Fluorescence-Based Enzyme Activity Assay: Ascertaining the Activity and Inhibition of Endocannabinoid Hydrolytic Enzymes
Abstract
1. Introduction
2. Enzymes Involved in Endocannabinoid Hydrolysis
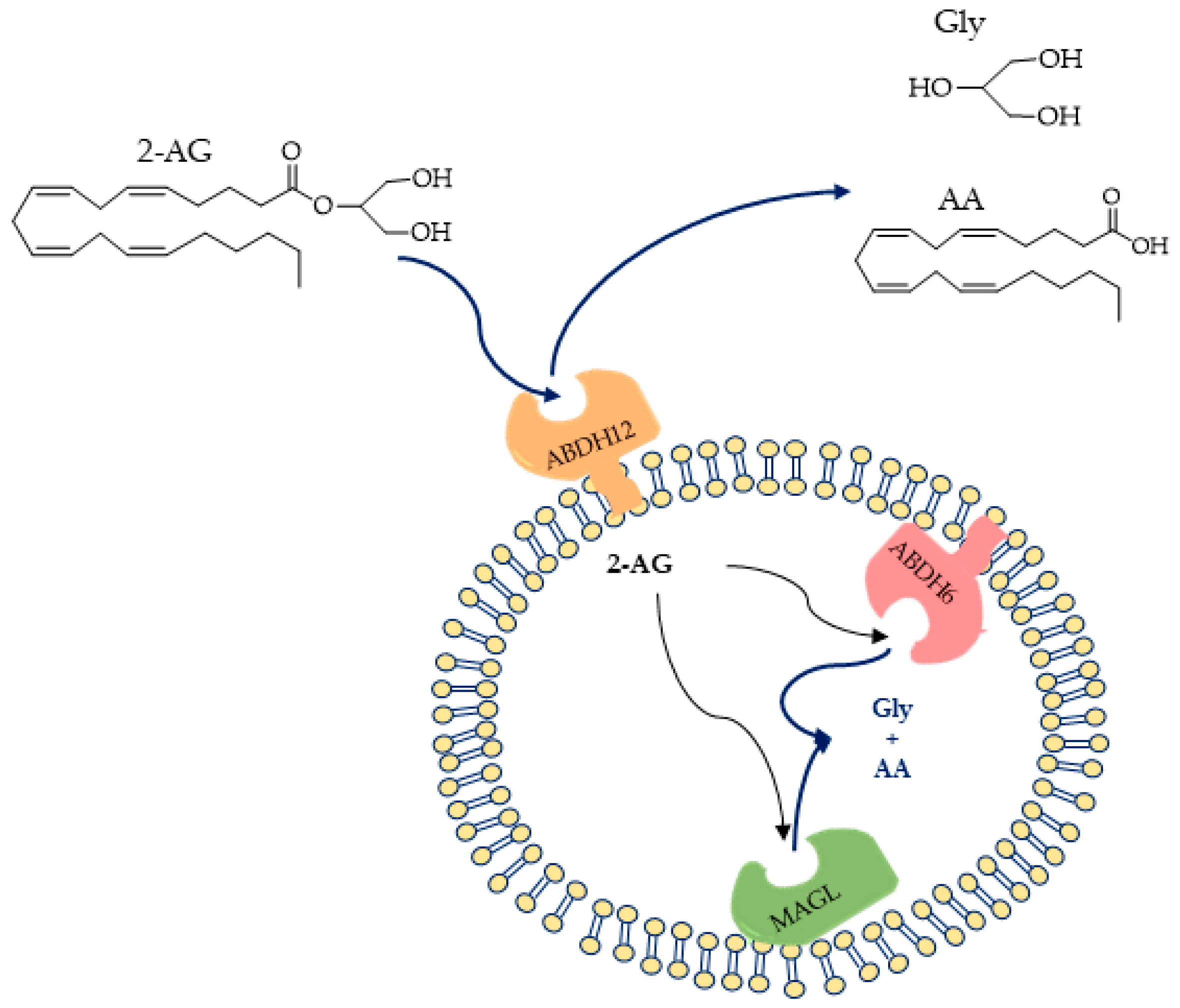
2.1. MAGL
2.2. ABHD6 and ABHD12
2.2.1. ABHD6
2.2.2. ABHD12
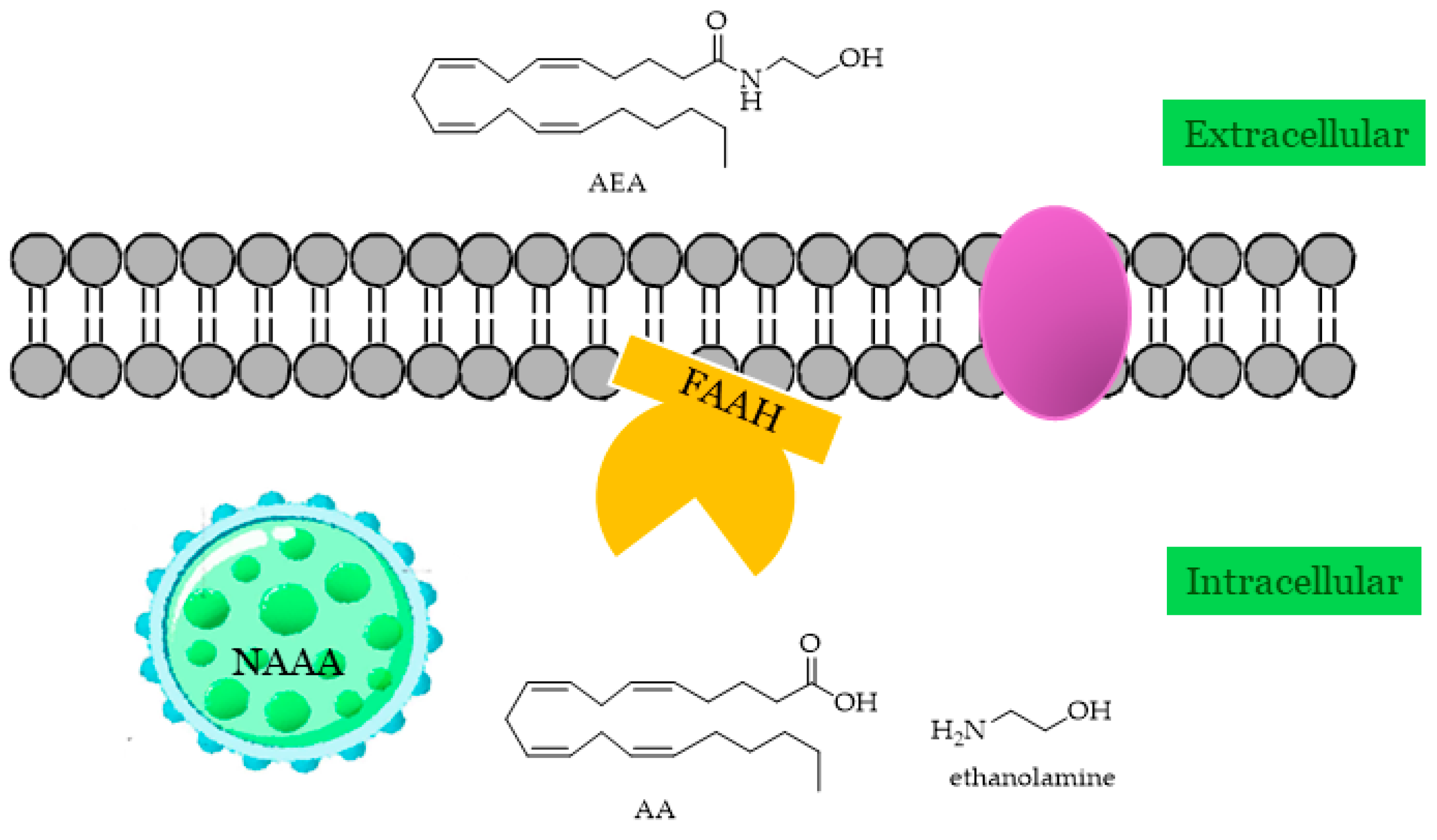
2.3. FAAH
2.4. NAAA
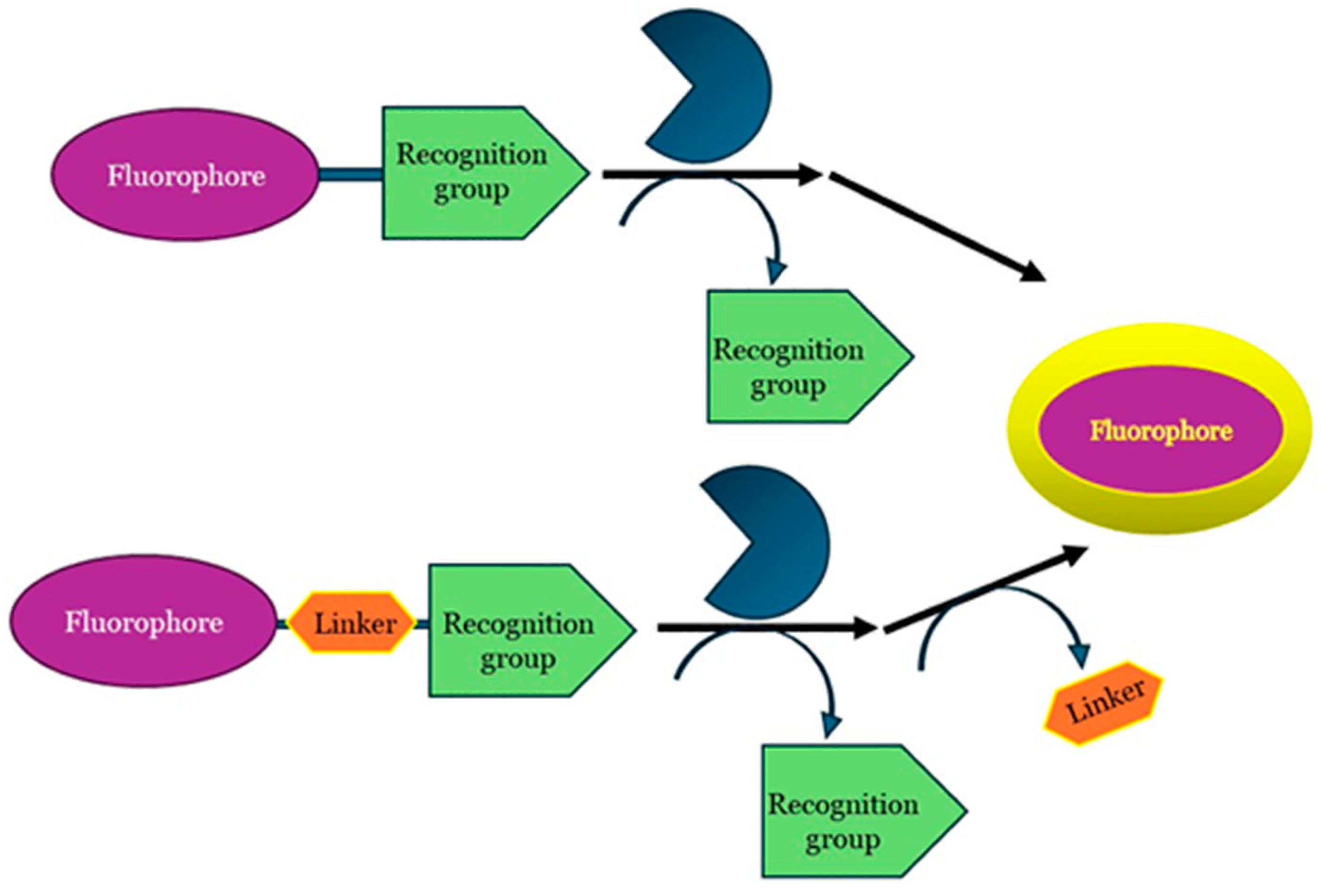
3. Fluorescence-Based Assay for Biological Applications
4. Fluorescence-Based Assays Used to Screen and Characterize Crucial Enzymes Involved in EC Hydrolysis
- They are more sensitive;
- Reaction kinetics can be monitored continuously;
- They require a relatively low amount of enzyme, usually between 0.005 and 0.5 mg/mL;
- They require a relatively low amount of substrate, usually between 0.0025 and 0.25 mM;
- They allow for the use of different buffers under different reaction conditions;
- They allow for micro-assay analysis, as they not only utilize a fluorometer (for one sample at a time) but also enable the use of a microplate reader with fluorimetric detection for the quantification of multiple samples simultaneously.
