Knockdown of DJ-1 Resulted in a Coordinated Activation of the Innate Immune Antiviral Response in HEK293 Cell Line
Abstract
1. Introduction
2. Results
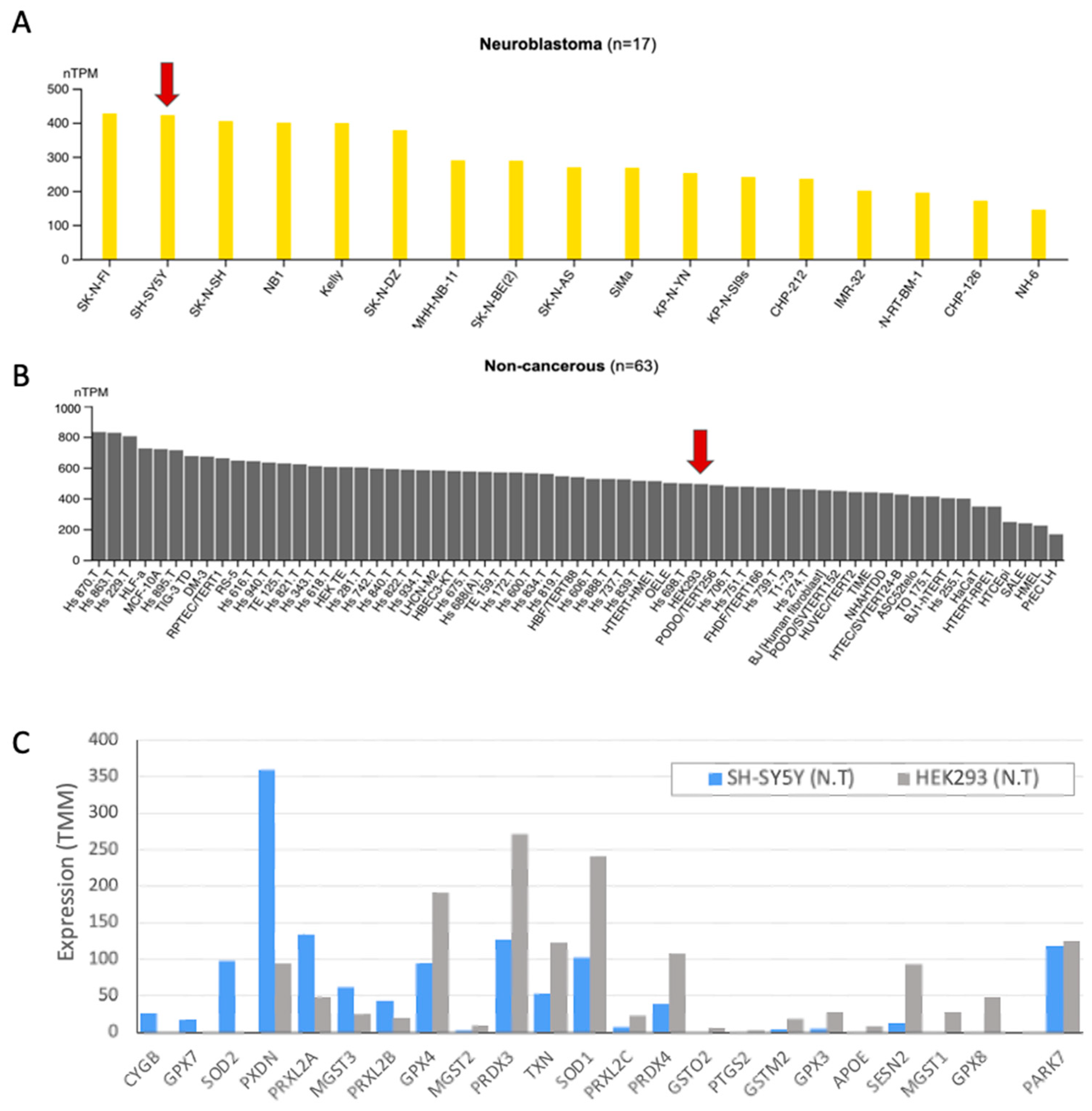
2.1. DJ-1 Response to Oxidative Stress by Translocation to Nucleus
2.2. Knockdown Resulted in 8-Fold Suppression of the Native PARK7/DJ-1 Transcript
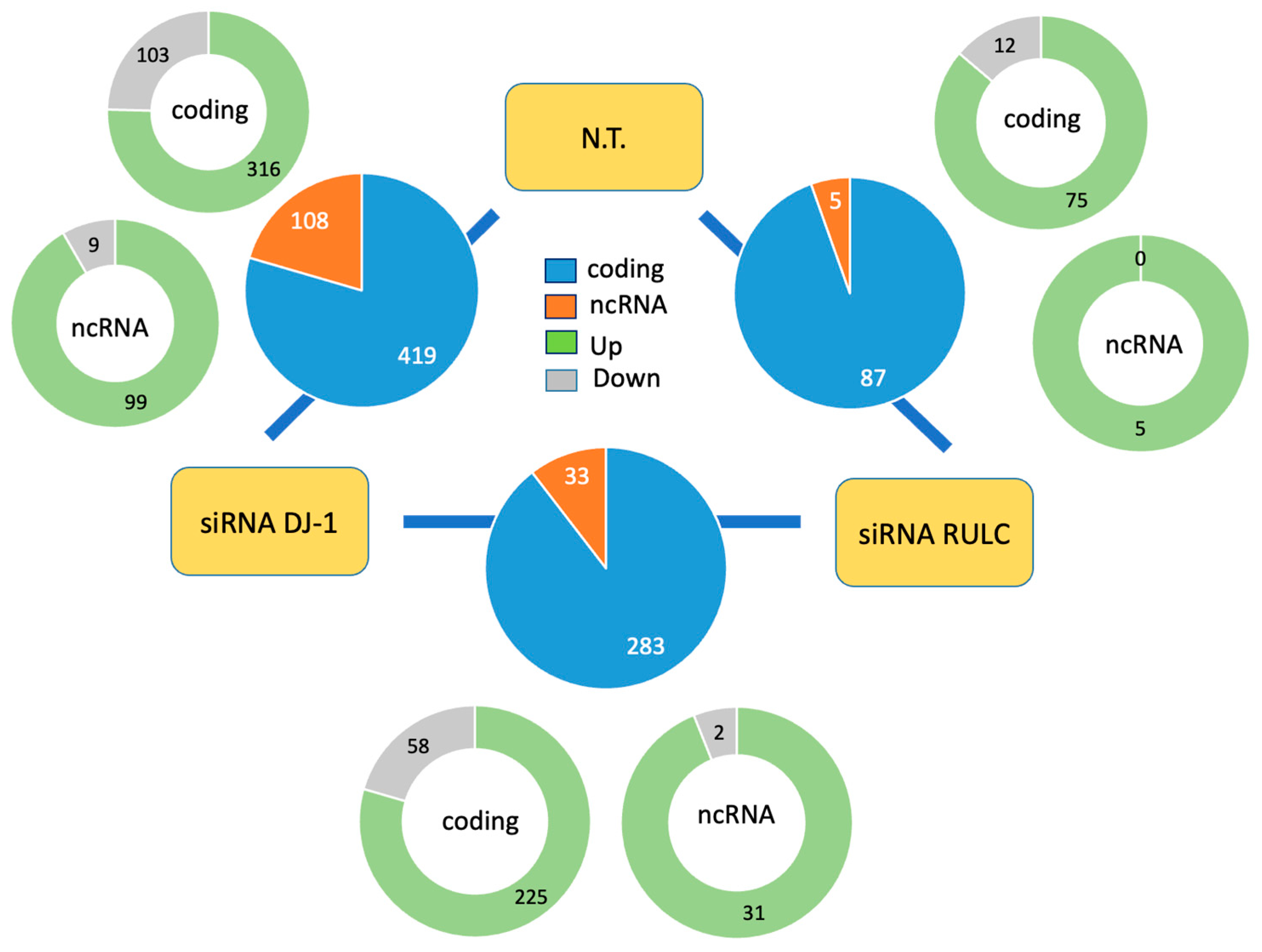
2.3. Knockdown of PARK7/DJ-1 Resulted in Coding and Non-Coding Differential Gene Expression
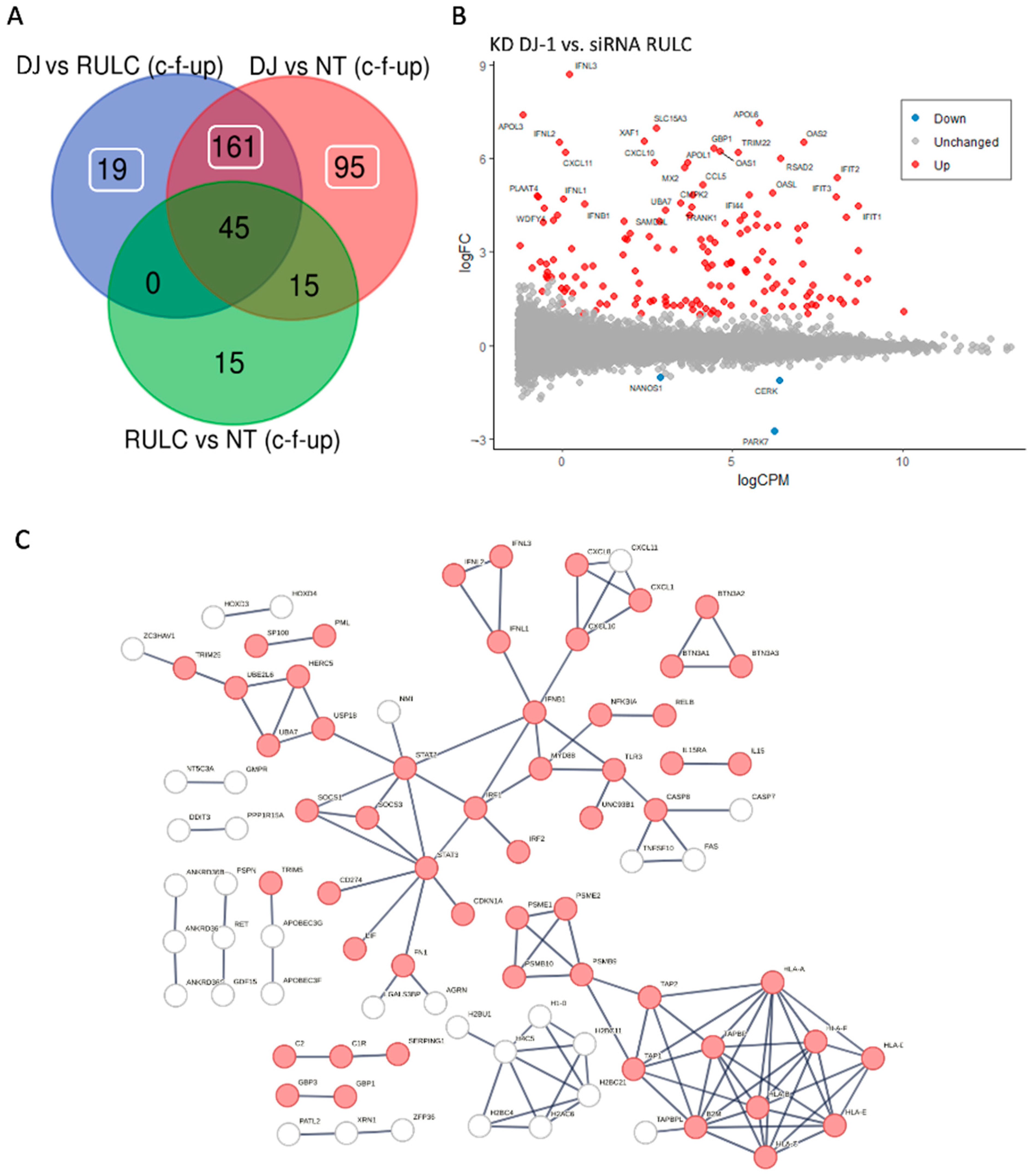
2.4. Non-Specific siRNA Induces Interferon Signaling and the Antiviral Response
2.5. Knockdown of PARK7/DJ-1 Strongly Boosted the Antiviral Response
2.6. Knockdown of PARK7/DJ-1 Exposed Downregulation of Genes with Membrane-Associated Functions
3. Methods
3.1. Cell Lines and siRNAs
3.2. Reverse Transcription PCR (RT-PCR) and PCR
3.3. RNA-Seq
3.4. Bioinformatic Analysis
3.5. Statistical Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| BSA | Bovine serum albumin |
| CNS | Central nervous system |
| DEG | Differentially expressed gene |
| DMEM | Dulbecco’s modified Eagle medium |
| FC | Fold change |
| FDR | False discovery rate |
| HPA | Human proteome atlas |
| GO | Gene ontology |
| IFN | Interferon |
| NT | Not treated |
| PCA | Principal component analysis |
| PCR | Polymerase chain reaction |
| PD | Parkinson’s disease |
| RNA-seq | RNA sequencing analysis |
| RT | Reverse transcription |
| TMM | Trimmed mean of means |
References
- Biosa, A.; Sandrelli, F.; Beltramini, M.; Greggio, E.; Bubacco, L.; Bisaglia, M. Recent findings on the physiological function of DJ-1: Beyond Parkinson’s disease. Neurobiol. Dis. 2017, 108, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Ishimori, C.; Takahashi-Niki, K.; Taira, T.; Kim, Y.-C.; Maita, H.; Maita, C.; Ariga, H.; Iguchi-Ariga, S.M. DJ-1 binds to mitochondrial complex I and maintains its activity. Biochem. Biophys. Res. Commun. 