Metformin Prevents Tumor Cell Growth and Invasion of Human Hormone Receptor-Positive Breast Cancer (HR+ BC) Cells via FOXA1 Inhibition
Abstract
:1. Introduction
2. Results
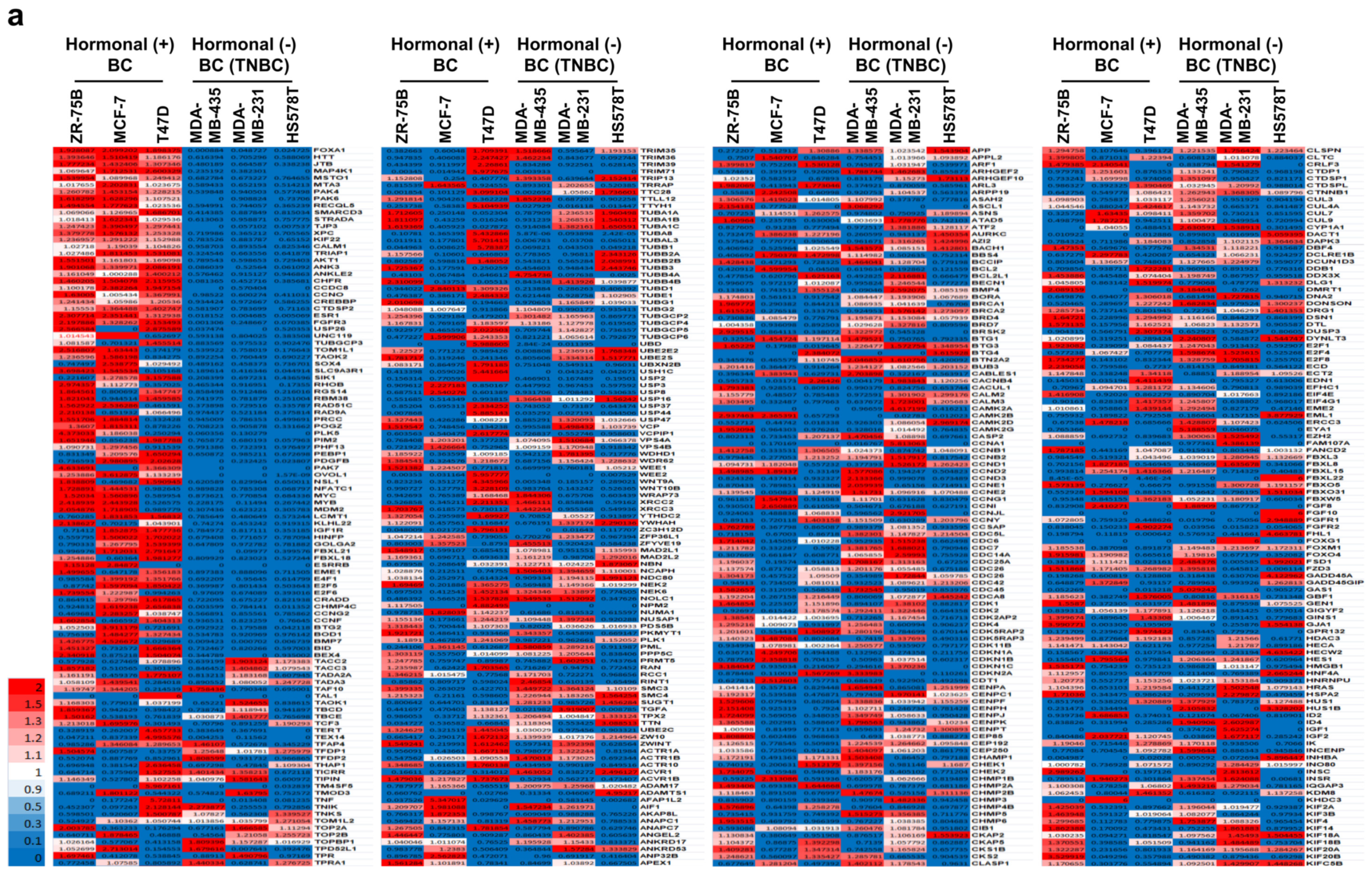
2.1. Fourteen Diabetes-Related Genes Are Significantly Expressed in Hormone-Receptor-Positive Breast Cancer (HR+ BC) Cell Lines from the NCBI GEO Dataset
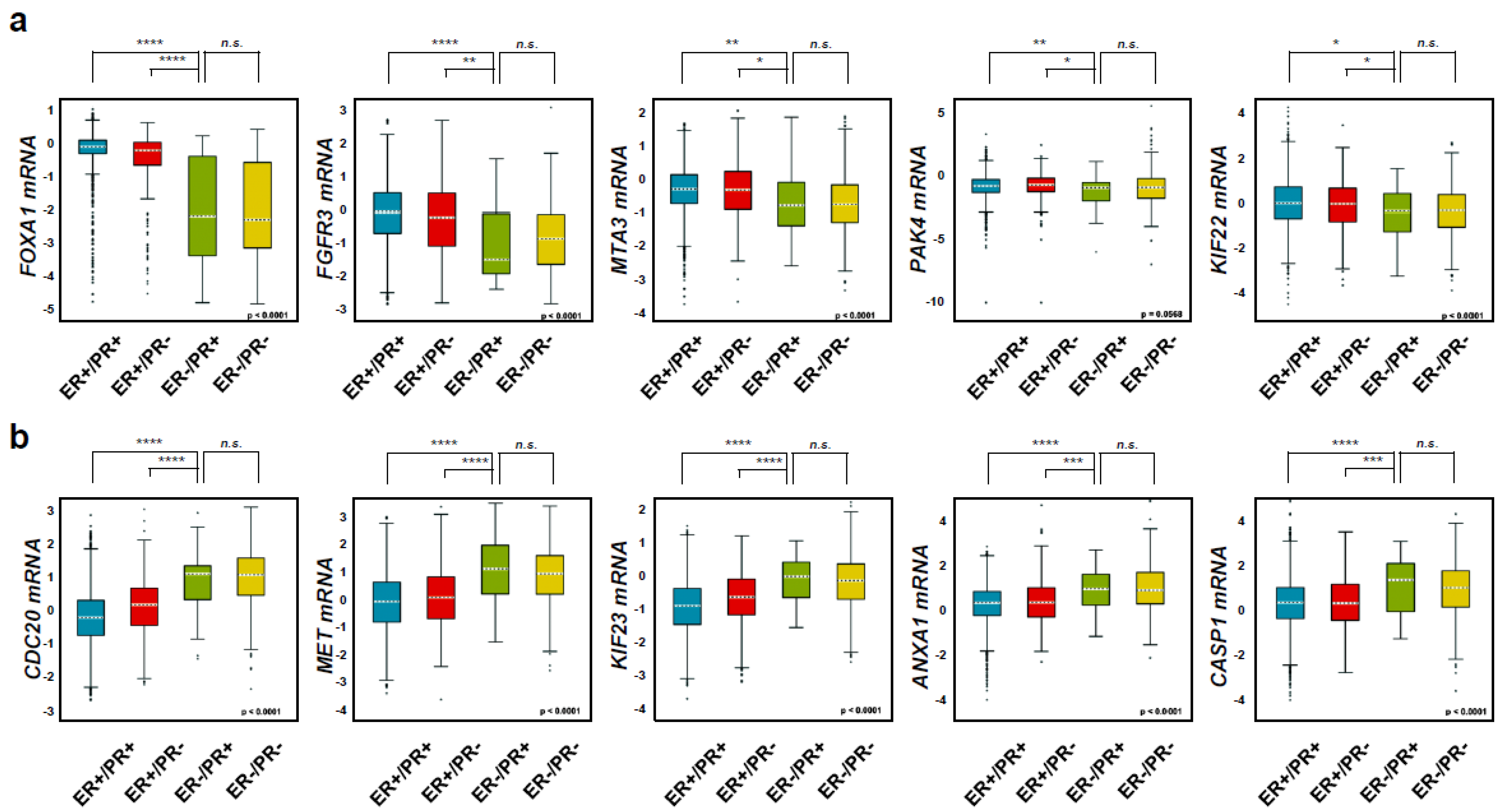
2.2. Diabetes-Related Genes Are Highly Expressed in Hormone-Receptor-Positive Breast Cancer (HR+ BC) Patients
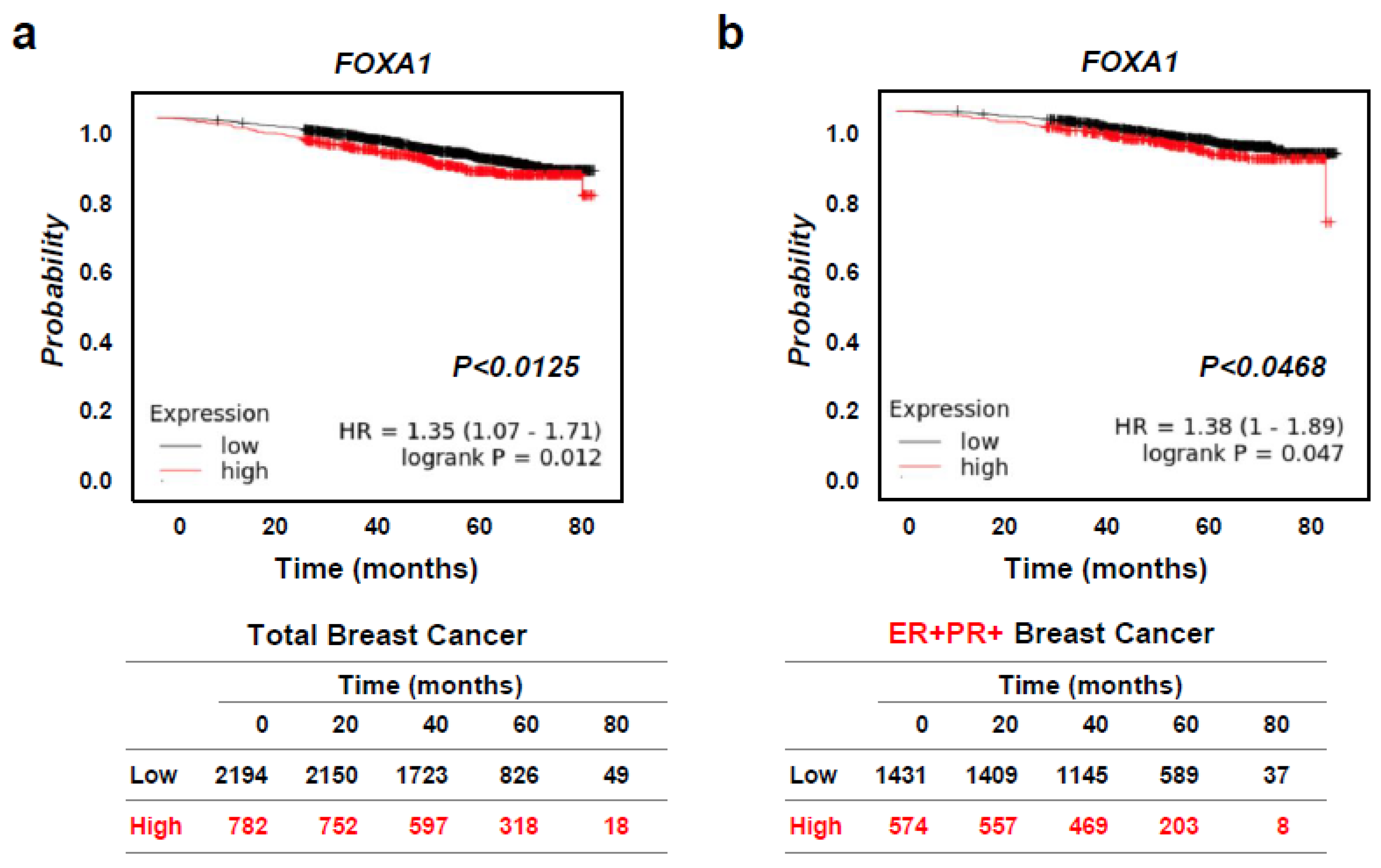
2.3. High FOXA1 Expression Leads to Worse Overall Survival in Hormone-Receptor-Positive Breast Cancer (HR+ BC) Patients
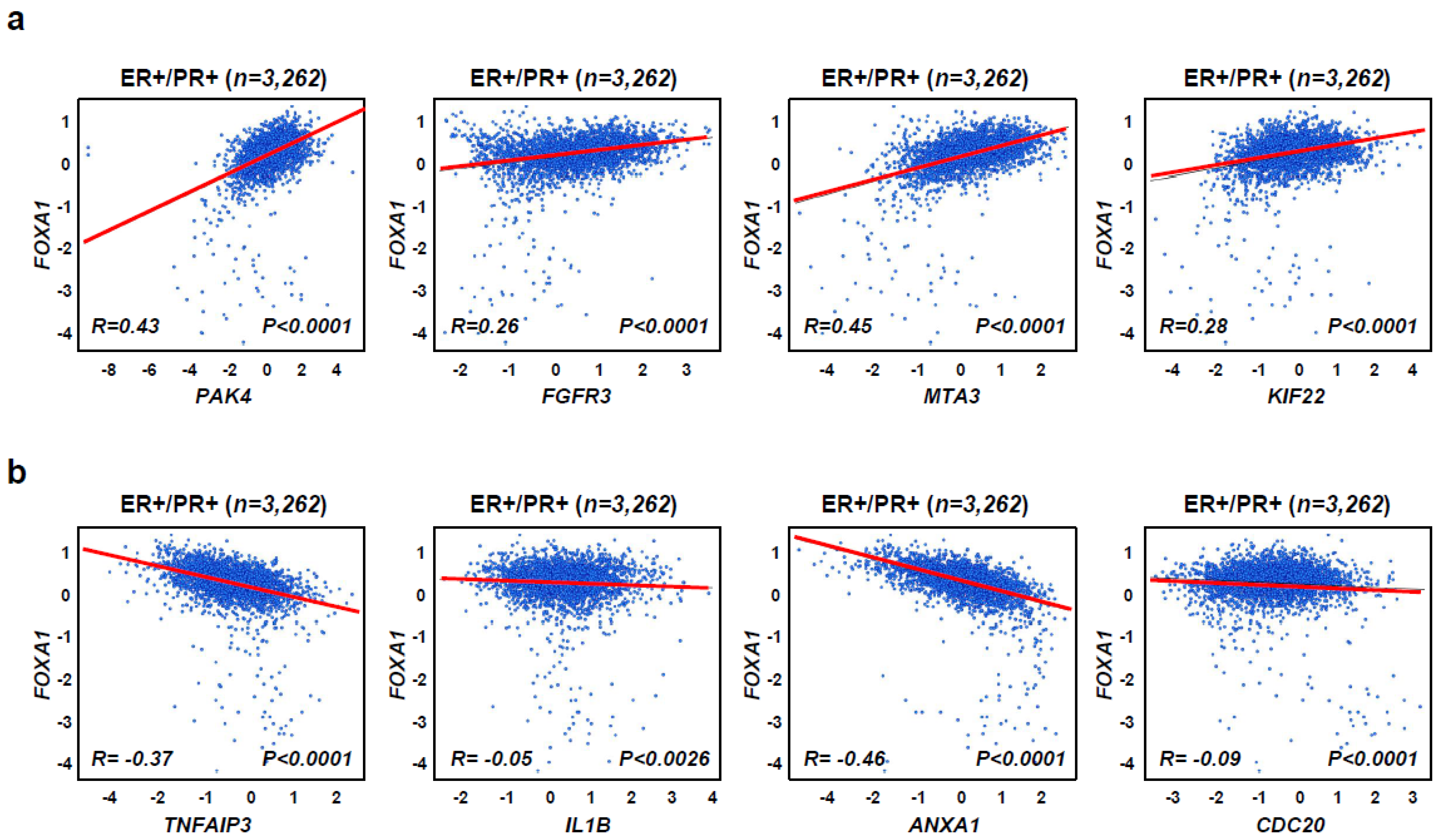
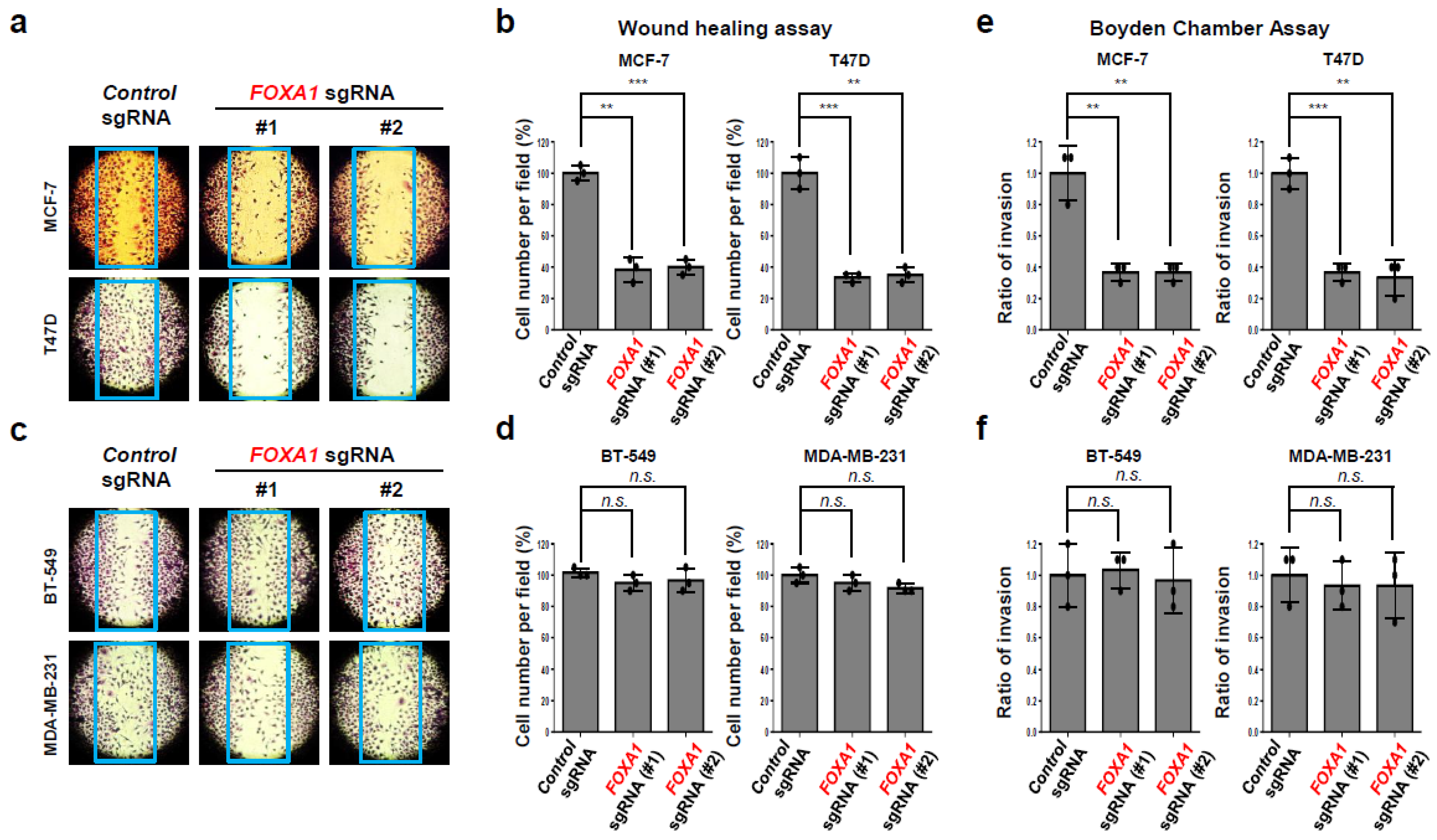
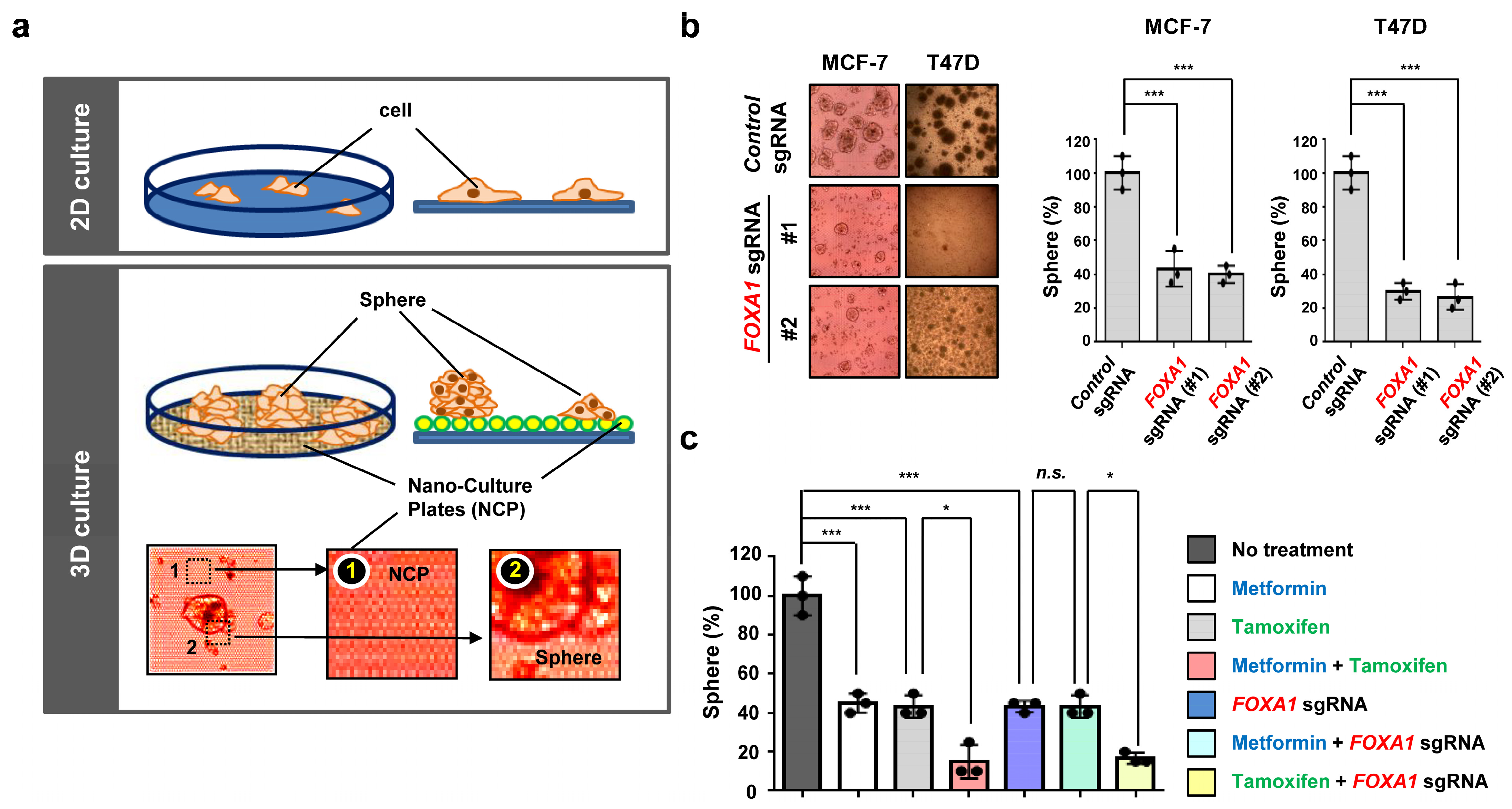
2.4. FOXA1 Is Highly Expressed in Hormone-Receptor-Positive Breast Cancer (HR+ BC) Cell Lines and Controls the HR+ Breast Cancer Cell Proliferation and Metastasis
2.5. Metformin and FOXA1 Deletion Enhance Tamoxifen-Mediated Tumor Cell Growth Inhibition in Hormone-Receptor-Positive Breast Cancer (HR+ BC) Cells
2.6. Metformin Is Effective in Treating Tamoxifen-Treated Hormone-Receptor-Positive Breast Cancer (HR+ BC) Cells in an Ex Vivo Model
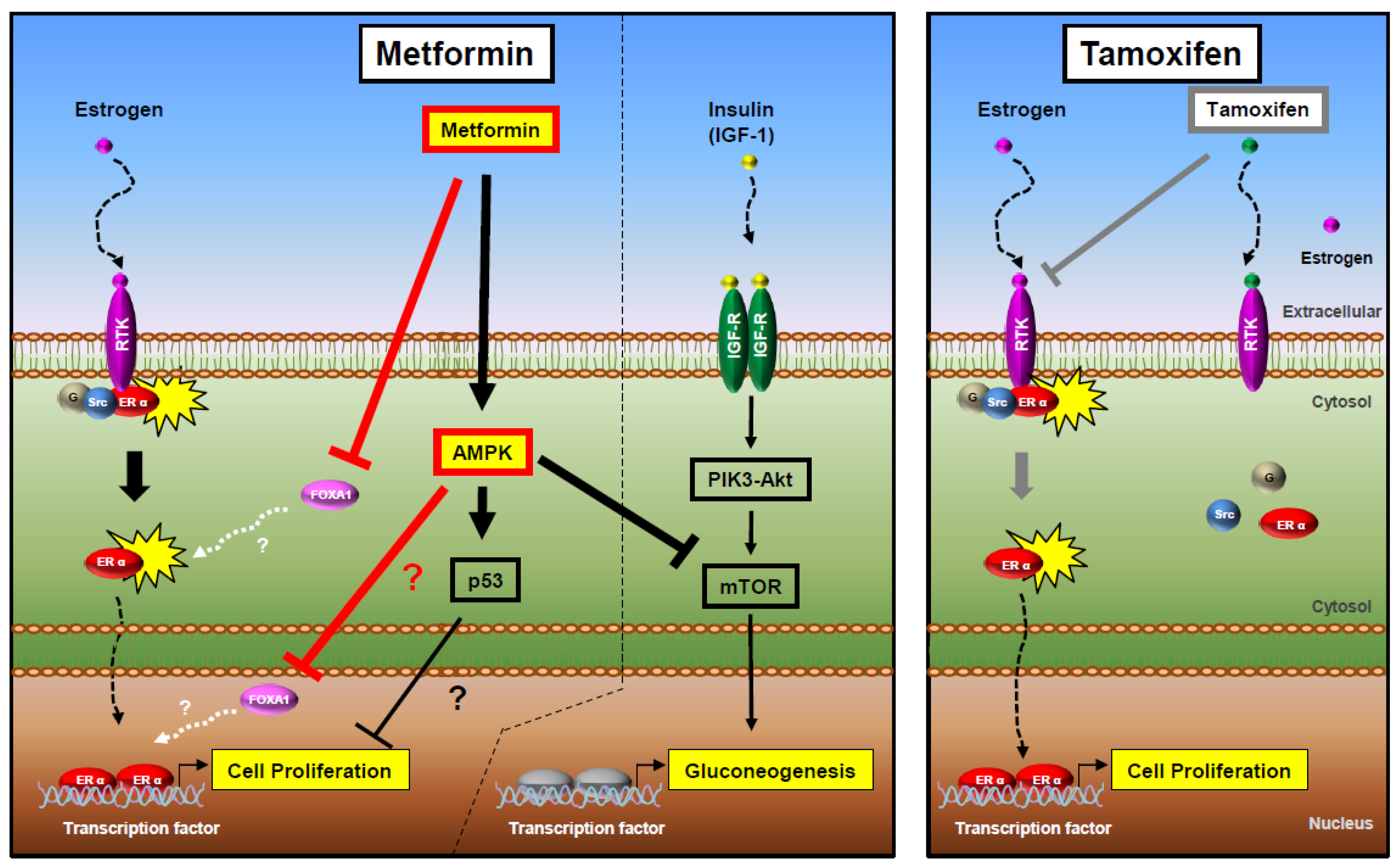
3. Discussion
4. Materials and Methods
4.1. Cancer Data Collection and Processing
4.2. Cells and Cell Lines, Reagents, and Gene Silencing
4.3. Western Blot and Antibodies
4.4. Colony-Forming Assay
4.5. Migration and Invasion Assays
4.6. Three-Dimensional Organoid Assay
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kennecke, H.; Yerushalmi, R.; Woods, R.; Cheang, M.C.; Voduc, D.; Speers, C.H.; Nielsen, T.O.; Gelmon, K. Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 2010, 28, 3271–3277. [Google Scholar] [CrossRef] [PubMed]