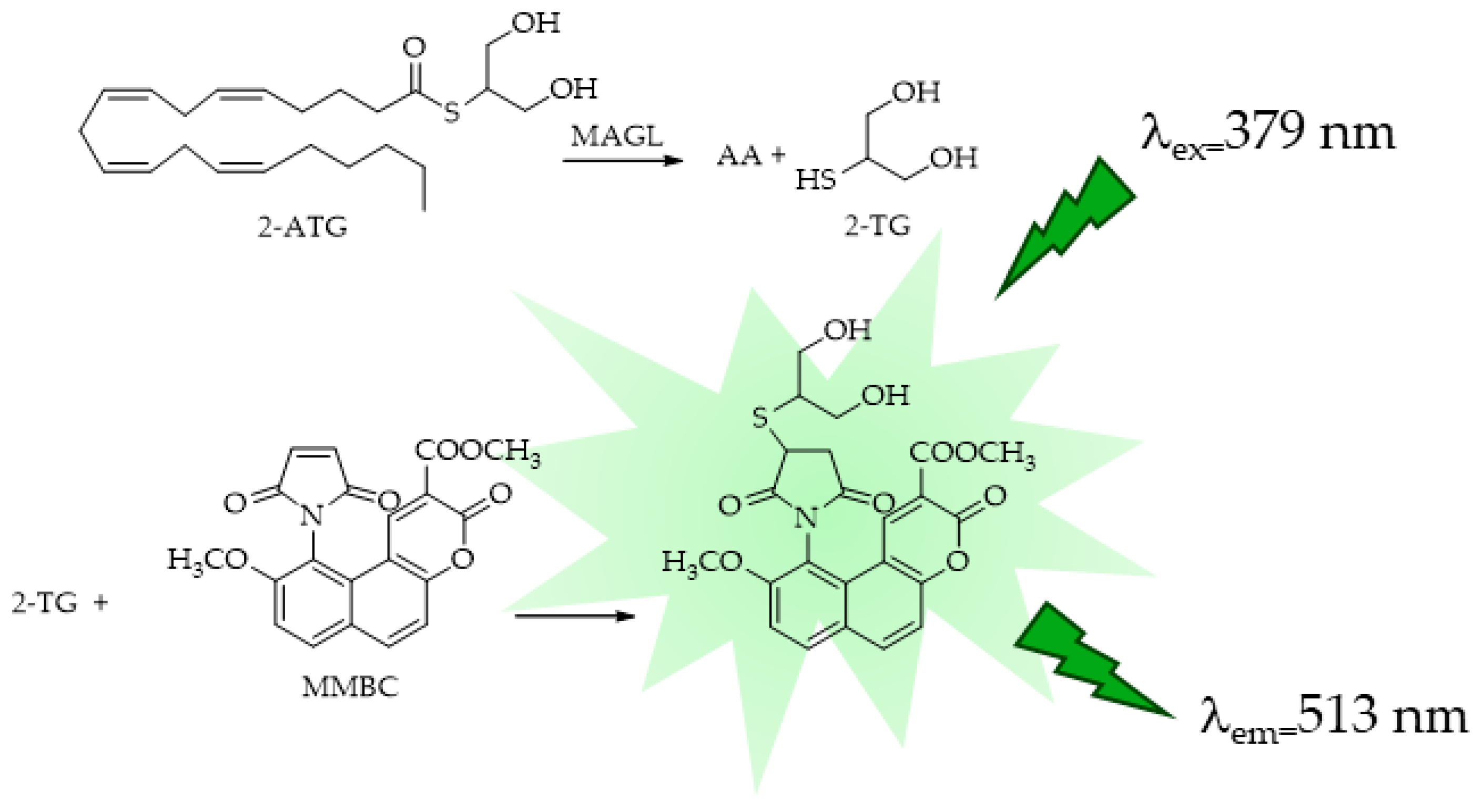
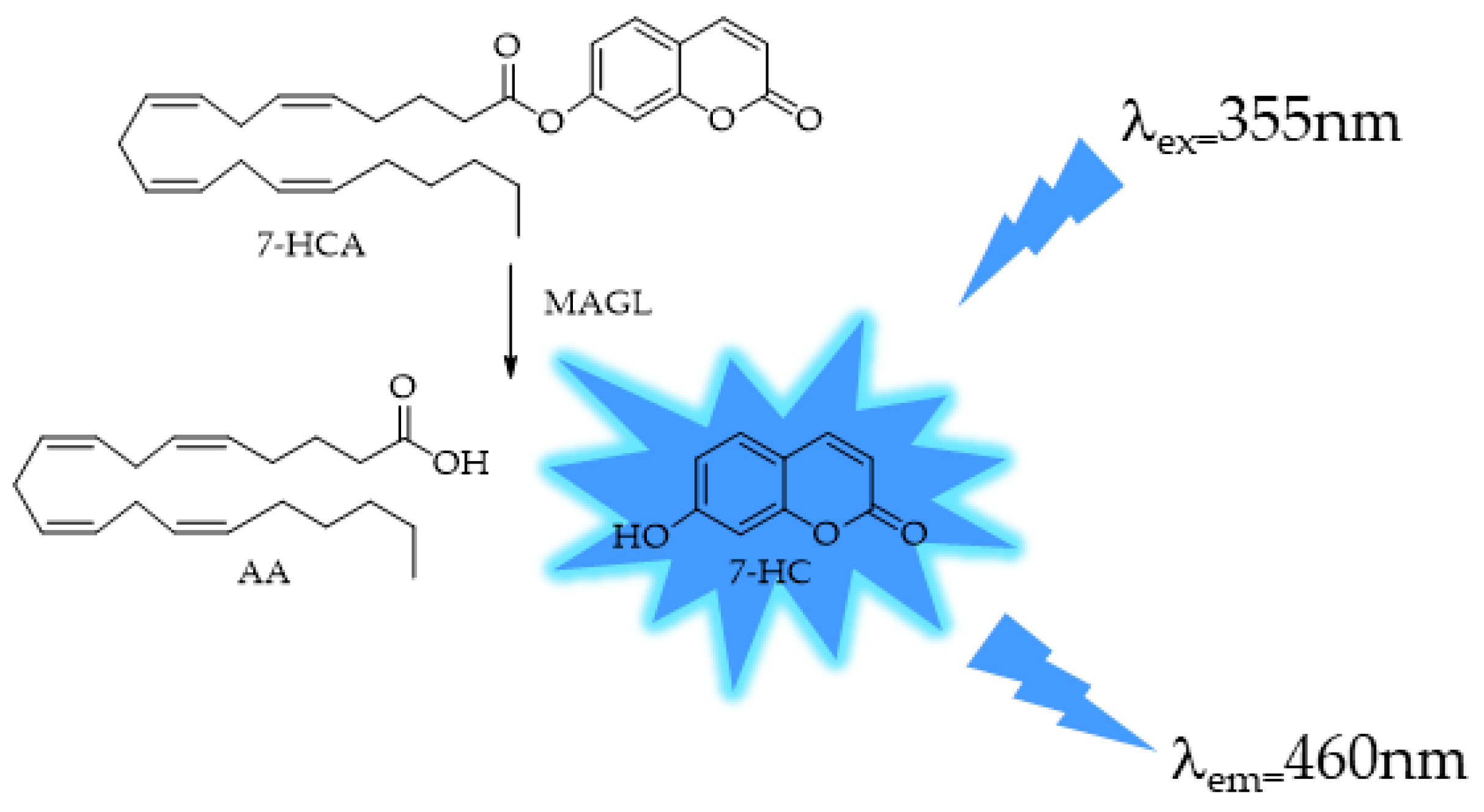
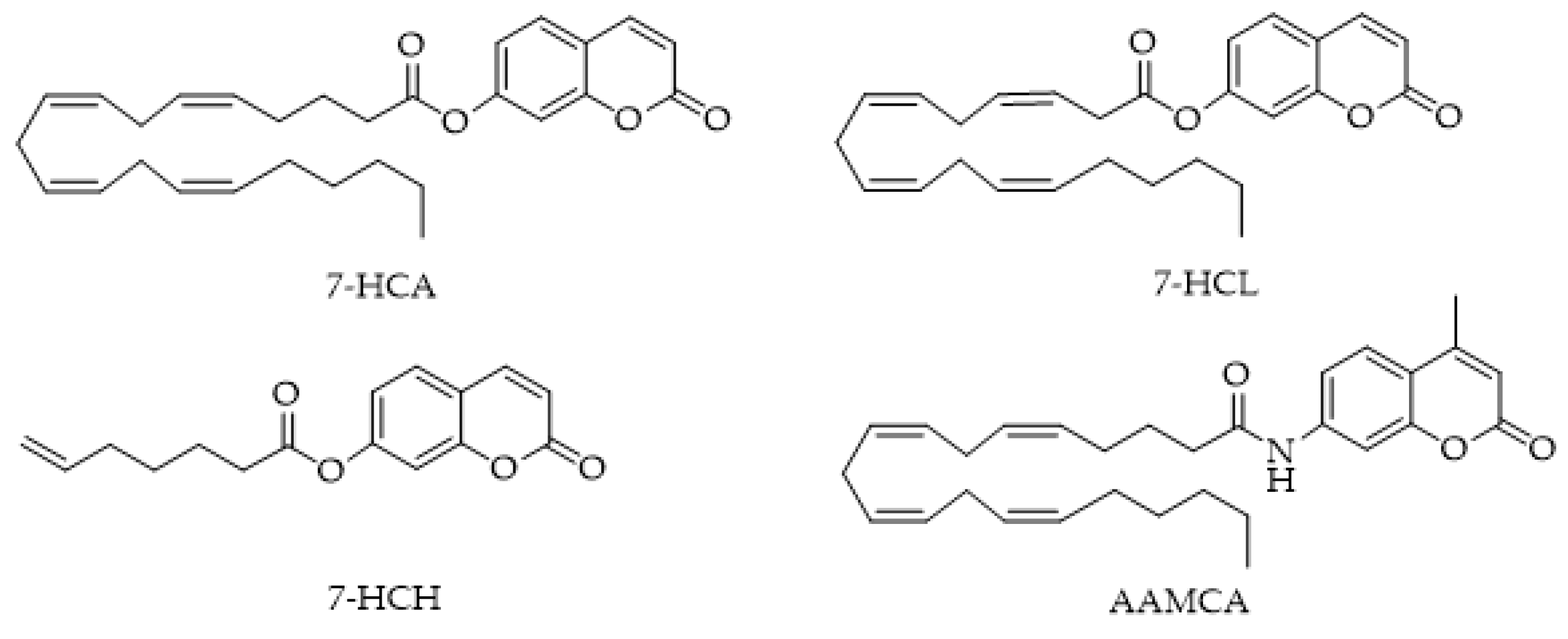
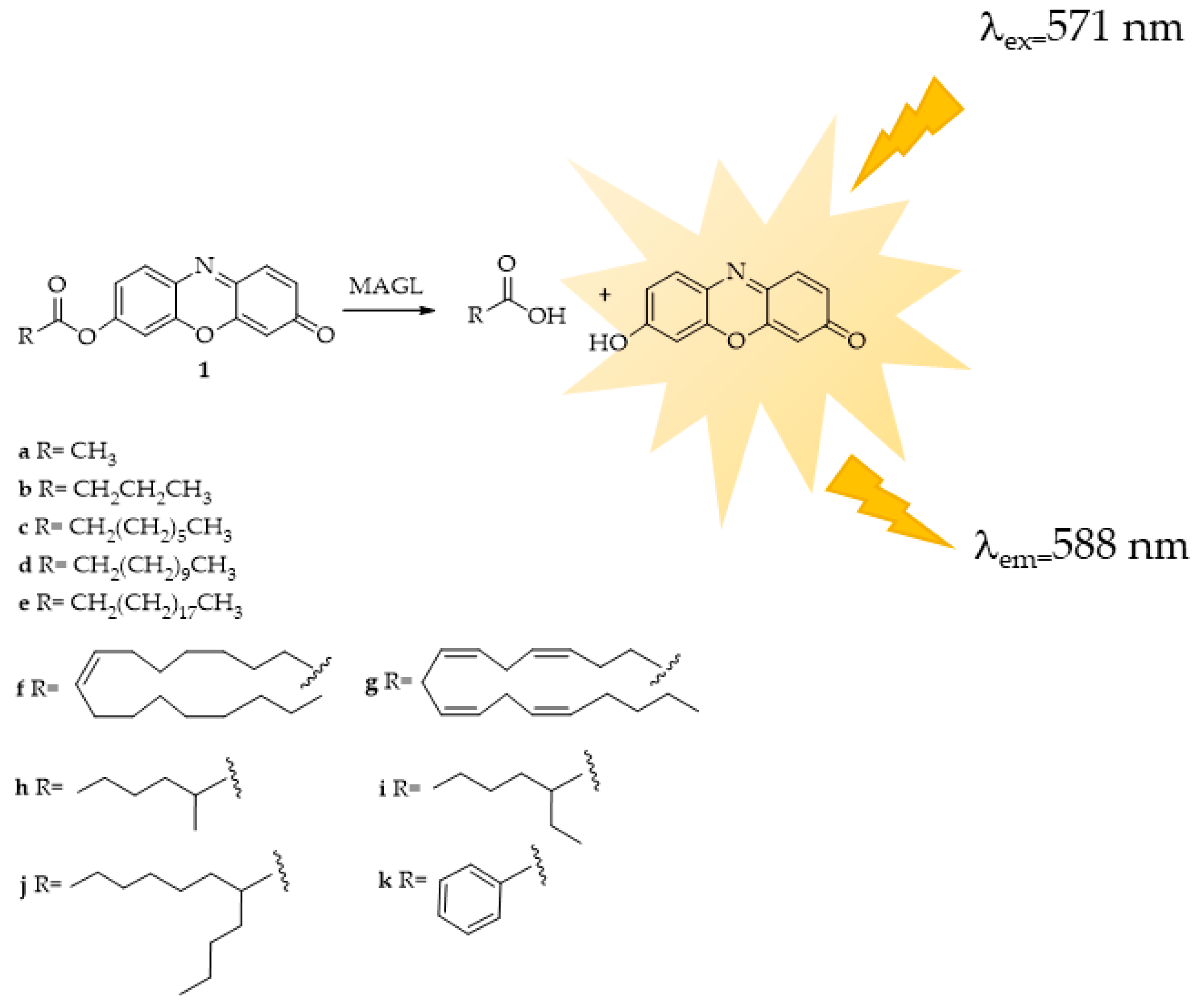
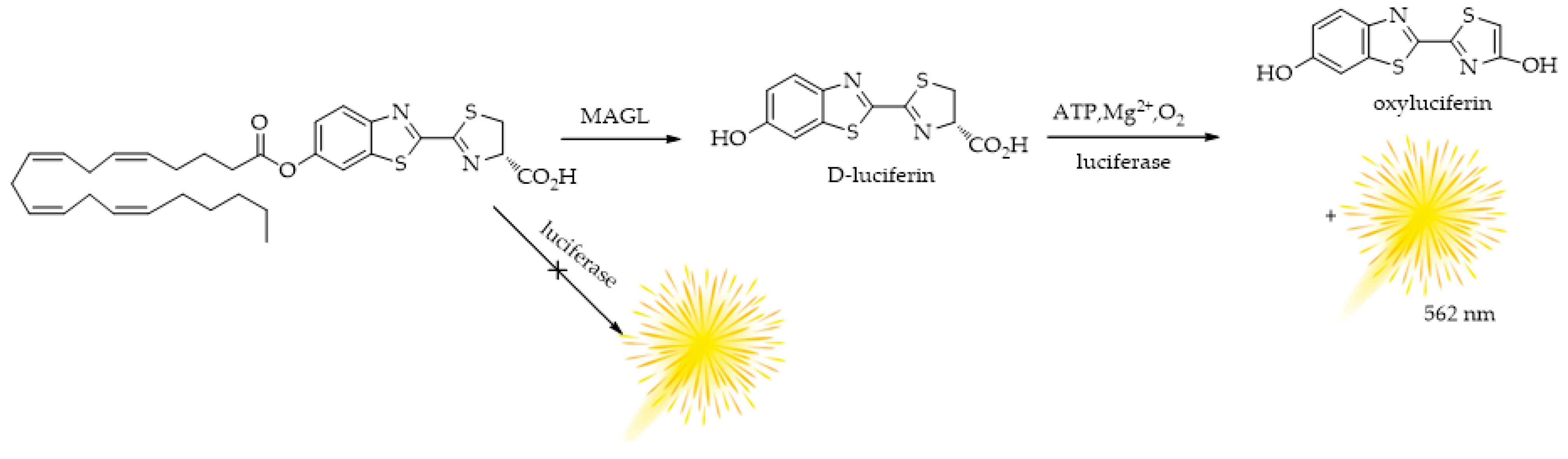
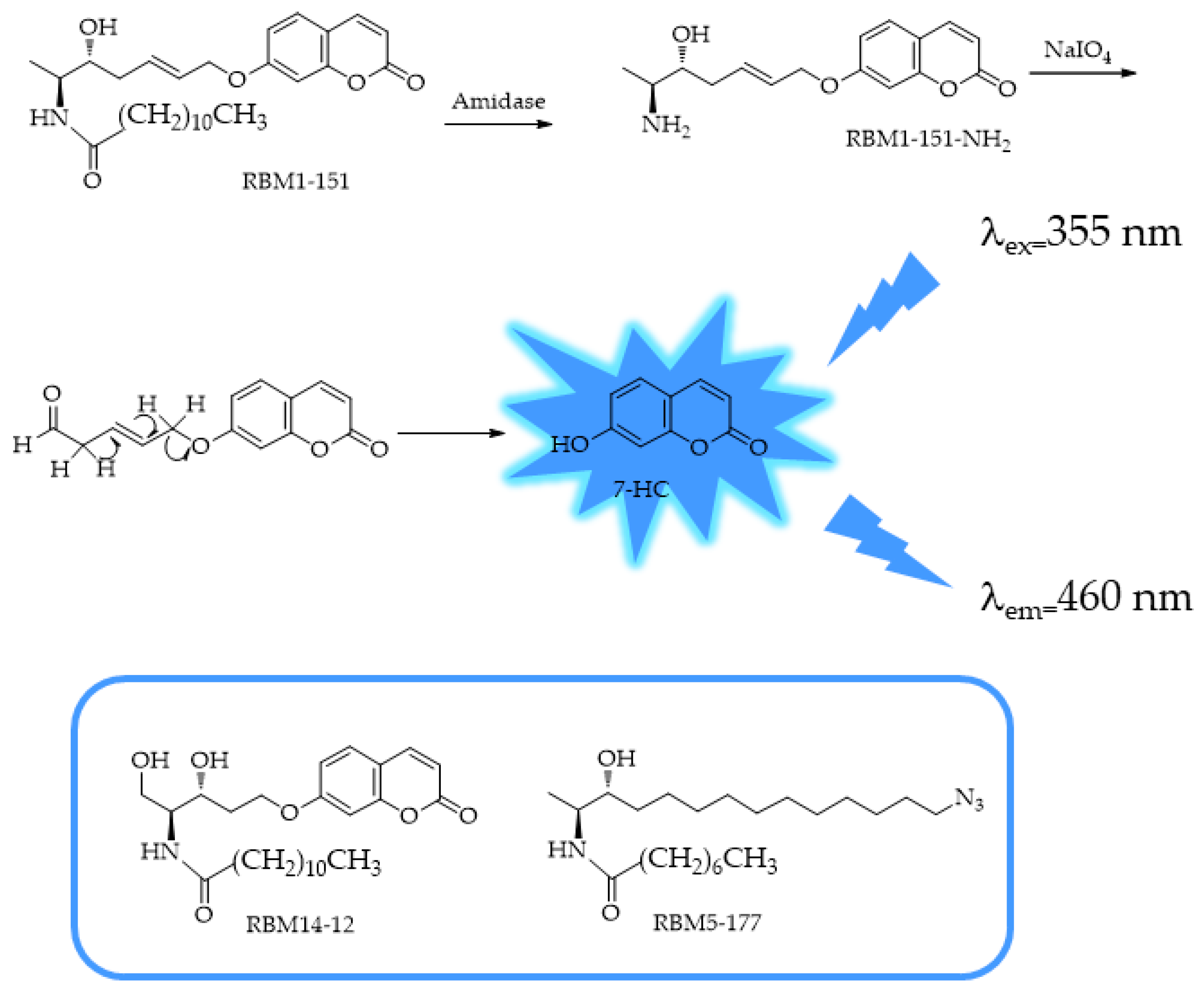
4.1. MAGL
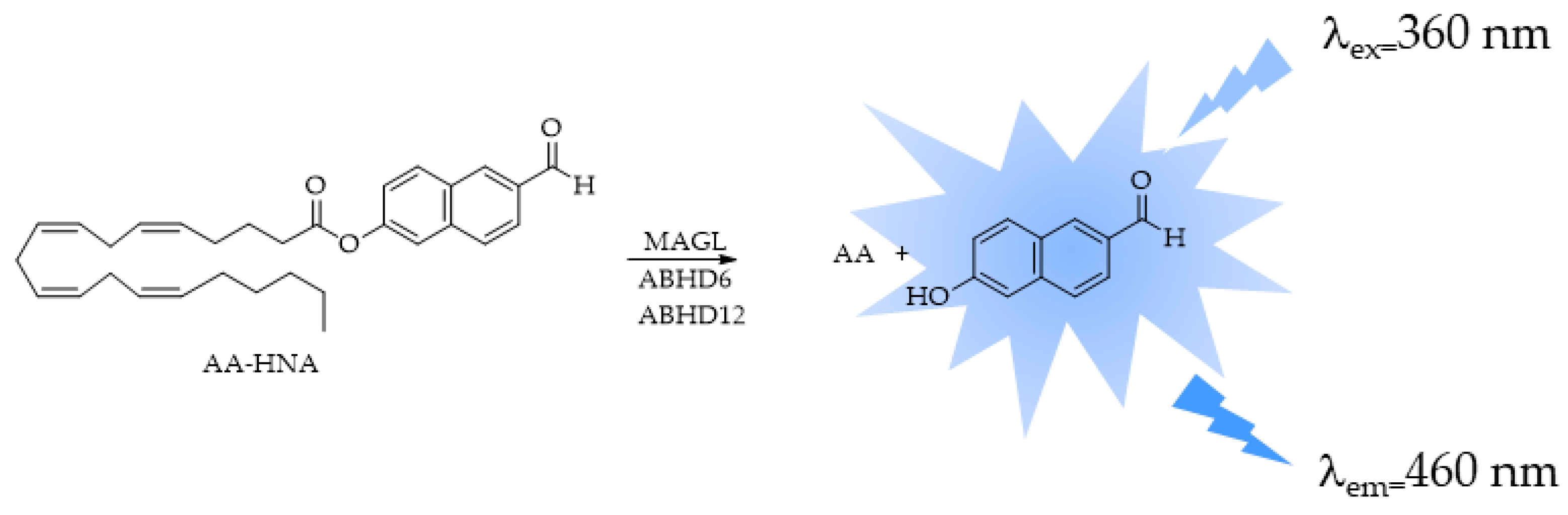
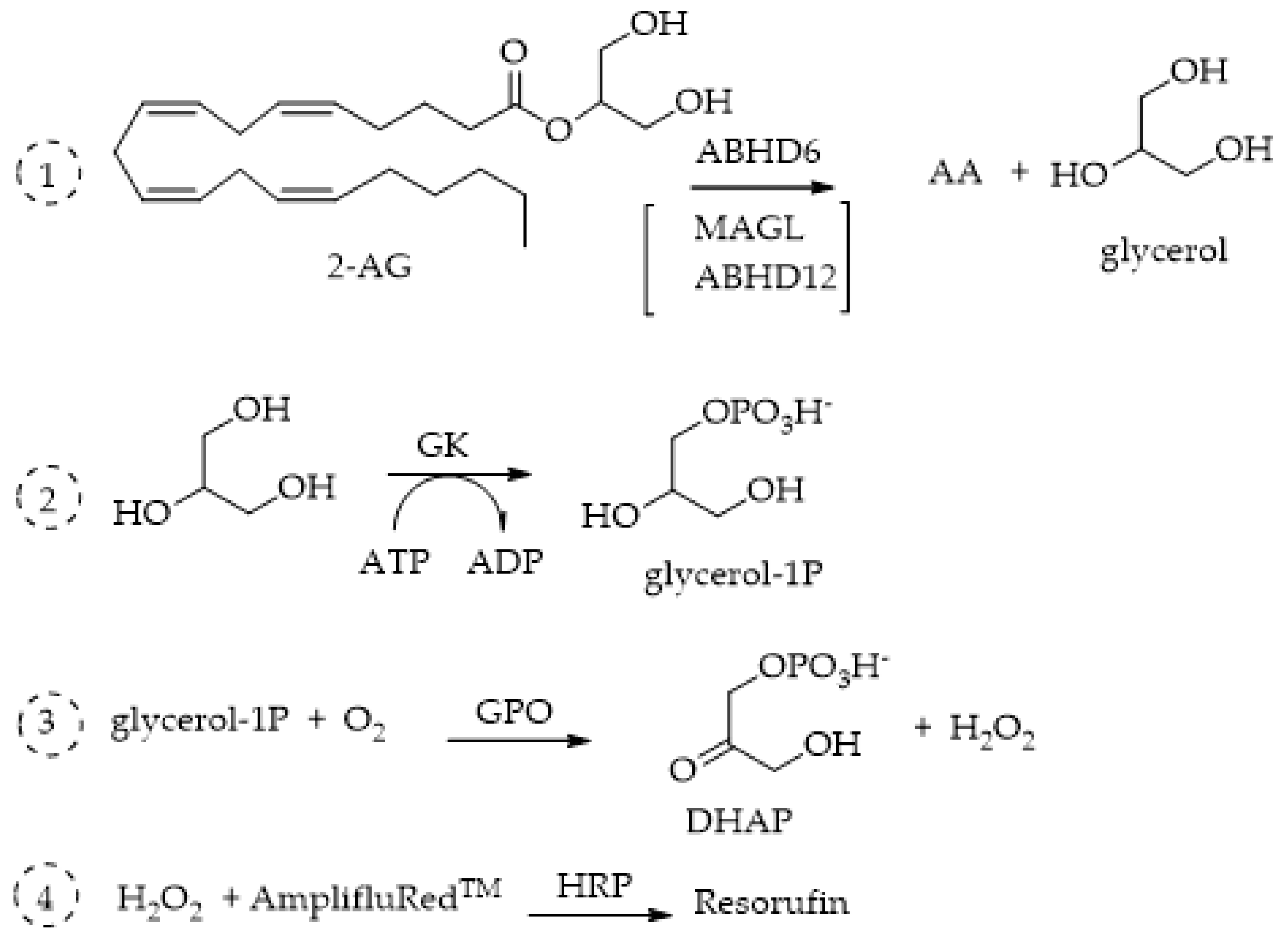
4.2. ABHD6 and ABHD12
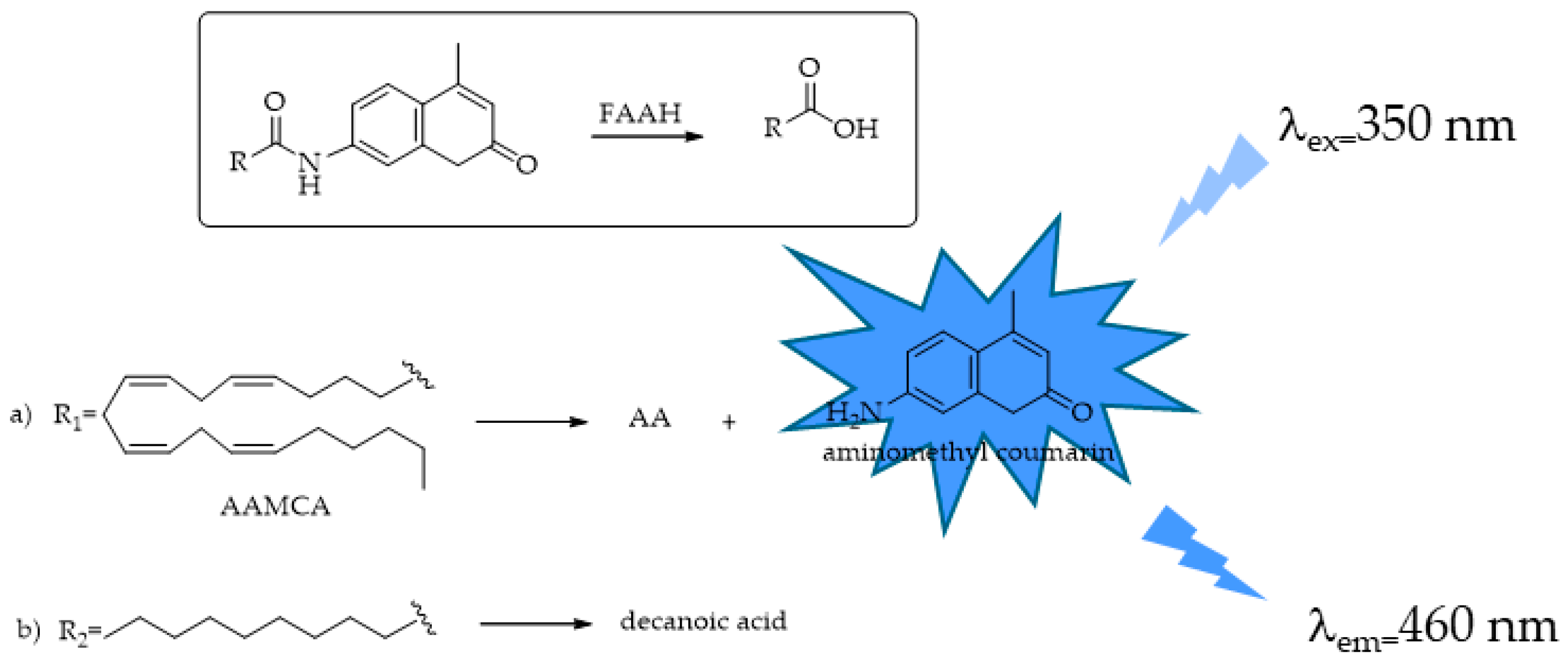
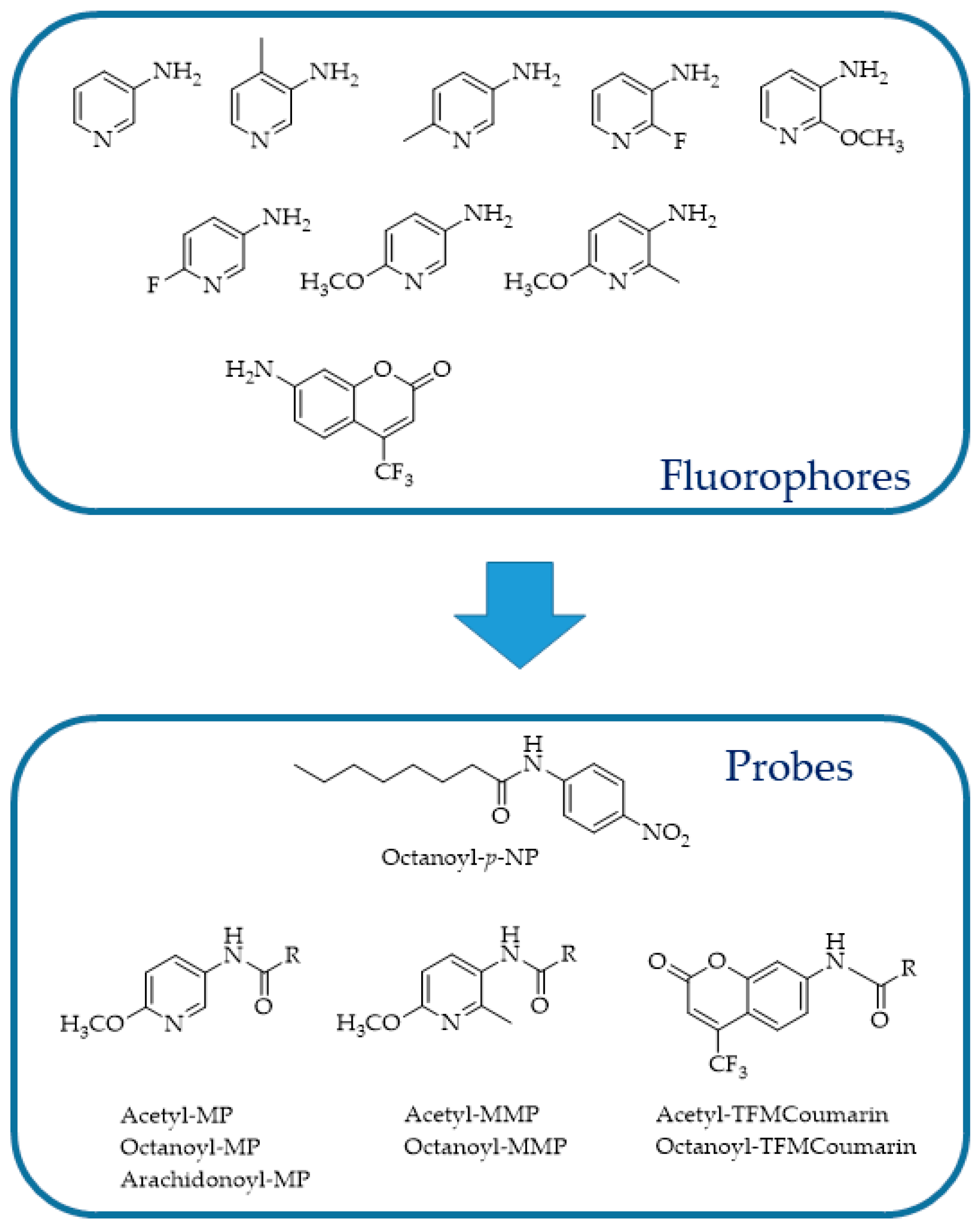
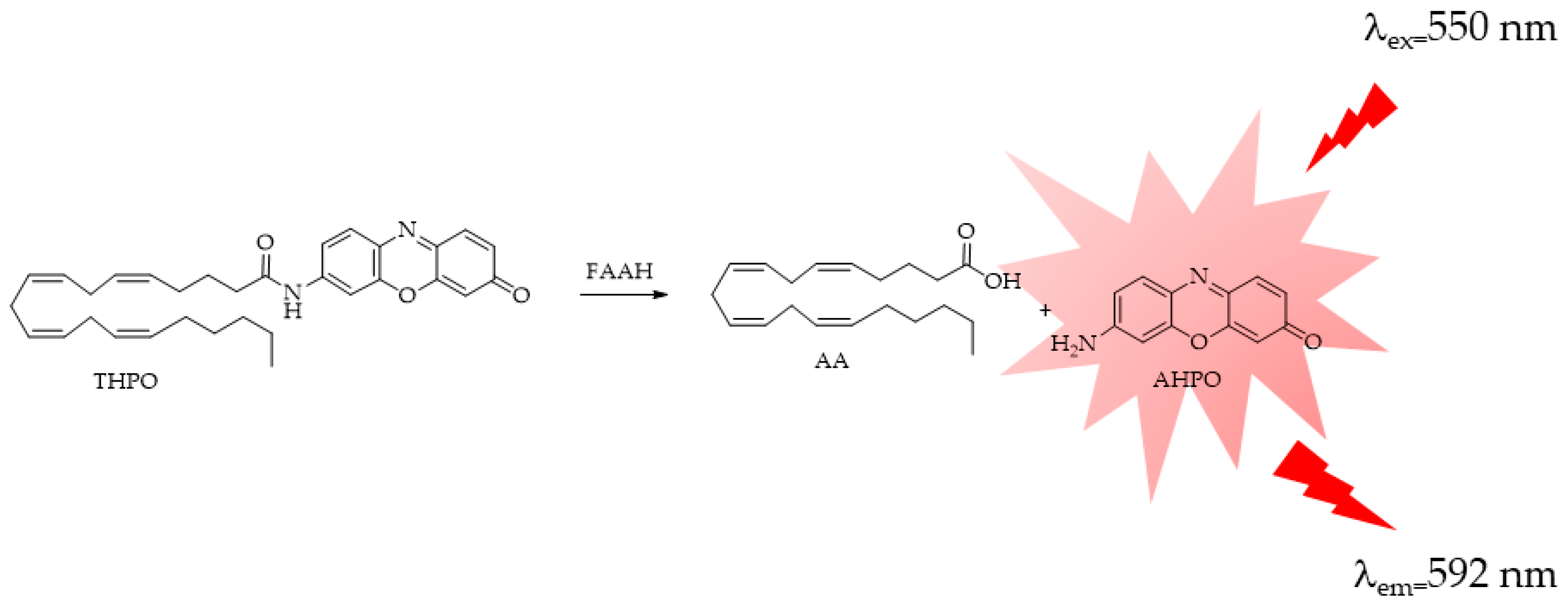
4.3. FAAH
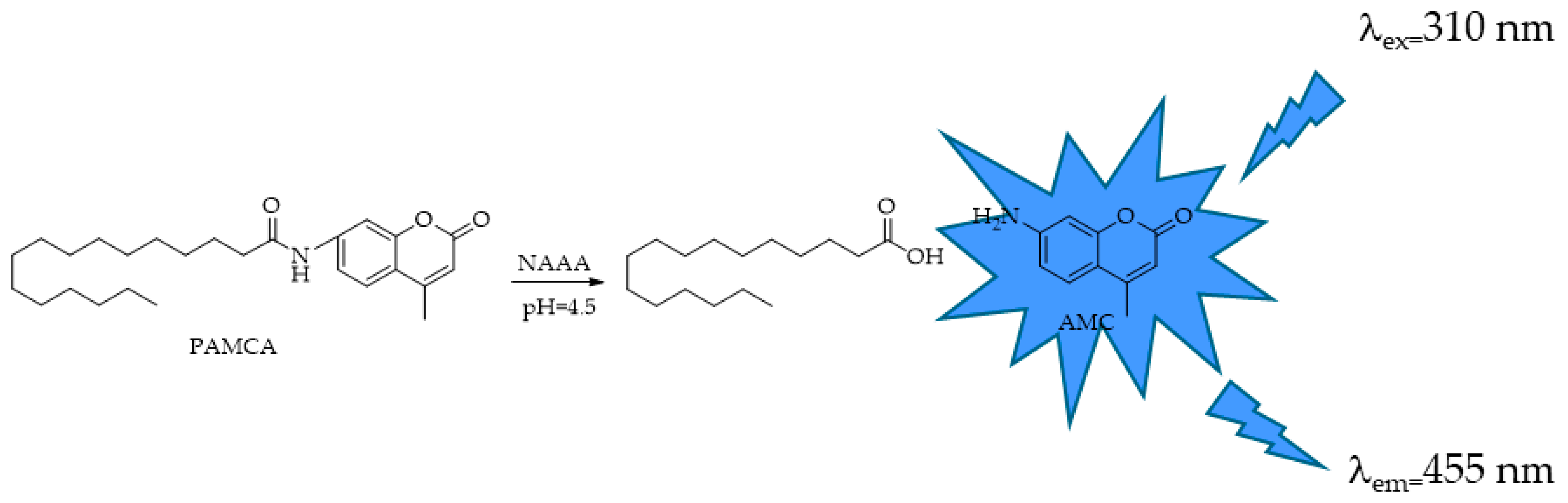
4.4. NAAA
5. Application of Fluorescent Probes in Living Cells
6. Advantages and Disadvantages
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maccarrone, M.; Finazzi-Agró, A. The endocannabinoid system, anandamide and the regulation of mammalian cell apoptosis. Cell Death Differ. 2003, 10, 946–955. [Google Scholar] [CrossRef] [PubMed]
- Mock, E.D.; Gagestein, B.; van der Stelt, M. Anandamide and other N-acylethanolamines: A class of signaling lipids with therapeutic opportunities. Prog. Lipid Res. 2023, 89, 101194. [Google Scholar] [CrossRef] [PubMed]
- Teplick, J.G.; Wallner, R.J.; Levine, A.H.; Haskin, M.E.; Teplick, S.K. Isolated dextrogastria: Report of two cases. AJR Am. J. Roentgenol. 1979, 132, 124–126. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Russell, L.; Condo, K.; DeFlorville, T. Nutrition, endocannabinoids, and the use of cannabis: An overview for the nutrition clinician. Nutr. Clin. Pract. 2024, 39, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Ottria, R.; Della Porta, M.; Xynomilakis, O.; Casati, S.; Cazzola, R.; Ciuffreda, P. Lipids and lipid signaling molecules in human milk and infant formula, a chemical characterization of relevant biochemical components. J. Nutr. Biochem. 2024, 126, 109580. [Google Scholar] [CrossRef] [PubMed]
- Ellermann, M. Emerging mechanisms by which endocannabinoids and their derivatives modulate bacterial populations within the gut microbiome. Adv. Drug Alcohol Res. 2023, 3, 11359. [Google Scholar] [CrossRef] [PubMed]
- Kurlyandchik, I.; Lauche, R.; Tiralongo, E.; Warne, L.N.; Schloss, J. Plasma and interstitial levels of endocannabinoids and N-acylethanolamines in patients with chronic widespread pain and fibromyalgia: A systematic review and meta-analysis. Pain Rep. 2022, 7, e1045. [Google Scholar] [CrossRef]
- Wu, Y.; Han, C.; Luo, R.; Cai, W.; Xia, Q.; Jiang, R.; Ferdek, P.E.; Liu, T.; Huang, W. Molecular mechanisms of pain in acute pancreatitis: Recent basic research advances and therapeutic implications. Front. Mol. Neurosci. 2023, 16, 1331438. [Google Scholar] [CrossRef] [PubMed]
- Pezzilli, R.; Ciuffreda, P.; Ottria, R.; Ravelli, A.; Melzi d’Eril, G.; Barassi, A. Serum endocannabinoids in assessing pain in patients with chronic pancreatitis and in those with pancreatic ductal adenocarcinoma. Scand. J. Gastroenterol. 2017, 52, 1133–1139. [Google Scholar] [CrossRef]
- Clouse, G.; Penman, S.; Hadjiargyrou, M.; Komatsu, D.E.; Thanos, P.K. Examining the role of cannabinoids on osteoporosis: A review. Arch. Osteoporos. 2022, 17, 146. [Google Scholar] [CrossRef]
- Casati, S.; Giannasi, C.; Minoli, M.; Niada, S.; Ravelli, A.; Angeli, I.; Mergenthaler, V.; Ottria, R.; Ciuffreda, P.; Orioli, M.; et al. Quantitative Lipidomic Analysis of Osteosarcoma Cell-Derived Products by UHPLC-MS/MS. Biomolecules 2020, 10, 1302. [Google Scholar] [CrossRef] [PubMed]
- Carnovali, M.; Ottria, R.; Pasqualetti, S.; Banfi, G.; Ciuffreda, P.; Mariotti, M. Effects of bioactive fatty acid amide derivatives in zebrafish scale model of bone metabolism and disease. Pharmacol. Res. 2016, 104, 1–8. [Google Scholar] [CrossRef]
- Ottria, R.; Cappelletti, L.; Ravelli, A.; Mariotti, M.; Gigli, F.; Romagnoli, S.; Ciuffreda, P.; Banfi, G.; Drago, L. Plasma endocannabinoid behaviour in total knee and hip arthroplasty. J. Biol. Regul. Homeost. Agents 2016, 30, 1147–1152. [Google Scholar]
- Di Marzo, V.; Fontana, A.; Cadas, H.; Schinelli, S.; Cimino, G.; Schwartz, J.C.; Piomelli, D. Formation and inactivation of endogenous cannabinoid anandamide in central neurons. Nature 1994, 372, 686–691. [Google Scholar] [CrossRef]
- Piomelli, D.; Beltramo, M.; Glasnapp, S.; Lin, S.Y.; Goutopoulos, A.; Xie, X.Q.; Makriyannis, A. Structural determinants for recognition and translocation by the anandamide transporter. Proc. Natl. Acad. Sci. USA 1999, 96, 5802–5807. [Google Scholar] [CrossRef]
- Cravatt, B.F.; Giang, D.K.; Mayfield, S.P.; Boger, D.L.; Lerner, R.A.; Gilula, N.B. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature 1996, 384, 83–87. [Google Scholar] [CrossRef]
- Dinh, T.P.; Carpenter, D.; Leslie, F.M.; Freund, T.F.; Katona, I.; Sensi, S.L.; Kathuria, S.; Piomelli, D. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc. Natl. Acad. Sci. USA 2002, 99, 10819–10824. [Google Scholar] [CrossRef]