2009, 390, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Hijioka, M.; Inden, M.; Yanagisawa, D.; Kitamura, Y. DJ-1/PARK7: A new therapeutic target for neurodegenerative disorders. Biol. Pharm. Bull. 2017, 40, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Yokota, T.; Sugawara, K.; Ito, K.; Takahashi, R.; Ariga, H.; Mizusawa, H. Down regulation of DJ-1 enhances cell death by oxidative stress, ER stress, and proteasome inhibition. Biochem. Biophys. Res. Commun. 2003, 312, 1342–1348. [Google Scholar] [CrossRef] [PubMed]
- Takahashi-Niki, K.; Niki, T.; Iguchi-Ariga, S.M.; Ariga, H. Transcriptional regulation of DJ-1. In DJ-1/PARK7 Protein: Parkinson’s Disease, Cancer and Oxidative Stress-Induced Diseases; Springer: Singapore, 2017; pp. 89–95. [Google Scholar]
- Tsoporis, J.N.; Drosatos, I.-A.; Gupta, S.; Amatullah, H.; Izhar, S.; Dos Santos, C.C.; Salpeas, V.; Rigopoulos, A.G.; Toumpoulis, I.K.; Triantafyllis, A.S. Cytoprotective mechanisms of DJ-1: Implications in cardiac pathophysiology. Molecules 2021, 26, 3795. [Google Scholar] [CrossRef] [PubMed]
- Neves, M.; Grãos, M.; Anjo, S.I.; Manadas, B. Modulation of signaling pathways by DJ-1: An updated overview. Redox Biol. 2022, 51, 102283. [Google Scholar] [CrossRef] [PubMed]
- Raninga, P.V.; Di Trapani, G.; Tonissen, K.F. The multifaceted roles of DJ-1 as an antioxidant. In DJ-1/PARK7 Protein: Parkinson’s Disease, Cancer and Oxidative Stress-Induced Diseases; Springer: Singapore, 2017; pp. 67–87. [Google Scholar]
- Liu, F.; Nguyen, J.L.; Hulleman, J.D.; Li, L.; Rochet, J.C. Mechanisms of DJ-1 neuroprotection in a cellular model of Parkinson’s disease. J. Neurochem. 2008, 105, 2435–2453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-L.; Wang, Z.-Z.; Shao, Q.-H.; Zhang, Z.; Li, L.; Guo, Z.-Y.; Sun, H.-M.; Zhang, Y.; Chen, N.-H. RNAi-mediated knockdown of DJ-1 leads to mitochondrial dysfunction via Akt/GSK-3ß and JNK signaling pathways in dopaminergic neuron-like cells. Brain Res. Bull. 2019, 146, 228–236. [Google Scholar] [CrossRef]