- Voduc, K.D.; Cheang, M.C.; Tyldesley, S.; Gelmon, K.; Nielsen, T.O.; Kennecke, H. Breast cancer subtypes and the risk of local and regional relapse. J. Clin. Oncol. 2010, 28, 1684–1691. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Wiechmann, L.; Sampson, M.; Stempel, M.; Jacks, L.M.; Patil, S.M.; King, T.; Morrow, M. Presenting features of breast cancer differ by molecular subtype. Ann. Surg. Oncol. 2009, 16, 2705–2710. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Linden, H.M.; Anderson, B.O.; Li, C.I. Trends in 5-year survival rates among breast cancer patients by hormone receptor status and stage. Breast Cancer Res. Treat. 2014, 147, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, A.; Tanimoto, T.; Saji, S. Palbociclib in Hormone-Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2015, 373, 1672–1673. [Google Scholar] [PubMed]
- Baselga, J.; Smith, B.L.; Rafferty, E.A.; Bombonati, A. Case records of the Massachusetts General Hospital. Case 16-2012. A 32-year-old woman with HER2-positive breast cancer. N. Engl. J. Med. 2012, 366, 2018–2026. [Google Scholar] [CrossRef]
- Hernandez-Aya, L.F.; Gonzalez-Angulo, A.M. Targeting the phosphatidylinositol 3-kinase signaling pathway in breast cancer. Oncologist 2011, 16, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Nunnery, S.E.; Mayer, I.A. Targeting the PI3K/AKT/mTOR Pathway in Hormone-Positive Breast Cancer. Drugs 2020, 80, 1685–1697. [Google Scholar] [CrossRef]
- Fritsch, M.; Jordan, V.C. Long-term Tamoxifen Therapy for the Treatment of Breast Cancer. Cancer Control 1994, 1, 356–366. [Google Scholar]
- Zheng, Y.; Ley, S.H.; Hu, F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat. Rev. Endocrinol. 2018, 14, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Lotfy, M.; Adeghate, J.; Kalasz, H.; Singh, J.; Adeghate, E. Chronic Complications of Diabetes Mellitus: A Mini Review. Curr. Diabetes Rev. 2017, 13, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Demissie, S.; Silliman, R.A.; Lash, T.L. Adjuvant tamoxifen: Predictors of use, side effects, and discontinuation in older women. J. Clin. Oncol. 2001, 19, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group; Davies, C.; Godwin, J.; Gray, R.; Clarke, M.; Cutter, D.; Darby, S.; McGale, P.; Pan, H.C.; Taylor, C.; et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomised trials. Lancet 2011, 378, 771–784. [Google Scholar] [CrossRef] [PubMed]
