Gadus morhua, broodstock. Aquac. Res. 2010, 41, 1791–1798. [Google Scholar] [CrossRef]
- Clark, M.S.; Edwards, Y.J.; Peterson, D.; Clifton, S.W.; Thompson, A.J.; Sasaki, M.; Suzuki, Y.; Kikuchi, K.; Watabe, S.; Kawakami, K.; et al. ESTs: New resources for transcription analysis and genome annotation. Genome Res. 2003, 13, 2747–2753. [Google Scholar] [CrossRef]
- Lo, J.; Lee, S.; Xu, M.; Liu, F.; Ruan, H.; Eun, A.; He, Y.; Ma, W.; Wang, W.; Wen, Z.; et al. 15,000 unique zebrafish EST clusters and their future use in microarray for profiling gene expression patterns during embryogenesis. Genom. Res. 2003, 13, 455–466. [Google Scholar] [CrossRef]
- Adzhubei, A.A.; Vlasova, A.V.; Hagen-Larsen, H.; Ruden, T.A.; Laerdahl, J.K.; Høyheim, B. Annotated expressed sequence tags (ESTs) from pre-smolt Atlantic salmon (Salmo salar) in a searchable data resource. BMC Genom. 2007, 8, 209. [Google Scholar] [CrossRef]
- Caballero-Solares, A.; Xue, X.; Parrish, C.C.; Foroutani, M.B.; Taylor, R.G.; Rise, M.L. Changes in the liver transcriptome of farmed Atlantic salmon (Salmo salar) fed experimental diets based on terrestrial alternatives to fish meal and fish oil. BMC Genom. 2018, 19, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Vallejos-Vidal, E.; Reyes-Cerpa, S.; Rivas-Pardo, J.A.; Maisey, K.; Yáñez, J.M.; Valenzuela, H.; Cea, P.A.; Castro-Fernandez, V.; Tort, L.; Sandino, A.M.; et al. Single-nucleotide polymorphisms (SNP) mining and their effect on the tridimensional protein structure prediction in a set of immunity-related expressed sequence tags (EST) in Atlantic salmon (Salmo salar). Front. Genet. 2020, 10, 1406. [Google Scholar] [CrossRef]
- Douglas, S.E.; Knickle, L.C.; Kimball, J.; Reith, M.E. Comprehensive EST analysis of Atlantic halibut (Hippoglossus hippoglossus), a commercially relevant aquaculture species. BMC Genom. 2007, 8, 144. [Google Scholar] [CrossRef]
- Radonić, I.; Trumbić, Ž.; Šegvić-Bubić, T.; Grubišić, L.; Mladineo, I. Development and potential application of new set of Atlantic bluefin tuna EST-SSRs in the survival success during farming cycle. Mediterr. Mar. Sci. 2020, 21, 298–307. [Google Scholar] [CrossRef]
- Booman, M.; Borza, T.; Feng, C.Y.; Hori, T.S.; Higgins, B.; Culf, A.; Léger, D.; Chute, I.C.; Belkaid, A.; Rise, M.; et al. Development and experimental validation of a 20K Atlantic cod (Gadus morhua) oligonucleotide microarray based on a collection of over 150,000 ESTs. Mar. Biotech. 2011, 13, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Edvardsen, R.B.; Malde, K.; Mittelholzer, C.; Taranger, G.L.; Nilsen, F. EST resources and establishment and validation of a 16 k cDNA microarray from Atlantic cod (Gadus morhua). Comp. Biochem. Physiol. Part D Genom. Proteom. 2011, 6, 23–30. [Google Scholar]