- Zohar, K.; Lezmi, E.; Eliyahu, T.; Linial, M. Ladostigil attenuates induced oxidative stress in human neuroblast-like SH-SY5Y cells. Biomedicines 2021, 9, 1251. [Google Scholar] [CrossRef]
- Im, J.-Y.; Lee, K.-W.; Woo, J.-M.; Junn, E.; Mouradian, M.M. DJ-1 induces thioredoxin 1 expression through the Nrf2 pathway. Hum. Mol. Genet. 2012, 21, 3013–3024. [Google Scholar] [CrossRef]
- Yanagida, T.; Tsushima, J.; Kitamura, Y.; Yanagisawa, D.; Takata, K.; Shibaike, T.; Yamamoto, A.; Taniguchi, T.; Yasui, H.; Taira, T. Oxidative stress induction of DJ-1 protein in reactive astrocytes scavenges free radicals and reduces cell injury. Oxidative Med. Cell. Longev. 2009, 2, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wu, R.; Zhang, Q.; Wu, J.-B.; Lou, J.; Zheng, Z.; Ding, J.-Q.; Yuan, Z. DJ-1 interacts with RACK1 and protects neurons from oxidative-stress-induced apoptosis. Biochem. J. 2014, 462, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Bahmed, K.; Boukhenouna, S.; Karim, L.; Andrews, T.; Lin, J.; Powers, R.; Wilson, M.A.; Lin, C.-R.; Messier, E.; Reisdorph, N. The effect of cysteine oxidation on DJ-1 cytoprotective function in human alveolar type II cells. Cell Death Dis. 2019, 10, 638. [Google Scholar] [CrossRef]
- Taira, T.; Saito, Y.; Niki, T.; Iguchi-Ariga, S.M.; Takahashi, K.; Ariga, H. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004, 5, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Inberg, A.; Linial, M. Protection of pancreatic beta-cells from various stress conditions is mediated by DJ-1. J. Biol. Chem. 2010, 285, 25686–25698. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Liao, S.-D.; Shi, J.-J.; Chang, L.-L.; Tong, Y.-G.; Cao, J.; Fu, Y.-Y.; Chen, X.-P.; Ying, M.-D.; Yang, B. DJ-1 mediates the resistance of cancer cells to dihydroartemisinin through reactive oxygen species removal. Free. Radic. Biol. Med. 2014, 71, 121–132. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, B.R.; Rocha, E.M.; Bai, Q.; El Ayadi, A.; Hinkle, D.; Burton, E.A.; Greenamyre, J.T. Astrocyte-specific DJ-1 overexpression protects against rotenone-induced neurotoxicity in a rat model of Parkinson’s disease. Neurobiol. Dis. 2018, 115, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.; Ambrosi, G.; Mullett, S.; Berman, S.; Hinkle, D. DJ-1 knock-down impairs astrocyte mitochondrial function. Neuroscience 2011, 196, 251–264. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cao, J.; Lou, S.; Ying, M.; Yang, B. DJ-1 as a human oncogene and potential therapeutic target. Biochem. Pharmacol. 2015, 93, 241–250. [Google Scholar] [CrossRef]
- Ren, H.; Fu, K.; Mu, C.; Li, B.; Wang, D.; Wang, G. DJ-1, a cancer and Parkinson’s disease associated protein, regulates autophagy through JNK pathway in cancer cells. Cancer Lett. 2010, 297, 101–108. [Google Scholar] [CrossRef]
- McCoy, M.K.; Cookson, M.R. DJ-1 regulation of mitochondrial function and autophagy through oxidative stress. Autophagy 2011, 7, 531–532. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Lu, A.; Ke, M.; Zhu, W.; Ye, X.; Wang, G.; Weng, G. DJ-1 inhibits autophagy activity of prostate cancer cells by repressing JNK–Bcl2–Beclin1 signaling. Cell Biol. Int. 2020, 44, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Luo, C.; Lin, K.; Bu, F.; Ye, F.; Huang, C.; Luo, H.; Huang, J.; Zhu, Z. Overexpression of DJ-1 enhances colorectal cancer cell proliferation through the cyclin-D1/MDM2-p53 signaling pathway. Biosci. Trends 2020, 14, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhou, C.; Lu, X.; Liu, Q.; Liu, M.; Chen, G.; Chen, W.; Wang, S.; Qiu, Y. DJ-1 promotes survival of human colon cancer cells under hypoxia by modulating HIF-1α expression through the PI3K-AKT pathway. Cancer Manag. Res. 2018, 10, 4615–4629. [Google Scholar] [CrossRef] [PubMed]