- Ottria, R.; Ravelli, A.; Gigli, F.; Ciuffreda, P. Simultaneous ultra-high performance liquid chromathograpy-electrospray ionization-quadrupole-time of flight mass spectrometry quantification of endogenous anandamide and related N-acylethanolamides in bio-matrices. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2014, 958, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Marchioni, C.; de Souza, I.D.; Acquaro, V.R.; de Souza Crippa, J.A.; Tumas, V.; Queiroz, M.E.C. Recent advances in LC-MS/MS methods to determine endocannabinoids in biological samples: Application in neurodegenerative diseases. Anal. Chim. Acta 2018, 1044, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Vago, R.; Ravelli, A.; Bettiga, A.; Casati, S.; Lavorgna, G.; Benigni, F.; Salonia, A.; Montorsi, F.; Orioli, M.; Ciuffreda, P.; et al. Urine Endocannabinoids as Novel Non-Invasive Biomarkers for Bladder Cancer at Early Stage. Cancers 2020, 12, 870. [Google Scholar] [CrossRef]
- Zoerner, A.A.; Gutzki, F.-M.; Batkai, S.; May, M.; Rakers, C.; Engeli, S.; Jordan, J.; Tsikas, D. Quantification of endocannabinoids in biological systems by chromatography and mass spectrometry: A comprehensive review from an analytical and biological perspective. Biochim. Biophys. Acta 2011, 1811, 706–723. [Google Scholar] [CrossRef] [PubMed]
- Ottria, R.; Casati, S.; Rota, P.; Ciuffreda, P. 2-Arachidonoylglycerol Synthesis: Facile and Handy Enzymatic Method That Allows to Avoid Isomerization. Molecules 2022, 27, 5190. [Google Scholar] [CrossRef] [PubMed]
- Ottria, R.; Casati, S.; Ciuffreda, P. Optimized synthesis and characterization of N-acylethanolamines and O-acylethanolamines, important family of lipid-signalling molecules. Chem. Phys. Lipids 2012, 165, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Piomelli, D.; Giuffrida, A.; Calignano, A.; Rodríguez de Fonseca, F. The endocannabinoid system as a target for therapeutic drugs. Trends Pharmacol. Sci. 2000, 21, 218–224. [Google Scholar] [CrossRef]
- Ramer, R.; Wittig, F.; Hinz, B. The Endocannabinoid System as a Pharmacological Target for New Cancer Therapies. Cancers 2021, 13, 5701. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.; Ayyannan, S.R. Anticancer Potential of Small-Molecule Inhibitors of Fatty Acid Amide Hydrolase and Monoacylglycerol Lipase. Chem. Med. Chem. 2021, 16, 2172–2187. [Google Scholar] [CrossRef] [PubMed]
- van Egmond, N.; Straub, V.M.; van der Stelt, M. Targeting Endocannabinoid Signaling: FAAH and MAG Lipase Inhibitors. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 441–463. [Google Scholar] [CrossRef] [PubMed]
- Lauria, S.; Perrotta, C.; Casati, S.; Di Renzo, I.; Ottria, R.; Eberini, I.; Palazzolo, L.; Parravicini, C.; Ciuffreda, P. Design, synthesis, molecular modelling and in vitro cytotoxicity analysis of novel carbamate derivatives as inhibitors of Monoacylglycerol lipase. Bioorg. Med. Chem. 2018, 26, 2561–2572. [Google Scholar] [CrossRef]
- Vago, R.; Bettiga, A.; Salonia, A.; Ciuffreda, P.; Ottria, R. Development of new inhibitors for N-acylethanolamine-hydrolyzing acid amidase as promising tool against bladder cancer. Bioorg. Med. Chem. 2017, 25, 1242–1249. [Google Scholar] [CrossRef]
- Lazarević, J.; Šmelcerović, A.; Zvezdanović, J.; Yancheva, D.; Casati, S.; Ottria, R.; Ciuffreda, P. Lipid peroxidation inhibition study: A promising case of 1,3-di(1,1′-biphenyl-3-yl)urea. Chem. Biol. Interact. 2020, 326, 109137. [Google Scholar] [CrossRef]
- Jain, S.; Bisht, A.; Verma, K.; Negi, S.; Paliwal, S.; Sharma, S. The role of fatty acid amide hydrolase enzyme inhibitors in Alzheimer’s disease. Cell Biochem. Funct. 2022, 40, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, A.; Kumar, S.; Dutt, R. A Review on Structurally Diversified Synthesized Molecules as Monoacylglycerol Lipase Inhibitors and their Therapeutic uses. Curr. Drug Res. Rev. 2022, 14, 96–115. [Google Scholar] [CrossRef] [PubMed]
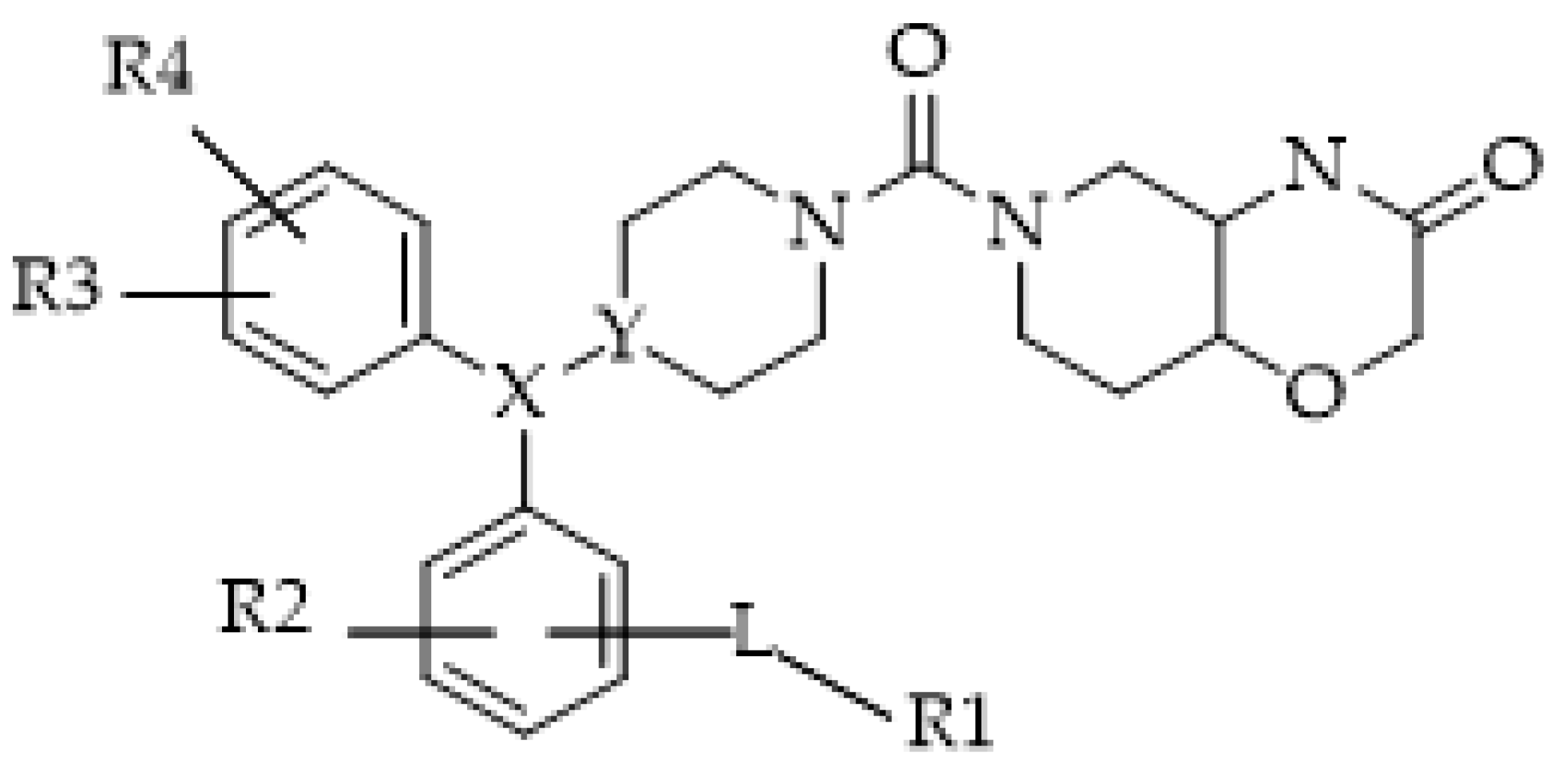
- Bononi, G.; Poli, G.; Rizzolio, F.; Tuccinardi, T.; Macchia, M.; Minutolo, F.; Granchi, C. An updated patent review of monoacylglycerol lipase (MAGL) inhibitors (2018-present). Expert Opin. Ther. Pat. 2021, 31, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Kicman, A.; Pędzińska-Betiuk, A.; Kozłowska, H. The potential of cannabinoids and inhibitors of endocannabinoid degradation in respiratory diseases. Eur. J. Pharmacol. 2021, 911, 174560. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, J.D.; Aripaka, S.S.; Egilmez, C.B.; Pazarlar, B.A. Binding of the monoacylglycerol lipase (MAGL) radiotracer 3HT-401 in the rat brain after status epilepticus. Neurochem. Int. 2024, 175, 105717. [Google Scholar] [CrossRef]
- Hou, L.; Rong, J.; Haider, A.; Ogasawara, D.; Varlow, C.; Schafroth, M.A.; Mu, L.; Gan, J.; Xu, H.; Fowler, C.J.; et al. Positron Emission Tomography Imaging of the Endocannabinoid System: Opportunities and Challenges in Radiotracer Development. J. Med. Chem. 2021, 64, 123–149. [Google Scholar] [CrossRef]
- Cécyre, B.; Monette, M.; Beudjekian, L.; Casanova, C.; Bouchard, J.-F. Localization of diacylglycerol lipase alpha and monoacylglycerol lipase during postnatal development of the rat retina. Front. Neuroanat. 2014, 8, 150. [Google Scholar] [CrossRef]