- Johansen, S.D.; Karlsen, B.O.; Furmanek, T.; Andreassen, M.; Jørgensen, T.E.; Bizuayehu, T.T.; Breines, R.; Emblem, Å.; Kettunen, P.; Luukko, K.; et al. RNA deep sequencing of the Atlantic cod transcriptome. Comp. Biochem. Physiol. Part D Genom. Proteom. 2011, 6, 18–22. [Google Scholar] [CrossRef]
- Guslund, N.; Solbakken, M.H.; Jakobsen, K.S.; Qiao, S.W. Single cell transcriptome profiling of the Atlantic cod immune system. bioRxiv 2020. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35 (Suppl. S2), W71–W74. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, S.; Furuichi, M.; Maruyama, K.; Matsuyama, M. Daily spawning and quality of eggs in one female red sea bream Pagrus major. Aquac. Sci. 1988, 36, 33–39. [Google Scholar]
- Fauvel, C.; Omnès, M.H.; Suquet, M.; Normant, Y. Reliable assessment of overripening in turbot (Scophthalmus maximus) by a simple pH measurement. Aquaculture 1993, 117, 107–113. [Google Scholar] [CrossRef]
- Kjørsvik, E.; Lønning, S. Effects of egg quality on normal fertilisation and early development of the cod, Gadus morhua L. J. Fish Biol. 1983, 23, 1–12. [Google Scholar] [CrossRef]
- Bromage, N.; Bruce, M.; Basavaraja, N.; Rana, K.; Shields, R.; Young, C.; Dye, J.; Smith, P.; Gillespie, M.; Gamble, J. Egg quality determinants in finfish the role of overripening with special reference to the timing of stripping in the Atlantic halibut Hippoglossus hippoglossus. J. World Aquac. Soc. 1994, 25, 13–21. [Google Scholar] [CrossRef]
- Pavlov, D.A.; Moksness, E. Production and quality of eggs obtained from wolffish (Anarhichas lupus L.) reared in captivity. Aquaculture 1994, 122, 295–312. [Google Scholar] [CrossRef]
- Lahnsteiner, F.; Soares, F.; Ribeiro, L.; Dinis, M.T. Egg quality determination in teleost fish. In Methods in Reproductive Aquaculture; CRC Press: Boca Raton, FL, USA, 2008; pp. 171–202. [Google Scholar]
- McEvoy, L.A. Ovulatory rhythms and over-ripening of eggs in cultivated turbot, Scophthalmus maximus L. J. Fish Biol. 1984, 24, 437–448. [Google Scholar]
- Dinis, M.T.; Ribeiro, L.; Soares, F.; Sarasquete, C. A review on the cultivation potential of Solea senegalensis in Spain and in Portugal. Aquaculture 1999, 176, 27–38. [Google Scholar] [CrossRef]
- Kjørsvik, E.; Mangor-Jensen, A.; Holmefjord, I. Egg quality in fishes. Adv. Mar. Biol. 1990, 26, 71–113. [Google Scholar]
- Brown, N.P.; Shields, R.J.; Bromage, N.R. The influence of water temperature on spawning patterns and egg quality in the Atlantic halibut (Hippoglossus hippoglossus L.). Aquaculture 2006, 261, 993–1002. [Google Scholar] [CrossRef]
- García Fernández, C. Maternal Effects on Oocyte Dynamic and Production in European Hake. Ph.D. Thesis, Institute of Marine Research (IIM-CSIC), Vigo, Spain, 2017. [Google Scholar]
- Bobe, J.; Labbé, C. Egg and sperm quality in fish. Gen. Comp. Endocrinol. 2010, 165, 535–548. [Google Scholar] [CrossRef]
- Bromage, N. Broodstock Management and Seed Quality-General Considerations. In Broodstock Management and Egg and Larval Quality; Blackwell Science Ltd.: Oxford, UK, 1995; pp. 1–24. [Google Scholar]
