Comprehensive Review of COVID-19: Epidemiology, Pathogenesis, Advancement in Diagnostic and Detection Techniques, and Post-Pandemic Treatment Strategies
Abstract
:1. Introduction
2. Epidemiology
2.1. Coronavirus
2.2. Origin of COVID-19
2.3. Routes of Transmission
2.4. SARS-CoV-2 Variant
2.5. SARS-CoV-2 Updated Infection Rate and Mortality across Different Regions
2.6. SARS-CoV-2 New Transmission Pattern
2.7. SARS-CoV-2 Reinfection Rate
- United States
- United Kingdom
- Austria
- Mexico
3. Pathogenesis
3.1. Infection Stage of SARS-CoV-2
3.2. Long COVID
3.2.1. Definition of Long COVID
3.2.2. Epidemiology of Long COVID
3.2.3. Pathogenesis of Long COVID
3.2.4. Treatments for Long COVID
4. Diagnostic Methods
4.1. Nucleic Acid Amplification Test (NAAT)
4.1.1. RT-qPCR
4.1.2. Multiplex RT-qPCR Assays for Detecting SARS-CoV-2 and Other Respiratory Pathogens
4.1.3. RT-dPCR
4.1.4. Other NAATs
4.2. Serological Tests
4.2.1. RAT
4.2.2. ELISA
4.2.3. Shortcomings of Antigen Tests
5. Ancillary Tests for COVID-19
5.1. Sequencing
5.2. Antibody Serology Tests
6. Treatment Strategies
6.1. Antiviral Therapy
6.1.1. Remdesivir
6.1.2. Nirmatrelvir–Ritonavir (Paxlovid)
6.2. Immunomodulators
6.2.1. Baricitinib (JAK Inhibitor)
6.2.2. Tocilizumab (IL-6 Inhibitor)
6.3. Cell Therapy
6.3.1. T Cell Therapy
6.3.2. Mesenchymal Stem Cell (MSC) Therapy
7. Vaccines
7.1. Impact of Vaccines on Epidemiology of COVID-19
7.2. Disparities in COVID-19 Response between High-Income and Low-Income Countries
7.3. Pfizer-BioNTech COVID-19 Vaccine (mRNA Vaccine)
7.4. Novavax COVID-19 Vaccine (Protein Subunit Vaccine)
8. Conclusions
Funding
Conflicts of Interest
References
- Centers for Disease Control and Prevention. CDC COVID Data Tracker. [Online]. Centers for Disease Control and Prevention. 2024. Available online: https://covid.cdc.gov/covid-data-tracker/#variant-proportions (accessed on 26 June 2024).
- Tsang, H.F.; Chan, L.W.C.; Cho, W.C.S.; Yu, A.C.S.; Yim, A.K.Y.; Chan, A.K.C.; Ng, L.P.W.; Wong, Y.K.E.; Pei, X.M.; Li, M.J.W.; et al. An update on COVID-19 pandemic: The epidemiology, pathogenesis, prevention and treatment strategies. Expert Rev. Anti-Infect. Ther. 2020, 19, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Huang, Y.; Yuen, K.-Y. Coronavirus Diversity, Phylogeny and Interspecies Jumping. Exp. Biol. Med. 2009, 234, 1117–1127. [Google Scholar] [CrossRef]
- Myint, S.H. Human coronavirus infections. In The Coronaviridae; Plenum Press: New York, NY, USA, 1995; pp. 389–401. [Google Scholar]
- Vabret, A.; Mourez, T.; Dina, J.; Van Der Hoek, L.; Gouarin, S.; Petitjean, J.; Brouard, J.; Freymuth, F. Human coronavirus NL63, France. Emerg. Infect. Dis. 2005, 11, 1225–1229. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Chu, C.-M.; Chan, K.-H.; Tsoi, H.-W.; Huang, Y.; Wong, B.H.L.; Poon, R.W.S.; Cai, J.J.; Luk, W.-K.; et al. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 2005, 79, 884–895. [Google Scholar] [CrossRef] [PubMed]
- Ogimi, C.; Kim, Y.J.; Martin, E.T.; Huh, H.J.; Chiu, C.H.; Englund, J.A. What’s new with the old coronaviruses? J. Pediatr. Infect. Dis. Soc. 2020, 9, 210–217. [Google Scholar] [CrossRef]
- De Groot, R.J.; Baker, S.C.; Baric, R.S.; Brown, C.S.; Drosten, C.; Enjuanes, L.; Fouchier, R.A.; Galiano, M.; Gorbalenya, A.E.; Memish, Z.A.; et al. Commentary: Middle East Respiratory Syndrome Coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. J. Virol. 2013, 87, 7790–7792. [Google Scholar] [CrossRef] [PubMed]
- Peiris, J.S.M.; Lai, S.T.; Poon, L.L.M.; Guan, Y.; Yam, L.Y.C.; Lim, W.; Nicholls, J.; Yee, W.K.S.; Yan, W.W.; Cheung, M.T.; et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003, 361, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Gao, S.-J. Global health concerns stirred by emerging viral infections. J. Med. Virol. 2020, 92, 399–400. [Google Scholar] [CrossRef]
- Ludwig, S.; Zarbock, A. Coronaviruses and SARS-CoV-2: A Brief Overview. Anesth. Analg. 2020, 131, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Fouchier, R.A.; Kuiken, T.; Schutten, M.; Van Amerongen, G.; Van Doornum, G.J.; Van Den Hoogen, B.G.; Peiris, M.; Lim, W.; Stöhr, K.; Osterhaus, A.D. Koch’s postulates fulfilled for SARS virus. Nature 2003, 423, 240. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, J.S.; Smith, D.W. COVID-19: A novel zoonotic disease caused by a coronavirus from China: What we know and what we don’t. Microbiol. Aust. 2020, 41, 45. [Google Scholar] [CrossRef]
- Lu, H.; Stratton, C.W.; Tang, Y.-W. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J. Med. Virol. 2020, 92, 401–402. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.R.; Cao, Q.D.; Hong, Z.S.; Tan, Y.Y.; Chen, S.D.; Jin, H.J.; Tan, K.S.; Wang, D.Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef]
- Horton, R. Offline: 2019-nCoV outbreak-early lessons. Lancet 2020, 395, 322. [Google Scholar] [CrossRef]
- World Health Organization. Why Is COVID-19 Data Being Presented as Weekly Statistics? [Online]. World Health Organization. 2024. Available online: https://data.who.int/dashboards/covid19/cases?m49=156&n=c (accessed on 13 April 2024).
- Belser, J.A.; Rota, P.A.; Tumpey, T.M. Ocular tropism of respiratory viruses. Microbiol. Mol. Biol. Rev. 2013, 77, 144–156. [Google Scholar] [CrossRef]
- Lu, Q.; Shi, Y. Coronavirus disease (COVID-19) and neonate: What neonatologist need to know. J. Med. Virol. 2020, 92, 564–567. [Google Scholar] [CrossRef]
- Li, H.; Leong, F.Y.; Xu, G.; Kang, C.W.; Lim, K.H.; Tan, B.H.; Loo, C.M. Airborne dispersion of droplets during coughing: A physical model of viral transmission. Sci. Rep. 2021, 11, 4617. [Google Scholar] [CrossRef]
- Jiang, F.; Deng, L.; Zhang, L.; Cai, Y.; Cheung, C.W.; Xia, Z. Review of the clinical characteristics of coronavirus disease 2019 (COVID-19). J. Gen. Intern. Med. 2020, 35, 1545–1549. [Google Scholar] [CrossRef]
- Chen, T. Fomites and the COVID-19 Pandemic: An Evidence Review on Its Role in Viral Transmission; National Collaborating Centre for Environmental Health: Vancouver, BC, Canada, 2021; pp. 1–24. [Google Scholar]
- Wu, D.; Wu, T.; Liu, Q.; Yang, Z. The SARS-CoV-2 outbreak: What we know. Int. J. Infect. Dis. 2020, 94, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Koenig, K.L.; Bey, C.K.; McDonald, E.C. 2019-nCoV: The identify-isolate-inform (3I) tool applied to a novel emerging coronavirus. West. J. Emerg. Med. 2020, 21, 184–190. [Google Scholar] [CrossRef]
- Tellier, R. COVID-19: The case for aerosol transmission. Interface Focus 2022, 12, 20210072. [Google Scholar] [CrossRef]
- Zhou, C. Evaluating new evidence in the early dynamics of the novel coronavirus COVID-19 outbreak in Wuhan, China with real time domestic traffic and potential asymptomatic transmissions. MedRxiv 2020. [Google Scholar] [CrossRef]
- Morawska, L.J.G.R.; Johnson, G.R.; Ristovski, Z.D.; Hargreaves, M.; Mengersen, K.; Corbett, S.; Chao, C.Y.H.; Li, Y.; Katoshevski, D. Size distribution and sites of origin of droplets expelled from the human respiratory tract during expiratory activities. J. Aerosol Sci. 2009, 40, 256–269. [Google Scholar] [CrossRef]
- Cortellessa, G.; Stabile, L.; Arpino, F.; Faleiros, D.E.; Van Den Bos, W.; Morawska, L.; Buonanno, G. Close proximity risk assessment for SARS-CoV-2 infection. Sci. Total Environ. 2021, 794, 148749. [Google Scholar] [CrossRef]
- Denison, M.R.; Graham, R.L.; Donaldson, E.F.; Eckerle, L.D.; Baric, R.S. Coronaviruses: An RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011, 8, 270–279. [Google Scholar] [CrossRef]
- Mlcochova, P.; Kemp, S.; Dhar, M.S.; Papa, G.; Meng, B.; Mishra, S.; Whittaker, C.; Mellan, T.; Ferreira, I.; Datir, R.; et al. SARS-CoV-2 B. 1.617. 2 Delta variant emergence, replication and sensitivity to neutralising antibodies. BioRxiv 2021. [Google Scholar] [CrossRef]
- Kandeel, M.; Mohamed, M.E.M.; Abd El-Lateef, H.M.A.; Venugopala, K.N.; El-Beltagi, H.S. Omicron variant genome evolution and phylogenetics. J. Med. Virol. 2022, 94, 1627–1632. [Google Scholar] [CrossRef]
- Planas, D.; Staropoli, I.; Michel, V.; Lemoine, F.; Donati, F.; Prot, M.; Porrot, F.; Guivel-Benhassine, F.; Jeyarajah, B.; Brisebarre, A.; et al. Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion. Nat. Commun. 2024, 15, 2254. [Google Scholar] [CrossRef] [PubMed]
- Stehlik, P.; Dowsett, C.; Camacho, X.; Falster, M.O.; Lim, R.; Nasreen, S.; Pratt, N.L.; Pearson, S.A.; Henry, D. Evolution of the data and methods in real-world COVID-19 vaccine effectiveness studies on mortality: A scoping review protocol. BMJ Open 2024, 14, e079071. [Google Scholar] [CrossRef] [PubMed]
- Haider, N.; Hasan, M.N.; Guitian, J.; Khan, R.A.; McCoy, D.; Ntoumi, F.; Dar, O.; Ansumana, R.; Uddin, J.; Zumla, A.; et al. The disproportionate case-fatality ratio of COVID-19 between countries with the highest vaccination rates and the rest of the world. IJID Reg. 2023, 6, 159–166. [Google Scholar] [CrossRef]
- World Health Organization. Statement on the Fifteenth Meeting of the IHR (2005) Emergency Committee on the COVID-19 Pandemic. [Online]. World Health Organization. 2023. Available online: https://www.who.int/news/item/05-05-2023-statement-on-the-fifteenth-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-coronavirus-disease-(covid-19)-pandemic (accessed on 26 June 2024).
- World Health Organization. Statement on the Update of Who’s Working Definitions and Tracking System for SARS-COV-2 Variants of Concern and Variants of Interest. [Online]. World Health Organization. 2023. Available online: https://www.who.int/news/item/16-03-2023-statement-on-the-update-of-who-s-working-definitions-and-tracking-system-for-sars-cov-2-variants-of-concern-and-variants-of-interest (accessed on 26 June 2024).
- Rodriguez, H.; Hartert, T.V.; Gebretsadik, T.; Carroll, K.N.; Larkin, E.K. A simple respiratory severity score that may be used in evaluation of acute respiratory infection. BMC Res. Notes 2016, 9, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Kaku, Y.; Yo, M.S.; Tolentino, J.E.; Uriu, K.; Okumura, K.; Ito, J.; Sato, K. Virological characteristics of the SARS-CoV-2 KP. 3, LB. 1, and KP. 2.3 variants. Lancet Infect. Dis. 2024. [Google Scholar] [CrossRef]
- World Health Organization. Coronavirus Disease (COVID-19) Situation Reports. [Online]. World Health Organization. 2024. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 26 June 2024).
- World Health Organization. COVID-19 Deaths WHO COVID-19 Dashboard. [Online]. World Health Organization. 2024. Available online: https://data.who.int/dashboards/covid19/deaths (accessed on 27 June 2024).
- Centers for Disease Control and Prevention. Vaccination Trends-Adults. [Online]. Centers for Disease Control and Prevention. 2024. Available online: https://www.cdc.gov/respiratory-viruses/data-research/dashboard/vaccination-trends-adults.html (accessed on 28 June 2024).
- European Centre for Disease Prevention and Control. Interim COVID-19 Vaccination Coverage in the EU/EEA during the 2023–2024 Season Campaigns 2024. [Online]. European Centre for Disease Prevention and Control. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/COVID-19-vaccination-coverage-2023%E2%80%9324.pdf (accessed on 28 June 2024).
- World Health Organization. COVID-19 Cases WHO COVID-19 Dashboard. [Online]. World Health Organization. 2024. Available online: https://data.who.int/dashboards/covid19/cases?m49=620&n=o (accessed on 26 June 2024).
- Centers for Disease Control and Prevention. COVID-19 Wastewater Data—National Trends. Centers for Disease Control and Prevention. 2024. Available online: https://www.cdc.gov/nwss/rv/COVID19-nationaltrend.html (accessed on 26 June 2024).
- Pulliam, J.R.; van Schalkwyk, C.; Govender, N.; von Gottberg, A.; Cohen, C.; Groome, M.J.; Dushoff, J.; Mlisana, K.; Moultrie, H. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science 2022, 376, eabn4947. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhu, W.; Han, X.; Chen, M.; Li, X.; Huang, H.; Zhang, M.; Wei, R.; Zhang, H.; Yang, C.; et al. How does the SARS-CoV-2 reinfection rate change over time? The global evidence from systematic review and meta-analysis. BMC Infect. Dis. 2024, 24, 339. [Google Scholar] [CrossRef]
- Guedes, A.R.; Oliveira, M.S.; Tavares, B.M.; Luna-Muschi, A.; Lazari, C.d.S.; Montal, A.C.; de Faria, E.; Maia, F.L.; Barboza, A.d.S.; Leme, M.D.; et al. Reinfection rate in a cohort of healthcare workers over 2 years of the COVID-19 pandemic. Sci. Rep. 2023, 13, 712. [Google Scholar] [CrossRef]
- Department of Health. COVID-19 Reinfection Data. [Online]. Department of Health. 2023. Available online: https://coronavirus.health.ny.gov/covid-19-reinfection-data (accessed on 26 June 2024).
