A Comparative Analysis of Two Automated Quantification Methods for Regional Cerebral Amyloid Retention: PET-Only and PET-and-MRI-Based Methods
Abstract
1. Introduction
2. Results
2.1. Demographics and Clinical Characteristics
2.2. Consistency of Amyloid PET SUVR between Two Quantification Methods
2.3. Difference in Amyloid PET SUVR between Two Quantification Methods
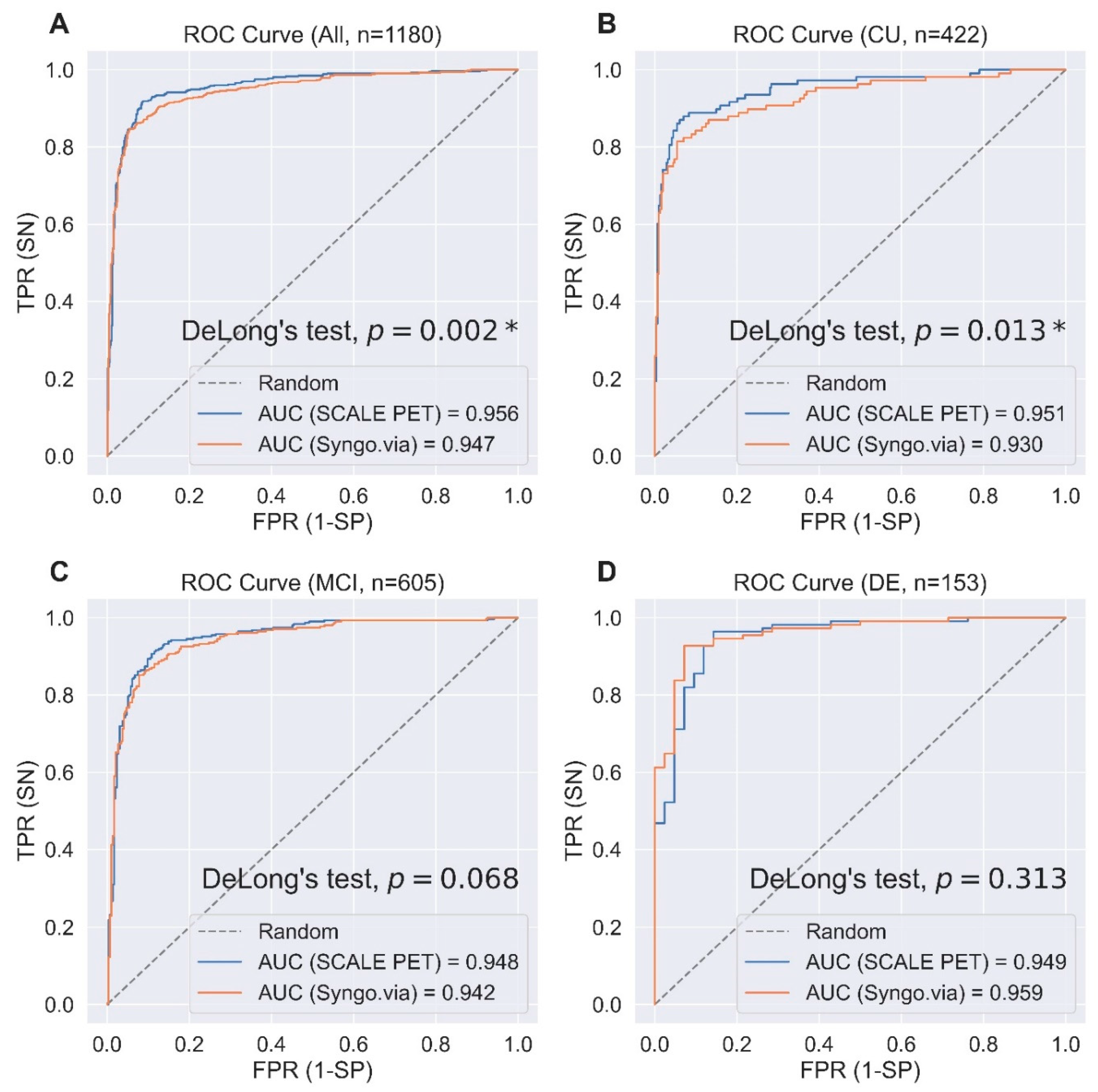
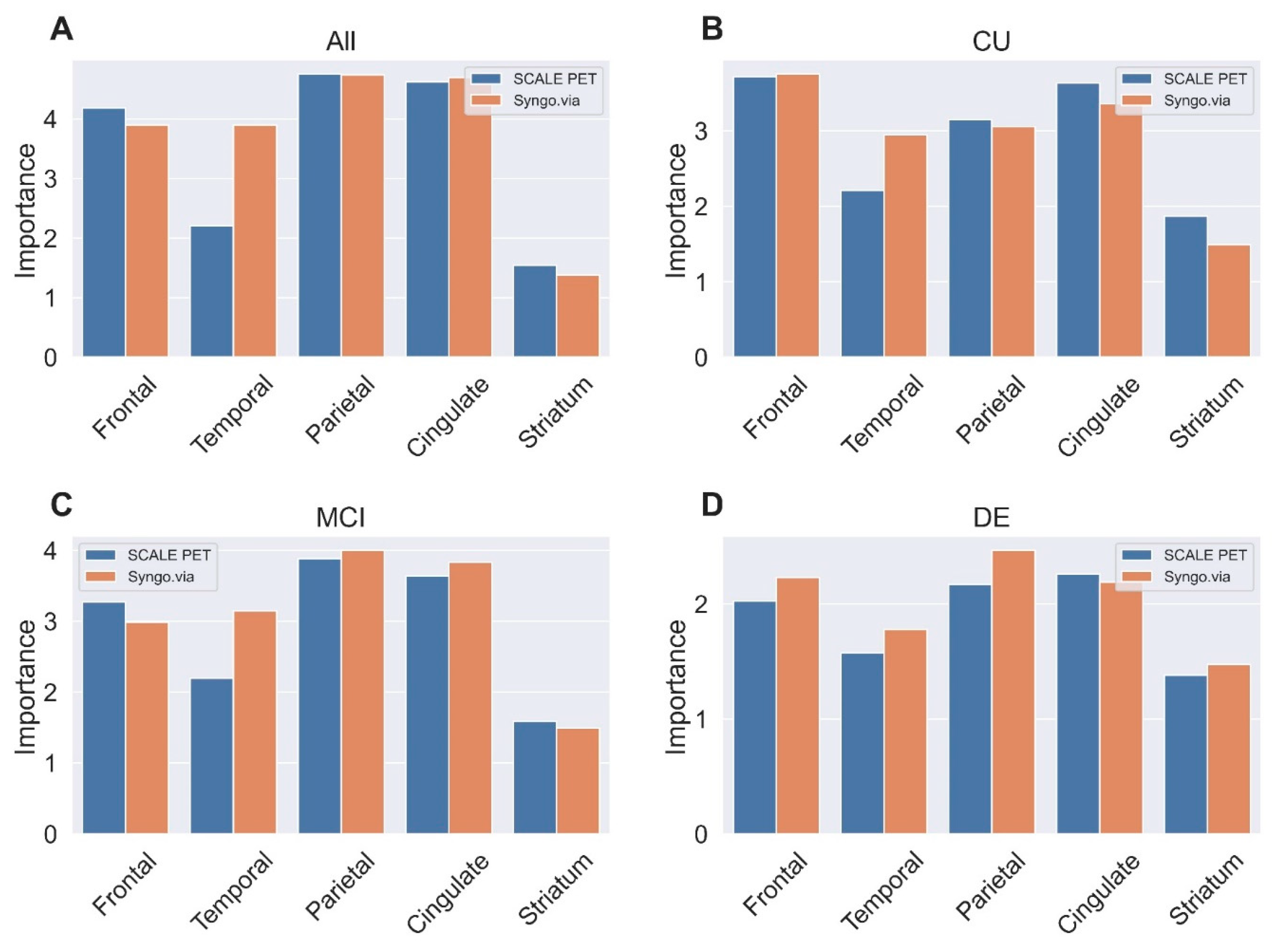
2.4. Predicting Visual Reads for Aβ-Positivity Using Regional SUVRs
3. Discussion
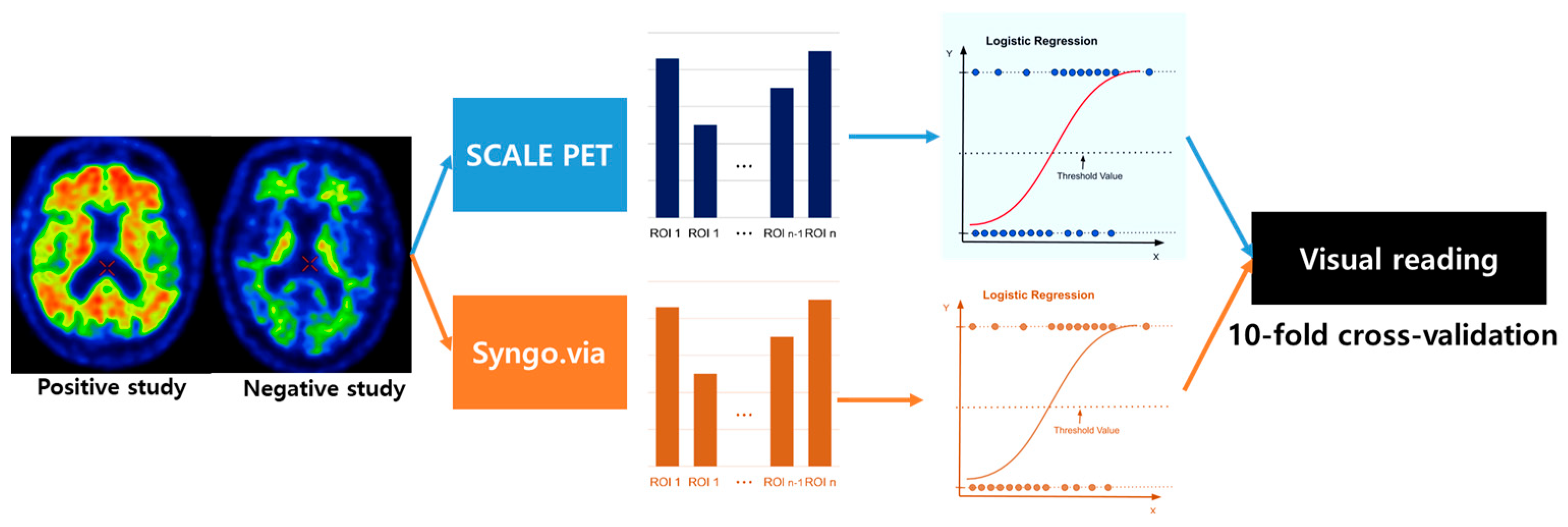
4. Materials and Methods
4.1. Study Participants
4.2. Amyloid PET and MRI Image Acquisition
4.3. Visual Analysis of Amyloid PET
4.4. Quantitative Assessment of Aβ Deposition
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- GBD 2019 Collaborators. Global mortality from dementia: Application of a new method and results from the Global Burden of Disease Study 2019. Alzheimer’s Dement. 2021, 7, e12200. [Google Scholar] [CrossRef]
- Joe, E.; Ringman, J.M. Cognitive symptoms of Alzheimer’s disease: Clinical management and prevention. BMJ 2019, 367, l6217. [Google Scholar] [CrossRef]
- Lyketsos, C.G.; Carrillo, M.C.; Ryan, J.M.; Khachaturian, A.S.; Trzepacz, P.; Amatniek, J.; Cedarbaum, J.; Brashear, R.; Miller, D.S. Neuropsychiatric Symptoms in Alzheimer’s Disease; Elsevier: Amsterdam, The Netherlands, 2011; Volume 7, pp. 532–539. [Google Scholar]
- Dubois, B.; Hampel, H.; Feldman, H.H.; Scheltens, P.; Aisen, P.; Andrieu, S.; Bakardjian, H.; Benali, H.; Bertram, L.; Blennow, K. Preclinical Alzheimer’s disease: Definition, natural history, and diagnostic criteria. Alzheimer’s Dement. 2016, 12, 292–323. [Google Scholar] [CrossRef]
- Zetterberg, H.; Bendlin, B.B. Biomarkers for Alzheimer’s disease—Preparing for a new era of disease-modifying therapies. Mol. Psychiatry 2021, 26, 296–308. [Google Scholar] [CrossRef]
- Dubois, B.; von Arnim, C.A.F.; Burnie, N.; Bozeat, S.; Cummings, J. Biomarkers in Alzheimer’s disease: Role in early and differential diagnosis and recognition of atypical variants. Alzheimer’s Res. Ther. 2023, 15, 175. [Google Scholar] [CrossRef]
- Jack Jr, C.R.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J. NIA-AA research framework: Toward a biological definition of Alzheimer’s disease. Alzheimer’s Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Um, Y.H.; Choi, W.H.; Jung, W.S.; Park, Y.H.; Lee, C.-U.; Lim, H.K. Whole brain voxel-wise analysis of cerebral retention of Beta-amyloid in cognitively normal older adults using 18F-florbetaben. Psychiatry Investig. 2017, 14, 883. [Google Scholar] [CrossRef]