- Kjørsvik, E.; Hoehne-Reitan, K.; Reitan, K. Egg and larval quality criteria as predictive measures for juvenile production in turbot (Scophthalmus maximus L.). Aquaculture 2003, 227, 9–20. [Google Scholar] [CrossRef]
- Da Silva, F.F.; Jacobsen, C.; Kjørsvik, E.; Støttrup, J.G.; Tomkiewicz, J. Oocyte and egg quality indicators in European eel: Lipid droplet coalescence and fatty acid composition. Aquaculture 2018, 496, 30–38. [Google Scholar] [CrossRef]
- Kjørsvik, E. Morphological, physiological and genetical studies of egg quality in cod Gadus morhua L. Flodevigen Rapp. 1984, 1, 67–86. [Google Scholar]
- Shields, R.J.; Brown, N.P.; Bromage, N.R. Blastomere morphology as a predictive measure of fish egg viability. Aquaculture 1997, 155, 1–12. [Google Scholar] [CrossRef]
- Kjørsvik, E. Egg quality in wild and broodstock cod Gadus morhua L. J. World Aquac. Soc. 1994, 25, 22–29. [Google Scholar] [CrossRef]
- Vallin, L.; Nissling, A. Cell morphology as an indicator of viability of cod eggs–results from an experimental study. Fish. Res. 1998, 38, 247–255. [Google Scholar] [CrossRef]
- Bromley, P. Evidence for density-dependent growth in North Sea gadoids. J. Fish Biol. 1989, 35, 117–123. [Google Scholar] [CrossRef]
- Ienaga, N.; Higuchi, K.; Takashi, T.; Gen, K.; Tsuda, K.; Terayama, K. Vision-based egg quality prediction in Pacific bluefin tuna (Thunnus orientalis) by deep neural network. Sci. Rep. 2021, 11, 6. [Google Scholar] [CrossRef]
- Kottmann, J.S.; Jørgensen, M.G.; Bertolini, F.; Loh, A.; Tomkiewicz, J. Differential impacts of carp and salmon pituitary extracts on induced oogenesis, egg quality, molecular ontogeny and embryonic developmental competence in European eel. PLoS ONE 2020, 15, e0235617. [Google Scholar] [CrossRef]
- Sullivan, C.V.; Chapman, R.W.; Reading, B.J.; Anderson, P.E. Transcriptomics of mRNA and egg quality in farmed fish: Some recent developments and future directions. Gen. Comp. Endocrinol. 2015, 221, 23–30. [Google Scholar] [CrossRef]
- Qiu, G.F.; Ramachandra, R.K.; Rexroad III, C.E.; Yao, J. Molecular characterisation and expression profiles of cyclin B1, B2 and Cdc2 kinase during oogenesis and spermatogenesis in rainbow trout (Oncorhynchus mykiss). Anim. Reprod. Sci. 2008, 105, 209–225. [Google Scholar] [CrossRef] [PubMed]
- Hamoutene, D.; Lush, L.; Drover, D.; Walsh, A. Investigation of the temporal effects of spawning season and maternal and paternal differences on egg quality in Atlantic cod Gadus morhua L. broodstock. Aquac. Res. 2009, 40, 1668–1669. [Google Scholar] [CrossRef]
- Balsa, E.; Marco, R.; Perales-Clemente, E.; Szklarczyk, R.; Calvo, E.; Landázuri, M.O.; Enríquez, J.A. NDUFA4 is a subunit of complex IV of the mammalian electro transport chain. Cell Metab. 2012, 16, 378–386. [Google Scholar] [CrossRef]