- Wei, J.; Stoesser, N.; Matthews, P.C.; Khera, T.; Gethings, O.; Diamond, I.; Studley, R.; Taylor, N.; Peto, T.E.; Walker, A.S.; et al. Risk of SARS-CoV-2 reinfection during multiple Omicron variant waves in the UK general population. Nat. Commun. 2024, 15, 1008. [Google Scholar] [CrossRef]
- Hoeggerl, A.D.; Nunhofer, V.; Weidner, L.; Lauth, W.; Zimmermann, G.; Badstuber, N.; Grabmer, C.; Kartal, O.; Jungbauer, C.; Neureiter, H.; et al. Dissecting the dynamics of SARS-CoV-2 reinfections in blood donors with pauci-or asymptomatic COVID-19 disease course at initial infection. Infect. Dis. 2024, 1–11. [Google Scholar] [CrossRef]
- de Anda-Jáuregui, G.; Gómez-Romero, L.; Cañas, S.; Campos-Romero, A.; Alcántar-Fernández, J.; Cedro-Tanda, A. COVID-19 reinfections in Mexico City: Implications for public health. Front. Public Health 2024, 11, 1321283. [Google Scholar] [CrossRef]
- Antia, R.; Halloran, M.E. Transition to endemicity: Understanding COVID-19. Immunity 2021, 54, 2172–2176. [Google Scholar] [CrossRef] [PubMed]
- Akkiz, H. Implications of the novel mutations in the SARS-CoV-2 genome for transmission, disease severity, and the vaccine development. Front. Med. 2021, 8, 636532. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Hillyer, C.; Du, L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020, 41, 355–359. [Google Scholar] [CrossRef] [PubMed]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-prior, P. Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection. J. Biol. Chem. 2021, 296, 100135. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Deng, W.; Li, S.; Yang, X. Advances in research on ACE2 as a receptor for 2019-nCoV. Cell. Mol. Life Sci. 2020, 78, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Hanff, T.C.; Harhay, M.O.; Brown, T.S.; Cohen, J.B.; Mohareb, A.M. Is there an association between COVID-19 mortality and the renin-angiotensin system? A call for epidemiologic investigations. Clin. Infect. Dis. 2020, 71, 870–874. [Google Scholar] [CrossRef]
- Shieh, W.-J.; Hsiao, C.-H.; Paddock, C.D.; Guarner, J.; Goldsmith, C.S.; Tatti, K.; Packard, M.; Mueller, L.; Wu, M.-Z.; Rollin, P.; et al. Immunohistochemical, in situ hybridization, and ultrastructural localization of SARS-associated coronavirus in lung of a fatal case of severe acute respiratory syndrome in Taiwan. Hum. Pathol. 2005, 36, 303–309. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, Y.; Wang, D.W. SARS-CoV-2: A potential novel etiology of fulminant myocarditis. Herz 2020, 45, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef] [PubMed]
- Ebbole, D.J. Magnaporthe as a model for understanding host-pathogen interactions. Annu. Rev. Phytopathol. 2007, 45, 437–456. [Google Scholar] [CrossRef] [PubMed]
- Streicher, F.; Jouvenet, N. Stimulation of innate immunity by host and viral RNAs. Trends Immunol. 2019, 40, 1134–1148. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Hackbart, M.; Mettelman, R.C.; O’brien, A.; Mielech, A.M.; Yi, G.; Kao, C.C.; Baker, S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. USA 2017, 114, E4251–E4260. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhao, B.; Qu, Y.; Chen, Y.; Xiong, J.; Feng, Y.; Men, D.; Huang, Q.; Liu, Y.; Yang, B.; et al. Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (RNAemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus disease 2019. Clin. Infect. Dis. 2020, 71, 1937–1942. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.D.; Lemle, M.D.; Komaroff, A.L.; Snyder, S.H. Redox imbalance links COVID-19 and myalgic encephalomyelitis/chronic fatigue syndrome. Proc. Natl. Acad. Sci. USA 2021, 118, e2024358118. [Google Scholar] [CrossRef]
- Channappanavar, R.; Perlman, S. Pathogenic human coronavirus infections: Causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017, 39, 529–539. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef]
- Shi, D.; Weng, T.; Wu, J.; Dai, C.; Luo, R.; Chen, K.; Zhu, M.; Lu, X.; Cheng, L.; Chen, Q.; et al. Dynamic characteristic analysis of antibodies in patients with COVID-19: A 13-month study. Front. Immunol. 2021, 12, 708184. [Google Scholar] [CrossRef] [PubMed]
- Zohar, T.; Alter, G. Dissecting antibody-mediated protection against SARS-CoV-2. Nat. Rev. Immunol. 2020, 20, 392–394. [Google Scholar] [CrossRef] [PubMed]
- Vetter, P.; Eberhardt, C.S.; Meyer, B.; Murillo, P.A.M.; Torriani, G.; Pigny, F.; Lemeille, S.; Cordey, S.; Laubscher, F.; Vu, D.-L.; et al. Daily viral kinetics and innate and adaptive immune response assessment in COVID-19: A case series. mSphere 2020, 5, e00827-20. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.M.; Bai, P.; He, W.; Wu, F.; Liu, X.F.; Han, D.M.; Liu, S.; Yang, J.K. Gender differences in patients with COVID-19: Focus on severity and mortality. Front. Public Health 2020, 8, 152. [Google Scholar] [CrossRef]
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J.; on behalf of the HLH Across Speciality Collaboration, UK. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Singh, M.K.; Mobeen, A.; Chandra, A.; Joshi, S.; Ramachandran, S. A meta-analysis of comorbidities in COVID-19: Which diseases increase the susceptibility of SARS-CoV-2 infection? Comput. Biol. Med. 2021, 130, 104219. [Google Scholar] [CrossRef]
- Agrawal, U.; Azcoaga-Lorenzo, A.; Fagbamigbe, A.F.; Vasileiou, E.; Henery, P.; Simpson, C.R.; Stock, S.J.; Shah, S.A.; Robertson, C.; Woolhouse, M.; et al. Association between multimorbidity and mortality in a cohort of patients admitted to hospital with COVID-19 in Scotland. J. R. Soc. Med. 2021, 115, 22–30. [Google Scholar] [CrossRef]
- Carfì, A.; Bernabei, R.; Landi, F. Persistent symptoms in patients after acute COVID-19. JAMA 2020, 324, 603–605. [Google Scholar] [CrossRef]
- Shah, W.; Hillman, T.; Playford, E.D.; Hishmeh, L. Managing the long term effects of COVID-19: Summary of NICE, SIGN, and RCGP rapid guideline. BMJ 2021, 372, n136. [Google Scholar] [CrossRef]
- Greenhalgh, T.; Knight, M.; A’court, C.; Buxton, M.; Husain, L. Management of post-acute COVID-19 in primary care. BMJ 2020, 370, m3026. [Google Scholar] [CrossRef] [PubMed]
- van Kampen, J.J.; van de Vijver, D.A.; Fraaij, P.L.; Haagmans, B.L.; Lamers, M.M.; Okba, N.; van den Akker, J.P.; Endeman, H.; Gommers, D.A.; Cornelissen, J.J.; et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat. Commun. 2021, 12, 267. [Google Scholar] [CrossRef]
- Raveendran, A. Long COVID-19: Challenges in the diagnosis and proposed diagnostic criteria. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 15, 145–146. [Google Scholar] [CrossRef] [PubMed]
- Ceban, F.; Ling, S.; Lui, L.M.; Lee, Y.; Gill, H.; Teopiz, K.M.; Rodrigues, N.B.; Subramaniapillai, M.; Di Vincenzo, J.D.; Cao, B.; et al. Fatigue and cognitive impairment in Post-COVID-19 Syndrome: A systematic review and meta-analysis. Brain Behav. Immun. 2021, 101, 93–135. [Google Scholar] [CrossRef]
- Ayoubkhani, D.; Bosworth, M.L.; King, S.; Pouwels, K.B.; Glickman, M.; Nafilyan, V.; Zaccardi, F.; Khunti, K.; Alwan, N.A.; Walker, A.S. Risk of Long Covid in people infected with SARS-CoV-2 after two doses of a COVID-19 vaccine: Community-based, matched cohort study. medRxiv 2022. [Google Scholar] [CrossRef]
- Subramanian, A.; Nirantharakumar, K.; Hughes, S.; Myles, P.; Williams, T.; Gokhale, K.M.; Taverner, T.; Chandan, J.S.; Brown, K.; Simms-Williams, N.; et al. Symptoms and risk factors for long COVID in non-hospitalized adults. Nat. Med. 2022, 28, 1706–1714. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Zheng, B.; Daines, L.; Sheikh, A. Long-term sequelae of COVID-19: A systematic review and meta-analysis of one-year follow-up studies on post-COVID symptoms. Pathogens 2022, 11, 269. [Google Scholar] [CrossRef] [PubMed]
- Singh, I.; Joseph, P.; Heerdt, P.M.; Cullinan, M.; Lutchmansingh, D.D.; Gulati, M.; Possick, J.D.; Systrom, D.M.; Waxman, A.B. Persistent exertional intolerance after COVID-19: Insights from invasive cardiopulmonary exercise testing. Chest 2022, 161, 54–63. [Google Scholar] [CrossRef]
- Fogarty, H.; Townsend, L.; Morrin, H.; Ahmad, A.; Comerford, C.; Karampini, E.; Englert, H.; Byrne, M.; Bergin, C.; O’sullivan, J.M.; et al. Persistent endotheliopathy in the pathogenesis of long COVID syndrome. J. Thromb. Haemost. 2021, 19, 2546–2553. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, Y.; Li, Y.; Wang, Z.; Ma, F.; Luo, R.; Xu, X.; Zhou, G.; Wang, J.; Niu, J.; et al. Platelets mediate inflammatory monocyte activation by SARS-CoV-2 spike protein. J. Clin. Investig. 2022, 132, e150101. [Google Scholar] [CrossRef]
- Grobbelaar, L.M.; Venter, C.; Vlok, M.; Ngoepe, M.; Laubscher, G.J.; Lourens, P.J.; Steenkamp, J.; Kell, D.B.; Pretorius, E. SARS-CoV-2 spike protein S1 induces fibrin (ogen) resistant to fibrinolysis: Implications for microclot formation in COVID-19. Biosci. Rep. 2021, 41, BSR20210611. [Google Scholar] [CrossRef]
- Ajaz, S.; McPhail, M.J.; Singh, K.K.; Mujib, S.; Trovato, F.M.; Napoli, S.; Agarwal, K. Mitochondrial metabolic manipulation by SARS-CoV-2 in peripheral blood mononuclear cells of patients with COVID-19. Am. J. Physiol.-Cell Physiol. 2021, 320, C57–C65. [Google Scholar] [CrossRef] [PubMed]
- Tsivgoulis, G.; Fragkou, P.C.; Lachanis, S.; Palaiodimou, L.; Lambadiari, V.; Papathanasiou, M.; Sfikakis, P.P.; Voumvourakis, K.I.; Tsiodras, S. Olfactory bulb and mucosa abnormalities in persistent COVID-19-induced anosmia: A magnetic resonance imaging study. Eur. J. Neurol. 2020, 28, E6–E8. [Google Scholar] [CrossRef]
- Brann, D.H.; Tsukahara, T.; Weinreb, C.; Lipovsek, M.; Van den Berge, K.; Gong, B.; Chance, R.; Macaulay, I.C.; Chou, H.J.; Fletcher, R.B.; et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 2020, 6, eabc5801. [Google Scholar] [CrossRef]
- Vaira, L.A.; Hopkins, C.; Sandison, A.; Manca, A.; Machouchas, N.; Turilli, D.; Lechien, J.R.; Barillari, M.R.; Salzano, G.; Cossu, A.; et al. Olfactory epithelium histopathological findings in long-term coronavirus disease 2019 related anosmia. J. Laryngol. Otol. 2020, 134, 1123–1127. [Google Scholar] [CrossRef]
- Davis, H.E.; Mccorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Català, M.; Mercadé-Besora, N.; Kolde, R.; Trinh, N.T.; Roel, E.; Burn, E.; Rathod-Mistry, T.; Kostka, K.; Man, W.Y.; Delmestri, A.; et al. The effectiveness of COVID-19 vaccines to prevent long COVID symptoms: Staggered cohort study of data from the UK, Spain, and Estonia. Lancet Respir. Med. 2024, 12, 225–236. [Google Scholar] [CrossRef]
- Watanabe, A.; Iwagami, M.; Yasuhara, J.; Takagi, H.; Kuno, T. Protective effect of COVID-19 vaccination against long COVID syndrome: A systematic review and meta-analysis. Vaccine 2023, 41, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Mccarthy, M.W. Metformin as a potential treatment for COVID-19. Expert Opin. Pharmacother. 2023, 24, 1199–1203. [Google Scholar] [CrossRef]
- Mccarthy, M.W. Paxlovid as a potential treatment for long COVID. Expert Opin. Pharmacother. 2023, 24, 1839–1843. [Google Scholar] [CrossRef]
- Knopman, D.S.; Laskowitz, D.T.; Koltai, D.C.; Charvet, L.E.; Becker, J.H.; Federman, A.D.; Wisnivesky, J.; Mahncke, H.; Van Vleet, T.M.; Bateman, L.; et al. RECOVER-NEURO: Study protocol for a multi-center, multi-arm, phase 2, randomized, active comparator trial evaluating three interventions for cognitive dysfunction in post-acute sequelae of SARS-CoV-2 infection (PASC). Trials 2024, 25, 326. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, K. RECOVER-AUTONOMIC: A Platform Protocol for Evaluation of Interventions for Autonomic Dysfunction in Post-Acute Sequelae of SARS-CoV-2 Infection (PASC). [Online]. 2024. Available online: https://trials.recovercovid.org/documents/RECOVER_AUTONOMIC_Protocol_V3.0.pdf (accessed on 30 June 2024).
- Zimmerman, K. RECOVER-SLEEP: A Platform Protocol for Evaluation of Interventions for Sleep Disturbances in Post-Acute Sequelae of SARS-CoV-2 Infection (PASC). [Online]. 2024. Available online: https://trials.recovercovid.org/documents/RECOVER_SLEEP_Protocol_V3.0.pdf (accessed on 30 June 2024).
- Gao, W.; Lv, J.; Pang, Y.; Li, L.-M. Role of asymptomatic and pre-symptomatic infections in COVID-19 pandemic. BMJ 2021, 375, n2342. [Google Scholar] [CrossRef]
- Ravindra, K.; Malik, V.; Padhi, B.; Goel, S.; Gupta, M. Asymptomatic infection and transmission of COVID-19 among clusters: Systematic review and meta-analysis. Public Health 2021, 203, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Jayaweera, M.; Perera, H.; Gunawardana, B.; Manatunge, J. Transmission of Null COVID-19 virus by droplets and aerosols: A critical review on the unresolved dichotomy. Environ. Res. 2020, 188, 109819. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. In Vitro Diagnostics for COVID-19 [Online]. Available online: https://www.who.int/teams/health-product-policy-and-standards/assistive-and-medical-technology/medical-devices/priority-medical-devices-for-covid/diagnostics-for-covid-19 (accessed on 6 July 2024).
- Food and Drug Administration. COVID-19 Test Basics [Online]. 2023. Available online: https://www.fda.gov/consumers/consumer-updates/covid-19-test-basics (accessed on 6 July 2024).
- Castellanos, M.; Somoza, Á. Emerging clinically tested detection methods for COVID-19. FEBS J. 2022, 290, 3089–3104. [Google Scholar] [CrossRef] [PubMed]
- Maia, R.; Carvalho, V.; Faria, B.; Miranda, I.; Catarino, S.; Teixeira, S.; Lima, R.; Minas, G.; Ribeiro, J. Diagnosis Methods for COVID-19: A Systematic Review. Micromachines 2022, 13, 1349. [Google Scholar] [CrossRef]
- Dong, H.; Zhang, K.; Zhang, J.; Xiao, Y.; Zhang, F.; Wang, M.; Wang, H.; Zhao, G.; Xie, S.; Xie, X.; et al. A fast RT-qPCR system significantly shortens the time for SARS-CoV-2 nucleic acid test. Drug Discov. Ther. 2023, 17, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Tsang, H.F.; Leung, W.M.S.; Chan, L.W.C.; Cho, W.C.S.; Wong, S.C.C. Performance comparison of the Cobas® Liat® and Cepheid® GeneXpert® systems on SARS-CoV-2 detection in nasopharyngeal swab and posterior oropharyngeal saliva. Expert Rev. Mol. Diagn. 2021, 21, 515–518. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tan, S.H.; Allicock, O.; Armstrong-Hough, M.; Wyllie, A.L. Saliva as a gold-standard sample for SARS-CoV-2 detection. Lancet Respir. Med. 2021, 9, 562–564. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, X.; Sun, J.; Ye, J.; Wang, F.; Hua, J.; Zhang, H.; Shi, T.; Li, Q.; Wu, X. Differences of Severe Acute Respiratory Syndrome Coronavirus 2 Shedding Duration in Sputum and Nasopharyngeal Swab Specimens among Adult Inpatients with Coronavirus Disease 2019. Chest 2020, 158, 1876–1884. [Google Scholar] [CrossRef]
- Akowuah, E.; Acheampong, G.; Ayisi-Boateng, N.K.; Amaniampong, A.; Agyapong, F.O.; Senyo Kamasah, J.; Agyei, G.; Owusu, D.O.; Nkrumah, B.; Mutocheluh, M.; et al. Comparable Detection of SARS-CoV-2 in Sputum and Oropharyngeal Swab Samples of Suspected COVID-19 Patients. COVID 2022, 2, 858–866. [Google Scholar] [CrossRef]
- Ahmadzadeh, M.; Vahidi, H.; Mahboubi, A.; Hajifathaliha, F.; Nematollahi, L.; Mohit, E. Different Respiratory Samples for COVID-19 Detection by Standard and Direct Quantitative RT-PCR: A Literature Review. Iran. J. Pharm. Res. IJPR 2021, 20, 285–299. [Google Scholar]
- Ali, D.Y.; Hussein, R.A.; Elshafie, S.M.; Mohamed, R.A.; El Reheem, F.A. Comparable detection of nasopharyngeal swabs and induced sputum specimens for viral nucleic acid detection of suspected novel coronavirus (SARS-Cov-2) patients in Fayoum governorate, Egypt. Beni-Suef Univ. J. Basic Appl. Sci. 2023, 12, 43. [Google Scholar] [CrossRef]
- Alexandersen, S.; Chamings, A.; Bhatta, T.R. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 2020, 11, 6059. [Google Scholar] [CrossRef] [PubMed]
- Maniruzzaman, M.; Islam, M.M.; Ali, M.H.; Mukerjee, N.; Maitra, S.; Kamal, M.A.; Ghosh, A.; Castrosanto, M.A.; Alexiou, A.; Ashraf, G.M.; et al. COVID-19 diagnostic methods in developing countries. Environ. Sci. Pollut. Res. Int. 2022, 29, 51384–51397. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, T.; Spencer, E.A.; Conly, J.M.; Rosca, E.C.; Maltoni, S.; Brassey, J.; Onakpoya, I.J.; Evans, D.H.; Heneghan, C.J.; Plüddemann, A. Viral cultures, cycle threshold values and viral load estimation for assessing SARS-CoV-2 infectiousness in haematopoietic stem cell and solid organ transplant patients: A systematic review. J. Hosp. Infect. 2023, 132, 62–72. [Google Scholar] [CrossRef]
- Benevides Lima, L.; Mesquita, F.P.; Brasil de Oliveira, L.L.; Andréa da Silva Oliveira, F.; Elisabete Amaral de Moraes, M.; Souza, P.F.; Montenegro, R.C. True or false: What are the factors that influence COVID-19 diagnosis by RT-qPCR? Expert Rev. Mol. Diagn. 2022, 22, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.-R. Eleven golden rules of quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef]
- Panchali, M.J.L.; Oh, H.J.; Lee, Y.M.; Kim, C.-M.; Tariq, M.; Seo, J.-W.; Kim, D.Y.; Yun, N.R.; Kim, D.-M. Accuracy of Real-Time Polymerase Chain Reaction in COVID-19 Patients. Microbiol. Spectr. 2022, 10, e0059121. [Google Scholar] [CrossRef]
- Bello-Lemus, Y.; Anaya-Romero, M.; Gómez-Montoya, J.; Árquez, M.; González-Torres, H.J.; Navarro-Quiroz, E.; Pacheco-Londoño, L.; Pacheco-Lugo, L.; Acosta-Hoyos, A.J. Comparative Analysis of In-House RT-qPCR Detection of SARS-CoV-2 for Resource-Constrained Settings. Diagnostics 2022, 12, 2883. [Google Scholar] [CrossRef]
- Walsh, K.A.; Jordan, K.; Clyne, B.; Rohde, D.; Drummond, L.; Byrne, P.; Ahern, S.; Carty, P.G.; O’Brien, K.K.; O’Murchu, E.; et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J. Infect. 2020, 81, 357–371. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Yi, S.; Zhang, J.; Lv, Z.; Zhu, C.; Zhang, Y. Viral Load Dynamics in Sputum and Nasopharyngeal Swab in Patients with COVID-19. J. Dent. Res. 2020, 99, 1239–1244. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January–March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration. In Vitro Diagnostics EUAs—Molecular Diagnostic Tests for SARS-CoV-2 [Online]. 2024. Available online: https://www.fda.gov/medical-devices/covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-molecular-diagnostic-tests-sars-cov-2 (accessed on 1 July 2024).
- Johns Hopkins Center for Health Security. Antigen and Molecular Tests for COVID-19 [Online]. 2022. Available online: https://covid19testingtoolkit.centerforhealthsecurity.org/testing-trackers/antigen-and-molecular-tests-for-covid-19#lab (accessed on 3 July 2024).
- Phan, T.; Valeriano, P.; Boes, S.; McCullough, M.; Gribschaw, J.; Wells, A. Evaluation of the ePlex Respiratory pathogen panel 2 to detect viral and bacterial pathogens, including SARS-CoV-2 Omicron in nasopharyngeal swabs. J. Clin. Virol. Plus 2022, 2, 100072. [Google Scholar] [CrossRef]
- Sahajpal, N.S.; Mondal, A.K.; Ananth, S.; Njau, A.; Jones, K.; Ahluwalia, P.; Oza, E.; Ross, T.M.; Kota, V.; Kothandaraman, A.; et al. Clinical validation of a multiplex PCR-based detection assay using saliva or nasopharyngeal samples for SARS-Cov-2, influenza A and B. Sci. Rep. 2022, 12, 3480. [Google Scholar] [CrossRef]
- Hashemi, S.A.; Safamanesh, S.; Ghasemzadeh-moghaddam, H.; Ghafouri, M.; Azimian, A. High prevalence of SARS-CoV-2 and influenza A virus (H1N1) coinfection in dead patients in Northeastern Iran. J. Med. Virol. 2021, 93, 1008–1012. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Zhang, Y.; Hang, C.; Ai, J.; Li, S.; Zhang, W. Comparisons of viral shedding time of SARS-CoV-2 of different samples in ICU and non-ICU patients. J. Infect. 2020, 81, 147–178. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Zhou, J.; Niu, C.; Wang, Q.; Pan, Y.; Sheng, S.; Wang, X.; Zhang, Y.; Yang, J.; Liu, M.; et al. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. Talanta 2021, 224, 121726. [Google Scholar] [CrossRef] [PubMed]
- Suo, T.; Liu, X.; Feng, J.; Guo, M.; Hu, W.; Guo, D.; Ullah, H.; Yang, Y.; Zhang, Q.; Wang, X.; et al. ddPCR: A more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020, 9, 1259–1268. [Google Scholar] [CrossRef]
- Dhar, B.C. Diagnostic assay and technology advancement for detecting SARS-CoV-2 infections causing the COVID-19 pandemic. Anal. Bioanal. Chem. 2022, 414, 2903–2934. [Google Scholar] [CrossRef]
- Shafie, M.H.; Antony Dass, M.; Ahmad Shaberi, H.S.; Zafarina, Z. Screening and confirmation tests for SARS-CoV-2: Benefits and drawbacks. Beni Suef Univ. J. Basic Appl. Sci. 2023, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.; Moehling, T.J.; Meagher, R.J. Advances in RT-LAMP for COVID-19 testing and diagnosis. Expert Rev. Mol. Diagn. 2023, 23, 9–28. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ma, Y.; Song, Z.; Sun, W.; Liu, Y.; Shu, C.; Hua, H.; Yang, M.; Liang, Q. Evaluation of an automated CRISPR-based diagnostic tool for rapid detection of COVID-19. Heliyon 2023, 9, e13190. [Google Scholar] [CrossRef] [PubMed]
- New England Biolabs. Loop-Mediated Isothermal Amplification [Online]. Available online: https://www.neb.com/en/applications/dna-amplification-pcr-and-qpcr/isothermal-amplification/loop-mediated-isothermal-amplification-lamp (accessed on 15 May 2024).