- DeFronzo, R.A.; Ferrannini, E.; Groop, L.; Henry, R.R.; Herman, W.H.; Holst, J.J.; Hu, F.B.; Kahn, C.R.; Raz, I.; Shulman, G.I.; et al. Type 2 diabetes mellitus. Nat. Rev. Dis. Primers 2015, 1, 15019. [Google Scholar] [CrossRef] [PubMed]
- Olokoba, A.B.; Obateru, O.A.; Olokoba, L.B. Type 2 diabetes mellitus: A review of current trends. Oman Med. J. 2012, 27, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Bronsveld, H.K.; Jensen, V.; Vahl, P.; De Bruin, M.L.; Cornelissen, S.; Sanders, J.; Auvinen, A.; Haukka, J.; Andersen, M.; Vestergaard, P.; et al. Diabetes and Breast Cancer Subtypes. PLoS ONE 2017, 12, e0170084. [Google Scholar] [CrossRef] [PubMed]
- Wolf, I.; Sadetzki, S.; Catane, R.; Karasik, A.; Kaufman, B. Diabetes mellitus and breast cancer. Lancet Oncol. 2005, 6, 103–111. [Google Scholar] [CrossRef]
- Peairs, K.S.; Barone, B.B.; Snyder, C.F.; Yeh, H.C.; Stein, K.B.; Derr, R.L.; Brancati, F.L.; Wolff, A.C. Diabetes mellitus and breast cancer outcomes: A systematic review and meta-analysis. J. Clin. Oncol. 2011, 29, 40–46. [Google Scholar] [CrossRef]
- Chomanicova, N.; Gazova, A.; Adamickova, A.; Valaskova, S.; Kyselovic, J. The role of AMPK/mTOR signaling pathway in anticancer activity of metformin. Physiol. Res. 2021, 70, 501–508. [Google Scholar] [CrossRef]
- Hadad, S.M.; Fleming, S.; Thompson, A.M. Targeting AMPK: A new therapeutic opportunity in breast cancer. Crit. Rev. Oncol. Hematol. 2008, 67, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Augello, M.A.; Hickey, T.E.; Knudsen, K.E. FOXA1: Master of steroid receptor function in cancer. EMBO J. 2011, 30, 3885–3894. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, G.M.; Keri, R.A. FOXA1: A transcription factor with parallel functions in development and cancer. Biosci. Rep. 2012, 32, 113–130. [Google Scholar] [CrossRef]
