The Role of the Gut and Airway Microbiota in Chronic Rhinosinusitis with Nasal Polyps: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Search Strategy
2.2. Selection Criteria
2.3. Review Process and Data Extraction
3. Results
3.1. Search Results
3.2. Study Analysis
3.2.1. Microbiota Composition in CRSwNP
| Ref. | Study Type | Objective | Sample Size | Country | Age (Average) | Sex (n with CRSwNP) | Other Disease (n with CRSwNP) | Sample Type | Methodology | Results/Conclusion | Observ. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| [73] | CSS | To compare the predominant bacteriological profiles in the middle meatus of Chinese CRSwNP and CRSsNP patients and DC subjects. | 537 (285 completed the study) CRSwNP: 165 CRSsNP: 76 DC: 44 | China | 18–80 CRSwNP: 18–78 (45.2) | M: 185 (109 with CRSwNP) F: 100 (56) | AS: 14 (13 with CRSwNP) AL: 11 (10) AsS: 1 (1) | MMSS collected during ESS | SMT | The most common bacteria were Coagulase-negative staphylococci (CoNS), Streptococcus, Corynebacterium spp., Staphylococcus aureus, and Haemophilus influenzae. No significant differences in the species or the number of different bacterial isolates between CRSwNP and DC. Very few fungi were isolated: one case each of Penicillium notatum, Verticillium, Aspergillus, and Schizophyllum. | No antibiotics or corticosteroids in the previous two weeks |
| [80] | CS | To evaluate human herpesviruses (HHV) 1–6 and community-acquired respiratory viruses (CARVS) prevalence in CRSwNP patients undergoing FESS | 35 CRSwNP: 35 | Italy | CRSwNP: 23–77 (50.3) | M: (25) F: (10) | AS: (8) AL: (15) | Bioptic samples of NP collected during FESS | MT (PCR and qPCR) | 60% of patients (21/35) were positive for at least one virus in at least one specimen studied. The highest prevalence was found for herpesviruses 6 (HHV-6), Epstein–Barr virus (EBV), cytomegalovirus (CMV), and Herpes simplex virus type 1 (HSV-1). | NA |
| [96] | RCT | To evaluate the efficacy of long-term antibiotic therapy to prevent recurrence of NP | 66 CRSwNP: 66 (55 completed the study) | Russia | 18–77 (48.7) | M: 36 F: 30 | AS: 35 AT: 19 AERD: 27 | MMSS pre-FESS and post-FESS | SMT (system Microscan walk away 40) | The most common bacteria were S. aureus, Streptococcus haemolyticus, Escherichia coli, Pseudomonas aeruginosa, and Enterobacter aerogenes. The bacterial spectrum changed significantly after surgery. | No corticosteroids |
| [66] | CS | To evaluate the association between bacterial infection and surgical outcomes following FEES | 71 CRSwNP: 71 | Republic of Korea | NA | M: 42 F: 29 | * Patients with a history of asthma were excluded | Maxillary sinus samples after MM antrostomy | SMT | Most of the patients (55/71, 77.5%) showed positive culture results. The most common bacteria were S. epidermidis, Propionibacterium acne, Corynebacterium, and E. aerogenes. The “normal flora group” (S. epidermidis and Corynebacterium) had the worst prognosis of postoperation, while the “culture-negative group” had the best prognosis. | NA |
| [99] | CSS | To compare the diversity of nasal microbiota and their secreted extracellular vesicles between CRSwNP patients and DCs | 11 CRSwNP: 5 CRSsNP: 3 DC: 3 | Republic of Korea | 19–67 (43.1) CRSwNP: 23–67, (45.6) | M: 8 (4) F: 3 (1) | AT: 3 (1) | Nasal lavage fluid samples during ESS | MT (sequencing, OTUs) | The major bacterial genera were Pseudomonas, Haemophilus, Staphylococcus, Moraxella, Enterobacter, and Fusobacterium *. | No antibiotics or corticosteroids in the previous month |