- Centers for Disease Control and Prevention. Interim Guidelines for COVID-19 Antibody Testing in Clinical and Public Health Settings [Online]. 2022. Available online: https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing/antibody-tests-guidelines.html (accessed on 10 April 2024).
- Food and Drug Administration. Antibody (Serology) Testing for COVID-19: Information for Patients and Consumers [Online]. 2023. Available online: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/antibody-serology-testing-covid-19-information-patients-and-consumers (accessed on 10 April 2024).
- Centers for Disease Control and Prevention. Considerations for SARS-CoV-2 Antigen Testing for Healthcare Providers Testing Individuals in the Community [Online]. 2023. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html (accessed on 10 April 2024).
- American Society for Microbiology. How the SARS-CoV-2 EUA Antigen Tests Work [Online]. 2020. Available online: https://asm.org/articles/2020/august/how-the-sars-cov-2-eua-antigen-tests-work (accessed on 10 April 2024).
- Hashim, I.A. (Ed.) Chapter 17—Analytical methods and special considerations. In Tutorials in Clinical Chemistry; Elsevier: Amsterdam, The Netherlands, 2024. [Google Scholar]
- Food and Drug Administration. At Home OTC COVID-19 Diagnostic Tests [Online]. 2024. Available online: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/home-otc-covid-19-diagnostic-tests (accessed on 10 April 2024).
- Katzenschlager, S.; Bruemmer, L.E.; Schmitz, S.; Tolle, H.; Manten, K.; Gaeddert, M.; Erdmann, C.; Lindner, A.; Tobian, F.; Grilli, M.; et al. Comparing SARS-CoV-2 antigen-detection rapid diagnostic tests for COVID-19 self-testing/self-sampling with molecular and professional-use tests: A systematic review and meta-analysis. Sci. Rep. 2023, 13, 21913. [Google Scholar] [CrossRef] [PubMed]
- Yadegari, H.; Mohammadi, M.; Maghsood, F.; Ghorbani, A.; Bahadori, T.; Golsaz-Shirazi, F.; Zarnani, A.H.; Salimi, V.; Jeddi-Tehrani, M.; Amiri, M.M.; et al. Diagnostic performance of a novel antigen-capture ELISA for the detection of SARS-CoV-2. Anal. Biochem. 2023, 666, 115079. [Google Scholar] [CrossRef] [PubMed]
- Adnan, N.; Khandker, S.S.; Haq, A.; Chaity, M.A.; Khalek, A.; Nazim, A.Q.; Kaitsuka, T.; Tomizawa, K.; Mie, M.; Kobatake, E.; et al. Detection of SARS-CoV-2 by antigen ELISA test is highly swayed by viral load and sample storage condition. Expert Rev. Anti-Infect. Ther. 2022, 20, 473–481. [Google Scholar] [CrossRef]
- Weiss, A.; Jellingsø, M.; Sommer, M.O.A. Spatial and temporal dynamics of SARS-CoV-2 in COVID-19 patients: A systematic review and meta-analysis. eBioMedicine 2020, 58, 102916. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Li, Y.; Wu, P.; Li, Z.; Lau, E.H.Y.; Qin, Y.; Wang, L.; Cowling, B.J.; Tsang, T.K.; Li, Z. Estimating the Latent Period of Coronavirus Disease 2019 (COVID-19). Clin. Infect. Dis. 2021, 74, 1678–1681. [Google Scholar] [CrossRef] [PubMed]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef]
- Hagag, I.T.; Pyrc, K.; Weber, S.; Balkema-Buschmann, A.; Groschup, M.H.; Keller, M. Mutations in SARS-CoV-2 nucleocapsid in variants of concern impair the sensitivity of SARS-CoV-2 detection by rapid antigen tests. Front. Virol. 2022, 2, 971862. [Google Scholar] [CrossRef]
- Johnson, B.A.; Zhou, Y.; Lokugamage, K.G.; Vu, M.N.; Bopp, N.; Crocquet-Valdes, P.A.; Kalveram, B.; Schindewolf, C.; Liu, Y.; Scharton, D.; et al. Nucleocapsid mutations in SARS-CoV-2 augment replication and pathogenesis. PLoS Pathog. 2022, 18, e1010627. [Google Scholar] [CrossRef] [PubMed]
- El-Daly, M.M. Advances and Challenges in SARS-CoV-2 Detection: A Review of Molecular and Serological Technologies. Diagnostics 2024, 14, 519. [Google Scholar] [CrossRef] [PubMed]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Holmes, E.C. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell 2020, 181, 223–227. [Google Scholar] [CrossRef]
- World Health Organization. Genomic Sequencing of SARS-CoV-2: A Guide to Implementation for Maximum Impact on Public Health. 8 January 2021. Available online: https://www.who.int/publications/i/item/9789240018440 (accessed on 24 July 2024).
- ImmunoDiagnostics. SARS-CoV-2 NP Ab ELISA Kit [CE-IVD] [Online]. Available online: https://www.immunodiagnostics.com.hk/product-page/sars-cov-2-np-ab-elisa-kit-ce-ivd (accessed on 3 July 2024).
- Centers for Disease Control and Prevention. Understanding How COVID-19 Vaccines Work [Online]. 2023. Available online: https://www.cdc.gov/coronavirus/2019-ncov/vaccines/different-vaccines/how-they-work.html (accessed on 3 July 2024).
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; Peacock, S.J.; Barclay, W.S.; De Silva, T.I.; Towers, G.J.; Robertson, D.L. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef]
- Papa Mze, N.; Kacel, I.; Beye, M.; Tola, R.; Sarr, M.; Basco, L.; Bogreau, H.; Colson, P.; Fournier, P.E. High Throughput SARS-CoV-2 Genome Sequencing from 384 Respiratory Samples Using the Illumina COVIDSeq Protocol. Genes 2023, 14, 681. [Google Scholar] [CrossRef]
- Fox, T.; Geppert, J.; Dinnes, J.; Scandrett, K.; Bigio, J.; Sulis, G.; Hettiarachchi, D.; Mathangasinghe, Y.; Weeratunga, P.; Wickramasinghe, D.; et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst. Rev. 2022, 6, cd013652. [Google Scholar] [CrossRef]
- Kontou, P.I.; Braliou, G.G.; Dimou, N.L.; Nikolopoulos, G.; Bagos, P.G. Antibody Tests in Detecting SARS-CoV-2 Infection: A Meta-Analysis. Diagnostics 2020, 10, 319. [Google Scholar] [CrossRef]
- Matinfar, S.; Mortezagholi, S.; Amiri, D.; Pashaiefar, H.; Eskandarian, M.; Ghadimi, S.; Nazari, M.F.; Tavakoli, S.; Valizadeh, M.; Namaki, S.; et al. Investigating the Seroconversion Patterns of Specific Antibodies against Various Antigens of SARS-CoV-2 in Hospitalized COVID-19 Patients and Vaccinated Individuals. Arch. Clin. Infect. Dis. 2024, 19, e140414. [Google Scholar] [CrossRef]
- Imai, K.; Kitagawa, Y.; Tabata, S.; Kubota, K.; Nagura-Ikeda, M.; Matsuoka, M.; Miyoshi, K.; Sakai, J.; Ishibashi, N.; Tarumoto, N.; et al. Antibody response patterns in COVID-19 patients with different levels of disease severity in Japan. J. Med. Virol. 2021, 93, 3211–3218. [Google Scholar] [CrossRef]
- Nakano, Y.; Kurano, M.; Morita, Y.; Shimura, T.; Yokoyama, R.; Qian, C.; Xia, F.; He, F.; Kishi, Y.; Okada, J.; et al. Time course of the sensitivity and specificity of anti-SARS-CoV-2 IgM and IgG antibodies for symptomatic COVID-19 in Japan. Sci. Rep. 2021, 11, 2776. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.X.; Liu, B.Z.; Deng, H.J.; Wu, G.C.; Deng, K.; Chen, Y.K.; Liao, P.; Qiu, J.F.; Lin, Y.; Cai, X.F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- Bhattacharjee, B.; Rynjah, D.; Ahmed, A.B.; Newar, A.; Sengupta, S.; Chakrabarty, S.; Sahu, R.K.; Khan, J. Overview of diagnostic tools and nano-based therapy of SARS-CoV-2 infection. Chem. Pap. 2024, 78, 2123–2154. [Google Scholar] [CrossRef]
- Food and Drug Administration. In Vitro Diagnostics Emergency Use Authorizations (EUAs)—Serology and Other Adaptive Immune Response Tests for SARS-CoV-2 [Online]. 2024. Available online: https://www.fda.gov/medical-devices/covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-emergency-use-authorizations-euas-serology-and-other-adaptive-immune-response (accessed on 3 July 2024).
- Augustine, R.; Das, S.; Hasan, A.; Abdul Salam, S.; Augustine, P.; Dalvi, Y.B.; Varghese, R.; Primavera, R.; Yassine, H.M.; Thakor, A.S.; et al. Rapid antibody-based COVID-19 mass surveillance: Relevance, challenges, and prospects in a pandemic and post-pandemic world. J. Clin. Med. 2020, 9, 3372. [Google Scholar] [CrossRef]
- Dimech, W.; Curley, S.; Subissi, L.; Ströher, U.; Perkins, M.D.; Cunningham, J. Comprehensive, Comparative Evaluation of 35 Manual SARS-CoV-2 Serological Assays. Microbiol. Spectr. 2023, 11, e05101-22. [Google Scholar] [CrossRef]
- Dimech, W.; Curley, S.; Cai, J.J. Comprehensive, comparative evaluation of 25 automated SARS-CoV-2 serology assays. Microbiol. Spectr. 2024, 12, e03228-23. [Google Scholar] [CrossRef]
- Li, G.; Hilgenfeld, R.; Whitley, R.; De Clercq, E. Therapeutic strategies for COVID-19: Progress and lessons learned. Nat. Rev. Drug Discov. 2023, 22, 449–475. [Google Scholar] [CrossRef] [PubMed]
- Eastman, R.T.; Roth, J.S.; Brimacombe, K.R.; Simeonov, A.; Shen, M.; Patnaik, S.; Hall, M.D. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Central Sci. 2020, 6, 672–683. [Google Scholar] [CrossRef]
- Barghash, R.F.; Fawzy, I.M.; Chandrasekar, V.; Singh, A.V.; Katha, U.; Mandour, A.A. In Silico Modeling as a Perspective in Developing Potential Vaccine Candidates and Therapeutics for COVID-19. Coatings 2021, 11, 1273. [Google Scholar] [CrossRef]
- Gottlieb, R.L.; Vaca, C.E.; Paredes, R.; Mera, J.; Webb, B.J.; Perez, G.; Oguchi, G.; Ryan, P.; Nielsen, B.U.; Brown, M.; et al. Early Remdesivir to Prevent Progression to Severe Covid-19 in Outpatients. N. Engl. J. Med. 2021, 386, 305–315. [Google Scholar] [CrossRef]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of COVID-19—Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef]
- Xie, J.; Wang, Z. Can remdesivir and its parent nucleoside GS-441524 be potential oral drugs? An in vitro and in vivo DMPK assessment. Acta Pharm. Sin. B 2021, 11, 1607–1616. [Google Scholar] [CrossRef]
- Ader, F.; Bouscambert-Duchamp, M.; Hites, M.; Peiffer-Smadja, N.; Poissy, J.; Belhadi, D.; Diallo, A.; Lê, M.-P.; Peytavin, G.; Staub, T.; et al. Remdesivir plus standard of care versus standard of care alone for the treatment of patients admitted to hospital with COVID-19 (DisCoVeRy): A phase 3, randomised, controlled, open-label trial. Lancet Infect. Dis. 2021, 22, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Peto, R.; Restrepo, A.M.H.; Preziosi, M.-P.; Sathiyamoorthy, V.; Karim, Q.A.; Alejandria, M.; García, C.H.; Kieny, M.-P.; Malekzadeh, R.; et al. Remdesivir and three other drugs for hospitalised patients with COVID-19: Final results of the WHO Solidarity randomised trial and updated meta-analyses. Lancet 2022, 399, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Kalil, A.C.; Patterson, T.F.; Mehta, A.K.; Tomashek, K.M.; Wolfe, C.R.; Ghazaryan, V.; Marconi, V.C.; Ruiz-Palacios, G.M.; Hsieh, L.; Kline, S.; et al. Baricitinib plus Remdesivir for Hospitalized Adults with COVID-19. N. Engl. J. Med. 2021, 384, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Kalil, A.C.; Mehta, A.K.; Patterson, T.F.; Erdmann, N.; Gomez, C.A.; Jain, M.K.; Wolfe, C.R.; Ruiz-Palacios, G.M.; Kline, S.; Pineda, J.R.; et al. Efficacy of interferon beta-1a plus remdesivir compared with remdesivir alone in hospitalised adults with COVID-19: A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Respir. Med. 2021, 9, 1365–1376. [Google Scholar] [CrossRef]
- Focosi, D.; Maggi, F.; McConnell, S.; Casadevall, A. Very low levels of remdesivir resistance in SARS-CoV-2 genomes after 18 months of massive usage during the COVID19 pandemic: A GISAID exploratory analysis. Antivir. Res. 2022, 198, 105247. [Google Scholar] [CrossRef]
- Hedskog, C.; Rodriguez, L.; Roychoudhury, P.; Huang, M.-L.; Jerome, K.R.; Hao, L.; Ireton, R.C.; Li, J.; Perry, J.K.; Han, D.; et al. Viral Resistance Analyses from the Remdesivir Phase 3 Adaptive COVID-19 Treatment Trial-1 (ACTT-1). J. Infect. Dis. 2023, 228, 1263–1273. [Google Scholar] [CrossRef]
- Vangeel, L.; Chiu, W.; De Jonghe, S.; Maes, P.; Slechten, B.; Raymenants, J.; André, E.; Leyssen, P.; Neyts, J.; Jochmans, D. Remdesivir, Molnupiravir and Nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antivir. Res. 2022, 198, 105252. [Google Scholar] [CrossRef]
- Uraki, R.; Ito, M.; Kiso, M.; Yamayoshi, S.; Iwatsuki-Horimoto, K.; Sakai-Tagawa, Y.; Imai, M.; Koga, M.; Yamamoto, S.; Adachi, E.; et al. Antiviral efficacy against and replicative fitness of an XBB.1.9.1 clinical isolate. iScience 2023, 26, 108147. [Google Scholar] [CrossRef]
- Pitts, J.; Li, J.; Perry, J.K.; Du Pont, V.; Riola, N.; Rodriguez, L.; Lu, X.; Kurhade, C.; Xie, X.; Camus, G.; et al. Remdesivir and GS-441524 Retain Antiviral Activity against Delta, Omicron, and Other Emergent SARS-CoV-2 Variants. Antimicrob. Agents Chemother. 2022, 66, e0022222. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Gao, W.; Bao, H.; Feng, H.; Mei, S.; Chen, P.; Gao, Y.; Cui, Z.; Zhang, Q.; Meng, X.; et al. VV116 versus Nirmatrelvir–Ritonavir for Oral Treatment of COVID-19. N. Engl. J. Med. 2023, 388, 406–417. [Google Scholar] [CrossRef]
- Fan, X.; Dai, X.; Ling, Y.; Wu, L.; Tang, L.; Peng, C.; Huang, C.; Liu, H.; Lu, H.; Shen, X.; et al. Oral VV116 versus placebo in patients with mild-to-moderate COVID-19 in China: A multicentre, double-blind, phase 3, randomised controlled study. Lancet Infect. Dis. 2024, 24, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Hashemian, S.M.; Sheida, A.; Taghizadieh, M.; Memar, M.Y.; Hamblin, M.R.; Baghi, H.B.; Nahand, J.S.; Asemi, Z.; Mirzaei, H. Paxlovid (Nirmatrelvir/Ritonavir): A new approach to COVID-19 therapy? Biomed. Pharmacother. 2023, 162, 114367. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Zheng, Y.; Zeng, X.; He, B.; Cheng, W. Structural biology of SARS-CoV-2: Open the door for novel therapies. Signal Transduct. Target. Ther. 2022, 7, 26. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef]
- Hammond, J.; Leister-Tebbe, H.; Gardner, A.; Abreu, P.; Bao, W.; Wisemandle, W.; Baniecki, M.; Hendrick, V.M.; Damle, B.; Simón-Campos, A.; et al. Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with COVID-19. N. Engl. J. Med. 2022, 386, 1397–1408. [Google Scholar] [CrossRef]
- Wong, C.K.H.; Lau, J.J.; Au, I.C.H.; Lau, K.T.K.; Hung, I.F.N.; Peiris, M.; Leung, G.M.; Wu, J.T. Optimal timing of nirmatrelvir/ritonavir treatment after COVID-19 symptom onset or diagnosis: Target trial emulation. Nat. Commun. 2023, 14, 8377. [Google Scholar] [CrossRef]
- Arbel, R.; Sagy, Y.W.; Hoshen, M.; Battat, E.; Lavie, G.; Sergienko, R.; Friger, M.; Waxman, J.G.; Dagan, N.; Balicer, R.; et al. Nirmatrelvir Use and Severe Covid-19 Outcomes during the Omicron Surge. N. Engl. J. Med. 2022, 387, 790–798. [Google Scholar] [CrossRef]
- Li, P.; Wang, Y.; Lavrijsen, M.; Lamers, M.M.; de Vries, A.C.; Rottier, R.J.; Bruno, M.J.; Peppelenbosch, M.P.; Haagmans, B.L.; Pan, Q. SARS-CoV-2 Omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination. Cell Res. 2022, 32, 322–324. [Google Scholar] [CrossRef]