- Mathavan, S.; Lee, S.G.P.; Mak, A.; Miller, L.D.; Murthy, K.R.K.; Govindarajan, K.R.; Tong, Y.; Wu, Y.L.; Lam, S.H.; Yang, H.; et al. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genetics 2005, 1, e29. [Google Scholar] [CrossRef]
- Fauvel, C.; Savoye, O.; Dreanno, C.; Cosson, J.; Suquet, M. Characteristics of sperm of captive seabass in relation to its fertilisation potential. J. Fish Biol. 1999, 54, 356–369. [Google Scholar] [CrossRef]
- Rouxel, C.; Suquet, M.; Cosson, J.; Severe, A.; Quemener, L.; Fauvel, C. Changes in Atlantic cod (Gadus morhua L.) sperm quality during the spawning season. Aquac. Res. 2008, 39, 434–440. [Google Scholar] [CrossRef]
- Hsiao, C.D.; Tsai, W.Y.; Horng, L.S.; Tsai, H.J. Molecular structure and developmental expression of three muscle-type troponin T genes in zebrafish. Dev. Dyn. 2003, 227, 266–279. [Google Scholar] [CrossRef]
- Leung, Y.F.; Dowling, J.E. Gene expression profiling of zebrafish embryonic retina. Zebrafish 2005, 2, 269–283. [Google Scholar] [CrossRef]
- Mommens, M.; Fernandes, J.M.; Bizuayehu, T.T.; Bolla, S.L.; Johnston, I.A.; Babiak, I. Maternal gene expression in Atlantic halibut (Hippoglossus hippoglossus L.) and its relation to egg quality. BMC Res. Notes 2010, 3, 138. [Google Scholar] [CrossRef]
- Rise, M.L.; Nash, G.W.; Hall, J.R.; Booman, M.; Hori, T.S.; Trippel, E.A.; Gamperl, A.K. Variation in embryonic mortality and maternal transcript expression among Atlantic cod (Gadus morhua) broodstock: A functional genomics study. Mar. Genom. 2014, 18, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, K.; Sarropoulou, E.; Edvardsen, V.; Fernandes, J.M. Substantial Downregulation of Myogenic Transcripts in Skeletal Muscle of Atlantic Cod during the Spawning Period. PLoS ONE 2016, 11, e0148374. [Google Scholar] [CrossRef]
- Hall, T.E.; Smith, P.; Johnston, I.A. Stages of embryonic development in the Atlantic cod Gadus morhua. J. Morphol. 2004, 259, 255–270. [Google Scholar] [CrossRef]
- Lush, L.; Hamoutene, D.; Drover, D.; Walsh, A.; Puvanendran, V. Gamete collection method and egg quality comparison in Atlantic cod, Gadus morhua L. mating pairs and its importance in selective breeding. Aquaculture 2011, 315, 407–409. [Google Scholar] [CrossRef]
- Hansen, Ø.J.; Puvanendran, V.; Bangera, R. Broodstock diet with water and astaxanthin improve condition and egg output of brood fish and larval survival in Atlantic cod, Gadus morhua L. Aquac. Res. 2016, 47, 819–829. [Google Scholar] [CrossRef]
- Mattson, N.S.; Riple, T.H. Metomidate, a better anesthetic for cod (Gadus morhua) in comparison with benzocaine, MS-222, chlorobutanol, and phenoxyethanol. Aquaculture 1989, 83, 89–94. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C T method. Nat. Protoc. 2008, 3, 1101. [Google Scholar] [CrossRef] [PubMed]