- Shou, J.; Lai, Y.; Xu, J.; Huang, J. Prognostic value of FOXA1 in breast cancer: A systematic review and meta-analysis. Breast 2016, 27, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Jozwik, K.M.; Chernukhin, I.; Serandour, A.A.; Nagarajan, S.; Carroll, J.S. FOXA1 Directs H3K4 Monomethylation at Enhancers via Recruitment of the Methyltransferase MLL3. Cell Rep. 2016, 17, 2715–2723. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Kendi, A.T.; Lowe, V.J.; Lee, S. The A20/TNFAIP3-CDC20-CASP1 Axis Promotes Inflammation-mediated Metastatic Disease in Triple-negative Breast Cancer. Anticancer Res. 2022, 42, 681–695. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Kendi, A.T.; Shim, J.Y.; Jung, D.; Kang, P.S.; Lowe, V.J.; Lee, S. ER-/PR+ breast cancer is controlled more effectively with an inflammatory inhibitor than hormonal inhibitor. Breast Cancer 2023, 30, 436–452. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Lowe, V.J.; Lee, S. Inhibition of Cdc20 suppresses the metastasis in triple negative breast cancer (TNBC). Breast Cancer 2021, 28, 1073–1086. [Google Scholar] [CrossRef] [PubMed]
- Arnaud-Dabernat, S.; Kritzik, M.; Kayali, A.G.; Zhang, Y.Q.; Liu, G.; Ungles, C.; Sarvetnick, N. FGFR3 is a negative regulator of the expansion of pancreatic epithelial cells. Diabetes 2007, 56, 96–106. [Google Scholar] [CrossRef]
- Berdeaux, R.; Hutchins, C. Anabolic and Pro-metabolic Functions of CREB-CRTC in Skeletal Muscle: Advantages and Obstacles for Type 2 Diabetes and Cancer Cachexia. Front. Endocrinol. 2019, 10, 535. [Google Scholar] [CrossRef]