| [100] | CSS | To determine whether smoking affects CT score, bacterial diversity of the upper airways, and distribution of inflammatory cells in nasal mucosa in CRS patients | 64 CRSwNP: 20 CRSsNP: 17 DC: 27 | Slovakia | CRSwNP: 22–76 (49.6) | NA | Patients with a history of asthma, atopy, and aspirin intolerance were excluded | MMSS | SMT | The most common bacteria were CoNS, S. aureus, and Corynebacterium species. | No antibiotics in the previous month |
| [70] | RCT | To compare the efficacy of topical nasal corticosteroids either as monotherapy or combined in eradicating the biofilm of NP | 44 CRSwNP: 44 (34 completed the study) | Turkey | NA | M: 26 F: 18 | NA | NP | Scanning electron microscopic (SEM) examination | In the initial baseline SEM examination, biofilm prevalence was 68.0% (23/34). | No antibiotics and/or corticosteroids in the previous month |
| [81] | CSS | To determine the presence of respiratory viruses in the paranasal sinuses of CRSwNP and CRSsNP patients compared to DCs | 35 CRSwNP: 13 CRSsNP: 8 DC: 14 | USA | CRSwNP: (55.3) | M: 19 (9) F: 16 (4) | AS: 15 (11) AR: 19 (11) AERD: 2 (2) | MMSS | MT (RT-PCR) | The presence of viruses (Coronavirus) was detected in only 7.7% of patients (1/13). None of the controls had a positive screen. | NA |
| [71] | CCS | To determine the presence of bacterial biofilm on the sinus mucosa of CRSwNP and CRSsNP patients and DCs | 100 CRwNP: 18 ACP: 12 CRSsNP: 20 DC: 50 | India | NA | NA | NA | Nasal–sinus tissue sample | SMT | Bacterial biofilm was detected in 66.7% of patients (12/18). | NA |
| [97] | CSS | To determine whether Gram-negative bacterial carriage impacted disease evolution and inflammatory profile in CRSwNP patients | 337 CRSwNP: 337 | Canada | NA (approx. 50) | NA | AS: aprox. 200 AL: 215 ASA: 101 (self-reported) | MMSS | SMT | The most common bacteria were CoNS, S. aureus, and C. diphtheriae. P. aeruginosa carriage was associated with a higher self-reported incidence of asthma. | NA |
| [95] | CSS | To evaluate the impact of saline irrigation and topical corticosteroids on the post-surgical sinonasal microbiota of CRSwNP patients and DCs | 42 CRSwNP: 14 DC: 28 | USA | 22–77 CRSwNP: 27–65 (52.0) | M: 15 (3) F: 27 (11) | NA | Nasal cavity and maxillary swab samples | MT (sequencing, 16S RNA gene sequences) | The most abundant bacteria were Propionibacterium, Corynebacterium, S. aureus, S. epidermis, and Staphylococcus pasteuri. A higher proportional abundance of Corynebacterium, Serratia, and Finegoldia was found in men than in women who used intranasal steroid spray. | No antibiotics or corticosteroids in the previous two months |
| [101] | CSS | To determine the association between smoking history and sinonasal microbiome alterations in both CRS patients and DCs | 101 CRSwNP: 22 CRSsNP: 48 DC: 31 | USA | >18 | M: 59 F: 42 | AS: 31 AR 60 | MMSS collected during ESS | MT (PCR and sequencing, OTUs) | The most abundant genera were Staphylococcus, Corynebacterium, Carnobacteriaceae, and Streptococcus. Smoking had a stronger effect on the microbiota at the phylum level (Bacteroides, Firmicutes, and Proteobacteria). Cigarette-smoking history was associated with less bacterial diversity, with a significant decrease in the phylum Firmicutes and an increase in the genera Carnobacteriaceae. | No systemic corticosteroids for at least one month |