- Olsvik, P.A.; Søfteland, L.; Lie, K.K. Selection of reference genes for qRT-PCR examination of wild populations of Atlantic cod Gadus morhua. BMC Res. Notes 2008, 1, 47. [Google Scholar] [CrossRef]
- Koedijk, R.M.; Le François, N.R.; Blier, P.U.; Foss, A.; Folkvord, A.; Ditlecadet, D.; Lamarre, S.G.; Stefansson, S.O.; Imsland, A.K. Ontogenetic effects of diet during early development on growth performance, myosin mRNA expression and metabolic enzyme activity in Atlantic cod juveniles reared at different salinities. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2010, 156, 102–109. [Google Scholar] [CrossRef]
- Breton, T.S.; Berlinsky, D.L. Characterizing ovarian gene expression during oocyte growth in Atlantic cod (Gadus morhua). Comp. Biochem. Physiol. Part D Genom. Proteom. 2014, 9, 1–10. [Google Scholar] [CrossRef]
- Kortner, T.M.; Arukwe, A. Effects of 17α-methyltestosterone exposure on steroidogenesis and cyclin-B mRNA expression in previtellogenic oocytes of Atlantic cod (Gadus morhua). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2007, 146, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Aursnes, I.A.; Rishovd, A.L.; Karlsen, H.E.; Gjøen, T. Validation of reference genes for quantitative RT-qPCR studies of gene expression in Atlantic cod (Gadus morhua L.) during temperature stress. BMC Res. Notes 2011, 4, 104. [Google Scholar] [CrossRef]
- Pérez-Casanova, J.C.; Hamoutene, D.; Samuelson, S.; Burt, K.; King, T.L.; Lee, K. The immune response of juvenile Atlantic cod (Gadus morhua L.) to chronic exposure to produced water. Mar. Environ. Res. 2010, 70, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.K.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalisation of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 0034.1–0034.11. [Google Scholar] [CrossRef]
- Xia, M.; Sherlock, J.; Hegerich, P.; You, X.; Lee, K.; Walworth, C.; Spier, E. DataAssist data analysis software for TaqMan real-time PCR data. In Proceedings of the International MultiConference of Engineers and Computer Scientists, Hong Kong, China, 17–19 March 2010; Volume 1, pp. 210–212. [Google Scholar]
- Hird, H.J.; Chisholm, J.; Kaye, J.; Colyer, A.; Hold, G.; Conyers, C.M.; Núñez, J.I.; Macarthur, R. Development of real-time PCR assays for the detection of Atlantic cod (Gadus morhua), Atlantic salmon (Salmo salar) and European plaice (Pleuronectes platessa) in complex food samples. Eur. Food Res. Technol. 2012, 234, 127–136. [Google Scholar] [CrossRef]
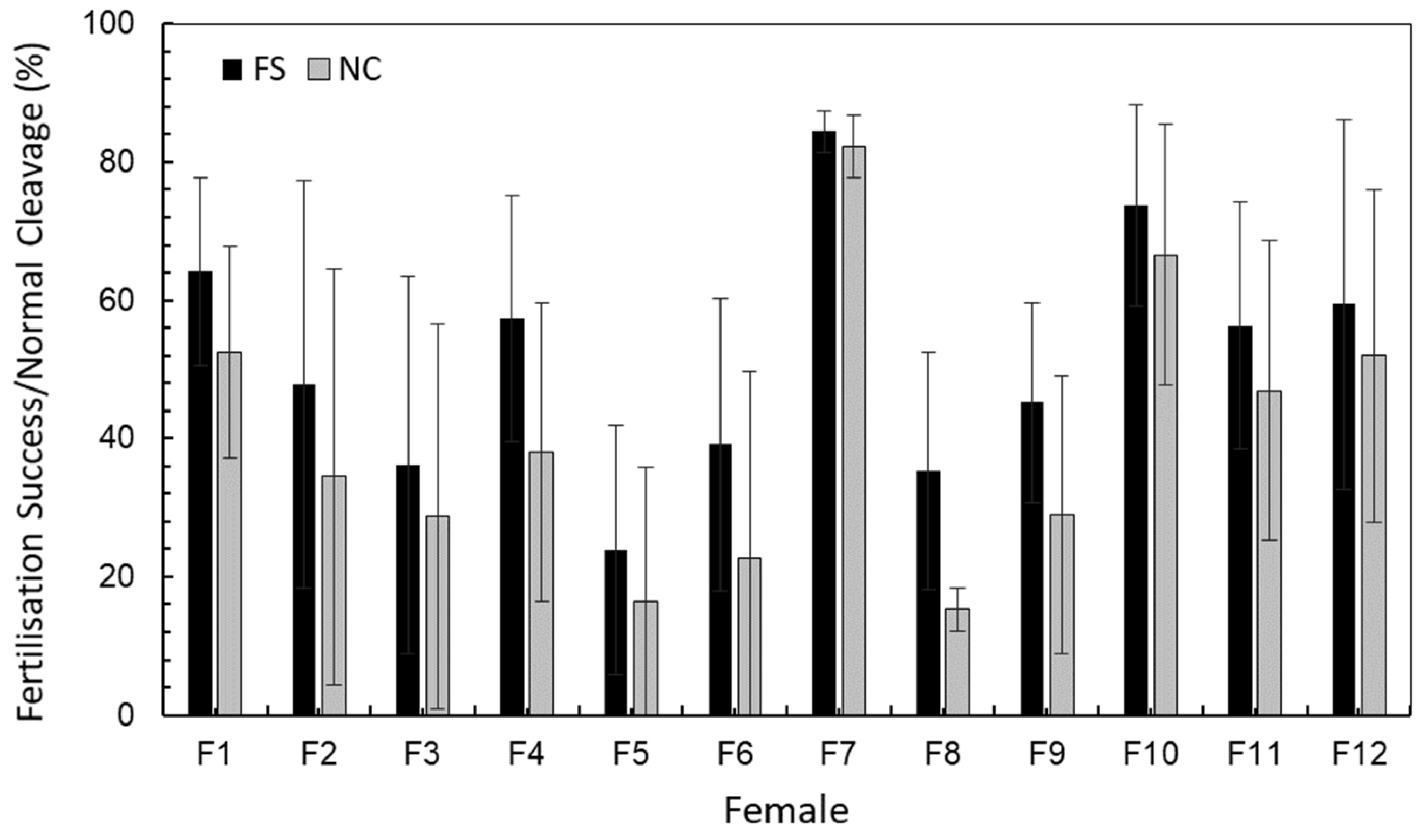

| SN | Mean | S.D. | S.E. | p | Tukey HSD—Q Statistic | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FS (%) | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 | F10 | F11 | F12 | |||||
| Female 1 | 7 | 64.13 | 13.55 | 5.12 | <0.001 | 2.13 | 3.81 | 0.79 | 5.48 * | 3.27 | 2.36 | 3.92 | 2.58 | 1.19 | 1.06 | 0.58 |
| Female 2 | 6 | 47.85 | 29.52 | 12.05 | 1.53 | 1.07 | 3.14 | 1.10 | 4.12 | 1.64 | 0.35 | 3.10 | 1.09 | 1.37 | ||