- Greasley, S.E.; Noell, S.; Plotnikova, O.; Ferre, R.; Liu, W.; Bolanos, B.; Fennell, K.; Nicki, J.; Craig, T.; Zhu, Y.; et al. Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants. J. Biol. Chem. 2022, 298, 101972. [Google Scholar] [CrossRef] [PubMed]
- Takashita, E.; Kinoshita, N.; Yamayoshi, S.; Sakai-Tagawa, Y.; Fujisaki, S.; Ito, M.; Iwatsuki-Horimoto, K.; Halfmann, P.; Watanabe, S.; Maeda, K.; et al. Efficacy of Antiviral Agents against the SARS-CoV-2 Omicron Subvariant BA.2. N. Engl. J. Med. 2022, 386, 1475–1477. [Google Scholar] [CrossRef] [PubMed]
- Takashita, E.; Yamayoshi, S.; Simon, V.; van Bakel, H.; Sordillo, E.M.; Pekosz, A.; Fukushi, S.; Suzuki, T.; Maeda, K.; Halfmann, P.; et al. Efficacy of Antibodies and Antiviral Drugs against Omicron BA.2.12.1, BA.4, and BA.5 Subvariants. N. Engl. J. Med. 2022, 387, 468–470. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Tamura, T.; Zahradnik, J.; Deguchi, S.; Tabata, K.; Anraku, Y.; Kimura, I.; Ito, J.; Yamasoba, D.; Nasser, H.; et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2.75 variant. Cell Host Microbe 2022, 30, 1540–1555.e15. [Google Scholar] [CrossRef] [PubMed]
- Imai, M.; Ito, M.; Kiso, M.; Yamayoshi, S.; Uraki, R.; Fukushi, S.; Watanabe, S.; Suzuki, T.; Maeda, K.; Sakai-Tagawa, Y.; et al. Efficacy of Antiviral Agents against Omicron Subvariants BQ.1.1 and XBB. N. Engl. J. Med. 2023, 388, 89–91. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, N.R.; Molina, K.C.; Beaty, L.E.; Bennett, T.D.; Carlson, N.E.; Mayer, D.A.; Peers, J.L.; Russell, S.; Wynia, M.K.; Ginde, A.A. Real-world use of nirmatrelvir–ritonavir in outpatients with COVID-19 during the era of omicron variants including BA.4 and BA.5 in Colorado, USA: A retrospective cohort study. Lancet Infect. Dis. 2023, 23, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Kelly, S.P.; Liang, C.; Li, L.; Shen, R.; Leister-Tebbe, H.K.; Terra, S.G.; Gaffney, M.; Russo, L. Real-World Effectiveness of Nirmatrelvir/Ritonavir in Preventing Hospitalization among Patients with COVID-19 at High Risk for Severe Disease in the United States: A Nationwide Population-Based Cohort Study. medRxiv 2022. [Google Scholar] [CrossRef]
- Cegolon, L.; Pol, R.; Simonetti, O.; Filon, F.L.; Luzzati, R. Molnupiravir, Nirmatrelvir/Ritonavir, or Sotrovimab for High-Risk COVID-19 Patients Infected by the Omicron Variant: Hospitalization, Mortality, and Time until Negative Swab Test in Real Life. Pharmaceuticals 2023, 16, 721. [Google Scholar] [CrossRef] [PubMed]
- Iketani, S.; Mohri, H.; Culbertson, B.; Hong, S.J.; Duan, Y.; Luck, M.I.; Annavajhala, M.K.; Guo, Y.; Sheng, Z.; Uhlemann, A.-C.; et al. Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir. Nature 2022, 613, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhou, H.; Liu, X.; Iketani, S.; Lin, M.; Zhang, X.; Bian, Q.; Wang, H.; Sun, H.; Hong, S.J.; et al. Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir. Nature 2023, 622, 376–382. [Google Scholar] [CrossRef]
- Charness, M.E.; Gupta, K.; Stack, G.; Strymish, J.; Adams, E.; Lindy, D.C.; Mohri, H.; Ho, D.D. Rebound of SARS-CoV-2 Infection after Nirmatrelvir–Ritonavir Treatment. N. Engl. J. Med. 2022, 387, 1045–1047. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Berger, N.A.; Davis, P.B.; Kaelber, D.C.; Volkow, N.D.; Xu, R. COVID-19 rebound after Paxlovid and Molnupiravir during January–June 2022. MedRxiv 2022. [Google Scholar] [CrossRef]
- Anderson, A.S.; Caubel, P.; Rusnak, J.M. Nirmatrelvir–Ritonavir and Viral Load Rebound in COVID-19. N. Engl. J. Med. 2022, 387, 1047–1049. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.K.H.; Lau, K.T.K.; Au, I.C.H.; Lau, E.H.Y.; Poon, L.L.M.; Hung, I.F.N.; Cowling, B.J.; Leung, G.M. Viral burden rebound in hospitalised patients with COVID-19 receiving oral antivirals in Hong Kong: A population-wide retrospective cohort study. Lancet Infect. Dis. 2023, 23, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.L.-H.; Yip, T.C.-F.; Lai, M.S.-M.; Wong, V.W.-S.; Hui, D.S.-C.; Lui, G.C.-Y. Incidence of Viral Rebound after Treatment with Nirmatrelvir-Ritonavir and Molnupiravir. JAMA Netw. Open 2022, 5, e2245086. [Google Scholar] [CrossRef] [PubMed]
- Perelson, A.S.; Ribeiro, R.M.; Phan, T. An explanation for SARS-CoV-2 rebound after Paxlovid treatment. medRxiv 2023. [Google Scholar] [CrossRef]
- Boucau, J.; Uddin, R.; Marino, C.; Regan, J.; Flynn, J.P.; Choudhary, M.C.; Chen, G.; Stuckwisch, A.M.; Mathews, J.; Liew, M.Y.; et al. Characterization of Virologic Rebound Following Nirmatrelvir-Ritonavir Treatment for Coronavirus Disease 2019 (COVID-19). Clin. Infect. Dis. 2022, 76, e526–e529. [Google Scholar] [CrossRef] [PubMed]
- Carlin, A.F.; Clark, A.E.; Chaillon, A.; Garretson, A.F.; Bray, W.; Porrachia, M.; Santos, A.T.; Rana, T.M.; Smith, D.M. Virologic and Immunologic Characterization of Coronavirus Disease 2019 Recrudescence After Nirmatrelvir/Ritonavir Treatment. Clin. Infect. Dis. 2022, 76, e530–e532. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhou, C.; Deng, J.; Zhou, J. JAK inhibition as a new treatment strategy for patients with COVID-19. Biochem. Pharmacol. 2022, 202, 115162. [Google Scholar] [CrossRef]
- Satarker, S.; Tom, A.A.; Shaji, R.A.; Alosious, A.; Luvis, M.; Nampoothiri, M. JAK-STAT Pathway Inhibition and their Implications in COVID-19 Therapy. Postgrad. Med. 2020, 133, 489–507. [Google Scholar] [CrossRef]
- Tanaka, Y. A review of Janus kinase inhibitors for the treatment of COVID-19 pneumonia. Inflamm. Regen. 2023, 43, 3. [Google Scholar] [CrossRef]
- Abani, O.; Abbas, A.; Abbas, F.; Abbas, J.; Abbas, K.; Abbas, M.; Abbasi, S.; Abbass, H.; Abbott, A.; Abbott, A.; et al. Baricitinib in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial and updated meta-analysis. Lancet 2022, 400, 359–368. [Google Scholar]
- Marconi, V.C.; Ramanan, A.V.; de Bono, S.; Kartman, C.E.; Krishnan, V.; Liao, R.; Piruzeli, M.L.B.; Alatorre-Alexander, J.; Pellegrini, R.d.C.; Estrada, V.; et al. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): A randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet Respir. Med. 2021, 9, 1407–1418. [Google Scholar] [CrossRef] [PubMed]
- Abizanda, P.; Mayo, J.M.C.; Romero, M.M.; Zamora, E.B.C.; Sahuquillo, M.T.T.; Rizos, L.R.; Sánchez-Jurado, P.M.; Sánchez-Nievas, G.; Escolano, C.C.; Serrano, A.O.; et al. Baricitinib reduces 30-day mortality in older adults with moderate-to-severe COVID-19 pneumonia. J. Am. Geriatr. Soc. 2021, 69, 2752–2758. [Google Scholar] [CrossRef] [PubMed]
- Stebbing, J.; Sánchez Nievas, G.; Falcone, M.; Youhanna, S.; Richardson, P.; Ottaviani, S.; Shen, J.X.; Sommerauer, C.; Tiseo, G.; Ghiadoni, L.; et al. JAK inhibition reduces SARS-CoV-2 liver infectivity and modulates inflammatory responses to reduce morbidity and mortality. Sci. Adv. 2021, 7, eabe4724. [Google Scholar] [CrossRef]
- Wolfe, C.R.; Tomashek, K.M.; Patterson, T.F.; Gomez, C.A.; Marconi, V.C.; Jain, M.K.; Yang, O.O.; Paules, C.I.; Palacios, G.M.R.; Grossberg, R.; et al. Baricitinib versus dexamethasone for adults hospitalised with COVID-19 (ACTT-4): A randomised, double-blind, double placebo-controlled trial. Lancet Respir. Med. 2022, 10, 888–899. [Google Scholar] [CrossRef] [PubMed]
- Troseid, M.; Arribas, J.R.; Assoumou, L.; Holten, A.R.; Poissy, J.; Terzic, V.; Mazzaferri, F.; Rodriguez-Bano, J.; Eustace, J.; Hites, M.; et al. Efficacy and safety of baricitinib in hospitalized adults with severe or critical COVID-19 (Bari-SolidAct): A randomised, double-blind, placebo-controlled phase 3 trial. Crit. Care 2023, 27, 9. [Google Scholar] [CrossRef]
- Ferro, F.; La Rocca, G.; Elefante, E.; Italiano, N.; Moretti, M.; Talarico, R.; Pelati, E.; Valentini, K.; Baldini, C.; Mozzo, R.; et al. Baricitinib and Pulse Steroids Combination Treatment in Hyperinflammatory COVID-19: A Rheumatological Approach in the Intensive Care Unit. Int. J. Mol. Sci. 2024, 25, 7273. [Google Scholar] [CrossRef]
- Rezabakhsh, A.; Mojtahedi, F.; Tekantapeh, S.T.; Mahmoodpoor, A.; Ala, A.; Soleimanpour, H. Therapeutic Impact of Tocilizumab in the Setting of Severe COVID-19; an Updated and Comprehensive Review on Current Evidence. Arch. Acad. Emerg. Med. 2024, 12, e47. [Google Scholar] [CrossRef]
- Zhang, C.; Wu, Z.; Li, J.-W.; Zhao, H.; Wang, G.Q. The cytokine release syndrome (CRS) of severe COVID-19 and Interleukin-6 receptor (IL-6R) antagonist Tocilizumab may be the key to reduce the mortality. Int. J. Antimicrob. Agents 2020, 55, 105954. [Google Scholar] [CrossRef]
- Le, R.Q.; Li, L.; Yuan, W.; Shord, S.S.; Nie, L.; Habtemariam, B.A.; Przepiorka, D.; Farrell, A.T.; Pazdur, R. FDA Approval Summary: Tocilizumab for Treatment of Chimeric Antigen Receptor T Cell-Induced Severe or Life-Threatening Cytokine Release Syndrome. Oncologist 2018, 23, 943–947. [Google Scholar] [CrossRef] [PubMed]
- Steuber, T.D.; Rosandich, T.; Cadwallader, T.; Steil, L.; Belk, M.; Yendrapalli, U.; Hassoun, A.; Edwards, J. Dosing and Administration Strategies of Tocilizumab in Patients with COVID-19: A Retrospective Cohort Analysis. Ann. Pharmacother. 2023, 58, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Leung, E.; Crass, R.L.; Jorgensen, S.C.J.; Raybardhan, S.; Langford, B.J.; Moore, W.J.; Rhodes, N.J. Pharmacokinetic/Pharmacodynamic Considerations of Alternate Dosing Strategies of Tocilizumab in COVID-19. Clin. Pharmacokinet. 2021, 61, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Tay, M.Z.; Poh, C.M.; Rénia, L.; MacAry, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef] [PubMed]
- RECOVERY Collaborative Group. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial. Lancet 2021, 397, 1637–1645. [Google Scholar] [CrossRef]
- Gordon, A.C.; Mouncey, P.R.; Al-Beidh, F.; Rowan, K.M.; Nichol, A.D.; Arabi, Y.M.; Annane, D.; Beane, A.; van Bentum-Puijk, W.; Berry, L.R. Interleukin-6 Receptor Antagonists in Critically Ill Patients with COVID-19. N. Engl. J. Med. 2021, 384, 1491–1502. [Google Scholar] [PubMed]
- Alkhateeb, T.; Stollings, J.L.; Sohn, I.; Liu, D.; Fleenor, L.M.; Ely, E.W.; Lahiri, S. Tocilizumab is associated with reduced delirium and coma in critically ill patients with COVID-19. Sci. Rep. 2024, 14, 11738. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H.; Sparrow, N.A.; Anwar, F.; Guidry, G.; Covarrubias, A.E.; Pang, H.; Bogguri, C.; Karumanchi, S.A.; Lahiri, S. Interleukin-6 mediates delirium-like phenotypes in a murine model of urinary tract infection. J. Neuroinflamm. 2021, 18, 247. [Google Scholar] [CrossRef] [PubMed]
- Anwar, F.; Sparrow, N.A.; Rashid, M.H.; Guidry, G.; Gezalian, M.M.; Ley, E.J.; Koronyo-Hamaoui, M.; Danovitch, I.; Ely, E.W.; Karumanchi, S.A.; et al. Systemic interleukin-6 inhibition ameliorates acute neuropsychiatric phenotypes in a murine model of acute lung injury. Crit. Care 2022, 26, 274. [Google Scholar] [CrossRef] [PubMed]
- Sparrow, N.A.; Anwar, F.; Covarrubias, A.E.; Rajput, P.S.; Rashid, M.H.; Nisson, P.L.; Gezalian, M.M.; Toossi, S.; Ayodele, M.O.; Karumanchi, S.A.; et al. IL-6 Inhibition Reduces Neuronal Injury in a Murine Model of Ventilator-induced Lung Injury. Am. J. Respir. Cell Mol. Biol. 2021, 65, 403–412. [Google Scholar] [CrossRef]
- Knorr, J.P.; Colomy, V.; Mauriello, C.M.; Ha, S. Tocilizumab in patients with severe COVID-19: A single-center observational analysis. J. Med. Virol. 2020, 92, 2813–2820. [Google Scholar] [CrossRef]
- Hager, D.N.; Dinglas, V.D.; Subhas, S.; Rowden, A.M.; Neufeld, K.J.; Bienvenu, O.J.; Touradji, P.; Colantuoni, E.; Reddy, D.R.; Brower, R.G.; et al. Reducing Deep Sedation and Delirium in Acute Lung Injury Patients. Crit. Care Med. 2013, 41, 1435–1442. [Google Scholar] [CrossRef]
- Chan, K.H.; Patel, B.; Podel, B.; Szablea, M.E.; Shaaban, H.S.; Guron, G.; Slim, J. Tocilizumab and Thromboembolism in COVID-19: A Retrospective Hospital-Based Cohort Analysis. Cureus 2021, 13, e15208. [Google Scholar] [CrossRef]
- Hafez, W.; Ziade, M.A.; Arya, A.; Saleh, H.; Abdelshakor, M.; Alla, O.F.; Agrawal, P.; Ali, S.; Rao, S.R.; Gupta, S.; et al. Treatment Outcomes of Tocilizumab in Critically-Ill COVID-19 Patients, Single-Centre Retrospective Study. Antibiotics 2022, 11, 241. [Google Scholar] [CrossRef]
- Rosas, I.O.; Bräu, N.; Waters, M.; Go, R.C.; Hunter, B.D.; Bhagani, S.; Skiest, D.; Aziz, M.S.; Cooper, N.; Douglas, I.S.; et al. Tocilizumab in Hospitalized Patients with Severe Covid-19 Pneumonia. N. Engl. J. Med. 2021, 384, 1503–1516. [Google Scholar] [CrossRef] [PubMed]
- Aljuhani, O.; Al Sulaiman, K.; Korayem, G.B.; Altebainawi, A.F.; Alsohimi, S.; Alqahtani, R.; Alfaifi, S.; Alharbi, A.; AlKhayrat, A.; Hattan, A.; et al. The association between tocilizumab therapy and the development of thrombosis in critically ill patients with COVID-19: A multicenter, cohort study. Sci. Rep. 2024, 14, 3037. [Google Scholar] [CrossRef]
- Guaraldi, G.; Meschiari, M.; Cozzi-Lepri, A.; Milic, J.; Tonelli, R.; Menozzi, M.; Franceschini, E.; Cuomo, G.; Orlando, G.; Borghi, V.; et al. Tocilizumab in patients with severe COVID-19: A retrospective cohort study. Lancet Rheumatol. 2020, 2, e474–e484. [Google Scholar] [CrossRef] [PubMed]
- Hermine, O.; Mariette, X.; Tharaux, P.-L.; Resche-Rigon, M.; Porcher, R.; Ravaud, P.; Bureau, S.; Dougados, M.; Tibi, A.; CORIMUNO-19 Collaborative Group; et al. Effect of Tocilizumab vs Usual Care in Adults Hospitalized with COVID-19 and Moderate or Severe Pneumonia: A Randomized Clinical Trial. JAMA Intern. Med. 2021, 181, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.H.; Frigault, M.J.; Serling-Boyd, N.J.; Fernandes, A.D.; Harvey, L.; Foulkes, A.S.; Horick, N.K.; Healy, B.C.; Shah, R.; Bensaci, A.M.; et al. Efficacy of Tocilizumab in Patients Hospitalized with COVID-19. N. Engl. J. Med. 2020, 383, 2333–2344. [Google Scholar] [CrossRef]
- Lu, D.-E.; Ou, T.-Y.; Kang, J.-W.; Ong, J.Y.; Chen, I.-J.; Lee, C.-H.; Lee, M.-C. The association between tocilizumab and the secondary bloodstream infection maybe nonsignificant in hospitalized patients with SARS-CoV-2 infection: A cohort study. J. Microbiol. Immunol. Infect. 2024, 57, 38–47. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, Q.; Chen, F.; Zheng, J.; Chen, Y.; Chen, Z.; Li, R.; Li, X. Immune-Cell-Based Therapy for COVID-19: Current Status. Viruses 2023, 15, 2148. [Google Scholar] [CrossRef]
- Kállay, K.; Kassa, C.; Réti, M.; Karászi, É.; Sinkó, J.; Goda, V.; Stréhn, A.; Csordás, K.; Horváth, O.; Szederjesi, A.; et al. Early Experience with CliniMACS Prodigy CCS (IFN-gamma) System in Selection of Virus-specific T Cells from Third-party Donors for Pediatric Patients with Severe Viral Infections after Hematopoietic Stem Cell Transplantation. J. Immunother. 2018, 41, 158–163. [Google Scholar] [CrossRef]