| [76] | CSS | To determine the presence of specific fungal microbial species in CRS patients and DCss | 28 CRSwNP: 15 Allergic fungal rhinosinusitis: 3 Fungus ball: 3 DC: 7 | USA | CRSwNP: (49.0) | M: 17 (9) F: 11 (6) | AS: 12 (10) AsS: 4 (4) | Ethmoid and maxillary sinus brush samples | MT (qPCR) | Malassezia spp., M. restricta, and M. globosa were identified in ten, seven, and four CRSwNP patients, respectively. | NA |
| [67] | CSS | To define the bacteriology of CRSwNP and CRSsNP patients and DCs | 163 CRSwNP: 60 CRSsNP: 50 DC: 26 HC: 27 | Israel | CRSwNP: (51.2) | M: (37) F: (23) | NA | MMSS pre-FESS | SMT | Positive cultures (52/60) mainly for pathogenic bacteria (47/60) were found in most of the patients (86.7% and 78.3%, respectively). A higher rate of Gram-negative bacteria isolates than Gram-positive bacteria was found. The pathogenic bacteria most frequently isolated were S. aureus, Citrobacter diversus, Proteus mirabilis, Enterobacter, Pseudomonas, Klebsiella oxytoca, S. pneumoniae, and Klebsiella pneumoniae. | No antibiotics in the previous month |
| [68] | CSS | To define the bacteriology of CRSwNP and “sinonasal complication of dental disease or treatment” (SCDDT) patients | 44 CRSwNP: 16 SCDDT: 28 | Italy | CRSwNP: (49,4) | M: 26 (13) F: 18 (3) | NA | Endosinusal pus and biopsies from nasal polyps and fungus balls collected during surgery | SMT and MT (sequencing, 16S RNA gene sequences) | 56.3% of patients (9/16) did not show microbial growth. S. aureus and other staphylococci, Peptostreptococcus spp., E. coli, and Bacteroides spp., were identified. | NA |
| [93] | CSS | To compare the nasal microbiota in regard to health state, anatomical region, and sampling strategy | 79 CRSwNP: 15 CRSsNP: 27 DC: 37 | Germany | 18–79 CRSwNP: 20–77 (52.0) | M: 50 (13) F: 29 (2) | AS: 6 (4) AR: 23 (5) AsS: 3 (3) | Swab samples at four different regions along the nasal passage (anterior and posterior vestibules and inferior meatus and MM) and tissue biopsies | MT (sequencing, 16S RNA gene sequences) | No significant differences in global bacterial profiles at the sampling sites. Significant differences between the bacterial assemblages and diversity measured for different sample types (tissue biopsies and swabs). | No antibiotics at the time of sampling |
| [102] | CSS | To determine the association between the sino-nasal microbiota and toll-like receptor (TLR) activation in DCs and CRS patients | 36 CRSwNP: 11 CRSsNP: 9 CRS/CF: 6 DC: 10 | New Zealand | 18–84 (43.9) CRSwNP: 18–71 (41.1) | M: 14 (7) F: 22 (4) | AS: 2 (8) AsS: 2 (0) | Nasal lavage and sino-nasal mucus samples collected during ESS | MT (qPCR and sequencing, OTUs) | Pseudomonas, Haemophilus, Enterobacter, and Staphylococcus were the dominating genera. | No antibiotics and prednisone |
| [103] | CSS | To determine the association of distinct pathogenic sinus microbiota with specific innate and adaptive immune responses and the relative risk of NP | 76 (69 completed the study) CRSwNP: 32 CRSsNP: 27 DC: 10 | USA | 18–88 (46.6) CRSwNP: 19–88 (48.4) | M: 40 (21) F: 29 (11) | AS: 18 (12) CF: 9 (7) | Sinus brushing samples collected during ESS | MT (qPCR and sequencing, OTUs) | Most of the patients clustered into a subgroup typically dominated by Corynebacteriaceae | Most of patients (30/32, 93.8%) had taken pre-operative antibiotics (<3 months) |