- Jack, C.R.; Knopman, D.S.; Jagust, W.J.; Petersen, R.C.; Weiner, M.W.; Aisen, P.S.; Shaw, L.M.; Vemuri, P.; Wiste, H.J.; Weigand, S.D. Tracking pathophysiological processes in Alzheimer’s disease: An updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013, 12, 207–216. [Google Scholar] [CrossRef]
- Jack, C.R.; Knopman, D.S.; Jagust, W.J.; Shaw, L.M.; Aisen, P.S.; Weiner, M.W.; Petersen, R.C.; Trojanowski, J.Q. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010, 9, 119–128. [Google Scholar] [CrossRef]
- Jia, J.; Ning, Y.; Chen, M.; Wang, S.; Yang, H.; Li, F.; Ding, J.; Li, Y.; Zhao, B.; Lyu, J. Biomarker Changes during 20 Years Preceding Alzheimer’s Disease. N. Engl. J. Med. 2024, 390, 712–722. [Google Scholar] [CrossRef]
- Mintun, M.A.; Lo, A.C.; Duggan Evans, C.; Wessels, A.M.; Ardayfio, P.A.; Andersen, S.W.; Shcherbinin, S.; Sparks, J.; Sims, J.R.; Brys, M. Donanemab in early Alzheimer’s disease. N. Engl. J. Med. 2021, 384, 1691–1704. [Google Scholar] [CrossRef]
- Van Dyck, C.H.; Swanson, C.J.; Aisen, P.; Bateman, R.J.; Chen, C.; Gee, M.; Kanekiyo, M.; Li, D.; Reyderman, L.; Cohen, S. Lecanemab in early Alzheimer’s disease. N. Engl. J. Med. 2023, 388, 9–21. [Google Scholar] [CrossRef]
- Lowe, V.J.; Lundt, E.S.; Albertson, S.M.; Przybelski, S.A.; Senjem, M.L.; Parisi, J.E.; Kantarci, K.; Boeve, B.; Jones, D.T.; Knopman, D.; et al. Neuroimaging correlates with neuropathologic schemes in neurodegenerative disease. Alzheimer’s Dement. 2019, 15, 927–939. [Google Scholar] [CrossRef]
- Thal, D.R.; Beach, T.G.; Zanette, M.; Heurling, K.; Chakrabarty, A.; Ismail, A.; Smith, A.P.; Buckley, C. [(18)F]flutemetamol amyloid positron emission tomography in preclinical and symptomatic Alzheimer’s disease: Specific detection of advanced phases of amyloid-β pathology. Alzheimer’s Dement. 2015, 11, 975–985. [Google Scholar] [CrossRef]
- Olsson, B.; Lautner, R.; Andreasson, U.; Öhrfelt, A.; Portelius, E.; Bjerke, M.; Hölttä, M.; Rosén, C.; Olsson, C.; Strobel, G. CSF and blood biomarkers for the diagnosis of Alzheimer’s disease: A systematic review and meta-analysis. Lancet Neurol. 2016, 15, 673–684. [Google Scholar] [CrossRef]
- Ben Bouallègue, F.; Mariano-Goulart, D.; Payoux, P.; the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Comparison of CSF markers and semi-quantitative amyloid PET in Alzheimer’s disease diagnosis and in cognitive impairment prognosis using the ADNI-2 database. Alzheimer’s Res. Ther. 2017, 9, 32. [Google Scholar] [CrossRef]