- Naik, S.; Nicholas, S.K.; Martinez, C.A.; Leen, A.M.; Hanley, P.J.; Gottschalk, S.M.; Rooney, C.M.; Hanson, I.C.; Krance, R.A.; Shpall, E.J.; et al. Adoptive immunotherapy for primary immunodeficiency disorders with virus-specific T lymphocytes. J. Allergy Clin. Immunol. 2016, 137, 1498–1505. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Milner, J.; Lamb, M.; Maryamchik, E.; Rigot, O.; Ayello, J.; Harrison, L.; Shaw, R.; Behbehani, G.K.; Mardis, E.R.; et al. Manufacture and Characterization of Good Manufacturing Practice-Compliant SARS-CoV-2 Cytotoxic T Lymphocytes. J. Infect. Dis. 2022, 227, 788–799. [Google Scholar] [CrossRef]
- Bleakley, M.M.; Gooley, T.A.; Hilzinger, B.; Riddell, S.R.; Shlomchik, W.D. NaïVe T Cell Depletion of PBSC Grafts Results in Very Low Rates of Chronic Gvhd and High Survival. Blood 2016, 128, 668. [Google Scholar] [CrossRef]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological Memory to SARS-CoV-2 Assessed for Up to 8 Months after Infection. Science 2021, 371, eabf4063. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, A.; Karavalakis, G.; Papadopoulou, E.; Xochelli, A.; Bousiou, Z.; Vogiatzoglou, A.; Papayanni, P.G.; Georgakopoulou, A.; Giannaki, M.; Stavridou, F.; et al. SARS-CoV-2-specific T cell therapy for severe COVID-19: A randomized phase 1/2 trial. Nat. Med. 2023, 29, 2019–2029. [Google Scholar] [CrossRef]
- Ferreras, C.; Hernández-Blanco, C.; Martín-Quirós, A.; Al-Akioui-Sanz, K.; Mora-Rillo, M.; Ibáñez, F.; Díaz-Almirón, M.; Cano-Ochando, J.; Lozano-Ojalvo, D.; Jiménez-González, M.; et al. Results of phase 2 randomized multi-center study to evaluate the safety and efficacy of infusion of memory T cells as adoptive therapy in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pneumonia and/or lymphopenia (RELEASE NCT04578210). Cytotherapy 2024, 26, 25–35. [Google Scholar] [CrossRef]
- Ong, R.Y.L.; Seah, V.X.F.; Chong, C.Y.; Thoon, K.C.; Tan, N.W.H.; Li, J.; Nadua, K.D.; Soh, S.Y.; Seng, M.S.-F.; Pham, T.N.A.; et al. A cohort study of COVID-19 infection in pediatric oncology patients plus the utility and safety of remdesivir treatment. Acta Oncol. 2023, 62, 53–57. [Google Scholar] [CrossRef]
- Tarke, A.; Coelho, C.H.; Zhang, Z.; Dan, J.M.; Yu, E.D.; Methot, N.; Bloom, N.I.; Goodwin, B.; Phillips, E.; Mallal, S.; et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 2022, 185, 847–859. [Google Scholar] [CrossRef]
- Zavvar, M.; Yahyapoor, A.; Baghdadi, H.; Zargaran, S.; Assadiasl, S.; Abdolmohammadi, K.; Abooei, A.H.; Sattarian, M.R.; JalaliFarahani, M.; Zarei, N.; et al. COVID-19 immunotherapy: Treatment based on the immune cell-mediated approaches. Int. Immunopharmacol. 2022, 107, 108655. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Jiang, X.; Dai, X.; Li, B. The Dynamic Role of FOXP3+ Tregs and Their Potential Therapeutic Applications during SARS-CoV-2 Infection. Front. Immunol. 2022, 13, 916411. [Google Scholar] [CrossRef]
- Fransson, M.; Piras, E.; Burman, J.; Nilsson, B.; Essand, M.; Lu, B.; Harris, R.A.; Magnusson, P.U.; Brittebo, E.; Loskog, A.S. CAR/FoxP3-engineered T regulatory cells target the CNS and suppress EAE upon intranasal delivery. J. Neuroinflamm. 2012, 9, 112. [Google Scholar] [CrossRef]
- Blat, D.; Zigmond, E.; Alteber, Z.; Waks, T.; Eshhar, Z. Suppression of Murine Colitis and its Associated Cancer by Carcinoembryonic Antigen-Specific Regulatory T Cells. Mol. Ther. 2014, 22, 1018–1028. [Google Scholar] [CrossRef]
- Skuljec, J.; Chmielewski, M.; Happle, C.; Habener, A.; Busse, M.; Abken, H.; Hansen, G. Chimeric Antigen Receptor-Redirected Regulatory T Cells Suppress Experimental Allergic Airway Inflammation, a Model of Asthma. Front. Immunol. 2017, 8, 1125. [Google Scholar] [CrossRef]
- Guan, T.; Zhou, X.; Zhou, W.; Lin, H. Regulatory T cell and macrophage crosstalk in acute lung injury: Future perspectives. Cell Death Discov. 2023, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Yang, X.O. TH17 responses in cytokine storm of COVID-19: An emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020, 53, 368–370. [Google Scholar] [CrossRef]
- Gonçalves-Pereira, M.H.; Santiago, L.; Ravetti, C.G.; Vassallo, P.F.; de Andrade, M.V.M.; Vieira, M.S.; Oliveira, F.d.F.S.d.; Carobin, N.V.; Li, G.; Sabino, A.d.P.; et al. Dysfunctional phenotype of systemic and pulmonary regulatory T cells associate with lethal COVID-19 cases. Immunology 2022, 168, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Khesht, A.M.; Karpisheh, V.; Saeed, B.Q.; Zekiy, A.O.; Yapanto, L.M.; Afjadi, M.N.; Aksoun, M.; Esfahani, M.N.; Aghakhani, F.; Movahed, M.; et al. Different T cell related immunological profiles in COVID-19 patients compared to healthy controls. Int. Immunopharmacol. 2021, 97, 107828. [Google Scholar] [CrossRef]
- Gladstone, D.E.; D’Alessio, F.R.; Howard, C.; Lyu, M.-A.; Mock, J.R.; Gibbs, K.W.; Abrams, D.; Huang, M.; Zeng, K.; Herlihy, J.P.; et al. Randomized, double-blinded, placebo-controlled trial of allogeneic cord blood T-regulatory cells for treatment of COVID-19 ARDS. Blood Adv. 2023, 7, 3075–3079. [Google Scholar] [CrossRef]
- Buitrago-Molina, L.E.; Pietrek, J.; Noyan, F.; Schlue, J.; Manns, M.P.; Wedemeyer, H.; Hardtke-Wolenski, M.; Jaeckel, E. Treg-specific IL-2 therapy can reestablish intrahepatic immune regulation in autoimmune hepatitis. J. Autoimmun. 2020, 117, 102591. [Google Scholar] [CrossRef] [PubMed]
- Humrich, J.Y.; Riemekasten, G. Restoring regulation—IL-2 therapy in systemic lupus erythematosus. Expert Rev. Clin. Immunol. 2016, 12, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Boyman, O.; Sprent, J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat. Rev. Immunol. 2012, 12, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, M.; Rabaan, A.A.; Alwarthan, S.; Alhajri, M.; Halwani, M.A.; Alshengeti, A.; Najim, M.A.; Alwashmi, A.S.S.; Alshehri, A.A.; Alshamrani, S.A.; et al. Regulatory T Cells (Tregs) and COVID-19: Unveiling the Mechanisms, and Therapeutic Potentialities with a Special Focus on Long COVID. Vaccines 2023, 11, 699. [Google Scholar] [CrossRef] [PubMed]
- Alavi-Dana, S.M.M.; Gholami, Y.; Meghdadi, M.; Fadaei, M.S.; Askari, V.R. Mesenchymal stem cell therapy for COVID-19 infection. Inflammopharmacology 2023, 32, 319–334. [Google Scholar] [CrossRef]
- Cipriani, P.; Carubbi, F.; Liakouli, V.; Marrelli, A.; Perricone, C.; Perricone, R.; Alesse, E.; Giacomelli, R. Stem cells in autoimmune diseases: Implications for pathogenesis and future trends in therapy. Autoimmun. Rev. 2013, 12, 709–716. [Google Scholar] [CrossRef]
- Peng, Y.; Chen, X.; Liu, Q.; Zhang, X.; Huang, K.; Liu, L.; Li, H.; Zhou, M.; Huang, F.; Fan, Z.; et al. Mesenchymal stromal cells infusions improve refractory chronic graft versus host disease through an increase of CD5+ regulatory B cells producing interleukin 10. Leukemia 2014, 29, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-Y.; Lee, Y.-K.; Ran, X.; Liao, S.-Y.; Yang, J.; Au, K.-W.; Lai, W.-H.; Esteban, M.A.; Tse, H.-F. Generation of Induced Cardiospheres via Reprogramming of Skin Fibroblasts for Myocardial Regeneration. Stem Cells 2016, 34, 2693–2706. [Google Scholar] [CrossRef]
- Saleh, M.; Vaezi, A.A.; Aliannejad, R.; Sohrabpour, A.A.; Kiaei, S.Z.F.; Shadnoush, M.; Siavashi, V.; Aghaghazvini, L.; Khoundabi, B.; Abdoli, S.; et al. Cell therapy in patients with COVID-19 using Wharton’s jelly mesenchymal stem cells: A phase 1 clinical trial. Stem Cell Res. Ther. 2021, 12, 410. [Google Scholar] [CrossRef]
- Sengupta, V.; Sengupta, S.; Lazo, A.; Woods, P.; Nolan, A.; Bremer, N. Exosomes Derived from Bone Marrow Mesenchymal Stem Cells as Treatment for Severe COVID-19. Stem Cells Dev. 2020, 29, 747–754. [Google Scholar] [CrossRef]
- Adas, G.; Cukurova, Z.; Yasar, K.K.; Yilmaz, R.; Isiksacan, N.; Kasapoglu, P.; Yesilbag, Z.; Koyuncu, I.; Karaoz, E. The Systematic Effect of Mesenchymal Stem Cell Therapy in Critical COVID-19 Patients: A Prospective Double Controlled Trial. Cell Transplant. 2021, 30, 096368972110249. [Google Scholar] [CrossRef]
- Dilogo, I.H.; Aditianingsih, D.; Sugiarto, A.; Burhan, E.; Damayanti, T.; Sitompul, P.A.; Mariana, N.; Antarianto, R.D.; Liem, I.K.; Kispa, T.; et al. Umbilical Cord Mesenchymal Stromal Cells as Critical COVID-19 Adjuvant Therapy: A Randomized Controlled Trial. Stem Cells Transl. Med. 2021, 10, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Ringden, O.; Roshandel, E.; Pirsalehi, A.; Kazemi, S.; Sankanian, G.; Majidi, M.; Salimi, M.; Aghdami, N.; Sadrosadat, H.; Kochaksaraei, S.S.; et al. Conquering the cytokine storm in COVID-19-induced ARDS using placenta-derived decidua stromal cells. Biol. Blood Marrow Transplant. 2022, 28, S222. [Google Scholar] [CrossRef]
- Farkhad, N.K.; Sedaghat, A.; Reihani, H.; Moghadam, A.A.; Moghadam, A.B.; Ghaebi, N.K.; Khodadoust, M.A.; Ganjali, R.; Tafreshian, A.R.; Tavakol-Afshari, J. Mesenchymal stromal cell therapy for COVID-19-induced ARDS patients: A successful phase 1, control-placebo group, clinical trial. Stem Cell Res. Ther. 2022, 13, 283. [Google Scholar] [CrossRef]
- Monsel, A.; Hauw-Berlemont, C.; Mebarki, M.; Heming, N.; Mayaux, J.; Tchoumba, O.N.; Diehl, J.-L.; Demoule, A.; Annane, D.; Marois, C.; et al. Treatment of COVID-19-associated ARDS with mesenchymal stromal cells: A multicenter randomized double-blind trial. Crit. Care 2022, 26, 48. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Zheng, Y.; Cheng, Z.; Ji, N.; Niu, C.; Wang, Y.; Huang, T.; Li, R.; Huang, M.; Chen, X.; et al. One-year follow-up study after patients with severe COVID-19 received human umbilical cord mesenchymal stem cells treatment. Stem Cell Res. Ther. 2022, 13, 321. [Google Scholar] [CrossRef] [PubMed]
- Li, T.-T.; Li, T.-T.; Zhang, B.; Zhang, B.; Fang, H.; Fang, H.; Shi, M.; Shi, M.; Yao, W.-Q.; Yao, W.-Q.; et al. Human mesenchymal stem cell therapy in severe COVID-19 patients: 2-year follow-up results of a randomized, double-blind, placebo-controlled trial. eBioMedicine 2023, 92, 104600. [Google Scholar] [CrossRef] [PubMed]
- Yudintceva, N.; Mikhailova, N.; Fedorov, V.; Samochernych, K.; Vinogradova, T.; Muraviov, A.; Shevtsov, M. Mesenchymal Stem Cells and MSCs-Derived Extracellular Vesicles in Infectious Diseases: From Basic Research to Clinical Practice. Bioengineering 2022, 9, 662. [Google Scholar] [CrossRef]
- Lukomska, B.; Stanaszek, L.; Zuba-Surma, E.; Legosz, P.; Sarzynska, S.; Drela, K. Challenges and Controversies in Human Mesenchymal Stem Cell Therapy. Stem Cells Int. 2019, 2019, 9628536. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Dunkle, L.M.; Kotloff, K.L.; Gay, C.L.; Áñez, G.; Adelglass, J.M.; Barrat Hernández, A.Q.; Harper, W.L.; Duncanson, D.M.; McArthur, M.A.; Florescu, D.F.; et al. Efficacy and safety of NVX-CoV2373 in adults in the United States and Mexico. N. Engl. J. Med. 2022, 386, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.B.; Montefiori, D.C.; McDermott, A.B.; Fong, Y.; Benkeser, D.; Deng, W.; Zhou, H.; Houchens, C.R.; Martins, K.; Jayashankar, L.; et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science 2022, 375, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Collier, A.-R.Y.; Yu, J.; McMahan, K.; Liu, J.; Chandrashekar, A.; Maron, J.S.; Atyeo, C.; Martinez, D.R.; Ansel, J.L.; Aguayo, R.; et al. Differential kinetics of immune responses elicited by COVID-19 vaccines. N. Engl. J. Med. 2021, 385, 2010–2012. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yu, Y.; Xu, Y.; Jian, F.; Song, W.; Yisimayi, A.; Wang, P.; Wang, J.; Liu, J.; Yu, L.; et al. Fast evolution of SARS-CoV-2 BA. 2.86 to JN. 1 under heavy immune pressure. Lancet Infect. Dis. 2024, 24, e70–e72. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, Y.; Bowen, A.; Mellis, I.A.; Valdez, R.; Gherasim, C.; Gordon, A.; Liu, L.; Ho, D.D. XBB.1.5 monovalent mRNA vaccine booster elicits robust neutralizing antibodies against XBB subvariants and JN.1. Cell Host Microbe 2024, 32, 315–321. [Google Scholar] [CrossRef]
- Kojima, N.; Adams, K.; Self, W.H.; Gaglani, M.; McNeal, T.; Ghamande, S.; Steingrub, J.S.; Shapiro, N.I.; Duggal, A.; Busse, L.W.; et al. Changing severity and epidemiology of adults hospitalized with coronavirus disease 2019 (COVID-19) in the United States after introduction of COVID-19 vaccines, March 2021–August 2022. Clin. Infect. Dis. 2023, 77, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Tenforde, M.W.; Self, W.H.; Adams, K.; Gaglani, M.; Ginde, A.A.; McNeal, T.; Ghamande, S.; Douin, D.J.; Talbot, H.K.; Casey, J.D.; et al. Association between mRNA vaccination and COVID-19 hospitalization and disease severity. JAMA 2021, 326, 2043–2054. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, M.C.; Galvani, A.P. Optimizing age-specific vaccination. Science 2021, 371, 890–891. [Google Scholar] [CrossRef] [PubMed]
- Zaoui, S.; Foguem, C.; Tchuente, D.; Fosso-Wamba, S.; Kamsu-Foguem, B. The viability of supply chains with interpretable learning systems: The case of COVID-19 vaccine deliveries. Glob. J. Flex. Syst. Manag. 2023, 24, 633–657. [Google Scholar] [CrossRef]
- Gavi, the Vaccine Alliance. COVAX Explained. [Online]. Gavi, the Vaccine Alliance. 2023. Available online: https://www.gavi.org/vaccineswork/covax-explained (accessed on 30 June 2024).
- World Health Organization. COVID-19 Vaccinations Shift to Regular Immunization as COVAX Draws to a Close. [Online]. World Health Organization. 2023. Available online: https://www.who.int/news/item/19-12-2023-covid-19-vaccinations-shift-to-regular-immunization-as-covax-draws-to-a-close (accessed on 30 June 2024).
- Sign¢, L. Strategies for Effective Health Care for Africa in the Fourth Industrial Revolution, Bridging the Gap between the Promise and Delivery. [Online]. 2021. Available online: https://www.brookings.edu/wp-content/uploads/2021/10/Strategies-for-effective-health-care-delivery-in-Africa_FINAL.pdf (accessed on 2 July 2024).
- Hierink, F.; Okiro, E.A.; Flahault, A.; Ray, N. The winding road to health: A systematic scoping review on the effect of geographical accessibility to health care on infectious diseases in low- and middle-income countries. PLoS ONE 2021, 16, e0244921. [Google Scholar] [CrossRef]
- Med Aditus. Rwanda Welcomes Africa’s First Mobile Vaccine-Production Units. Med Aditus. [Online]. 2023. Available online: https://medaditus.org/news-articles/rwanda-welcomes-africas-first-mobile-vaccine-production-units/ (accessed on 30 June 2024).