- Olivo, E.; La Chimia, M.; Ceramella, J.; Catalano, A.; Chiaradonna, F.; Sinicropi, M.S.; Cuda, G.; Iacopetta, D.; Scumaci, D. Moving beyond the tip of the iceberg: DJ-1 implications in cancer metabolism. Cells 2022, 11, 1432. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, J.; Wang, J.; Yang, B.; He, Q.; Weng, Q. Role of DJ-1 in immune and inflammatory diseases. Front. Immunol. 2020, 11, 529121. [Google Scholar] [CrossRef]
- Mullett, S.J.; Hinkle, D.A. DJ-1 knock-down in astrocytes impairs astrocyte-mediated neuroprotection against rotenone. Neurobiol. Dis. 2009, 33, 28–36. [Google Scholar] [CrossRef]
- Kim, J.-H.; Choi, D.-J.; Jeong, H.-K.; Kim, J.; Kim, D.W.; Choi, S.Y.; Park, S.-M.; Suh, Y.H.; Jou, I.; Joe, E.-H. DJ-1 facilitates the interaction between STAT1 and its phosphatase, SHP-1, in brain microglia and astrocytes: A novel anti-inflammatory function of DJ-1. Neurobiol. Dis. 2013, 60, 1–10. [Google Scholar] [CrossRef]
- Milde, S.; van Tartwijk, F.W.; Vilalta, A.; Hornik, T.C.; Dundee, J.M.; Puigdellivol, M.; Brown, G.C. Inflammatory neuronal loss in the substantia nigra induced by systemic lipopolysaccharide is prevented by knockout of the P2Y(6) receptor in mice. J. Neuroinflamm. 2021, 18, 225. [Google Scholar] [CrossRef]
- Oh, S.E.; Mouradian, M.M. Regulation of Signal Transduction by DJ-1. Adv. Exp. Med. Biol. 2017, 1037, 97–131. [Google Scholar]
- Liu, T.; Castro, S.; Brasier, A.R.; Jamaluddin, M.; Garofalo, R.P.; Casola, A. Reactive oxygen species mediate virus-induced STAT activation: Role of tyrosine phosphatases. J. Biol. Chem. 2004, 279, 2461–2469. [Google Scholar] [CrossRef]
- Simon, A.R.; Rai, U.; Fanburg, B.L.; Cochran, B.H. Activation of the JAK-STAT pathway by reactive oxygen species. Am. J. Physiol. 1998, 275, C1640–C1652. [Google Scholar] [CrossRef]