| Female 3 | 7 | 36.13 | 27.29 | 10.32 | 2.46 | 1.67 | 0.39 | 5.60 ** | 0.11 | 1.23 | 4.66 | 2.72 | 2.87 | |||
| Female 4 | 4 | 57.35 | 17.84 | 8.92 | 3.89 | 2.06 | 2.79 | 2.56 | 1.41 | 1.77 | 0.12 | 0.22 | ||||
| Female 5 | 7 | 23.81 | 18.00 | 6.80 | 2.00 | 7.03 ** | 1.56 | 2.90 | 6.19 ** | 4.38 | 4.39 | |||||
| Female 6 | 6 | 39.10 | 21.11 | 8.62 | 5.11 * | 0.50 | 0.79 | 4.15 | 2.23 | 2.41 | ||||||
| Female 7 | 4 | 84.45 | 3.040 | 1.52 | 5.70 ** | 4.56 | 1.17 | 3.24 | 2.66 | |||||||
| Female 8 | 7 | 35.30 | 17.09 | 6.46 | 1.34 | 4.76 * | 2.83 | 2.97 | ||||||||
| Female 9 | 7 | 45.17 | 14.43 | 5.45 | 3.54 | 1.50 | 1.75 | |||||||||
| Female 10 | 5 | 73.68 | 14.53 | 6.50 | 2.12 | 1.60 | ||||||||||
| Female 11 | 7 | 56.29 | 17.90 | 6.77 | 0.27 | |||||||||||
| Female 12 | 6 | 59.40 | 26.78 | 10.93 | ||||||||||||
| NC (%) | ||||||||||||||||
| Female 1 | 7 | 52.61 | 15.43 | 5.83 | <0.001 | 2.19 | 3.00 | 1.56 | 4.58 | 3.62 | 3.21 | 4.71 * | 2.99 | 1.62 | 0.70 | 0.06 |
| Female 2 | 6 | 34.50 | 30.08 | 12.28 | 0.69 | 0.37 | 2.21 | 1.38 | 5.01 * | 2.33 | 0.68 | 3.58 | 1.50 | 2.00 | ||
| Female 3 | 7 | 28.79 | 27.83 | 10.52 | 1.00 | 1.58 | 0.74 | 5.77 ** | 1.71 | 0.02 | 4.36 | 2.27 | 2.76 | |||
| Female 4 | 4 | 38.03 | 21.50 | 10.75 | 2.34 | 1.60 | 4.23 | 2.45 | 0.98 | 2.88 | 0.95 | 1.43 | ||||
| Female 5 | 7 | 16.33 | 19.45 | 7.35 | 0.78 | 7.12 ** | 0.13 | 1.59 | 5.80 ** | 3.83 | 4.24 | |||||
| Female 6 | 6 | 22.72 | 26.99 | 11.02 | 6.24 ** | 0.90 | 0.75 | 4.90 * | 2.92 | 3.35 | ||||||
| Female 7 | 4 | 82.28 | 4.49 | 2.25 | 7.23 ** | 5.76 * | 1.58 | 3.77 | 3.10 | |||||||
| Female 8 | 7 | 15.30 | 3.13 | 1.18 | 1.72 | 5.92 ** | 3.96 | 4.36 | ||||||||
| Female 9 | 7 | 28.91 | 20.14 | 7.61 | 4.35 | 2.26 | 2.74 | |||||||||
| Female 10 | 5 | 66.56 | 18.90 | 8.45 | 2.25 | 1.59 | ||||||||||
| Female 11 | 7 | 46.96 | 21.61 | 8.17 | 0.59 | |||||||||||
| Female 12 | 6 | 51.98 | 24.09 | 9.83 | ||||||||||||
| Spawn Volume | MR | FS | NC | Length t90 | Weight t90 | |
|---|---|---|---|---|---|---|
| Female 1 | 261.71 ± 166.53 | 4.76 | 64.13 ± 13.55 | 52.61 ± 15.43 | 36.23 ± 3.33 | 0.40 ± 0.13 |
| Female 5 | 226.14 ± 103.40 | 5.94 | 23.81 ± 18.00 | 16.33 ± 19.45 | 40.51 ± 7.61 | 0.65 ± 0.29 |
| Female 6 | 171.50 ± 81.22 | 8.18 | 39.10 ± 21.11 | 22.72 ± 26.99 | 37.07 ± 3.52 | 0.50 ± 0.14 |
| Female 7 | 207.00 ± 109.18 | 2.34 | 84.45 ± 3.04 | 82.28 ± 4.49 | 37.04 ± 4.86 | 0.55 ± 0.21 |
| Female 11 | 209.43 ± 124.22 | 5.39 | 56.29 ± 17.90 | 46.96 ± 21.61 | 32.66 ± 4.25 | 0.36 ± 0.18 |
| Gene | Code | Function | Stages | 5′-Forward Primer-3′ | 5′-Reverse Primer-3′ | Size | Tm | E |
|---|---|---|---|---|---|---|---|---|
| Target genes | ||||||||
| Cyclin B2 | ccnb2 | Mitosis regulation | UE-GA | GGCCGGTAGTGCACCATGGC | TCAGGAGAGCCTCAAAGGCTGCA | 106 | 60 | 92.86 |
| Myosin | myhc | Muscle contraction | ES-HL | CAGAAGCTATAAAAGGTGTCCG | GCAGCCATTCTTCTTATCCTCCTC | 86 | 60 | 101.8 |