- Masresha, B.; Ruiz, M.A.S.; Atuhebwe, P.; Mihigo, R. The first year of COVID-19 vaccine roll-out in Africa: Challenges and lessons learned. Pan Afr. Med. J. 2022, 41, 2. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.B.; Kanevsky, I.; Che, Y.; Swanson, K.A.; Muik, A.; Vormehr, M.; Kranz, L.M.; Walzer, K.C.; Hein, S.; Güler, A.; et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 2021, 592, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Muik, A.; Lui, B.G.; Wallisch, A.-K.; Bacher, M.; Mühl, J.; Reinholz, J.; Ozhelvaci, O.; Beckmann, N.; Garcia, R.d.l.C.G.; Poran, A.; et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine–elicited human sera. Science 2022, 375, 678–680. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T-cell responses. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.J.; Moreira, E.D., Jr.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Polack, F.P.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine through 6 Months. N. Engl. J. Med. 2021, 385, 1761–1773. [Google Scholar] [CrossRef]
- Chemaitelly, H.; Tang, P.; Hasan, M.R.; AlMukdad, S.; Yassine, H.M.; Benslimane, F.M.; Al Khatib, H.A.; Coyle, P.; Ayoub, H.H.; Al Kanaani, Z.; et al. Waning of BNT162b2 Vaccine Protection against SARS-CoV-2 Infection in Qatar. N. Engl. J. Med. 2021, 385, e83. [Google Scholar] [CrossRef] [PubMed]
- Tartof, S.Y.; Slezak, J.M.; Fischer, H.; Hong, V.; Ackerson, B.K.; Ranasinghe, O.N.; Frankland, T.B.; Ogun, O.A.; Zamparo, J.M.; Gray, S.; et al. Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: A retrospective cohort study. Lancet 2021, 398, 1407–1416. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.; Hasan, M.R.; Chemaitelly, H.; Yassine, H.M.; Benslimane, F.M.; Al Khatib, H.A.; AlMukdad, S.; Coyle, P.; Ayoub, H.H.; Al Kanaani, Z.; et al. BNT162b2 and mRNA-1273 COVID-19 vaccine effectiveness against the SARS-CoV-2 Delta variant in Qatar. Nat. Med. 2021, 27, 2136–2143. [Google Scholar] [CrossRef]
- Abu-Raddad, L.J.; Chemaitelly, H.; Butt, A.A. Effectiveness of the BNT162b2 COVID-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl. J. Med. 2021, 385, 187–189. [Google Scholar] [CrossRef]
- Abu-Raddad, L.J.; Chemaitelly, H.; Ayoub, H.H.; AlMukdad, S.; Yassine, H.M.; Al-Khatib, H.A.; Smatti, M.K.; Tang, P.; Hasan, M.R.; Coyle, P.; et al. Effect of mRNA Vaccine Boosters against SARS-CoV-2 Omicron Infection in Qatar. N. Engl. J. Med. 2022, 386, 1804–1816. [Google Scholar] [CrossRef]
- Veneti, L.; Berild, J.D.; Watle, S.V.; Starrfelt, J.; Greve-Isdahl, M.; Langlete, P.; Bøås, H.; Bragstad, K.; Hungnes, O.; Meijerink, H. Effectiveness of BNT162b2 vaccine against SARS-CoV-2 Delta and Omicron infection in adolescents, Norway, August 2021 to January 2022. Int. J. Infect. Dis. 2023, 130, 182–188. [Google Scholar] [CrossRef]
- Chatzilena, A.; Hyams, C.; Challen, R.; Marlow, R.; King, J.; Adegbite, D.; Kinney, J.; Clout, M.; Maskell, N.; Oliver, J.; et al. Effectiveness of BNT162b2 COVID-19 vaccination in prevention of hospitalisations and severe disease in adults with SARS-CoV-2 Delta (B.1.617.2) and Omicron (B.1.1.529) variant between June 2021 and July 2022: A prospective test negative case–control study. Lancet Reg. Health—Eur. 2022, 25, 100552. [Google Scholar] [CrossRef]
- Moreira, E.D.; Kitchin, N.; Xu, X.; Dychter, S.S.; Lockhart, S.; Gurtman, A.; Perez, J.L.; Zerbini, C.; Dever, M.E.; Jennings, T.W.; et al. Safety and Efficacy of a Third Dose of BNT162b2 COVID-19 Vaccine. N. Engl. J. Med. 2022, 386, 1910–1921. [Google Scholar] [CrossRef]
- Pather, S.; Muik, A.; Rizzi, R.; Mensa, F. Clinical development of variant-adapted BNT162b2 COVID-19 vaccines: The early Omicron era. Expert Rev. Vaccines 2023, 22, 650–661. [Google Scholar] [CrossRef]
- US Centers for Disease Control and Prevention. Stay Up to Date with COVID-19 Vaccines. 2024. Available online: https://www.cdc.gov/coronavirus/2019-ncov/vaccines/stay-up-to-date.html#UTD (accessed on 7 July 2024).
- Winokur, P.; Gayed, J.; Fitz-Patrick, D.; Thomas, S.J.; Diya, O.; Lockhart, S.; Xu, X.; Zhang, Y.; Bangad, V.; Schwartz, H.I.; et al. Bivalent Omicron BA.1–Adapted BNT162b2 Booster in Adults Older than 55 Years. N. Engl. J. Med. 2023, 388, 214–227. [Google Scholar] [CrossRef]
- Muik, A.; Lui, B.G.; Bacher, M.; Wallisch, A.-K.; Toker, A.; Couto, C.I.C.; Güler, A.; Mampilli, V.; Schmitt, G.J.; Mottl, J.; et al. Exposure to BA.4/5 S protein drives neutralization of Omicron BA.1, BA.2, BA.2.12.1, and BA.4/5 in vaccine-experienced humans and mice. Sci. Immunol. 2022, 7, eade9888. [Google Scholar] [CrossRef]
- Muik, A.; Lui, B.G.; Bacher, M.; Wallisch, A.-K.; Toker, A.; Finlayson, A.; Krüger, K.; Ozhelvaci, O.; Grikscheit, K.; Hoehl, S.; et al. Omicron BA.2 breakthrough infection enhances cross-neutralization of BA.2.12.1 and BA.4/BA.5. Sci. Immunol. 2022, 7, eade2283. [Google Scholar] [CrossRef]
- Zou, J.; Kurhade, C.; Patel, S.; Kitchin, N.; Tompkins, K.; Cutler, M.; Cooper, D.; Yang, Q.; Cai, H.; Muik, A.; et al. Neutralization of BA.4–BA.5, BA.4.6, BA.2.75.2, BQ.1.1, and XBB.1 with Bivalent Vaccine. N. Engl. J. Med. 2023, 388, 854–857. [Google Scholar] [CrossRef]
- Andersson, N.W.; Thiesson, E.M.; Baum, U.; Pihlström, N.; Starrfelt, J.; Faksová, K.; Poukka, E.; Meijerink, H.; Ljung, R.; Hviid, A. Comparative effectiveness of bivalent BA.4-5 and BA.1 mRNA booster vaccines among adults aged ≥ 50 years in Nordic countries: Nationwide cohort study. BMJ 2023, 382, e075286. [Google Scholar] [CrossRef]
- Tartof, S.Y.; Slezak, J.M.; Puzniak, L.; Hong, V.; Frankland, T.B.; Ackerson, B.K.; Xie, F.; Takhar, H.; Ogun, O.A.; Simmons, S.; et al. Effectiveness of BNT162b2 BA.4/5 bivalent mRNA vaccine against a range of COVID-19 outcomes in a large health system in the USA: A test-negative case–control study. Lancet Respir. Med. 2023, 11, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Kurhade, C.; Zou, J.; Xia, H.; Liu, M.; Chang, H.C.; Ren, P.; Xie, X.; Shi, P. Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Nat. Med. 2022, 29, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Gayed, J.; Bangad, V.; Xu, X.; Mensa, F.; Cutler, M.; Türeci, Ö.; Şahin, U.; Modjarrad, K.; Swanson, K.A.; Anderson, A.S.; et al. Immunogenicity of the Monovalent Omicron XBB.1.5-Adapted BNT162b2 COVID-19 Vaccine against XBB.1.5, BA.2.86, and JN.1 Sublineages: A Phase 2/3 Trial. Vaccines 2024, 12, 734. [Google Scholar] [CrossRef]
- Gayed, J.; Diya, O.; Lowry, F.S.; Xu, X.; Bangad, V.; Mensa, F.; Zou, J.; Xie, X.; Hu, Y.; Lu, C.; et al. Safety and Immunogenicity of the Monovalent Omicron XBB.1.5-Adapted BNT162b2 COVID-19 Vaccine in Individuals ≥12 Years Old: A Phase 2/3 Trial. Vaccines 2024, 12, 118. [Google Scholar] [CrossRef]
- Hansen, C.H.; Hansen, C.H.; Moustsen-Helms, I.R.; Moustsen-Helms, I.R.; Rasmussen, M.; Rasmussen, M.; Søborg, B.; Søborg, B.; Ullum, H.; Ullum, H.; et al. Short-term effectiveness of the XBB.1.5 updated COVID-19 vaccine against hospitalisation in Denmark: A national cohort study. Lancet Infect. Dis. 2024, 24, e73–e74. [Google Scholar] [CrossRef] [PubMed]
- van Werkhoven, C.H.; Valk, A.-W.; Smagge, B.; de Melker, H.E.; Knol, M.J.; Hahné, S.J.; Hof, S.v.D.; de Gier, B. Early COVID-19 vaccine effectiveness of XBB.1.5 vaccine against hospitalisation and admission to intensive care, the Netherlands, 9 October to 5 December 2023. Eurosurveillance 2024, 29, 2300703. [Google Scholar] [CrossRef]
- Shrestha, N.K.; Burke, P.C.; Nowacki, A.S.; Gordon, S.M. Effectiveness of the 2023–2024 Formulation of the Coronavirus Disease 2019 Messenger RNA Vaccine. Clin. Infect. Dis. 2024, ciae132. [Google Scholar] [CrossRef]
- Link-Gelles, R.; Ciesla, A.A.; Mak, J.; Miller, J.D.; Silk, B.J.; Lambrou, A.S.; Paden, C.R.; Shirk, P.; Britton, A.; Smith, Z.R.; et al. Early Estimates of Updated 2023–2024 (Monovalent XBB.1.5) COVID-19 Vaccine Effectiveness Against Symptomatic SARS-CoV-2 Infection Attributable to Co-Circulating Omicron Variants Among Immunocompetent Adults—Increasing Community Access to Testing Program, United States, September 2023–January 2024. MMWR. Morb. Mortal. Wkly. Rep. 2024, 73, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Witberg, G.; Barda, N.; Hoss, S.; Richter, I.; Wiessman, M.; Aviv, Y.; Grinberg, T.; Auster, O.; Dagan, N.; Balicer, R.D.; et al. Myocarditis after Covid-19 Vaccination in a Large Health Care Organization. N. Engl. J. Med. 2021, 385, 2132–2139. [Google Scholar] [CrossRef]
- Comirnaty COVID-19 Vaccine. [Online] Summary of Product Characteristics. European Medicines Agency. 2022. Available online: https://www.ema.europa.eu/en/documents/product-information/comirnaty-epar-product-information_en.pdf (accessed on 21 July 2024).
- Goddard, K.; Lewis, N.; Fireman, B.; Weintraub, E.; Shimabukuro, T.; Zerbo, O.; Boyce, T.G.; Oster, M.E.; Hanson, K.E.; Donahue, J.G.; et al. Risk of myocarditis and pericarditis following BNT162b2 and mRNA-1273 COVID-19 vaccination. Vaccine 2022, 40, 5153–5159. [Google Scholar] [CrossRef]
- Husby, A.; Gulseth, H.L.; Hovi, P.; Hansen, J.V.; Pihlström, N.; Gunnes, N.; Härkänen, T.; Dahl, J.; Karlstad, Ø.; Heliö, T.; et al. Clinical outcomes of myocarditis after SARS-CoV-2 mRNA vaccination in four Nordic countries: Population based cohort study. BMJ Med. 2023, 2, e000373. [Google Scholar] [CrossRef]
- Fronza, M.; Thavendiranathan, P.; Chan, V.; Karur, G.R.; Udell, J.A.; Wald, R.M.; Hong, R.; Hanneman, K. Myocardial Injury Pattern at MRI in COVID-19 Vaccine–Associated Myocarditis. Radiology 2022, 304, 553–562. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. [Online] Selected Adverse Events Reported after COVID-19 Vaccination. 2023. Available online: https://www.cdc.gov/coronavirus/2019-ncov/vaccines/safety/adverse-events.html (accessed on 21 July 2024).
- Smadja, D.M.; Yue, Q.-Y.; Chocron, R.; Sanchez, O.; Louet, A.L.-L. Vaccination against COVID-19: Insight from arterial and venous thrombosis occurrence using data from VigiBase. Eur. Respir. J. 2021, 58, 2100956. [Google Scholar] [CrossRef] [PubMed]
- Elalamy, I.; Gerotziafas, G.; Alamowitch, S.; Laroche, J.-P.; Van Dreden, P.; Ageno, W.; Beyer-Westendorf, J.; Cohen, A.T.; Jimenez, D.; Brenner, B.; et al. SARS-CoV-2 Vaccine and Thrombosis: An Expert Consensus on Vaccine-Induced Immune Thrombotic Thrombocytopenia. Thromb. Haemost. 2021, 121, 982–991. [Google Scholar] [CrossRef]
- Fan, B.E.; Shen, J.Y.; Lim, X.R.; Tu, T.M.; Chang, C.C.R.; Khin, H.S.W.; Koh, J.S.; Rao, J.P.; Lau, S.L.; Tan, G.B.; et al. Cerebral venous thrombosis post BNT162b2 mRNA SARS-CoV-2 vaccination: A black swan event. Am. J. Hematol. 2021, 96, E357–E361. [Google Scholar] [CrossRef]
- Whiteley, W.N.; Ip, S.; Cooper, J.A.; Bolton, T.; Keene, S.; Walker, V.; Denholm, R.; Akbari, A.; Omigie, E.; Hollings, S.; et al. Association of COVID-19 vaccines ChAdOx1 and BNT162b2 with major venous, arterial, or thrombocytopenic events: A population-based cohort study of 46 million adults in England. PLoS Med. 2022, 19, e1003926. [Google Scholar] [CrossRef] [PubMed]
- Cari, L.; Fiore, P.; Alhosseini, M.N.; Sava, G.; Nocentini, G. Blood clots and bleeding events following BNT162b2 and ChAdOx1 nCoV-19 vaccine: An analysis of European data. J. Autoimmun. 2021, 122, 102685. [Google Scholar] [CrossRef] [PubMed]
- Sekulovski, M.; Mileva, N.; Vasilev, G.V.; Miteva, D.; Gulinac, M.; Peshevska-Sekulovska, M.; Chervenkov, L.; Batselova, H.; Vasilev, G.H.; Tomov, L.; et al. Blood Coagulation and Thrombotic Disorders following SARS-CoV-2 Infection and COVID-19 Vaccination. Biomedicines 2023, 11, 2813. [Google Scholar] [CrossRef]
- Grupper, A.; Rabinowich, L.; Schwartz, D.; Schwartz, I.F.; Ben-Yehoyada, M.; Shashar, M.; Katchman, E.; Halperin, T.; Turner, D.; Goykhman, Y.; et al. Reduced humoral response to mRNA SARS-CoV-2 BNT162b2 vaccine in kidney transplant recipients without prior exposure to the virus. Arab. Archaeol. Epigr. 2021, 21, 2719–2726. [Google Scholar] [CrossRef]
- Boyarsky, B.J.; Ou, M.T.B.; Greenberg, R.S.B.; Teles, A.T.B.; Werbel, W.A.; Avery, R.K.; Massie, A.B.; Segev, D.L.; Garonzik-Wang, J.M. Safety of the First Dose of SARS-CoV-2 Vaccination in Solid Organ Transplant Recipients. Transplantation 2021, 105, e56–e57. [Google Scholar] [CrossRef]
- Rahav, G.; Lustig, Y.; Lavee, J.; Benjamini, O.; Magen, H.; Hod, T.; Shem-Tov, N.; Shmueli, E.S.; Merkel, D.; Ben-Ari, Z.; et al. BNT162b2 mRNA COVID-19 vaccination in immunocompromised patients: A prospective cohort study. eClinicalMedicine 2021, 41, 101158. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, S.; Pascucci, D.; Laurenti, P. Humoral response after a fourth dose of SARS-CoV-2 vaccine in immunocompromised patients. Results of a systematic review. Front. Public Health 2023, 11, 1108546. [Google Scholar] [CrossRef]
- Novavax. Novavax and Serum Institute of India Announce World Health Organization Grants Emergency Use Listing for NVX-CoV2373 COVID-19 Vaccine [Online]. 2021. Available online: https://ir.novavax.com/press-releases/2021-12-17-Novavax-and-Serum-Institute-of-India-Announce-World-Health-Organization-Grants-Emergency-Use-Listing-for-NVX-CoV2373-COVID-19-Vaccine (accessed on 6 July 2024).
- Fix, J.; Mast, T.C.; Smith, K.; Baker, N. Benefit–risk assessment for the Novavax COVID-19 vaccine (NVX-CoV2373). Vaccine 2024, 42, 2161–2165. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Trost, J.F.; Guebre-Xabier, M.; Zhou, H.; Norton, J.; Jiang, D.; Cai, Z.; Zhu, M.; Marchese, A.M.; Greene, A.M.; et al. XBB.1.5 spike protein COVID-19 vaccine induces broadly neutralizing and cellular immune responses against EG.5.1 and emerging XBB variants. Sci. Rep. 2023, 13, 19176. [Google Scholar] [CrossRef]
- Food and Drug Administration. Novavax COVID-19 Vaccine, Adjuvanted [Online]. 2023. Available online: https://www.fda.gov/vaccines-blood-biologics/coronavirus-covid-19-cber-regulated-biologics/novavax-covid-19-vaccine-adjuvanted#additional (accessed on 6 July 2024).
- World Health Organization. Background Document on the Novavax NVX-CoV2373 Vaccine against COVID-19 [Online]. 2021. Available online: https://www.who.int/publications/i/item/WHO-2019-nCoV-vaccines-SAGE-recommendation-Novavax-NVX-CoV2373-background (accessed on 6 July 2024).
- Novavax. Novavax Submits Application to U.S. FDA for Updated Protein-Based 2024–2025 Formula COVID-19 Vaccine [Online]. 2024. Available online: https://ir.novavax.com/press-releases/2024-06-14-Novavax-Submits-Application-to-U-S-FDA-for-Updated-Protein-based-2024-2025-Formula-COVID-19-Vaccine (accessed on 6 July 2024).
- Hotez, P.J.; Bottazzi, M.E. Whole inactivated virus and protein-based COVID-19 vaccines. Annu. Rev. Med. 2022, 73, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Reimer, J.M.; Karlsson, K.H.; Lövgren-Bengtsson, K.; Magnusson, S.E.; Fuentes, A.; Stertman, L. Matrix-M™ adjuvant induces local recruitment, activation and maturation of central immune cells in absence of antigen. PLoS ONE 2012, 7, e41451. [Google Scholar] [CrossRef] [PubMed]
- Stertman, L.; Palm, A.K.; Zarnegar, B.; Carow, B.; Lunderius Andersson, C.; Magnusson, S.E.; Carnrot, C.; Shinde, V.; Smith, G.; Glenn, G.; et al. The Matrix-M™ adjuvant: A critical component of vaccines for the 21st century. Hum. Vaccines Immunother. 2023, 19, 2189885. [Google Scholar] [CrossRef]
- Heath, P.T.; Galiza, E.P.; Baxter, D.N.; Boffito, M.; Browne, D.; Burns, F.; Chadwick, D.R.; Clark, R.; Cosgrove, C.; Galloway, J.; et al. Safety and efficacy of NVX-CoV2373 COVID-19 vaccine. N. Engl. J. Med. 2021, 385, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Áñez, G.; Dunkle, L.M.; Gay, C.L.; Kotloff, K.L.; Adelglass, J.M.; Essink, B.; Campbell, J.D.; Cloney-Clark, S.; Zhu, M.; Plested, J.S.; et al. Safety, Immunogenicity, and Efficacy of the NVX-CoV2373 COVID-19 Vaccine in Adolescents: A Randomized Clinical Trial. JAMA Netw. Open 2023, 6, e239135. [Google Scholar] [CrossRef]
- Ahmad, S.; Yuson, C.; Le, A.; Hissaria, P. Myopericarditis following both BNT162b2 and NVX-CoV2373. Allergy Asthma Clin. Immunol. 2022, 18, 109. [Google Scholar] [CrossRef]
- Romanson, B. Notes from the Field: Safety Monitoring of Novavax COVID-19 Vaccine among Persons Aged ≥ 12 Years—United States, 13 July 2022–13 March 2023. MMWR. Morb. Mortal. Wkly. Rep. 2023, 72, 850–851. [Google Scholar] [CrossRef] [PubMed]
- Clothier, H.J.; Parker, C.; Mallard, J.H.; Effler, P.; Bloomfield, L.; Carcione, D.; Buttery, J.P. Real-world Nuvaxovid COVID-19 vaccine safety profile after first 100,000 doses in Australia, 2022–2023. medRxiv 2024. [Google Scholar] [CrossRef]
- Ogunjimi, O.B.; Tsalamandris, G.; Paladini, A.; Varrassi, G.; Zis, P. Guillain-Barré Syndrome Induced by Vaccination Against COVID-19: A Systematic Review and Meta-Analysis. Cureus 2023, 15, e37578. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-W.; Hu, W.; Shen, L.; Wang, F.-S. Safety and immunogenicity of COVID-19 vaccination in immunocompromised patients. Chin. Med. J. 2022, 135, 2656–2666. [Google Scholar] [CrossRef] [PubMed]
- HMA-EMA Catalogues of Real-World Data Sources and Studies. Safety Profile of the NVX-CoV2373 Vaccine in Individuals ≥ 12 Years of Age in the United States [Online]. HMA-EMA Catalogues of Real-World Data Sources and Studies. 2024. Available online: https://catalogues.ema.europa.eu/node/3713/methodological-aspects (accessed on 21 July 2024).