- Rivera, P.; Arrabal, S.; Cifuentes, M.; Grondona, J.M.; Pérez-Martín, M.; Rubio, L.; Vargas, A.; Serrano, A.; Pavón, F.J.; Suárez, J.; et al. Localization of the cannabinoid CB1 receptor and the 2-AG synthesizing (DAGLα) and degrading (MAGL, FAAH) enzymes in cells expressing the Ca(2+)-binding proteins calbindin, calretinin, and parvalbumin in the adult rat hippocampus. Front. Neuroanat. 2014, 8, 56. [Google Scholar] [CrossRef]
- Zhou, L.; Tian, M.; Zhang, B.; Cao, X.; Huo, X.; Yang, F.; Cao, P.; Feng, L.; Ma, X.; Tian, X. Lysosome targeting fluorescent probe for NAAA imaging and its applications in the drug development for anti-inflammatory. Int. J. Biol. Macromol. 2024, 263, 130307. [Google Scholar] [CrossRef]
- Yapa, U.; Prusakiewicz, J.J.; Wrightstone, A.D.; Christine, L.J.; Palandra, J.; Groeber, E.; Wittwer, A.J. High-performance liquid chromatography-tandem mass spectrometry assay of fatty acid amide hydrolase (FAAH) in blood: FAAH inhibition as clinical biomarker. Anal. Biochem. 2012, 421, 556–565. [Google Scholar] [CrossRef]
- Angelucci, C.B.; Giacominelli-Stuffler, R.; Maccarrone, M. Fluorimetric Assay of FAAH Activity. Methods Mol. Biol. 2023, 2576, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.-M.; Piomelli, D. Assay of Monoacylglycerol Lipase Activity. Methods Mol. Biol. 2023, 2576, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Blankman, J.L.; Simon, G.M.; Cravatt, B.F. A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2-arachidonoylglycerol. Chem. Biol. 2007, 14, 1347–1356. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, T.; Augé, F.; Houtmann, J.; Rak, A.; Vallée, F.; Mikol, V.; Berne, P.F.; Michot, N.; Cheuret, D.; Hoornaert, C.; et al. Structural basis for human monoglyceride lipase inhibition. J. Mol. Biol. 2010, 396, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Labar, G.; Bauvois, C.; Muccioli, G.G.; Wouters, J.; Lambert, D.M. Disulfiram is an inhibitor of human purified monoacylglycerol lipase, the enzyme regulating 2-arachidonoylglycerol signaling. ChemBioChem 2007, 8, 1293–1297. [Google Scholar] [CrossRef] [PubMed]
- Long, J.Z.; Cravatt, B.F. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem. Rev. 2011, 111, 6022–6063. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.; Contreras, J.A.; Hellman, U.; Tornqvist, H.; Holm, C. cDNA cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase. Evolutionary relationship to esterases, lysophospholipases, and haloperoxidases. J. Biol. Chem. 1997, 272, 27218–27223. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, T.; Kondo, S.; Sukagawa, A.; Nakane, S.; Shinoda, A.; Itoh, K.; Yamashita, A.; Waku, K. 2-Arachidonoylglycerol: A possible endogenous cannabinoid receptor ligand in brain. Biochem. Biophys. Res. Commun. 1995, 215, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Dinh, T.P.; Kathuria, S.; Piomelli, D. RNA interference suggests a primary role for monoacylglycerol lipase in the degradation of the endocannabinoid 2-arachidonoylglycerol. Mol. Pharmacol. 2004, 66, 1260–1264. [Google Scholar] [CrossRef]
- Deng, H.; Li, W. Therapeutic potential of targeting α/β-Hydrolase domain-containing 6 (ABHD6). Eur. J. Med. Chem. 2020, 198, 112353. [Google Scholar] [CrossRef]
- Vandevoorde, S.; Saha, B.; Mahadevan, A.; Razdan, R.K.; Pertwee, R.G.; Martin, B.R.; Fowler, C.J. Influence of the degree of unsaturation of the acyl side chain upon the interaction of analogues of 1-arachidonoylglycerol with monoacylglycerol lipase and fatty acid amide hydrolase. Biochem. Biophys. Res. Commun. 2005, 337, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Alhouayek, M.; Masquelier, J.; Muccioli, G.G. Controlling 2-arachidonoylglycerol metabolism as an anti-inflammatory strategy. Drug Discov. Today 2014, 19, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Nomura, D.K.; Long, J.Z.; Niessen, S.; Hoover, H.S.; Ng, S.-W.; Cravatt, B.F. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 2010, 140, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Muccioli, G.G.; Xu, C.; Odah, E.; Cudaback, E.; Cisneros, J.A.; Lambert, D.M.; López Rodríguez, M.L.; Bajjalieh, S.; Stella, N. Identification of a novel endocannabinoid-hydrolyzing enzyme expressed by microglial cells. J. Neurosci. 2007, 27, 2883–2889. [Google Scholar] [CrossRef] [PubMed]
- Kind, L.; Kursula, P. Structural properties and role of the endocannabinoid lipases ABHD6 and ABHD12 in lipid signalling and disease. Amino Acids 2019, 51, 151–174. [Google Scholar] [CrossRef] [PubMed]
- Baggelaar, M.P.; van Esbroeck, A.C.M.; van Rooden, E.J.; Florea, B.I.; Overkleeft, H.S.; Marsicano, G.; Chaouloff, F.; van der Stelt, M. Chemical Proteomics Maps Brain Region Specific Activity of Endocannabinoid Hydrolases. ACS Chem. Biol. 2017, 12, 852–861. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.; Betters, J.L.; Lord, C.C.; Brown, A.L.; Marshall, S.; Ferguson, D.; Sawyer, J.; Davis, M.A.; Melchior, J.T.; Blume, L.C.; et al. The Serine Hydrolase ABHD6 Is a Critical Regulator of the Metabolic Syndrome. Cell Rep. 2013, 5, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Navia-Paldanius, D.; Savinainen, J.R.; Laitinen, J.T. Biochemical and pharmacological characterization of human α/β-hydrolase domain containing 6 (ABHD6) and 12 (ABHD12). J. Lipid Res. 2012, 53, 2413–2424. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.K.; Kaplan, J.; Stella, N. ABHD6: Its Place in Endocannabinoid Signaling and Beyond. Trends Pharmacol. Sci. 2019, 40, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Alhouayek, M.; Masquelier, J.; Cani, P.D.; Lambert, D.M.; Muccioli, G.G. Implication of the anti-inflammatory bioactive lipid prostaglandin D2-glycerol ester in the control of macrophage activation and inflammation by ABHD6. Proc. Natl. Acad. Sci. USA 2013, 110, 17558–17563. [Google Scholar] [CrossRef]