- O’Bryant, S.E.; Petersen, M.; Hall, J.; Johnson, L.A.; Team, H.-H.S. Medical comorbidities and ethnicity impact plasma Alzheimer’s disease biomarkers: Important considerations for clinical trials and practice. Alzheimer’s Dement. 2023, 19, 36–43. [Google Scholar] [CrossRef]
- Bonomi, S.; Lu, R.; Schindler, S.E.; Bui, Q.; Lah, J.J.; Wolk, D.; Gleason, C.E.; Sperling, R.; Roberson, E.D.; Levey, A.I. Relationships of cognitive measures with cerebrospinal fluid but not imaging biomarkers of Alzheimer disease vary between Black and White individuals. Ann. Neurol. 2024, 95, 495–506. [Google Scholar] [CrossRef]
- Cullen, N.C.; Janelidze, S.; Mattsson-Carlgren, N.; Palmqvist, S.; Bittner, T.; Suridjan, I.; Jethwa, A.; Kollmorgen, G.; Brum, W.S.; Zetterberg, H. Test-retest variability of plasma biomarkers in Alzheimer’s disease and its effects on clinical prediction models. Alzheimer’s Dement. 2023, 19, 797–806. [Google Scholar] [CrossRef]
- Pichet Binette, A.; Janelidze, S.; Cullen, N.; Dage, J.L.; Bateman, R.J.; Zetterberg, H.; Blennow, K.; Stomrud, E.; Mattsson-Carlgren, N.; Hansson, O. Confounding factors of Alzheimer’s disease plasma biomarkers and their impact on clinical performance. Alzheimer’s Dement. 2023, 19, 1403–1414. [Google Scholar] [CrossRef]
- Collij, L.E.; Salvadó, G.; Shekari, M.; Lopes Alves, I.; Reimand, J.; Wink, A.M.; Zwan, M.; Niñerola-Baizán, A.; Perissinotti, A.; Scheltens, P.; et al. Visual assessment of [18F]flutemetamol PET images can detect early amyloid pathology and grade its extent. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 2169–2182. [Google Scholar] [CrossRef] [PubMed]
- Haller, S.; Montandon, M.-L.; Lilja, J.; Rodriguez, C.; Garibotto, V.; Herrmann, F.R.; Giannakopoulos, P. PET amyloid in normal aging: Direct comparison of visual and automatic processing methods. Sci. Rep. 2020, 10, 16665. [Google Scholar] [CrossRef] [PubMed]
- Pontecorvo, M.J.; Arora, A.K.; Devine, M.; Lu, M.; Galante, N.; Siderowf, A.; Devadanam, C.; Joshi, A.D.; Heun, S.L.; Teske, B.F.; et al. Quantitation of PET signal as an adjunct to visual interpretation of florbetapir imaging. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 825–837. [Google Scholar] [CrossRef]
- Lopresti, B.J.; Klunk, W.E.; Mathis, C.A.; Hoge, J.A.; Ziolko, S.K.; Lu, X.; Meltzer, C.C.; Schimmel, K.; Tsopelas, N.D.; DeKosky, S.T. Simplified quantification of Pittsburgh Compound B amyloid imaging PET studies: A comparative analysis. J. Nucl. Med. 2005, 46, 1959–1972. [Google Scholar]