- Jomova, K.; Raptova, R.; Alomar, S.Y.; Alwasel, S.H.; Nepovimova, E.; Kuca, K.; Valko, M. Reactive oxygen species, toxicity, oxidative stress, and antioxidants: Chronic diseases and aging. Arch. Toxicol. 2023, 97, 2499–2574. [Google Scholar] [CrossRef]
- Liu, S.; Li, Z.; Lan, S.; Hao, H.; Baz, A.A.; Yan, X.; Gao, P.; Chen, S.; Chu, Y. The Dual Roles of Activating Transcription Factor 3 (ATF3) in Inflammation, Apoptosis, Ferroptosis, and Pathogen Infection Responses. Int. J. Mol. Sci. 2024, 25, 824. [Google Scholar] [CrossRef]
- Sheridan, M.; Ogretmen, B. The Role of Ceramide Metabolism and Signaling in the Regulation of Mitophagy and Cancer Therapy. Cancers 2021, 13, 2475. [Google Scholar] [CrossRef]
- Onishi, M.; Nagumo, S.; Iwashita, S.; Okamoto, K. The ER membrane insertase Get1/2 is required for efficient mitophagy in yeast. Biochem. Biophys. Res. Commun. 2018, 503, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wang, J.E.; Yu, L.; Cui, B.; Wang, H.; Xiao, X.; Zhang, Y.; Zheng, J.; Wang, J.; Hui, R. ANKRD36 is involved in hypertension by altering expression of ENaC genes. Circ. Res. 2021, 129, 1067–1081. [Google Scholar] [CrossRef]
- Iqbal, Z.; Absar, M.; Akhtar, T.; Aleem, A.; Jameel, A.; Basit, S.; Ullah, A.; Afzal, S.; Ramzan, K.; Rasool, M.; et al. Integrated Genomic Analysis Identifies ANKRD36 Gene as a Novel and Common Biomarker of Disease Progression in Chronic Myeloid Leukemia. Biology 2021, 10, 1182. [Google Scholar] [CrossRef] [PubMed]
- Azzam, T.; Raskin, A.; Makovitzki, A.; Brem, H.; Vierling, P.; Lineal, M.; Domb, A.J. Cationic polysaccharides for gene delivery. Macromolecules 2002, 35, 9947–9953. [Google Scholar] [CrossRef]
- Hunt, M.A.; Currie, M.J.; Robinson, B.A.; Dachs, G.U. Optimizing transfection of primary human umbilical vein endothelial cells using commercially available chemical transfection reagents. J. Biomol. Tech. 2010, 21, 66–72. [Google Scholar]