- Bergeron, V.; Ghislain, J.; Poitout, V. The P21-activated kinase PAK4 is implicated in fatty-acid potentiation of insulin secretion downstream of free fatty acid receptor 1. Islets 2016, 8, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, J.; Yang, W.; Chen, J.; Meng, Y.; Feng, B.; Chi, Y.; Geng, B.; Zhou, Y.; Cui, Q.; et al. FAM3C activates HSF1 to suppress hepatic gluconeogenesis and attenuate hyperglycemia of type 1 diabetic mice. Oncotarget 2017, 8, 106038–106049. [Google Scholar] [CrossRef] [PubMed]
- Chuang, H.C.; Wang, J.S.; Lee, I.T.; Sheu, W.H.; Tan, T.H. Epigenetic regulation of HGK/MAP4K4 in T cells of type 2 diabetes patients. Oncotarget 2016, 7, 10976–10989. [Google Scholar] [CrossRef] [PubMed]
- Dammann, K.; Khare, V.; Coleman, C.; Berdel, H.; Gasche, C. p-21 Activated Kinase as a Molecular Target for Chemoprevention in Diabetes. Geriatrics 2018, 3, 73. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, C.J.; Langefeld, C.D.; Gordon, C.J.; Campbell, J.K.; Mychaleckyj, J.C.; Bryer-Ash, M.; Rich, S.S.; Bowden, D.W.; Sale, M.M. Association of the estrogen receptor-alpha gene with the metabolic syndrome and its component traits in African-American families: The Insulin Resistance Atherosclerosis Family Study. Diabetes 2007, 56, 2135–2141. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Luo, T.; Yang, J.; Zhang, D.; Dai, M.; Jian, W.; Zheng, S.; Zhou, W.; Zhou, W.; Wu, Y.; et al. The -822G/A polymorphism in the promoter region of the MAP4K5 gene is associated with reduced risk of type 2 diabetes in Chinese Hans from Shanghai. J. Hum. Genet. 2006, 51, 605–610. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Shu, Y.; Wang, X.; Liu, X.; Liu, G.; Chen, Z.; Wang, Z.; Li, L.; Liu, R.; Zhou, H.; et al. Intensive Glucose Control Reduces the Risk Effect of TRIB3, SMARCD3, and ATF6 Genetic Variation on Diabetic Vascular Complications. Front. Pharmacol. 2018, 9, 1422. [Google Scholar] [CrossRef]
- Heddad Masson, M.; Poisson, C.; Guerardel, A.; Mamin, A.; Philippe, J.; Gosmain, Y. Foxa1 and Foxa2 regulate alpha-cell differentiation, glucagon biosynthesis, and secretion. Endocrinology 2014, 155, 3781–3792. [Google Scholar] [CrossRef]