- Fleisher, A.S.; Chen, K.; Liu, X.; Roontiva, A.; Thiyyagura, P.; Ayutyanont, N.; Joshi, A.D.; Clark, C.M.; Mintun, M.A.; Pontecorvo, M.J. Using positron emission tomography and florbetapir F 18 to image cortical amyloid in patients with mild cognitive impairment or dementia due to Alzheimer disease. Arch. Neurol. 2011, 68, 1404–1411. [Google Scholar] [CrossRef]
- Cho, S.H.; Choe, Y.S.; Park, S.; Kim, Y.J.; Kim, H.J.; Jang, H.; Kim, S.J.; Kim, J.P.; Jung, Y.H.; Kim, B.C.; et al. Appropriate reference region selection of 18F-florbetaben and 18F-flutemetamol beta-amyloid PET expressed in Centiloid. Sci. Rep. 2020, 10, 14950. [Google Scholar] [CrossRef]
- Klunk, W.E.; Koeppe, R.A.; Price, J.C.; Benzinger, T.L.; Devous Sr, M.D.; Jagust, W.J.; Johnson, K.A.; Mathis, C.A.; Minhas, D.; Pontecorvo, M.J. The Centiloid Project: Standardizing quantitative amyloid plaque estimation by PET. Alzheimer’s Dement. 2015, 11, 1–15.e4. [Google Scholar] [CrossRef]
- Jeong, Y.J.; Yoon, H.J.; Kang, D.-Y.; Park, K.W. Quantitative comparative analysis of amyloid PET images using three radiopharmaceuticals. Ann. Nucl. Med. 2023, 37, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Kolinger, G.D.; Vállez García, D.; Willemsen, A.T.; Reesink, F.E.; de Jong, B.M.; Dierckx, R.A.; De Deyn, P.P.; Boellaard, R. Amyloid burden quantification depends on PET and MR image processing methodology. PLoS ONE 2021, 16, e0248122. [Google Scholar] [CrossRef]
- Ciarmiello, A.; Giovannini, E.; Riondato, M.; Giovacchini, G.; Duce, V.; Ferrando, O.; De Biasi, M.; Passera, C.; Carabelli, E.; Mannironi, A.; et al. Longitudinal cognitive decline in mild cognitive impairment subjects with early amyloid-β neocortical deposition. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2090–2098. [Google Scholar] [CrossRef]
- Harn, N.R.; Hunt, S.L.; Hill, J.; Vidoni, E.; Perry, M.; Burns, J.M. Augmenting Amyloid PET Interpretations With Quantitative Information Improves Consistency of Early Amyloid Detection. Clin. Nucl. Med. 2017, 42, 577–581. [Google Scholar] [CrossRef]
- Akamatsu, G.; Ikari, Y.; Ohnishi, A.; Matsumoto, K.; Nishida, H.; Yamamoto, Y.; Japanese Alzheimer’s Disease Neuroimaging Initiative. Voxel-based statistical analysis and quantification of amyloid PET in the Japanese Alzheimer’s disease neuroimaging initiative (J-ADNI) multi-center study. EJNMMI Res. 2019, 9, 91. [Google Scholar] [CrossRef]
- Pemberton, H.G.; Collij, L.E.; Heeman, F.; Bollack, A.; Shekari, M.; Salvadó, G.; Alves, I.L.; Garcia, D.V.; Battle, M.; Buckley, C. Quantification of amyloid PET for future clinical use: A state-of-the-art review. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 3508–3528. [Google Scholar] [CrossRef]