- Del Toro, N.; Shrivastava, A.; Ragueneau, E.; Meldal, B.; Combe, C.; Barrera, E.; Perfetto, L.; How, K.; Ratan, P.; Shirodkar, G. The IntAct database: Efficient access to fine-grained molecular interaction data. Nucleic Acids Res. 2022, 50, D648–D653. [Google Scholar] [CrossRef] [PubMed]
- Digre, A.; Lindskog, C. The Human Protein Atlas—Spatial localization of the human proteome in health and disease. Protein Sci. 2021, 30, 218–233. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis; Taylor & Francis: Abingdon, UK, 2019. [Google Scholar]
- Reynolds, A.; Anderson, E.M.; Vermeulen, A.; Fedorov, Y.; Robinson, K.; Leake, D.; Karpilow, J.; Marshall, W.S.; Khvorova, A. Induction of the interferon response by siRNA is cell type–and duplex length–dependent. RNA 2006, 12, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, C.; Wang, Z.; Shen, J.; Xiang, P.; Chen, X.; Nan, J.; Lin, Y. Lipofectamine 2000/siRNA complexes cause endoplasmic reticulum unfolded protein response in human endothelial cells. J. Cell Physiol. 2019, 234, 21166–21181. [Google Scholar] [CrossRef] [PubMed]
- Forsbach, A.; Nemorin, J.-G.; Montino, C.; Muller, C.; Samulowitz, U.; Vicari, A.P.; Jurk, M.; Mutwiri, G.K.; Krieg, A.M.; Lipford, G.B. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J. Immunol. 2008, 180, 3729–3738. [Google Scholar] [CrossRef] [PubMed]
- Nowicka, H.; Sekrecka, A.; Blaszczyk, K.; Kluzek, K.; Chang, C.Y.; Wesoly, J.; Lee, C.K.; Bluyssen, H.A.R. ISGF3 and STAT2/IRF9 Control Basal and IFN-Induced Transcription through Genome-Wide Binding of Phosphorylated and Unphosphorylated Complexes to Common ISRE-Containing ISGs. Int. J. Mol. Sci. 2023, 24, 17635. [Google Scholar] [CrossRef] [PubMed]
- Sledz, C.A.; Holko, M.; de Veer, M.J.; Silverman, R.H.; Williams, B.R. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003, 5, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Irrcher, I.; Aleyasin, H.; Seifert, E.; Hewitt, S.; Chhabra, S.; Phillips, M.; Lutz, A.; Rousseaux, M.; Bevilacqua, L.; Jahani-Asl, A. Loss of the Parkinson’s disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum. Mol. Genet. 2010, 19, 3734–3746. [Google Scholar] [CrossRef]