- Zhao, S.; Poursharifi, P.; Mugabo, Y.; Levens, E.J.; Vivot, K.; Attane, C.; Iglesias, J.; Peyot, M.-L.; Joly, E.; Madiraju, S.R.M.; et al. α/β-Hydrolase domain-6 and saturated long chain monoacylglycerol regulate insulin secretion promoted by both fuel and non-fuel stimuli. Mol. Metab. 2015, 4, 940–950. [Google Scholar] [CrossRef]
- Zhao, S.; Mugabo, Y.; Ballentine, G.; Attane, C.; Iglesias, J.; Poursharifi, P.; Zhang, D.; Nguyen, T.A.; Erb, H.; Prentki, R.; et al. α/β-Hydrolase Domain 6 Deletion Induces Adipose Browning and Prevents Obesity and Type 2 Diabetes. Cell Rep. 2016, 14, 2872–2888. [Google Scholar] [CrossRef] [PubMed]
- Max, D.; Hesse, M.; Volkmer, I.; Staege, M.S. High expression of the evolutionarily conserved alpha/beta hydrolase domain containing 6 (ABHD6) in Ewing tumors. Cancer Sci. 2009, 100, 2383–2389. [Google Scholar] [CrossRef]
- Joshi, A.; Shaikh, M.; Singh, S.; Rajendran, A.; Mhetre, A.; Kamat, S.S. Biochemical characterization of the PHARC-associated serine hydrolase ABHD12 reveals its preference for very-long-chain lipids. J. Biol. Chem. 2018, 293, 16953–16963. [Google Scholar] [CrossRef] [PubMed]
- Saghatelian, A.; McKinney, M.K.; Bandell, M.; Patapoutian, A.; Cravatt, B.F. A FAAH-regulated class of N-acyl taurines that activates TRP ion channels. Biochemistry 2006, 45, 9007–9015. [Google Scholar] [CrossRef] [PubMed]
- Chebrou, H.; Bigey, F.; Arnaud, A.; Galzy, P. Study of the amidase signature group. Biochim. Biophys. Acta 1996, 1298, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Lichtman, A.H.; Hawkins, E.G.; Griffin, G.; Cravatt, B.F. Pharmacological activity of fatty acid amides is regulated, but not mediated, by fatty acid amide hydrolase in vivo. J. Pharmacol. Exp. Ther. 2002, 302, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Kathuria, S.; Gaetani, S.; Fegley, D.; Valiño, F.; Duranti, A.; Tontini, A.; Mor, M.; Tarzia, G.; La Rana, G.; Calignano, A.; et al. Modulation of anxiety through blockade of anandamide hydrolysis. Nat. Med. 2003, 9, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Cravatt, B.F.; Saghatelian, A.; Hawkins, E.G.; Clement, A.B.; Bracey, M.H.; Lichtman, A.H. Functional disassociation of the central and peripheral fatty acid amide signaling systems. Proc. Natl. Acad. Sci. USA 2004, 101, 10821–10826. [Google Scholar] [CrossRef] [PubMed]
- Boger, D.L.; Sato, H.; Lerner, A.E.; Hedrick, M.P.; Fecik, R.A.; Miyauchi, H.; Wilkie, G.D.; Austin, B.J.; Patricelli, M.P.; Cravatt, B.F. Exceptionally potent inhibitors of fatty acid amide hydrolase: The enzyme responsible for degradation of endogenous oleamide and anandamide. Proc. Natl. Acad. Sci. USA 2000, 97, 5044–5049. [Google Scholar] [CrossRef]
- Tsuboi, K.; Sun, Y.-X.; Okamoto, Y.; Araki, N.; Tonai, T.; Ueda, N. Molecular characterization of N-acylethanolamine-hydrolyzing acid amidase, a novel member of the choloylglycine hydrolase family with structural and functional similarity to acid ceramidase. J. Biol. Chem. 2005, 280, 11082–11092. [Google Scholar] [CrossRef]
- Rossocha, M.; Schultz-Heienbrok, R.; von Moeller, H.; Coleman, J.P.; Saenger, W. Conjugated bile acid hydrolase is a tetrameric N-terminal thiol hydrolase with specific recognition of its cholyl but not of its tauryl product. Biochemistry 2005, 44, 5739–5748. [Google Scholar] [CrossRef]
- Zhao, L.-Y.; Tsuboi, K.; Okamoto, Y.; Nagahata, S.; Ueda, N. Proteolytic activation and glycosylation of N-acylethanolamine-hydrolyzing acid amidase, a lysosomal enzyme involved in the endocannabinoid metabolism. Biochim. Biophys. Acta 2007, 1771, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Saturnino, C.; Petrosino, S.; Ligresti, A.; Palladino, C.; de Martino, G.; Bisogno, T.; Di Marzo, V. Synthesis and biological evaluation of new potential inhibitors of N-acylethanolamine hydrolyzing acid amidase. Bioorg. Med. Chem. Lett. 2010, 20, 1210–1213. [Google Scholar] [CrossRef] [PubMed]
- Solorzano, C.; Antonietti, F.; Duranti, A.; Tontini, A.; Rivara, S.; Lodola, A.; Vacondio, F.; Tarzia, G.; Piomelli, D.; Mor, M. Synthesis and structure-activity relationships of N-(2-oxo-3-oxetanyl)amides as N-acylethanolamine-hydrolyzing acid amidase inhibitors. J. Med. Chem. 2010, 53, 5770–5781. [Google Scholar] [CrossRef][Green Version]
- Liu, H.-W.; Chen, L.; Xu, C.; Li, Z.; Zhang, H.; Zhang, X.-B.; Tan, W. Recent progresses in small-molecule enzymatic fluorescent probes for cancer imaging. Chem. Soc. Rev. 2018, 47, 7140–7180. [Google Scholar] [CrossRef]
- Wang, R.; Chen, J.; Gao, J.; Chen, J.-A.; Xu, G.; Zhu, T.; Gu, X.; Guo, Z.; Zhu, W.-H.; Zhao, C. A molecular design strategy toward enzyme-activated probes with near-infrared I and II fluorescence for targeted cancer imaging. Chem. Sci. 2019, 10, 7222–7227. [Google Scholar] [CrossRef]