- Bourgeat, P.; Villemagne, V.L.; Dore, V.; Brown, B.; Macaulay, S.L.; Martins, R.; Masters, C.L.; Ames, D.; Ellis, K.; Rowe, C.C. Comparison of MR-less PiB SUVR quantification methods. Neurobiol. Aging 2015, 36, S159–S166. [Google Scholar] [CrossRef]
- Dale, A.M.; Fischl, B.; Sereno, M.I. Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage 1999, 9, 179–194. [Google Scholar] [CrossRef]
- Fischl, B.; Sereno, M.I.; Dale, A.M. Cortical surface-based analysis: II: Inflation, flattening, and a surface-based coordinate system. Neuroimage 1999, 9, 195–207. [Google Scholar] [CrossRef]
- Fischl, B. FreeSurfer. Neuroimage 2012, 62, 774–781. [Google Scholar] [CrossRef]
- D’agostino, E.; Maes, F.; Vandermeulen, D.; Suetens, P. A viscous fluid model for multimodal non-rigid image registration using mutual information. Med. Image Anal. 2003, 7, 565–575. [Google Scholar] [CrossRef]
- Lee, J.; Ha, S.; Kim, R.E.; Lee, M.; Kim, D.; Lim, H.K. Development of Amyloid PET Analysis Pipeline Using Deep Learning-Based Brain MRI Segmentation—A Comparative Validation Study. Diagnostics 2022, 12, 623. [Google Scholar] [CrossRef]
- Payoux, P.; Delrieu, J.; Gallini, A.; Adel, D.; Salabert, A.S.; Hitzel, A.; Cantet, C.; Tafani, M.; De Verbizier, D.; Darcourt, J.; et al. Cognitive and functional patterns of nondemented subjects with equivocal visual amyloid PET findings. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 1459–1468. [Google Scholar] [CrossRef]
- Son, H.J.; Oh, J.S.; Oh, M.; Kim, S.J.; Lee, J.-H.; Roh, J.H.; Kim, J.S. The clinical feasibility of deep learning-based classification of amyloid PET images in visually equivocal cases. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 332–341. [Google Scholar] [CrossRef]
- Choi, H.; Lee, D.S.; Alzheimer’s Disease Neuroimaging Initiative. Generation of Structural MR Images from Amyloid PET: Application to MR-Less Quantification. J. Nucl. Med. 2018, 59, 1111–1117. [Google Scholar] [CrossRef]
- Kim, J.S.; Cho, H.; Choi, J.Y.; Lee, S.H.; Ryu, Y.H.; Lyoo, C.H.; Lee, M.S. Feasibility of computed tomography-guided methods for spatial normalization of dopamine transporter positron emission tomography image. PLoS ONE 2015, 10, e0132585. [Google Scholar] [CrossRef]
- Tahmi, M.; Bou-Zeid, W.; Razlighi, Q.R. A Fully Automatic Technique for Precise Localization and Quantification of Amyloid-beta PET Scans. J. Nucl. Med. 2019, 60, 1771–1779. [Google Scholar] [CrossRef]