- Health Canada. Novavax Nuvaxovid COVID-19 Vaccine [Online]. 2024. Available online: https://www.canada.ca/en/health-canada/services/drugs-health-products/covid19-industry/drugs-vaccines-treatments/vaccines/novavax.html (accessed on 6 July 2024).
- Mallory, R.M.; Formica, N.; Pfeiffer, S.; Wilkinson, B.; Marcheschi, A.; Albert, G.; McFall, H.; Robinson, M.; Plested, J.S.; Zhu, M.; et al. Safety and immunogenicity following a homologous booster dose of a SARS-CoV-2 recombinant spike protein vaccine (NVX-CoV2373): A secondary analysis of a randomised, placebo-controlled, phase 2 trial. Lancet Infect. Dis. 2022, 22, 1565–1576. [Google Scholar] [CrossRef] [PubMed]
- Munro, A.P.S.; Janani, L.; Cornelius, V.; Aley, P.K.; Babbage, G.; Baxter, D.; Bula, M.; Cathie, K.; Chatterjee, K.; Dodd, K.; et al. Safety and immunogenicity of seven COVID-19 vaccines as a third dose (booster) following two doses of ChAdOx1 nCov-19 or BNT162b2 in the UK (COV-BOOST): A blinded, multicentre, randomised, controlled, phase 2 trial. Lancet 2021, 398, 2258–2276. [Google Scholar] [CrossRef] [PubMed]
- Raiser, F.; Davis, M.; Adelglass, J.; Cai, M.R.; Chau, G.; Cloney-Clark, S.; Eickhoff, M.; Kalkeri, R.; McKnight, I.; Plested, J.; et al. Immunogenicity and safety of NVX-CoV2373 as a booster: A phase 3 randomized clinical trial in adults. Vaccine 2023, 41, 5965–5973. [Google Scholar] [CrossRef] [PubMed]
- Kutikova, L.; Brash, J.T.; Helme, K.; Brewster, J.; Brand, M.; Adam, A.; Seager, S.; Kostev, K.; Schelling, J. Characteristics and Outcomes for Recipients of NVX-CoV2373: A Real-World Retrospective Study in Germany. Vaccines 2024, 12, 387. [Google Scholar] [CrossRef] [PubMed]
- Mateo-Urdiales, A.; Sacco, C.; Petrone, D.; Bella, A.; Riccardo, F.; Del Manso, M.; Bressi, M.; Siddu, A.; Brusaferro, S.; Palamara, A.T.; et al. Estimated Effectiveness of a Primary Cycle of Protein Recombinant Vaccine NVX-CoV2373 Against COVID-19. JAMA Netw. Open 2023, 6, e2336854. [Google Scholar] [CrossRef]
- Gwak, E.; Choe, S.A.; Bolormaa, E.; Choe, Y.J.; Wang, C.; Fix, J.; Vadivale, M.; Rousculp, M.D. Short-Term Relative Effectiveness of Homologous NVX-CoV2373 and BNT162b2 COVID-19 Vaccinations in South Korea. medRxiv 2024. [Google Scholar] [CrossRef]
- Liu, B.; Stepien, S.; Qian, J.; Gidding, H.; Nicolopoulos, K.; Amin, J.; Cheng, A.; Macartney, K. Comparative effectiveness of four COVID-19 vaccines, BNT162b2 mRNA, mRNA-1273, ChAdOx1 nCov-19 and NVX-CoV2373 against SARS-CoV-2 B.1.1.529 (Omicron) infection. Vaccine 2023, 41, 5587–5591. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.S.M.; Choe, Y.J.; Choe, S.A.; Gwak, E.S.; Kwon, D.D. Relative Effectiveness of the NVX-CoV2373 Vaccine Compared with the BNT162b2 Vaccine in Adolescents. Pediatr. Infect. Dis. J. 2024. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Updated (2023–24 Formula) Novavax COVID-19 Vaccine [Online]. 2024. Available online: https://www.cdc.gov/vaccines/covid-19/info-by-product/novavax/downloads/novavax-standing-orders.pdf (accessed on 29 June 2024).
- ClinicalTrials.gov. A Study to Evaluate the Safety and Immunogenicity of an (Omicron Subvariant) COVID-19 Vaccine Booster Dose in Previously Vaccinated Participants and Unvaccinated Participants. (COVID-19) [Online]. 2024. Available online: https://clinicaltrials.gov/study/NCT05975060?intr=NVX-CoV2601&rank=3 (accessed on 6 July 2024).
- ClinicalTrials.gov. Phase 3 Adolescent Study for SARS-CoV-2 rS Variant Vaccines (COVID-19) [Online]. 2024. Available online: https://clinicaltrials.gov/study/NCT05973006?intr=NVX-CoV2601&rank=2 (accessed on 6 July 2024).