- Fernández-Ruiz, R. TXRF spectrometry in the bioanalytical sciences: A brief review. X-ray Spectrom. 2022, 51, 279–293. [Google Scholar] [CrossRef]
- Hua, L.; Wang, D.; Wang, K.; Wang, Y.; Gu, J.; Zhang, Q.; You, Q.; Wang, L. Design of Tracers in Fluorescence Polarization Assay for Extensive Application in Small Molecule Drug Discovery. J. Med. Chem. 2023, 66, 10934–10958. [Google Scholar] [CrossRef]
- Fang, B.; Shen, Y.; Peng, B.; Bai, H.; Wang, L.; Zhang, J.; Hu, W.; Fu, L.; Zhang, W.; Li, L.; et al. Small-Molecule Quenchers for Förster Resonance Energy Transfer: Structure, Mechanism, and Applications. Angew. Chem. Int. Ed. Engl. 2022, 61, e202207188. [Google Scholar] [CrossRef]
- Kundu, S.; Das, S.; Patra, A. Fluorescence correlation spectroscopy and fluorescence lifetime imaging microscopy for deciphering the morphological evolution of supramolecular self-assembly. Chem. Commun. 2023, 59, 8017–8031. [Google Scholar] [CrossRef]
- Millar, D.P. Time-resolved fluorescence spectroscopy. Curr. Opin. Struct. Biol. 1996, 6, 637–642. [Google Scholar] [CrossRef]
- Krichevsky, O.; Bonnet, G. Fluorescence correlation spectroscopy: The technique and its applications. Rep. Prog. Phys. 2002, 65, 251–297. [Google Scholar] [CrossRef]
- Kask, P.; Palo, K.; Ullmann, D.; Gall, K. Fluorescence-intensity distribution analysis and its application in biomolecular detection technology. Proc. Natl. Acad. Sci. USA 1999, 96, 13756–13761. [Google Scholar] [CrossRef]
- Ning, J.; Liu, T.; Dong, P.; Wang, W.; Ge, G.; Wang, B.; Yu, Z.; Shi, L.; Tian, X.; Huo, X.; et al. Molecular Design Strategy to Construct the Near-Infrared Fluorescent Probe for Selectively Sensing Human Cytochrome P450 2J2. J. Am. Chem. Soc. 2019, 141, 1126–1134. [Google Scholar] [CrossRef]
- Tian, Z.; Yan, F.; Tian, X.; Feng, L.; Cui, J.; Deng, S.; Zhang, B.; Xie, T.; Huang, S.; Ma, X. A NIR fluorescent probe for Vanin-1 and its applications in imaging, kidney injury diagnosis, and the development of inhibitor. Acta Pharm. Sin. B 2022, 12, 316–325. [Google Scholar] [CrossRef]
- Zhou, H.; Tang, J.; Zhang, J.; Chen, B.; Kan, J.; Zhang, W.; Zhou, J.; Ma, H. A red lysosome-targeted fluorescent probe for carboxylesterase detection and bioimaging. J. Mater. Chem. B 2019, 7, 2989–2996. [Google Scholar] [CrossRef]
- Wu, X.; Shi, W.; Li, X.; Ma, H. Recognition Moieties of Small Molecular Fluorescent Probes for Bioimaging of Enzymes. Acc. Chem. Res. 2019, 52, 1892–1904. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, J.; Ma, S.; Liu, Q.; Huang, L.; Chen, X.; Lou, K.; Wang, W. Recent developments in multimodality fluorescence imaging probes. Acta Pharm. Sin. B 2018, 8, 320–338. [Google Scholar] [CrossRef]
- Wang, P.; Yang, H.; Liu, C.; Qiu, M.; Ma, X.; Mao, Z.; Sun, Y.; Liu, Z. Recent advances in the development of activatable multifunctional probes for in vivo imaging of caspase-3. Chin. Chem. Lett. 2021, 32, 168–178. [Google Scholar] [CrossRef]
- Saario, S.M.; Savinainen, J.R.; Laitinen, J.T.; Järvinen, T.; Niemi, R. Monoglyceride lipase-like enzymatic activity is responsible for hydrolysis of 2-arachidonoylglycerol in rat cerebellar membranes. Biochem. Pharmacol. 2004, 67, 1381–1387. [Google Scholar] [CrossRef]
- King, A.R.; Lodola, A.; Carmi, C.; Fu, J.; Mor, M.; Piomelli, D. A critical cysteine residue in monoacylglycerol lipase is targeted by a new class of isothiazolinone-based enzyme inhibitors. Br. J. Pharmacol. 2009, 157, 974–983. [Google Scholar] [CrossRef]
- Ghafouri, N.; Tiger, G.; Razdan, R.K.; Mahadevan, A.; Pertwee, R.G.; Martin, B.R.; Fowler, C.J. Inhibition of monoacylglycerol lipase and fatty acid amide hydrolase by analogues of 2-arachidonoylglycerol. Br. J. Pharmacol. 2004, 143, 774–784. [Google Scholar] [CrossRef][Green Version]
- Brengdahl, J.; Fowler, C.J. A novel assay for monoacylglycerol hydrolysis suitable for high-throughput screening. Anal. Biochem. 2006, 359, 40–44. [Google Scholar] [CrossRef]
- Muccioli, G.G.; Labar, G.; Lambert, D.M. CAY10499, a novel monoglyceride lipase inhibitor evidenced by an expeditious MGL assay. ChemBioChem 2008, 9, 2704–2710. [Google Scholar] [CrossRef]
- Kanaoka, Y.; Sekine, T.; Machida, M.; Soma, Y.; Tanizawa, K.; Ban, Y. Studies on protein-sulfhydryl reagents. I. Synthesis of benzimidazole derivatives of maleimide; fluorescent labeling of maleimide. Chem. Pharm. Bull. 1964, 12, 127–134. [Google Scholar] [CrossRef][Green Version]
- Casida, J.E.; Gulevich, A.G.; Sarpong, R.; Bunnelle, E.M. S-Arachidonoyl-2-thioglycerol synthesis and use for fluorimetric and colorimetric assays of monoacylglycerol lipase. Bioorg. Med. Chem. 2010, 18, 1942–1947. [Google Scholar] [CrossRef]
- Yang, J.-R.; Langmuir, M.E. Synthesis and properties of a maleimide fluorescent thiol reagent derived from a naphthopyranone. J. Heterocycl. Chem. 1991, 28, 1177–1180. [Google Scholar] [CrossRef]
- Wang, Y.; Chanda, P.; Jones, P.G.; Kennedy, J.D. A fluorescence-based assay for monoacylglycerol lipase compatible with inhibitor screening. Assay Drug Dev. Technol. 2008, 6, 387–393. [Google Scholar] [CrossRef]