- Thal, D.R.; Rub, U.; Orantes, M.; Braak, H. Phases of A beta-deposition in the human brain and its relevance for the development of AD. Neurology 2002, 58, 1791–1800. [Google Scholar] [CrossRef]
- Dickerson, B.C.; Bakkour, A.; Salat, D.H.; Feczko, E.; Pacheco, J.; Greve, D.N.; Grodstein, F.; Wright, C.I.; Blacker, D.; Rosas, H.D. The cortical signature of Alzheimer’s disease: Regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb. Cortex 2009, 19, 497–510. [Google Scholar] [CrossRef]
- Lecy, E.E.; Min, H.K.; Apgar, C.J.; Maltais, D.D.; Lundt, E.S.; Albertson, S.M.; Senjem, M.L.; Schwarz, C.G.; Botha, H.; Graff-Radford, J.; et al. Patterns of Early Neocortical Amyloid-beta Accumulation: A PET Population-Based Study. J. Nucl. Med. 2024, 65, 1122–1128. [Google Scholar] [CrossRef]
- Collij, L.E.; Salvado, G.; Wottschel, V.; Mastenbroek, S.E.; Schoenmakers, P.; Heeman, F.; Aksman, L.; Wink, A.M.; Berckel, B.N.M.; van de Flier, W.M.; et al. Spatial-Temporal Patterns of beta-Amyloid Accumulation: A Subtype and Stage Inference Model Analysis. Neurology 2022, 98, e1692–e1703. [Google Scholar] [CrossRef]
- Park, C.J.; Seo, Y.; Choe, Y.S.; Jang, H.; Lee, H.; Kim, J.P.; Alzheimer’s Disease Neuroimaging Initiative. Predicting conversion of brain β-amyloid positivity in amyloid-negative individuals. Alzheimer’s Res. Ther. 2022, 14, 129. [Google Scholar] [CrossRef]
- Lim, H.K.; Nebes, R.; Snitz, B.; Cohen, A.; Mathis, C.; Price, J.; Weissfeld, L.; Klunk, W.; Aizenstein, H.J. Regional amyloid burden and intrinsic connectivity networks in cognitively normal elderly subjects. Brain 2014, 137, 3327–3338. [Google Scholar] [CrossRef]
- Zhukovsky, P.; Coughlan, G.; Buckley, R.; Grady, C.; Voineskos, A.N. Connectivity between default mode and frontoparietal networks mediates the association between global amyloid-β and episodic memory. Hum. Brain Mapp. 2023, 44, 1147–1157. [Google Scholar] [CrossRef]
- Ranganath, C.; Ritchey, M. Two cortical systems for memory-guided behaviour. Nat. Rev. Neurosci. 2012, 13, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Márquez, F.; Yassa, M.A. Neuroimaging Biomarkers for Alzheimer’s Disease. Mol. Neurodegener. 2019, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Berron, D.; van Westen, D.; Ossenkoppele, R.; Strandberg, O.; Hansson, O. Medial temporal lobe connectivity and its associations with cognition in early Alzheimer’s disease. Brain 2020, 143, 1233–1248. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. BLA 761269 Accelerated Approval Letter. 7 June 2023. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/appletter/2023/761269Orig1s000ltr.pdf (accessed on 6 July 2024).