- Huang, X.; Liu, G.; Guo, J.; Su, Z. The PI3K/AKT pathway in obesity and type 2 diabetes. Int. J. Biol. Sci. 2018, 14, 1483–1496. [Google Scholar] [CrossRef]
- Li, Q.; Li, Z.; Lin, Y.; Che, H.; Hu, Y.; Kang, X.; Zhang, Y.; Wang, L.; Zhang, Y. High glucose promotes hepatic fibrosis via miR32/MTA3mediated epithelialtomesenchymal transition. Mol. Med. Rep. 2019, 19, 3190–3200. [Google Scholar]
- Li, L.; Yang, K.; Ye, F.; Xu, Y.; Cao, L.; Sheng, J. Abnormal expression of TRIAP1 and its role in gestational diabetes mellitus-related pancreatic beta cells. Exp. Ther. Med. 2021, 21, 187. [Google Scholar] [CrossRef]
- Zhang, D.; Xu, X.L.; Li, F.; Sun, H.C.; Cui, Y.Q.; Liu, S.; Xu, P.Y. Upregulation of the checkpoint protein CHFR is associated with tumor suppression in pancreatic cancers. Oncol. Lett. 2017, 14, 8042–8050. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, Y.; Gao, H.; Jia, Y.; Xu, Y.; Wan, X.; Zhang, Z.; Yu, H.; Yan, S. Identification of Key Genes Involved in Pancreatic Ductal Adenocarcinoma with Diabetes Mellitus Based on Gene Expression Profiling Analysis. Pathol. Oncol. Res. 2021, 27, 604730. [Google Scholar] [CrossRef]
- Kim, J.J.; Lee, S.B.; Yi, S.Y.; Han, S.A.; Kim, S.H.; Lee, J.M.; Tong, S.Y.; Yin, P.; Gao, B.; Zhang, J.; et al. WSB1 overcomes oncogene-induced senescence by targeting ATM for degradation. Cell Res. 2017, 27, 274–293. [Google Scholar] [CrossRef]
- Wood, L.D.; Ewald, A.J. Organoids in cancer research: A review for pathologist-scientists. J. Pathol. 2021, 254, 395–404. [Google Scholar] [CrossRef]
- Mehta, R.J.; Jain, R.K.; Leung, S.; Choo, J.; Nielsen, T.; Huntsman, D.; Nakshatri, H.; Badve, S. FOXA1 is an independent prognostic marker for ER-positive breast cancer. Breast Cancer Res. Treat. 2012, 131, 881–890. [Google Scholar] [CrossRef]
- Hardie, D.G.; Hawley, S.A. AMP-activated protein kinase: The energy charge hypothesis revisited. Bioessays 2001, 23, 1112–1119. [Google Scholar] [CrossRef]