| [90] | CSS | To compare the microbiota of the MM and inferior meatus in HCs, AR, and CRS patients and characterize intra- and inter-subject and inter-group differences | 65 (48 for mapping intrasubject microbiota diversity and composition) CRSwNP: 18 CRSsNP: 15 AR: 11 HC: 4 | USA | NA * | NA * | NA | MM and inferior meatus swab samples | MT (sequencing, OTUs) | No differences in phylogenetic diversity or Shannon diversity between MM and inferior meatus-associated microbiota. No differences in beta diversity across all subjects. Taxa enriched included Staphylococcus and Alloiococcus, as well as low-abundance Corynebacterium, Haemophilus, Prevotella, and Porphyromonas compared to HC. | No antibiotics and/or corticosteroids in the previous month |
| [74] | CSS | To analyze and quantify the sinonasal mycobiome in HC and CRS patients in an attempt to better elucidate its role in sinus disease | 90 CRSwNP: 31 CRSsNP: 32 DC: 27 | Australia | NA | NA | AS and AL | MMSS intra-operatively collected | SMT and MT (sequencing, OTUs) | Fungi were detected in 12.9% of patients (4/31) (Aspergillus, Fusarium, and without fungus identified). | NA |
| [104] | CSS | To study the association between inflammatory cells and signaling markers of CRS endotypes and the sinonasal bacterial community patterns | 110 CRSwNP: 46 CRSsNP: 46 CRS/CF: 7 DC: 17 | New Zealand (Most of the patients were European (31/38)) | 18–84 CRSwNP: 18–71 (48.0) | M: 55 (25) F: 55 (14) | AS: 48 (28) AsS: 14 (40) AERD: 11 (11) | Tissue biopsies collected from the bulla ethmoidal | MT (qPCR and sequencing, OTUs) | Patients were grouped in subject clusters mainly associated with Staphylococcus, Corynebacterium, Streptococcus, and Propionibacterium. | Most of the patients (35/39, 89.7%) had not taken pre-operative antibiotics and corticosteroids (<3 months) |
| [105] | RCT | To describe the effect of oral antibiotics and corticosteroids on the bacterial microbiome within the paranasal mucus in CRS patients | 26 CRSwNP: 13 CRSsNP: 13 | New Zealand | 22–67 (48.9) CRSwNP: 29–64 (49.5) | M: 13 (7) F: 13 (6) | NA | MMSS | MT (qPCR and sequencing, OTUs) | Bacterial communities were typically dominated by Corynebacterium and Staphylococcus and at lower abundance by Streptococcus, Dolosigranulum, Haemophilus, and Moraxella. | No antibiotics and systemic corticosteroids in the previous month |
| [106] | CCS | To detect bacteria in culture-negative cases of CRS using 16S rRNA gene PCR and sequencing | 20 CRSwNP: 15 CRSsNP: 5 | India | NA | NA | NA | Discharge from the sinus and mucosal biopsies from the MM region collected during FESS | SMT and MT (qPCR and sequencing) | Two patients with a history of a previous sinus surgery yielded Staphylococcus spp. by qPCR (2/15, 13.3%). MRSA was isolated from one of them. | Cases with a history of a previous sinus surgery also had history of prior treatment with corticosteroids |