- Rafii, M.S.; Sperling, R.A.; Donohue, M.C.; Zhou, J.; Roberts, C.; Irizarry, M.C.; Dhadda, S.; Sethuraman, G.; Kramer, L.D.; Swanson, C.J.; et al. The AHEAD 3-45 Study: Design of a prevention trial for Alzheimer’s disease. Alzheimer’s Dement 2023, 19, 1227–1233. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Guo, J.L.; McBride, J.D.; Narasimhan, S.; Kim, H.; Changolkar, L.; Zhang, B.; Gathagan, R.J.; Yue, C.; Dengler, C.; et al. Amyloid-beta plaques enhance Alzheimer’s brain tau-seeded pathologies by facilitating neuritic plaque tau aggregation. Nat. Med. 2018, 24, 29–38. [Google Scholar] [CrossRef]
- Sperling, R.; Mormino, E.; Johnson, K. The evolution of preclinical Alzheimer’s disease: Implications for prevention trials. Neuron 2014, 84, 608–622. [Google Scholar] [CrossRef]
- Verger, A.; Yakushev, I.; Albert, N.L.; van Berckel, B.; Brendel, M.; Cecchin, D.; Fernandez, P.A.; Fraioli, F.; Guedj, E.; Morbelli, S.; et al. FDA approval of lecanemab: The real start of widespread amyloid PET use?—The EANM Neuroimaging Committee perspective. Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 1553–1555. [Google Scholar] [CrossRef]
- Zhang, L.; Geisler, T.; Ray, H.; Xie, Y. Improving logistic regression on the imbalanced data by a novel penalized log-likelihood function. J. Appl. Stat. 2022, 49, 3257–3277. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, K.U.; Lee, D.Y.; Kim, K.W.; Jhoo, J.H.; Kim, J.H.; Lee, K.H.; Kim, S.Y.; Han, S.H.; Woo, J.I. Development of the Korean version of the Consortium to Establish a Registry for Alzheimer’s Disease Assessment Packet (CERAD-K): Clinical and neuropsychological assessment batteries. J. Gerontol. B Psychol. Sci. Soc. Sci. 2002, 57, P47–P53. [Google Scholar] [CrossRef]
- Tay, L.; Lim, W.S.; Chan, M.; Ali, N.; Mahanum, S.; Chew, P.; Lim, J.; Chong, M.S. New DSM-V neurocognitive disorders criteria and their impact on diagnostic classifications of mild cognitive impairment and dementia in a memory clinic setting. Am. J. Geriatr. Psychiatry 2015, 23, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Morgan, P.S.; Ashburner, J.; Smith, J.; Rorden, C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods 2016, 264, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Siemens. Quantitative Software Evaluation of Beta-Amyloid Brain Pet Imaging in Dementia: White Paper; Siemens Medical Solutions Inc.: Erlangen, Germany, 2015. [Google Scholar]
- Desikan, R.S.; Ségonne, F.; Fischl, B.; Quinn, B.T.; Dickerson, B.C.; Blacker, D.; Buckner, R.L.; Dale, A.M.; Maguire, R.P.; Hyman, B.T. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006, 31, 968–980. [Google Scholar] [CrossRef] [PubMed]
- Landau, S.; Jagust, W. Florbetapir Processing Methods. 2015. Available online: https://adni.bitbucket.io/reference/docs/UCBERKELEYAV45/ADNI_AV45_Methods_JagustLab_06.25.15.pdf (accessed on 10 June 2024).