| Actin alpha cardiac muscle 1 | actc1 | Muscle contraction | ES-HL | CTTCCCTGTCCACCTTCCAG | ACGGAGACGACGATGGAGAA | 122 | 62 | 97.88 |
| Actin alpha skeletal muscle | acta | Muscle contraction | LS-HL | TGTTCACAGTTCGTTCTCCGA | TCGTCTCCGTCGTCATCATC | 200 | 62 | 86.04 |
| NADH ubiquinone reductase 4l | ndufa4l | Respiratory chain | BL-GA | CTTTTTCATCGGTGGAGGCG | TTGTTCTTGCGATCCCAGCT | 90 | 60 | 97.22 |
| Troponin T skeletal | tnnt3 | Muscle contraction | LS-HL | ACATGGGCTCCAACTACAGC | TTGCGTCTTCCAGCCAGAAT | 112 | 62 | 96.97 |
| Parvalbumin 1 | pvalb1 | Calcium signalling | LS-HL | CAGAGCGGCTTCATTGAGGA | CTCCGATCATGCCATCACCA | 136 | 62 | 83.21 |
| Reference genes | ||||||||
| Ribosomal protein S9 | rps9 | 40S ribosomal protein | BL-HL | TCTTTGAAGGTAATGCTCTGTTGAGA | CGAGGATGTAATCCAACTTCATCTT | 84 | 62 | 85.26 |
| Acidic ribosomal protein | arp | 60 S ribosomal protein | BL-HL | TGATCCTCCACGACGATGAG | CAGGGCCTTGGCGAAGA | 113 | 62 | 87.11 |
| Ubiquitin | ubi | Protein transport and degradation | BL-HL | GGCCGCAAAGATGCAGAT | CTGGGCTCGACCTCAAGAGT | 69 | 58 | 98.32 |
| Heat Shock Protein 90-Beta | hsp90β | Chaperone, polypeptides stabilisation | UE-HL | CGTGGCGTGGTGGACTCT | GACTATGTTCTTGCGGATGACCTT | 96 | 58 | 94.45 |
| FS | NC | MR | acta | actc1 | ccnb2 | myhc | ndufa4l | pvalb1 | tnnt3 | |
|---|---|---|---|---|---|---|---|---|---|---|
| FS | - | |||||||||
| NC | 0.912 * | - | ||||||||
| MR | −0.336 | −0.379 | - | |||||||
| acta | 0.290 * | 0.295 * | −0.167 | - | ||||||
| actc1 | 0.307 * | 0.262 | −0.161 | 0.592 | - | |||||
| ccnb2 | 0.090 | 0.094 | 0.281 | 0.117 | −0.085 | - | ||||
| myhc | −0.002 | 0.066 | −0.153 | 0.529 * | 0.360 * | −0.017 | - | |||
| ndufa4l | 0.123 | 0.197 | 0.024 | 0.148 | −0.074 | −0.088 | −0.029 | - | ||
| pvalb1 | 0.211 | 0.156 | −0.353 | 0.589 * | 0.612 * | −0.114 | 0.272 | 0.138 | - | |
| tnnt3 | −0.044 | −0.048 | 0.049 | 0.313 | 0.464 | −0.017 | 0.308 | −0.139 | 0.387 * | - |
| F1 | F3 | F5 | F6 | F7 | F8 | F9 | F11 | |
|---|---|---|---|---|---|---|---|---|
| Fertilisation success | ||||||||
| FS/NC | 0.919 * | 0.984 * | 0.984 * | 0.786 | 0.991 * | 0.744 * | 0.931 * | 0.865 * |
| FS/MR | −0.219 | −0.394 | 0.417 | −0.187 | −0.568 | −0.997 | 0.344 | −0.954 * |
| FS/acta | 0.487 | 0.319 | 0.777 * | 0.684 | 0.908 | 0.334 | −0.065 | 0.717 |
| FS/actc1 | 0.280 | 0.876* | 0.565 | 0.457 | 0.881 | 0.757 * | 0.240 | 0.799 * |
| FS/ccnb2 | 0.658 | −0.135 | 0.674 | 0.401 | 0.609 | −0.417 | −0.078 | −0.236 |
| FS/myhc | −0.050 | −0.310 | −0.010 | −0.091 | 0.879 | 0.431 | 0.358 | 0.482 |
| FS/ndufa4l | −0.126 | 0.052 | −0.353 | 0.588 | −0.805 | −0.090 | 0.053 | 0.029 |
| FS/pvalb1 | −0.738 * | 0.698 | 0.198 | 0.304 | 0.825 | 0.664 | −0.648 | 0.739 |
| FS/tnnt3 | 0.003 | −0.476 | 0.472 | 0.271 | 0.697 | 0.395 | −0.462 | 0.691 |
| Normal cleavage | ||||||||
| NC/MR | −0.073 | −0.248 | 0.316 | −0.283 | −0.666 | −0.738 | 0.195 | −0.929 * |