| [38] | CCS | To compare the microbiological features in middle meatus samples from CRSwNP and CRSsNP patients and DCs | 251 CRSwNP: 136 CRSsNP: 66 DC: 49 | China | CRSwNP: (45.4) | M: 154 (89) F: 96 (47) | AS: 41 (37) AR: 85 (61) | MMSS pre-ESS | SMT | Most patients (120/136, 88.4%) showed a positive culture result. The most abundant bacteria were CoNS, Corynebacterium, and S. epidermidis. The isolate rate of fungi was very low (3.7%). The strains were mainly Gram-positive aerobic and facultative anaerobic bacteria (69.8%). Patients with asthma showed a significantly lower isolation rate of Corynebacterium and P. aeruginosa. Patients with a history of ESS exhibited a significantly lower isolation rate of CoNS, and a significantly higher isolation rate of P. aeruginosa. A relatively high proportion of Citrobacter was observed compared with DCs. The isolation rate of S. aureus in the subgroup of patients with an increased percentage of eosinophils (>5%) in peripheral blood was higher than that in the subgroup with a standard percentage of eosinophils. | No antibiotics and glucocorticoids at least one month before surgery |
| [72] | CS | To investigate the relevance of the bitter-taste receptor TAS2R38 genetic variants in the susceptibility to bacterial infections associated with in vivo biofilm formation in CRSwNP patients | 100 CRSwNP: 100 | Italy | CRSwNP: (53.0) | M: (68) F: (32) | NA | Sinonasal mucosa samples pre-FESS | SMT and confocal laser scanning microscopy assay | 63.0% of patients (63/100) showed positive culture result. The most common bacteria were S. epidermidis, S. aureus, and Enterobacteriaceae (Klebsiella spp., Citrobacter koseri, and Serratia marcescens). Only in one sample was found a mixed microbiota composed of Candida albicans with S. aureus. 19 of 43 samples (44.2%) were biofilm-positive. Biofilms were associated with Klebsiella, Citrobacter, Haemophilus, Kocuria, S. aureus, and S. epidermidis. | No antibiotics or corticosteroids in the previous month |
| [85] | CSS | To evaluate the bacterial community composition on the distinct types of CRS compared to healthy bacterial communities | 18 CRSwNP: 5 CRSsNP: 5 CRS with unilateral purulent maxillary: 5 DC: 3 | Germany | 13–>70 CRSwNP: 13–29 | M: 8 F: 10 | NA | MMSS and tissue samples collected during ESS | SMT and MT (sequencing, OTUs) | Enterobacteria, Staphylococci, coryneform bacteria, Propionibacteria, Viridans streptococci, and Haemophilus were identified. The most common bacteria were S. epidermidis, P. acnes, and Corynebacterium spp. No significant differences were found in the microbiome between patients and DCs. | All patients had eosinophilia (>5%). No antibiotics in the previous month |
| [24] | CSS | To compare the bacterial communities of HCs with CRSwNP patients with (CRSwNP+A)/without (CRSwNP-A) comorbid asthma | 58 CRSwNP: 41 (CRSwNP+A: 20 CRSwNP-A: 21) HC *: 17 | Belgium | CRSwNP+A: 45.8 CRSwNP-A: 47.5 | M: 31 (22) F: 27 (19) | AS: (20) AT: (20) AaS: (7) | MMSS and tissue samples collected during ESS | MT (sequencing, OTUs) | HCs and CRSwNP patients had about the same total bacterial load, but the bacterial diversity and evenness were significantly lower in the CRSwNP group, especially the CRSwNP-A group (both evenness and Shannon’s diversity), compared with HCs. The phylum Proteobacteria and genus Haemophilus (H. influenzae) were more abundant than in HCs. In contrast, Staphylococcus xylosus and Bifidobacterium longum were less prevalent and abundant than in the HCs. The most abundant species in CRSwNP-A was S. aureus, and E. coli was found in high amounts in CRSwNP+A. | No antibiotics or systematic corticosteroids in the previous three months * |