- Ramarao, M.K.; Murphy, E.A.; Shen, M.W.H.; Wang, Y.; Bushell, K.N.; Huang, N.; Pan, N.; Williams, C.; Clark, J.D. A fluorescence-based assay for fatty acid amide hydrolase compatible with high-throughput screening. Anal. Biochem. 2005, 343, 143–151. [Google Scholar] [CrossRef]
- Savinainen, J.R.; Yoshino, M.; Minkkilä, A.; Nevalainen, T.; Laitinen, J.T. Characterization of binding properties of monoglyceride lipase inhibitors by a versatile fluorescence-based technique. Anal. Biochem. 2010, 399, 132–134. [Google Scholar] [CrossRef]
- Sun, W.C.; Gee, K.R.; Haugland, R.P. Synthesis of novel fluorinated coumarins: Excellent UV-light excitable fluorescent dyes. Bioorg. Med. Chem. Lett. 1998, 8, 3107–3110. [Google Scholar] [CrossRef]
- Clemente, J.C.; Nulton, E.; Nelen, M.; Todd, M.J.; Maguire, D.; Schalk-Hihi, C.; Kuo, L.C.; Zhang, S.-P.; Flores, C.M.; Kranz, J.K. Screening and characterization of human monoglyceride lipase active site inhibitors using orthogonal binding and functional assays. J. Biomol. Screen. 2012, 17, 629–640. [Google Scholar] [CrossRef]
- Lauria, S.; Casati, S.; Ciuffreda, P. Synthesis and characterization of a new fluorogenic substrate for monoacylglycerol lipase and application to inhibition studies. Anal. Bioanal. Chem. 2015, 407, 8163–8167. [Google Scholar] [CrossRef]
- Miceli, M.; Casati, S.; Ottria, R.; Di Leo, S.; Eberini, I.; Palazzolo, L.; Parravicini, C.; Ciuffreda, P. Set-Up and Validation of a High Throughput Screening Method for Human Monoacylglycerol Lipase (MAGL) Based on a New Red Fluorescent Probe. Molecules 2019, 24, 2241. [Google Scholar] [CrossRef]
- Miceli, M.; Casati, S.; Allevi, P.; Berra, S.; Ottria, R.; Rota, P.; Branchini, B.R.; Ciuffreda, P. A New Ultrasensitive Bioluminescence-Based Method for Assaying Monoacylglycerol Lipase. Int. J. Mol. Sci. 2021, 22, 6148. [Google Scholar] [CrossRef]
- Branchini, B.R.; Southworth, T.L.; Fontaine, D.M.; Kohrt, D.; Talukder, M.; Michelini, E.; Cevenini, L.; Roda, A.; Grossel, M.J. An enhanced chimeric firefly luciferase-inspired enzyme for ATP detection and bioluminescence reporter and imaging applications. Anal. Biochem. 2015, 484, 148–153. [Google Scholar] [CrossRef]
- Deng, H.; Zhang, Q.; Lei, Q.; Yang, N.; Yang, K.; Jiang, J.; Yu, Z. Discovering monoacylglycerol lipase inhibitors by a combination of fluorogenic substrate assay and activity-based protein profiling. Front. Pharmacol. 2022, 13, 941522. [Google Scholar] [CrossRef]
- Jiang, M.; Huizenga, M.C.W.; Wirt, J.L.; Paloczi, J.; Amedi, A.; van den Berg, R.J.B.H.N.; Benz, J.; Collin, L.; Deng, H.; Di, X.; et al. A monoacylglycerol lipase inhibitor showing therapeutic efficacy in mice without central side effects or dependence. Nat. Commun. 2023, 14, 8039. [Google Scholar] [CrossRef]
- Savinainen, J.R.; Navia-Paldanius, D.; Laitinen, J.T. A Sensitive and Versatile Fluorescent Activity Assay for ABHD6. Methods Mol. Biol. 2016, 1412, 169–178. [Google Scholar] [CrossRef]
- Savinainen, J.R.; Navia-Paldanius, D.; Laitinen, J.T. A Sensitive and Versatile Fluorescent Activity Assay for ABHD12. Methods Mol. Biol. 2016, 1412, 179–189. [Google Scholar] [CrossRef]
- Maccarrone, M.; Bari, M.; Agrò, A.F. A sensitive and specific radiochromatographic assay of fatty acid amide hydrolase activity. Anal. Biochem. 1999, 267, 314–318. [Google Scholar] [CrossRef]
- Omeir, R.L.; Chin, S.; Hong, Y.; Ahern, D.G.; Deutsch, D.G. Arachidonoyl ethanolamide-1,2-14C as a substrate for anandamide amidase. Life Sci. 1995, 56, 1999–2005. [Google Scholar] [CrossRef]
- Giang, D.K.; Cravatt, B.F. Molecular characterization of human and mouse fatty acid amide hydrolases. Proc. Natl. Acad. Sci. USA 1997, 94, 2238–2242. [Google Scholar] [CrossRef]
- Patricelli, M.P.; Lashuel, H.A.; Giang, D.K.; Kelly, J.W.; Cravatt, B.F. Comparative characterization of a wild type and transmembrane domain-deleted fatty acid amide hydrolase: Identification of the transmembrane domain as a site for oligomerization. Biochemistry 1998, 37, 15177–15187. [Google Scholar] [CrossRef]
- Thumser, A.E.; Voysey, J.; Wilton, D.C. A fluorescence displacement assay for the measurement of arachidonoyl ethanolamide (anandamide) and oleoyl amide (octadecenoamide) hydrolysis. Biochem. Pharmacol. 1997, 53, 433–435. [Google Scholar] [CrossRef]
- de Bank, P.A.; Kendall, D.A.; Alexander, S.P.H. A spectrophotometric assay for fatty acid amide hydrolase suitable for high-throughput screening. Biochem. Pharmacol. 2005, 69, 1187–1193. [Google Scholar] [CrossRef]
- Wang, Y.; Ramirez, F.; Krishnamurthy, G.; Gilbert, A.; Kadakia, N.; Xu, J.; Kalgaonkar, G.; Ramarao, M.K.; Edris, W.; Rogers, K.E.; et al. High-throughput screening for the discovery of inhibitors of fatty acid amide hydrolase using a microsome-based fluorescent assay. J. Biomol. Screen. 2006, 11, 519–527. [Google Scholar] [CrossRef][Green Version]
- Kage, K.L.; Richardson, P.L.; Traphagen, L.; Severin, J.; Pereda-Lopez, A.; Lubben, T.; Davis-Taber, R.; Vos, M.H.; Bartley, D.; Walter, K.; et al. A high throughput fluorescent assay for measuring the activity of fatty acid amide hydrolase. J. Neurosci. Methods 2007, 161, 47–54. [Google Scholar] [CrossRef]