| NC/acta | 0.222 | 0.206 | 0.780 * | 0.911 * | 0.870 | 0.689 | 0.229 | 0.715 |
| NC/actc1 | 0.076 | 0.797 * | 0.541 | 0.661 | 0.876 | 0.498 | 0.429 | 0.772 * |
| NC/ccnb2 | 0.563 * | −0.059 | 0.596 | 0.116 | 0.500 | −0.122 | 0.111 | −0.480 |
| NC/myhc | −0.075 | −0.380 | −0.089 | 0.440 | 0.834 | 0.732 | 0.598 | 0.546 |
| NC/ndufa4l | 0.064 | −0.024 | −0.358 | 0.936 * | −0.731 | 0.203 | 0.152 | −0.098 |
| NC/pvalb1 | −0.662 | 0.607 | 0.148 | 0.589 | 0.743 | 0.523 | −0.405 | 0.521 |
| NC/tnnt3 | 0.002 | −0.413 | 0.377 | 0.120 | 0.607 | 0.291 | −0.186 | 0.762 * |
| Mortality rate | ||||||||
| MR/acta | −0.637 | −0.402 | −0.054 | −0.125 | −0.248 | −0.021 | −0.191 | −0.626 |
| MR/actc1 | −0.661 | −0.561 | 0.035 | 0.150 | −0.400 | −0.703 | −0.213 | −0.795 |
| MR/ccnb2 | 0.208 | 0.375 | 0.637 | −0.599 | 0.291 | 0.709 | −0.150 | 0.464 |
| MR/myhc | −0.796 * | −0.276 | −0.114 | −0.026 | −0.181 | −0.286 | 0.328 | −0.383 |
| MR/ndufa4l | 0.834 * | −0.245 | 0.286 | −0.124 | 0.129 | −0.157 | 0.014 | 0.137 |
| MR/pvalb1 | −0.179 | −0.536 | −0.430 | −0.446 | −0.023 | −0.541 | −0.710 | −0.794 |
| MR/tnnt3 | −0.417 | 0.624 | 0.107 | −0.476 | 0.188 | −0.118 | 0.567 | −0.653 |
| Female | No. Batches | No. Samples |
|---|---|---|
| Female 1 *,+ | 7 | 49 |
| Female 2 | 6 | 42 |
| Female 3 + | 7 | 49 |
| Female 4 | 4 | 28 |
| Female 5 *,+ | 7 | 49 |
| Female 6 *,+ | 6 | 42 |
| Female 7 *,+ | 4 | 28 |
| Female 8 + | 7 | 49 |
| Female 9 + | 7 | 49 |
| Female 10 | 5 | 35 |
| Female 11 *,+ | 7 | 49 |
| Female 12 | 6 | 42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández Míguez, M.; Presa, P.; Puvanendran, V.; Tveiten, H.; Hansen, Ø.J.; Pérez, M. Gene Expression and Phenotypic Assessment of Egg Quality across Developmental Stages of Atlantic Cod throughout the Spawning Season. Int. J. Mol. Sci. 2024, 25, 7488. https://doi.org/10.3390/ijms25137488
Fernández Míguez M, Presa P, Puvanendran V, Tveiten H, Hansen ØJ, Pérez M. Gene Expression and Phenotypic Assessment of Egg Quality across Developmental Stages of Atlantic Cod throughout the Spawning Season. International Journal of Molecular Sciences. 2024; 25(13):7488. https://doi.org/10.3390/ijms25137488
Chicago/Turabian StyleFernández Míguez, María, Pablo Presa, Velmurugu Puvanendran, Helge Tveiten, Øyvind J. Hansen, and Montse Pérez. 2024. "Gene Expression and Phenotypic Assessment of Egg Quality across Developmental Stages of Atlantic Cod throughout the Spawning Season" International Journal of Molecular Sciences 25, no. 13: 7488. https://doi.org/10.3390/ijms25137488
APA StyleFernández Míguez, M., Presa, P., Puvanendran, V., Tveiten, H., Hansen, Ø. J., & Pérez, M. (2024). Gene Expression and Phenotypic Assessment of Egg Quality across Developmental Stages of Atlantic Cod throughout the Spawning Season. International Journal of Molecular Sciences, 25(13), 7488. https://doi.org/10.3390/ijms25137488