- Hardie, D.G.; Scott, J.W.; Pan, D.A.; Hudson, E.R. Management of cellular energy by the AMP-activated protein kinase system. FEBS Lett. 2003, 546, 113–120. [Google Scholar] [CrossRef]
- Goodwin, P.J.; Chen, B.E.; Gelmon, K.A.; Whelan, T.J.; Ennis, M.; Lemieux, J.; Ligibel, J.A.; Hershman, D.L.; Mayer, I.A.; Hobday, T.J.; et al. Effect of Metformin vs Placebo on Invasive Disease-Free Survival in Patients With Breast Cancer: The MA.32 Randomized Clinical Trial. JAMA 2022, 327, 1963–1973. [Google Scholar] [CrossRef] [PubMed]
- Kahn, B.B.; Alquier, T.; Carling, D.; Hardie, D.G. AMP-activated protein kinase: Ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005, 1, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Kemp, B.E.; Stapleton, D.; Campbell, D.J.; Chen, Z.P.; Murthy, S.; Walter, M.; Gupta, A.; Adams, J.J.; Katsis, F.; van Denderen, B.; et al. AMP-activated protein kinase, super metabolic regulator. Biochem. Soc. Trans. 2003, 31 Pt 1, 162–168. [Google Scholar] [CrossRef]
- Kearney, A.Y.; Fan, Y.H.; Giri, U.; Saigal, B.; Gandhi, V.; Heymach, J.V.; Zurita, A.J. 8-Chloroadenosine Sensitivity in Renal Cell Carcinoma Is Associated with AMPK Activation and mTOR Pathway Inhibition. PLoS ONE 2015, 10, e0135962. [Google Scholar] [CrossRef]
- Shi, W.Y.; Xiao, D.; Wang, L.; Dong, L.H.; Yan, Z.X.; Shen, Z.X.; Chen, S.J.; Chen, Y.; Zhao, W.L. Therapeutic metformin/AMPK activation blocked lymphoma cell growth via inhibition of mTOR pathway and induction of autophagy. Cell Death Dis. 2012, 3, e275. [Google Scholar] [CrossRef]
- Cerezo, M.; Tichet, M.; Abbe, P.; Ohanna, M.; Lehraiki, A.; Rouaud, F.; Allegra, M.; Giacchero, D.; Bahadoran, P.; Bertolotto, C.; et al. Metformin blocks melanoma invasion and metastasis development in AMPK/p53-dependent manner. Mol. Cancer Ther. 2013, 12, 1605–1615. [Google Scholar] [CrossRef]