- Huang, H.; Nishi, K.; Tsai, H.-J.; Hammock, B.D. Development of highly sensitive fluorescent assays for fatty acid amide hydrolase. Anal. Biochem. 2007, 363, 12–21. [Google Scholar] [CrossRef]
- Patricelli, M.P.; Cravatt, B.F. Characterization and manipulation of the acyl chain selectivity of fatty acid amide hydrolase. Biochemistry 2001, 40, 6107–6115. [Google Scholar] [CrossRef]
- Dato, F.M.; Maaßen, A.; Goldfuß, B.; Pietsch, M. Characterization of fatty acid amide hydrolase activity by a fluorescence-based assay. Anal. Biochem. 2018, 546, 50–57. [Google Scholar] [CrossRef]
- Forster, L.; Schulze Elfringhoff, A.; Lehr, M. High-performance liquid chromatographic assay with fluorescence detection for the evaluation of inhibitors against fatty acid amide hydrolase. Anal. Bioanal. Chem. 2009, 394, 1679–1685. [Google Scholar] [CrossRef]
- Tian, X.; Liu, T.; Li, L.; Shao, B.; Yao, D.; Feng, L.; Cui, J.; James, T.D.; Ma, X. Visual High-Throughput Screening for Developing a Fatty Acid Amide Hydrolase Natural Inhibitor Based on an Enzyme-Activated Fluorescent Probe. Anal. Chem. 2020, 92, 9493–9500. [Google Scholar] [CrossRef]
- Casasampere, M.; Ung, J.; Iñáñez, A.; Dufau, C.; Tsuboi, K.; Casas, J.; Tan, S.-F.; Feith, D.J.; Andrieu-Abadie, N.; Segui, B.; et al. A fluorogenic substrate for the detection of lipid amidases in intact cells. J. Lipid Res. 2024, 65, 100520. [Google Scholar] [CrossRef]
- Sanllehí, P.; Casasampere, M.; Abad, J.-L.; Fabriàs, G.; López, O.; Bujons, J.; Casas, J.; Delgado, A. The first fluorogenic sensor for sphingosine-1-phosphate lyase activity in intact cells. Chem. Commun. 2017, 53, 5441–5444. [Google Scholar] [CrossRef]
- Tsuboi, K.; Ueda, N. Assay of NAAA Activity. Methods Mol. Biol. 2023, 2576, 261–274. [Google Scholar] [CrossRef]
- Solorzano, C.; Zhu, C.; Battista, N.; Astarita, G.; Lodola, A.; Rivara, S.; Mor, M.; Russo, R.; Maccarrone, M.; Antonietti, F.; et al. Selective N-acylethanolamine-hydrolyzing acid amidase inhibition reveals a key role for endogenous palmitoylethanolamide in inflammation. Proc. Natl. Acad. Sci. USA 2009, 106, 20966–20971. [Google Scholar] [CrossRef]
- West, J.M.; Zvonok, N.; Whitten, K.M.; Wood, J.T.; Makriyannis, A. Mass spectrometric characterization of human N-acylethanolamine-hydrolyzing acid amidase. J. Proteome Res. 2012, 11, 972–981. [Google Scholar] [CrossRef]
- Yang, L.; Ji, C.; Li, Y.; Hu, F.; Zhang, F.; Zhang, H.; Li, L.; Ren, J.; Wang, Z.; Qiu, Y. Natural Potent NAAA Inhibitor Atractylodin Counteracts LPS-Induced Microglial Activation. Front. Pharmacol. 2020, 11, 577319. [Google Scholar] [CrossRef]
- West, J.M.; Zvonok, N.; Whitten, K.M.; Vadivel, S.K.; Bowman, A.L.; Makriyannis, A. Biochemical and mass spectrometric characterization of human N-acylethanolamine-hydrolyzing acid amidase inhibition. PLoS ONE 2012, 7, e43877. [Google Scholar] [CrossRef]
- Tian, M.; Tian, Z.; Yao, D.; Ning, J.; Deng, S.; Feng, L.; Huo, X.; Tian, X.; Zhang, B.; Wang, C.; et al. A NIR fluorescent probe for fatty acid amide hydrolase bioimaging and its application in development of inhibitors. J. Mater. Chem. B 2021, 9, 6460–6465. [Google Scholar] [CrossRef]
- Gu, X.; Wang, X.; Cai, W.; Han, Y.; Zhang, Q.-W. Monofluorophore-based Two-Photon Ratiometric Fluorescent Probe for the Quantitative Imaging of Fatty Acid Amide Hydrolase in Live Neurons and Mouse Brain Tissues. ACS Sens. 2024, 9, 3387–3393. [Google Scholar] [CrossRef]
- Xie, Y.; Fang, Y.; Huang, Z.; Tallon, A.M.; am Ende, C.W.; Fox, J.M. Divergent Synthesis of Monosubstituted and Unsymmetrical 3,6-Disubstituted Tetrazines from Carboxylic Ester Precursors. Angew. Chem. Int. Ed. Engl. 2020, 59, 16967–16973. [Google Scholar] [CrossRef]
- Mohammad, I.; Liebmann, K.L.; Miller, S.C. Firefly luciferin methyl ester illuminates the activity of multiple serine hydrolases. Chem. Commun. 2023, 59, 8552–8555. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciuffreda, P.; Xynomilakis, O.; Casati, S.; Ottria, R. Fluorescence-Based Enzyme Activity Assay: Ascertaining the Activity and Inhibition of Endocannabinoid Hydrolytic Enzymes. Int. J. Mol. Sci. 2024, 25, 7693. https://doi.org/10.3390/ijms25147693
Ciuffreda P, Xynomilakis O, Casati S, Ottria R. Fluorescence-Based Enzyme Activity Assay: Ascertaining the Activity and Inhibition of Endocannabinoid Hydrolytic Enzymes. International Journal of Molecular Sciences. 2024; 25(14):7693. https://doi.org/10.3390/ijms25147693
Chicago/Turabian StyleCiuffreda, Pierangela, Ornella Xynomilakis, Silvana Casati, and Roberta Ottria. 2024. "Fluorescence-Based Enzyme Activity Assay: Ascertaining the Activity and Inhibition of Endocannabinoid Hydrolytic Enzymes" International Journal of Molecular Sciences 25, no. 14: 7693. https://doi.org/10.3390/ijms25147693
APA StyleCiuffreda, P., Xynomilakis, O., Casati, S., & Ottria, R. (2024). Fluorescence-Based Enzyme Activity Assay: Ascertaining the Activity and Inhibition of Endocannabinoid Hydrolytic Enzymes. International Journal of Molecular Sciences, 25(14), 7693. https://doi.org/10.3390/ijms25147693