- Thurfjell, L.; Lilja, J.; Lundqvist, R.; Buckley, C.; Smith, A.; Vandenberghe, R.; Sherwin, P. Automated quantification of 18F-flutemetamol PET activity for categorizing scans as negative or positive for brain amyloid: Concordance with visual image reads. J. Nucl. Med. 2014, 55, 1623–1628. [Google Scholar] [CrossRef] [PubMed]
- Muller, E.G.; Stokke, C.; Stokmo, H.L.; Edwin, T.H.; Knapskog, A.B.; Revheim, M.E. Evaluation of semi-quantitative measures of (18)F-flutemetamol PET for the clinical diagnosis of Alzheimer’s disease. Quant. Imaging Med. Surg. 2022, 12, 493–509. [Google Scholar] [CrossRef]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Sun, X.; Xu, W. Fast implementation of DeLong’s algorithm for comparing the areas under correlated receiver operating characteristic curves. IEEE Signal Process. Lett. 2014, 21, 1389–1393. [Google Scholar] [CrossRef]
- Klunk, W.E. Imaging brain amyloid in Alzheimer’s disease with Pittsburgh Compound-B. Ann. Neurol. Off. J. Am. Neurol. Assoc. Child Neurol. Soc. 2004, 55, 306–319. [Google Scholar] [CrossRef]
- Villemagne, V.L.; Doré, V.; Burnham, S.C.; Masters, C.L.; Rowe, C.C. Imaging tau and amyloid-β proteinopathies in Alzheimer disease and other conditions. Nat. Rev. Neurol. 2018, 14, 225–236. [Google Scholar] [CrossRef]
- Hosmer, D.W., Jr.; Lemeshow, S.; Sturdivant, R.X. Applied Logistic Regression; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Vapnik, V.N. The support vector method. In International Conference on Artificial Neural Networks; Springer: Berlin/Heidelberg, Germany, 1997. [Google Scholar]


| Amyloid Negative (n = 651) | Amyloid Positive (n = 539) | Statistics (p-Value) | |
|---|---|---|---|
| Age, years | 72.84 ± 9.40 | 76.18 ± 7.99 | t = −6.49 (p < 0.001 *) |
| Education, years | 10.71 ± 5.19 | 10.21 ± 5.40 | t = 1.61 (p = 0.109) |
| Sex, n (%) | χ2 = 0.57 (p = 0.452) | ||
| Female | 461 (70.8%) | 363 (68.6%) | |
| Male | 190 (29.2%) | 166 (31.4%) | |
| Diagnosis, n (%) | χ2 = 120.73 (p < 0.001 *) | ||
| CU | 314 (48.2%) | 108 (20.4%) | |
| MCI | 295 (45.3%) | 310 (58.6%) | |
| DE | 42 (6.5%) | 111 (21.0%) | |
| APOE | χ2 = 134.18 (p < 0.001 *) | ||
| ε4 carrier, n (%) | 127 (19.5%) | 274 (51.8%) | |
| ε4 non-carrier, n (%) | 524 (80.5%) | 255 (48.2%) | |
| CDR | 0.31 ± 0.32 | 0.58 ± 0.42 | t = −12.44 (p < 0.001 *) |
| CDR-SB | 1.28 ± 1.86 | 2.83 ± 2.84 | t = −11.28 (p < 0.001 *) |
| Method | Accuracy | Sensitivity | Specificity | F1 Score | AUROC | |
|---|---|---|---|---|---|---|
| All | SCALE PET | 0.914 | 0.917 | 0.912 | 0.906 | 0.956 |
| Syngo.via | 0.900 | 0.845 | 0.945 | 0.883 | 0.947 | |
| CU | SCALE PET | 0.919 | 0.861 | 0.939 | 0.845 | 0.951 |
| Syngo.via | 0.910 | 0.806 | 0.946 | 0.821 | 0.930 | |
| MCI | SCALE PET | 0.898 | 0.903 | 0.892 | 0.900 | 0.948 |
| Syngo.via | 0.884 | 0.848 | 0.922 | 0.883 | 0.942 | |
| DE | SCALE PET | 0.928 | 0.955 | 0.857 | 0.951 | 0.949 |
| Syngo.via | 0.922 | 0.919 | 0.929 | 0.944 | 0.959 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Wang, S.-M.; Kang, D.W.; Um, Y.H.; Han, E.J.; Park, S.Y.; Ha, S.; Choe, Y.S.; Kim, H.W.; Kim, R.E.; et al. A Comparative Analysis of Two Automated Quantification Methods for Regional Cerebral Amyloid Retention: PET-Only and PET-and-MRI-Based Methods. Int. J. Mol. Sci. 2024, 25, 7649. https://doi.org/10.3390/ijms25147649
Kim S, Wang S-M, Kang DW, Um YH, Han EJ, Park SY, Ha S, Choe YS, Kim HW, Kim RE, et al. A Comparative Analysis of Two Automated Quantification Methods for Regional Cerebral Amyloid Retention: PET-Only and PET-and-MRI-Based Methods. International Journal of Molecular Sciences. 2024; 25(14):7649. https://doi.org/10.3390/ijms25147649
Chicago/Turabian StyleKim, Sunghwan, Sheng-Min Wang, Dong Woo Kang, Yoo Hyun Um, Eun Ji Han, Sonya Youngju Park, Seunggyun Ha, Yeong Sim Choe, Hye Weon Kim, Regina EY Kim, and et al. 2024. "A Comparative Analysis of Two Automated Quantification Methods for Regional Cerebral Amyloid Retention: PET-Only and PET-and-MRI-Based Methods" International Journal of Molecular Sciences 25, no. 14: 7649. https://doi.org/10.3390/ijms25147649
APA StyleKim, S., Wang, S.-M., Kang, D. W., Um, Y. H., Han, E. J., Park, S. Y., Ha, S., Choe, Y. S., Kim, H. W., Kim, R. E., Kim, D., Lee, C. U., & Lim, H. K. (2024). A Comparative Analysis of Two Automated Quantification Methods for Regional Cerebral Amyloid Retention: PET-Only and PET-and-MRI-Based Methods. International Journal of Molecular Sciences, 25(14), 7649. https://doi.org/10.3390/ijms25147649