- Bernardo, G.M.; Lozada, K.L.; Miedler, J.D.; Harburg, G.; Hewitt, S.C.; Mosley, J.D.; Godwin, A.K.; Korach, K.S.; Visvader, J.E.; Kaestner, K.H.; et al. FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development 2010, 137, 2045–2054. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, C.; Tutzauer, J.; Church, S.E.; Stal, O.; Ekholm, M.; Forsare, C.; Nordenskjold, B.; Ferno, M.; Bendahl, P.O.; Ryden, L. Tamoxifen-predictive value of gene expression signatures in premenopausal breast cancer: Data from the randomized SBII:2 trial. Breast Cancer Res. 2023, 25, 110. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, C.; Jung, D.; Kendi, A.T.; Rho, J.K.; Kim, E.-J.; Horn, I.; Curran, G.L.; Ghattamaneni, S.; Shim, J.Y.; Kang, P.S.; et al. Metformin Prevents Tumor Cell Growth and Invasion of Human Hormone Receptor-Positive Breast Cancer (HR+ BC) Cells via FOXA1 Inhibition. Int. J. Mol. Sci. 2024, 25, 7494. https://doi.org/10.3390/ijms25137494
Song C, Jung D, Kendi AT, Rho JK, Kim E-J, Horn I, Curran GL, Ghattamaneni S, Shim JY, Kang PS, et al. Metformin Prevents Tumor Cell Growth and Invasion of Human Hormone Receptor-Positive Breast Cancer (HR+ BC) Cells via FOXA1 Inhibition. International Journal of Molecular Sciences. 2024; 25(13):7494. https://doi.org/10.3390/ijms25137494
Chicago/Turabian StyleSong, Christine, Dawa Jung, Ayse Tuba Kendi, Jin Kyung Rho, Eun-Joo Kim, Ian Horn, Geoffry L. Curran, Sujala Ghattamaneni, Ji Yeon Shim, Pil Soo Kang, and et al. 2024. "Metformin Prevents Tumor Cell Growth and Invasion of Human Hormone Receptor-Positive Breast Cancer (HR+ BC) Cells via FOXA1 Inhibition" International Journal of Molecular Sciences 25, no. 13: 7494. https://doi.org/10.3390/ijms25137494
APA StyleSong, C., Jung, D., Kendi, A. T., Rho, J. K., Kim, E.-J., Horn, I., Curran, G. L., Ghattamaneni, S., Shim, J. Y., Kang, P. S., Kang, D., Thakkar, J. B., Dewan, S., Lowe, V. J., & Lee, S. B. (2024). Metformin Prevents Tumor Cell Growth and Invasion of Human Hormone Receptor-Positive Breast Cancer (HR+ BC) Cells via FOXA1 Inhibition. International Journal of Molecular Sciences, 25(13), 7494. https://doi.org/10.3390/ijms25137494