- ClinicalTrials.gov. Phase 2/3 Heterologous Boosting Study with Different Dose Levels of Monovalent SARS-CoV-2 rS Vaccines (COVID-19) [Online]. 2024. Available online: https://clinicaltrials.gov/study/NCT05925127?intr=NVX-CoV2601&rank=1 (accessed on 6 July 2024).
- Singh, A.V.; Kayal, A.; Malik, A.; Maharjan, R.S.; Dietrich, P.; Thissen, A.; Siewert, K.; Curato, C.; Pande, K.; Prahlad, D.; et al. Interfacial Water in the SARS Spike Protein: Investigating the Interaction with Human ACE2 Receptor and In Vitro Uptake in A549 Cells. Langmuir 2022, 38, 7976–7988. [Google Scholar] [CrossRef] [PubMed]
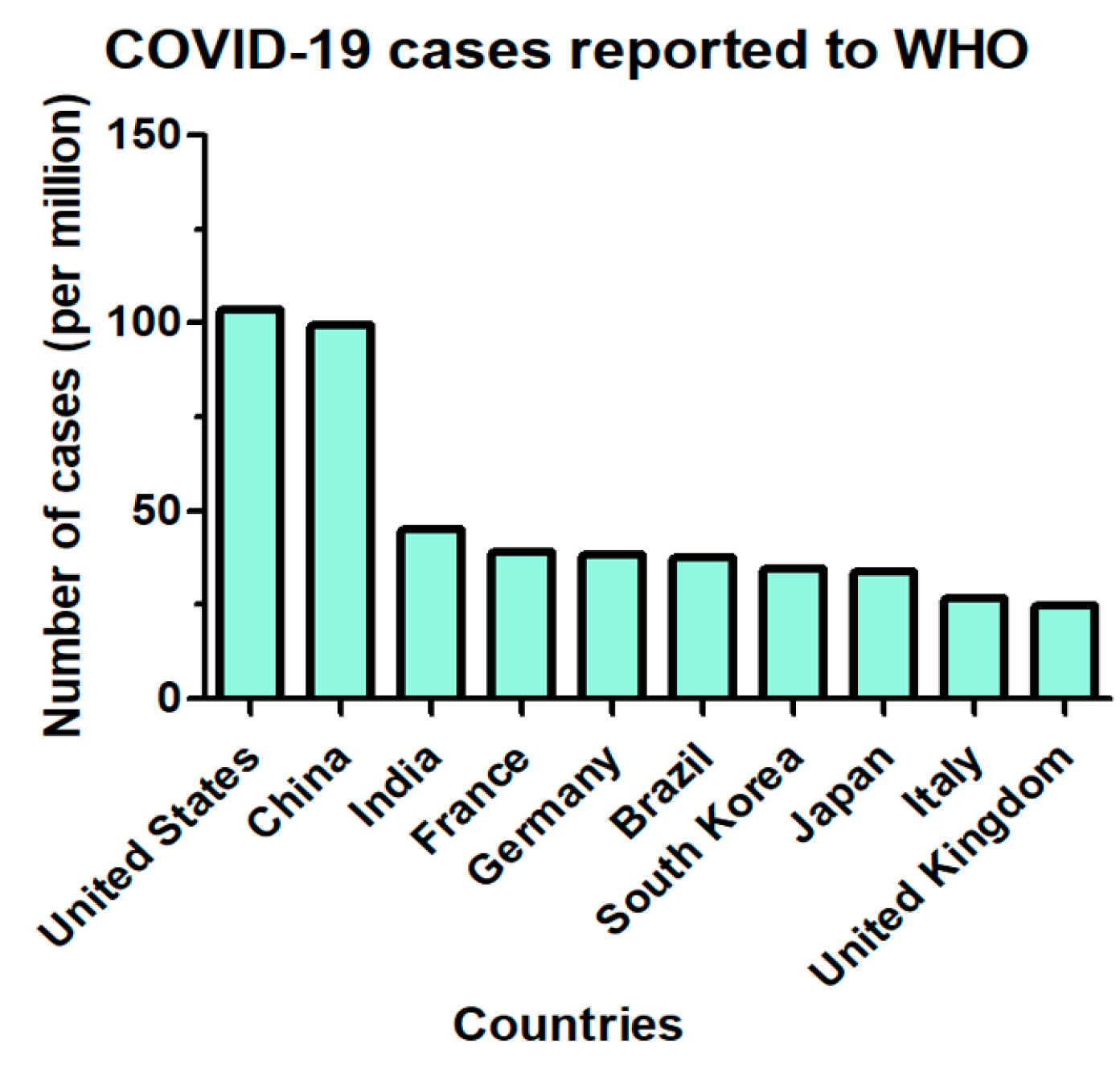
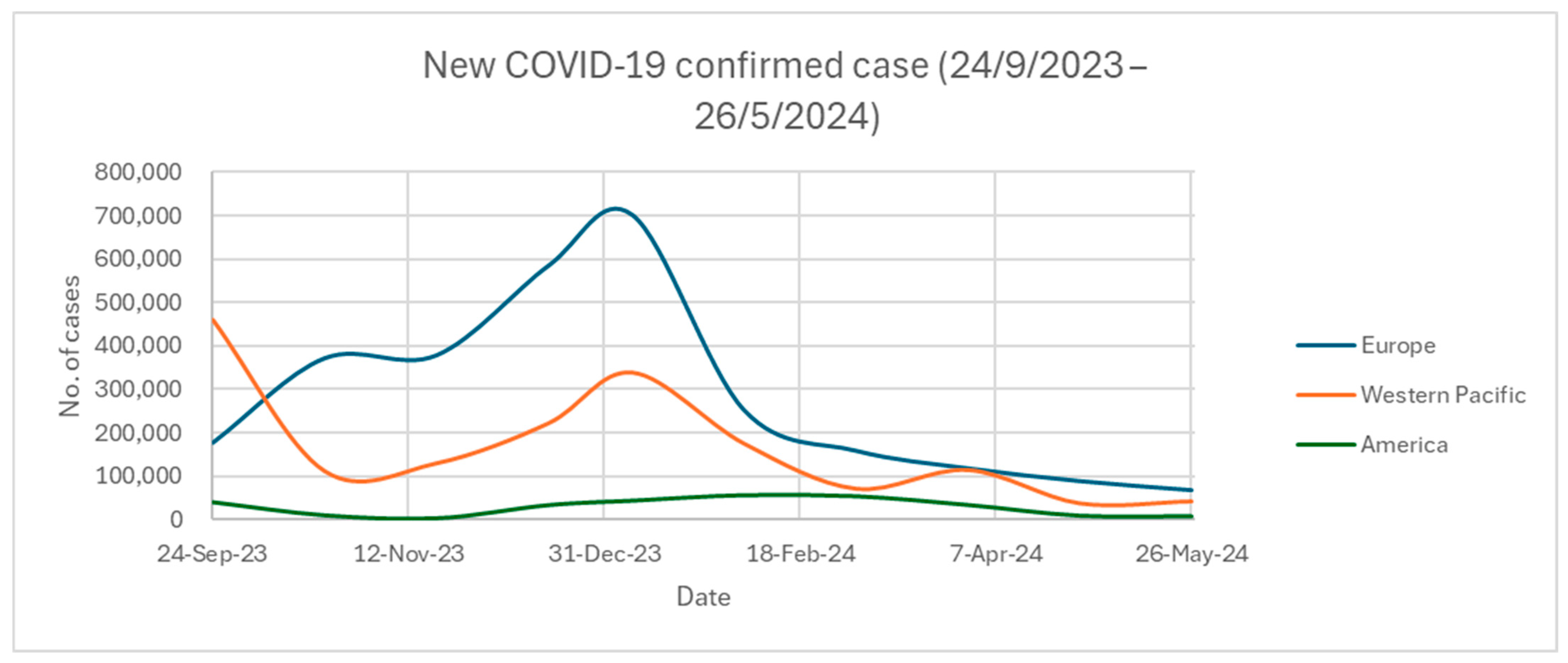
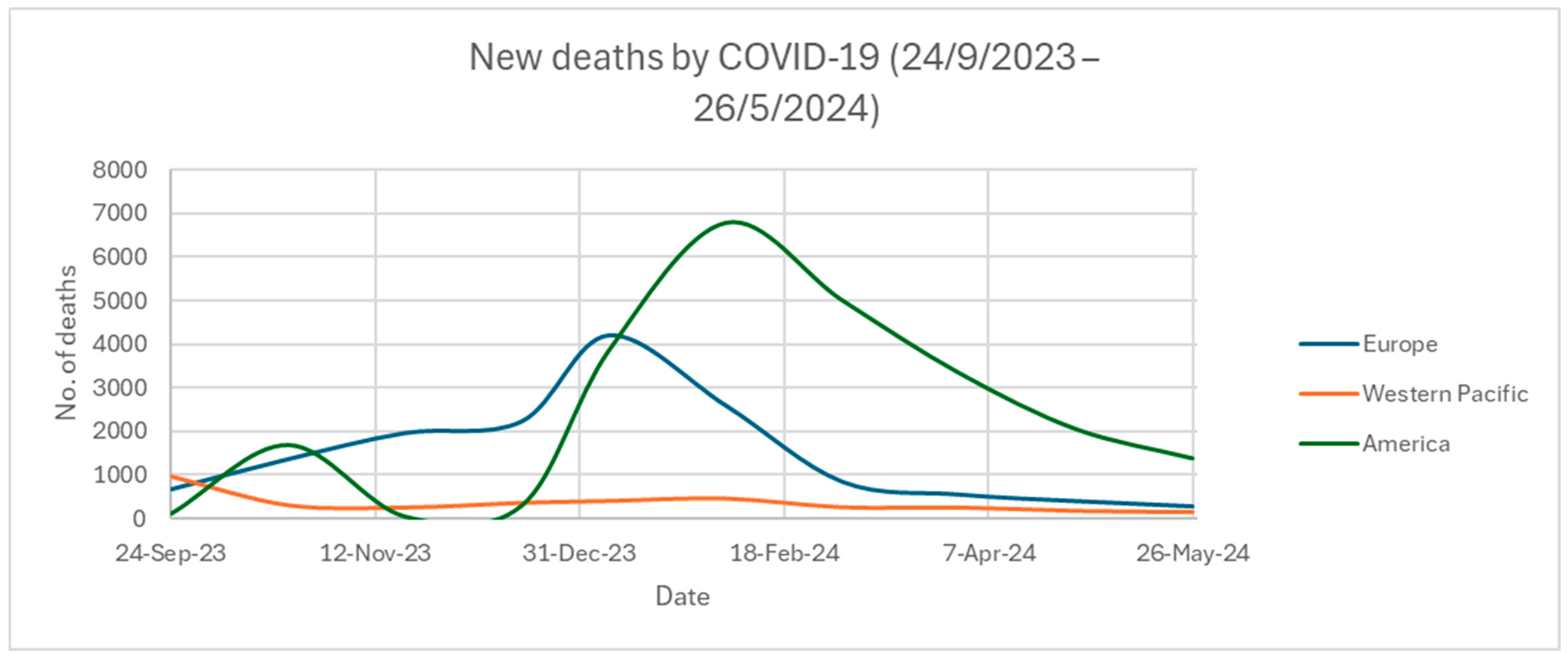
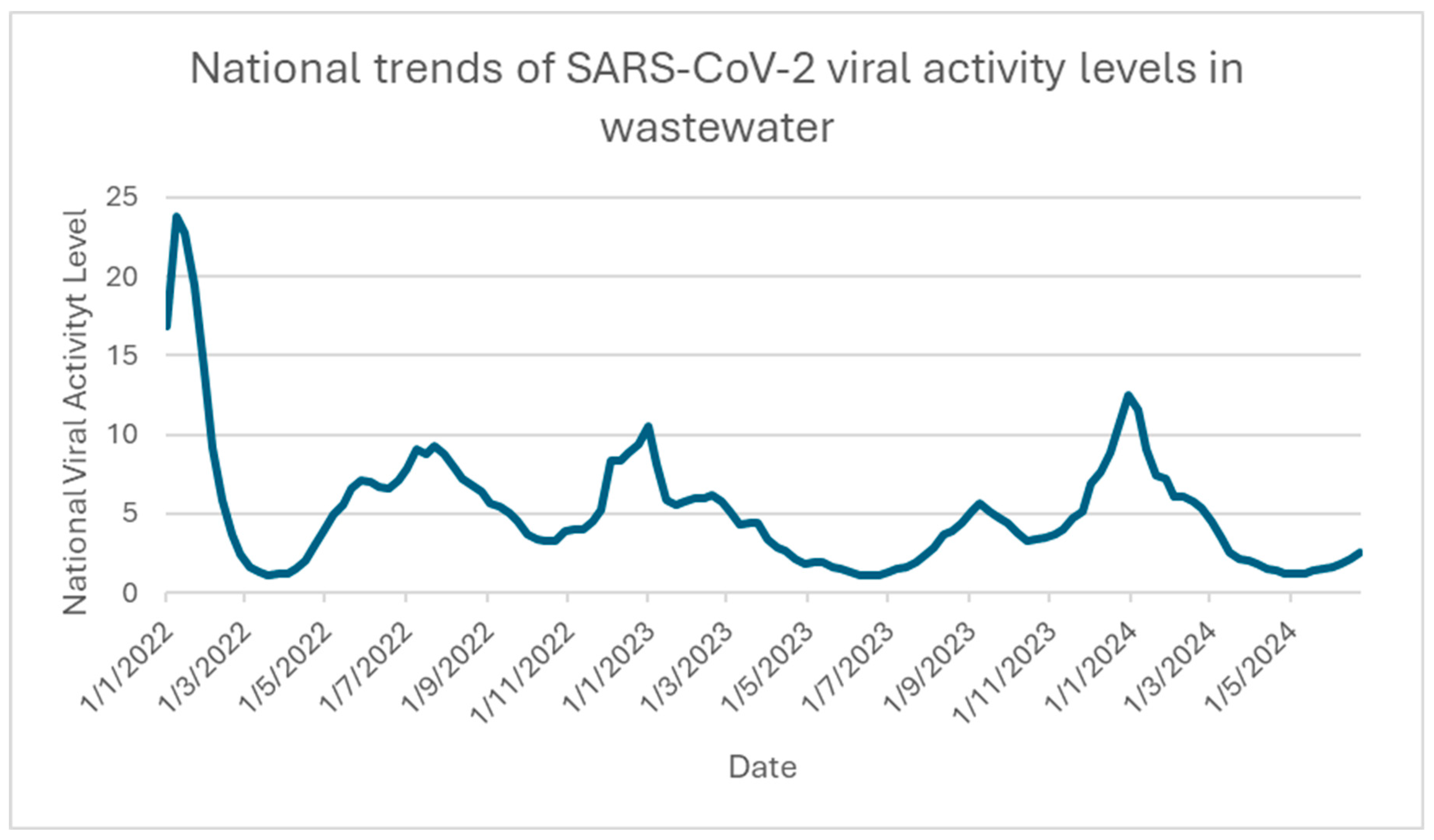
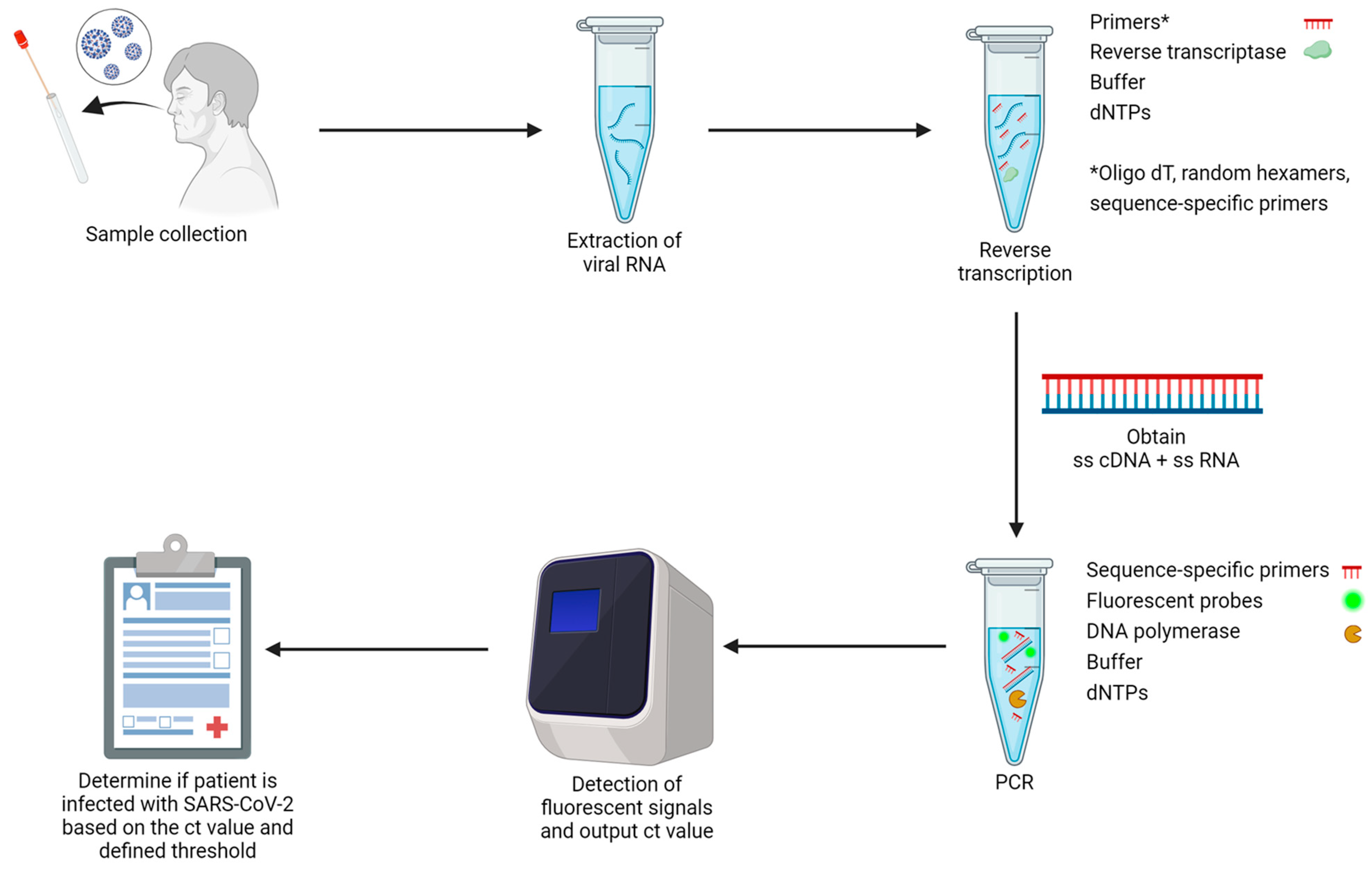
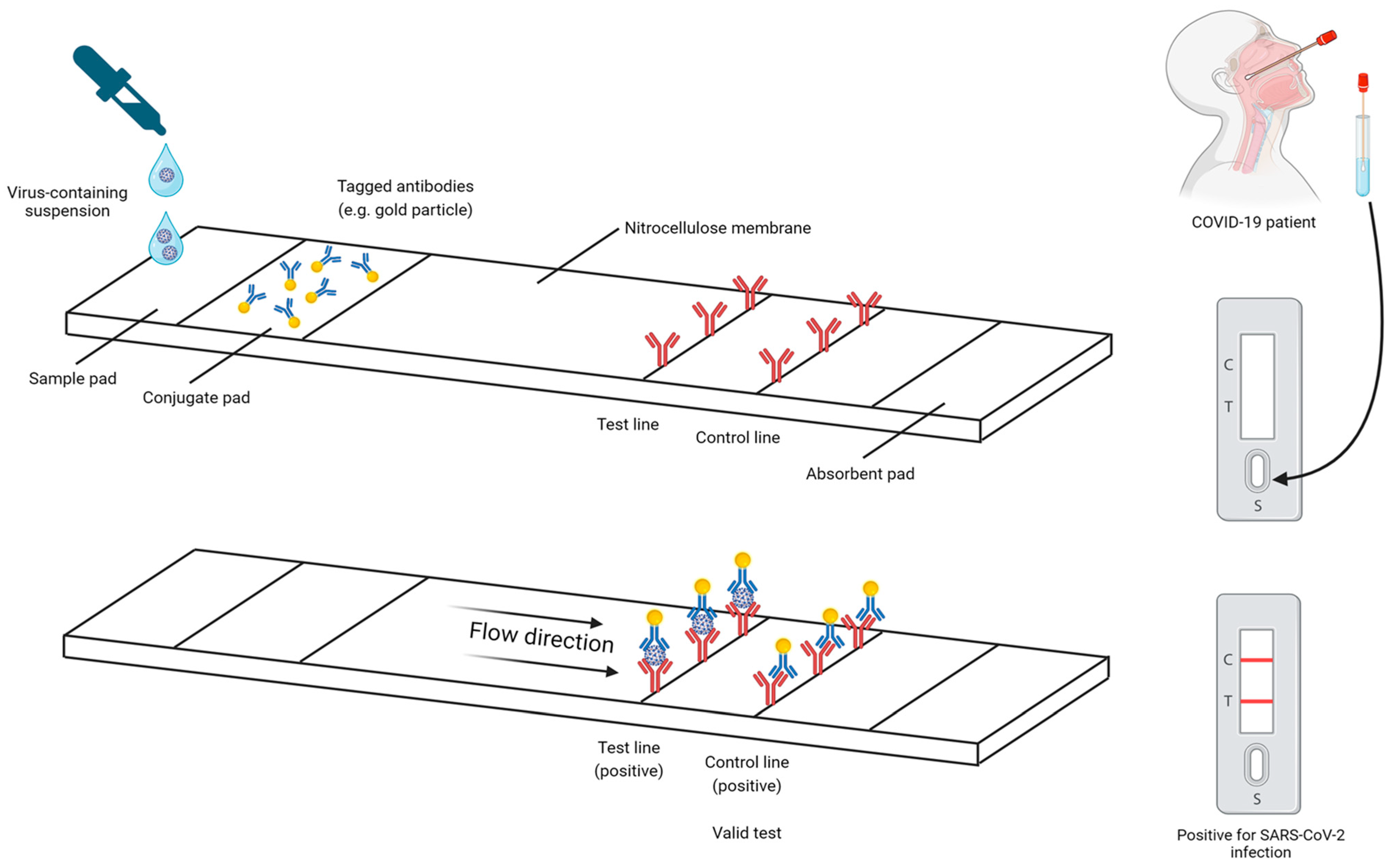
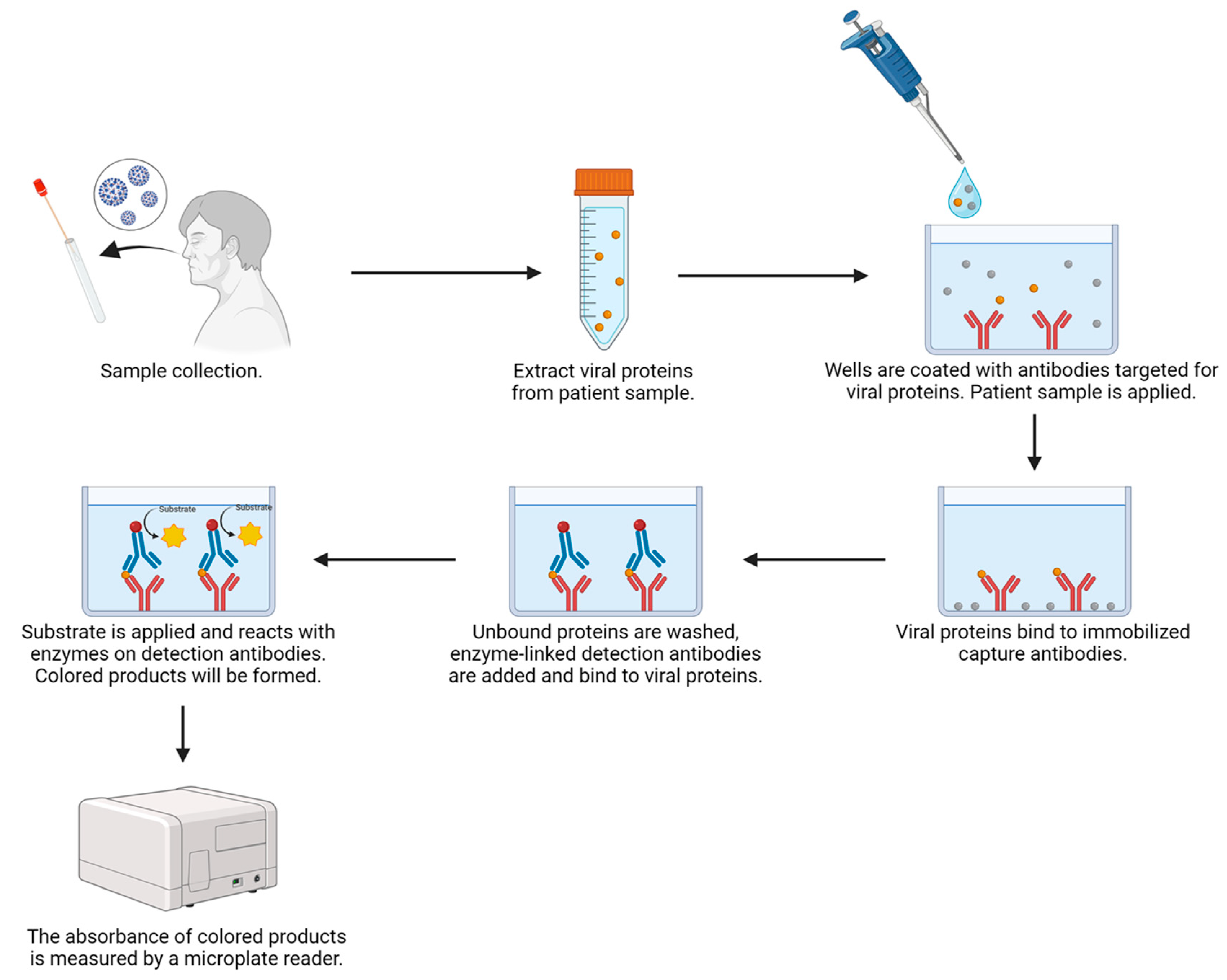

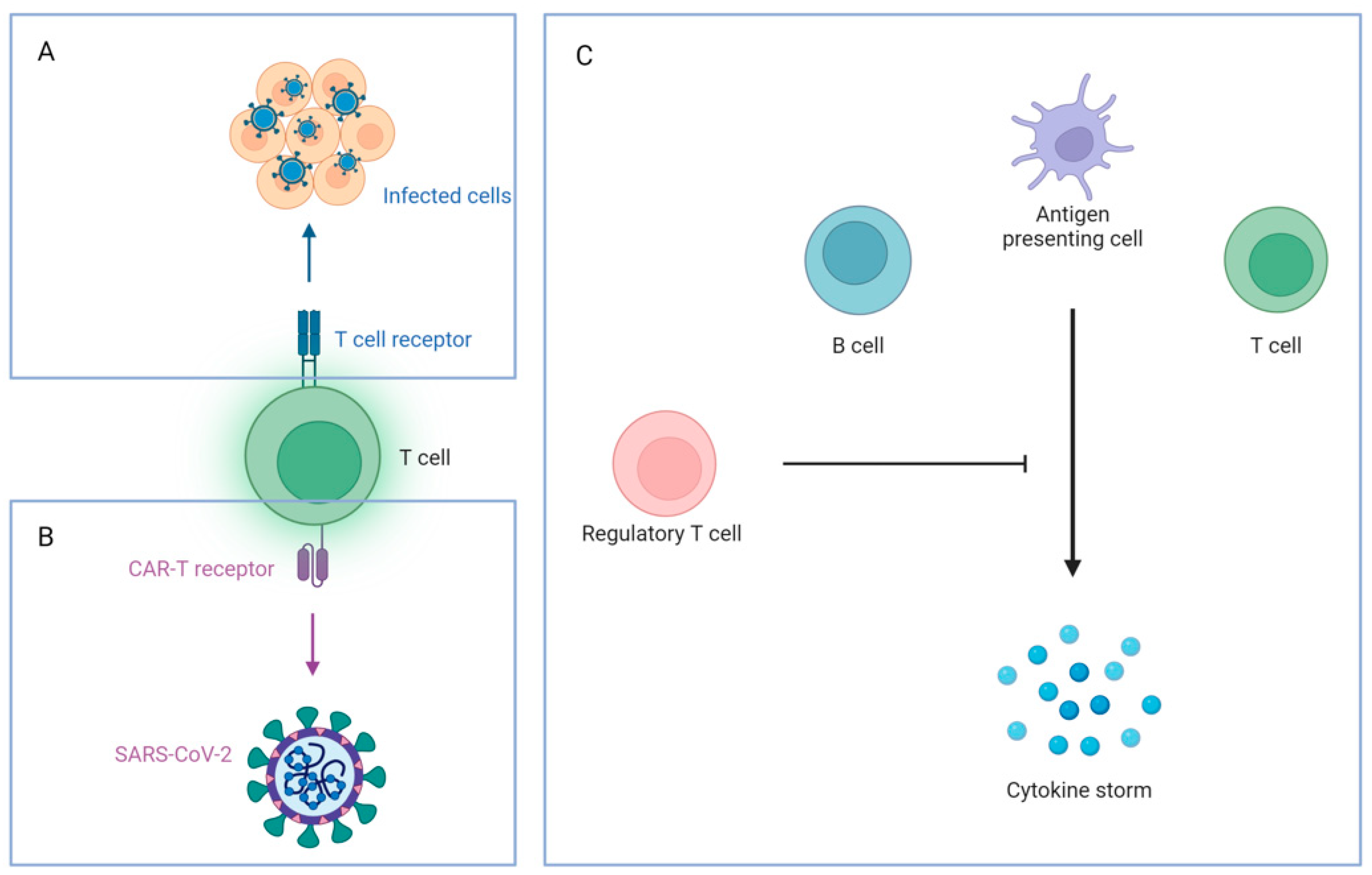
| WHO Label | Pango Lineage | First Outbreak | Number of Cases Worldwide |
|---|---|---|---|
| Alpha | B.1.1.7 | United Kingdom | 1,141,525 |
| Beta | B.1.351 | South Africa | 35,651 |
| Gamma | P.1 | Brazil | 73,495 |
| Delta | B.1.617.2 | India | 288,894 |
| Omicron | B.1.1.529 | South Africa | 1895 * |
| Method | RT-qPCR | ELISA (Antigen) | Rapid Antigen Test (RAT) |
|---|---|---|---|
| Category | Nucleic acid amplification test (NAAT) | Serological test | Serological test |
| Comment from important organizations | - Gold standard for COVID-19 diagnosis (WHO) | - Acceptable specificity - Can be set as a diagnosis criterion, together with clinical and epidemiological histories and case definition (WHO) | - Several FDA-approved over-the-counter COVID-19 diagnostic tests available in the market - Need to perform repeat testing to prevent obtaining a false-negative result (FDA) |
| Target for detection | Nucleic acids - Particular regions on the cDNA originated from the ssRNA of the virus | Viral proteins | Viral proteins |
| Specimens | Nasopharyngeal swab, oropharyngeal swab, saliva, sputum | Nasopharyngeal swab, oropharyngeal swab, saliva, sputum (depends on instructions) | Nasopharyngeal swab, oropharyngeal swab, saliva, sputum (depends on instructions) |
| Sensitivity | High | Limited | Limited |
| Specificity | High | Varies | Varies |
| Result generation time | Varies | Varies | 10–15 min |
| Cost | High | Moderate | Low |
| Trained operators | Yes | Yes | No |
| Affected by antigenically different variants | Not likely | Yes | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chung, Y.-S.; Lam, C.-Y.; Tan, P.-H.; Tsang, H.-F.; Wong, S.-C.C. Comprehensive Review of COVID-19: Epidemiology, Pathogenesis, Advancement in Diagnostic and Detection Techniques, and Post-Pandemic Treatment Strategies. Int. J. Mol. Sci. 2024, 25, 8155. https://doi.org/10.3390/ijms25158155
Chung Y-S, Lam C-Y, Tan P-H, Tsang H-F, Wong S-CC. Comprehensive Review of COVID-19: Epidemiology, Pathogenesis, Advancement in Diagnostic and Detection Techniques, and Post-Pandemic Treatment Strategies. International Journal of Molecular Sciences. 2024; 25(15):8155. https://doi.org/10.3390/ijms25158155
Chicago/Turabian StyleChung, Yiu-Sing, Ching-Yin Lam, Pak-Hei Tan, Hin-Fung Tsang, and Sze-Chuen Cesar Wong. 2024. "Comprehensive Review of COVID-19: Epidemiology, Pathogenesis, Advancement in Diagnostic and Detection Techniques, and Post-Pandemic Treatment Strategies" International Journal of Molecular Sciences 25, no. 15: 8155. https://doi.org/10.3390/ijms25158155
APA StyleChung, Y.-S., Lam, C.-Y., Tan, P.-H., Tsang, H.-F., & Wong, S.-C. C. (2024). Comprehensive Review of COVID-19: Epidemiology, Pathogenesis, Advancement in Diagnostic and Detection Techniques, and Post-Pandemic Treatment Strategies. International Journal of Molecular Sciences, 25(15), 8155. https://doi.org/10.3390/ijms25158155