| [77] | CSS | To characterize the sinonasal fungal communities (mycobiota) in CRS patients and DCs via the fungal ITS2 marker amplicon sequencing | 144 CRSwNP: 49 CRSsNP: 50 CRS/CF: 7 DC: 38 | New Zealand (Most of the patients were European (111/144, 77.1%)) | 18–84 CRSwNP: 18–71 (50.0) | M: 72 (29) F: 72 (19) | AS: 58 (35) AsS: 18 (15) | MMSS collected during ESS | MT (sequencing, ZOTUs) | At the phylum level, Basidiomycota and Ascomycota showed the highest RA. The most abundant fungi were Malassezia, followed by Davidiella. | Most of the patients had not taken antibiotics (41/49, 83.7%) and corticosteroids (44/49, 89.8%) in the previous month |
| [107] | RCT | To investigate the safety and preliminary efficacy of Manuka honey (MH) with augmented methylglyoxal (MGO) rinses in recalcitrant CRS | 25 CRSwNP: 20 (10 with useful information) CRSsNP: 5 | New Zealand | 27–86 CRSwNP: 49–69 | M: 14 F: 11 | AS: 10 AR: 12 AsS: 2 | Sinonasal swab samples post-EES | SMT | The most common and abundant bacteria were S. aureus and Pseudomonas. | NA |
| [108] | CSS | To describe the sinus microbiota of acute exacerbations in CRSwNP, CRSsNP, and allergic fungal rhinosinusitis (AFRS) patients | 143 CRSwNP: 55 CRSsNP: 65 AFRS: 14 | USA | (52.7) CRSwNP: (53.2) | M: 75 (33) F: 59 (22) | AS: 56 (38) AL: 66 (33) AsS: 9 (7) | Aspiration samples of purulent secretions from within the MM or previously opened sinus | MT (sequencing, OTUs) | The most common bacteria were Staphylococcus spp. (S. aureus and S. epidermidis), Pseudomonas spp., Haemophilus spp. Enterobacter spp., and Corynebacterium spp. Staphylococcus spp., Pseudomonas spp., and Streptococcus spp. showed the most RA. An average of 3 taxa per specimen isolated showed a low level of diversity in acute exacerbation CRSwNP | Some patients (no data) had taken antibiotics and corticosteroids |
| [69] | CS | To demonstrate differential expression of trefoil family factor (TFF) protein genes in CRSwNP patients and the impact of bacterial colonization on their expression. | 54 CRSwNP: 29 DC: 25 | Croatia | 21–69 CRSwNP: 26–69 (53.4) | M: 30 (16) F: 24 (13) | AERD: (3) | Nasal and sinus swab samples collected during FESS | SMT | Most of the patients (23/29, 79.3%) showed positive culture results, of which 15 had isolated pathogenic bacteria and 8 nonpathogenic bacteria (S. epidermidis). * Pathogenic bacteria isolated: S. aureus, E. coli, Group B S. haemolyticus, Morganella morganii, Enterobacter spp., Serratia marcescens, P. mirabilis, Enterobacter freundii, and K. oxytoca. | Patients had taken corticosteroids in the previous three weeks and some cases had also taken antibiotics (no data) |
| [109] | CCS | To identify a microbiome profile in CRSwNP and CRSsNP patients | 20 CRSwNP: 10 CRSsNP: 10 | Indonesia | 18–>50 | M: 9 F: 11 | NA | Nasal tissue sample collected during FESS | MT (sequencing, OTUs) | The most common bacteria (phyla) were Proteobacteria, Firmicutes, Cyanobacteria, Fusobacteria, Actinobacteria, and Bacteoidetes. | NA |
| [57] | CS | To identify trends in bacteria isolated and their antibiotic resistance from Korean adults with CRS | 510 CRSwNP: 376 (ECRSwNP: 36 NECRSwNP: 22) CRSsNP: 134 | Korea | >18 | NA | AS: (33) AT: (51) | Purulent discharge in the maxillary and ethmoid sinuses samples collected during ESS | SMT | 73.9% of patients (278/376) showed positive culture results. The most common bacteria were CoNS (S. epidermidis), Streptococcus spp., Corynebacterium, Propionibacterium spp., Haemophilus spp., S. aureus, and Klebsiella spp. S. epidermidis, Corynebacterium spp., and Enterobacter spp. were significantly associated with ECRSwNP, and Haemophilus spp., Klebsiella spp., and P. aeruginosa with NECRSwNP. | No antibiotics or systemic corticosteroids for at least the previous month |