- Dodson, M.W.; Guo, M. Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson’s disease. Curr. Opin. Neurobiol. 2007, 17, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Zohar, K.; Giladi, E.; Eliyahu, T.; Linial, M. Alteration in long noncoding RNAs in response to oxidative stress and ladostigil in SH-SY5Y cells. bioRxiv 2022. [Google Scholar] [CrossRef]
- Boros, F.A.; Maszlag-Török, R.; Vécsei, L.; Klivényi, P. Increased level of NEAT1 long non-coding RNA is detectable in peripheral blood cells of patients with Parkinson’s disease. Brain Res. 2020, 1730, 146672. [Google Scholar] [CrossRef]
- Schubert, M.; Klinger, B.; Klunemann, M.; Sieber, A.; Uhlitz, F.; Sauer, S.; Garnett, M.J.; Bluthgen, N.; Saez-Rodriguez, J. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 2018, 9, 20. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zohar, K.; Linial, M. Knockdown of DJ-1 Resulted in a Coordinated Activation of the Innate Immune Antiviral Response in HEK293 Cell Line. Int. J. Mol. Sci. 2024, 25, 7550. https://doi.org/10.3390/ijms25147550
Zohar K, Linial M. Knockdown of DJ-1 Resulted in a Coordinated Activation of the Innate Immune Antiviral Response in HEK293 Cell Line. International Journal of Molecular Sciences. 2024; 25(14):7550. https://doi.org/10.3390/ijms25147550
Chicago/Turabian StyleZohar, Keren, and Michal Linial. 2024. "Knockdown of DJ-1 Resulted in a Coordinated Activation of the Innate Immune Antiviral Response in HEK293 Cell Line" International Journal of Molecular Sciences 25, no. 14: 7550. https://doi.org/10.3390/ijms25147550
APA StyleZohar, K., & Linial, M. (2024). Knockdown of DJ-1 Resulted in a Coordinated Activation of the Innate Immune Antiviral Response in HEK293 Cell Line. International Journal of Molecular Sciences, 25(14), 7550. https://doi.org/10.3390/ijms25147550