| [110] | CSS | To stratify CRS patients based on their microbial community compositions using a probabilistic modeling approach and the traditional phenotypic approach | 31 CRSwNP: 8 CRSsNP: 8 CRS/CF: 7 DC: 8 | New Zealand | >18 | NA | NA | Ethmoidal sinus biopsy specimens collected during FESS | MT (sequencing, OTUs) | Staphylococcus, Streptococcus, Propionibacterium, and Corynebacterium were prevalent in a majority but at low RA. In CRSwNP, Moraxella and Stenotrophomonas were dominant but showed less Streptococcus and Veillonella than DC. | No antibiotics or systemic corticosteroids in the previous month |
| [111] | CSS | To identify reactive allergens of IgE antibodies produced locally in NPs | 51 ECRSwNP: 46 (4 finally included) DC: 15 | Japan | ECRSwNP: 39–77 (53.3) | M: (2) F: (2) | AS: (3) | Swab and NP samples | SMT | Moraxella catarrhalis, Corynebacterium spp., and S. aureus were identified. | NA |
| [86] | CSS | To characterize the differences in microbiome profiles between CRSwNP patients and DCs | 86 CRSwNP: 59 DC: 27 | China | CRSwNP: (46.4) | M: 53 (35) F: 33 (24) | AS: 12 (11) AR: 16 (14) | MMSS collected during FESS | MT (sequencing, OTUs) | The predominant bacterial phyla were Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes, and Fusobacteria. The predominant bacterial genera were Lactobacillus, Corynebacterium, Staphylococcus, Streptococcus, Erysipelotrichales, Escherichia–Shigella, Haemophilus, Enterobacter, Propionibacterium, and Pseudomonas. CRSwNP had a lower nasal microbiome richness than DC. The RA of Actinobacteria (predominantly Corynebacterium), and Dolosigranulum was significantly lower in CRSwNP than in DC. Lactobacillus, Escherichia–Shigella, Turicibacter, Clostridium, Enterococcus, and Romboutsia were positively correlated with the severity of CRSwNP (Lund–Mackay CT score). Smoking status, asthma, or allergic rhinitis did not change the microbiome distribution. | No antibiotics and corticosteroids in the previous month |
| [112] | CSS | To compare bacterial community composition and absolute abundances of S. aureus and S. epidermidis between CRS patients and DCs | 54 CRSwNP: 18 CRSsNP: 22 DC: 14 | New Zealand (Most of the patients were European (44/54, 81.5%)) | CRSwNP: (53.2) | M: 31 F: 23 | AS: 14 (10) | MMSS | MT (qPCR and sequencing, OTUs) | The most common bacteria were Corynebacterium, Haemophilus, Staphylococcus, and Dolosigranulum. CRSwNP had a significantly higher overall bacterial load than DCs. | Most of the patients had taken antibiotics and corticosteroids in the previous year |
| [58] | CSS | To investigate the expression of lipopolysaccharide (LPS) and its relationship with glucocorticoid receptors (GRs) in CRSwNP | 162 CRSwNP: 112 (ECRSwNP: 49 NECRSwNP: 63) | China | CRSwNP: 13–71 (ECRSwNP: 19–65 (46.0) NECRSwNP: 13–71 (44.0)) | M: 91 (65, ECRSwNP: 28, NECRSwNP: 37) F: 71 (47, ECRSwNP: 21, NECRSwNP: 26) | AS: (8, ECRSwNP: 5, NECRSwNP: 3) AT: (5, ECRSwNP: 4, NECRSwNP: 1) | Swab samples and specimens pre-ESS | SMT | 82.1% of patients (82.1%) showed positive culture. The positive rate of bacterial culture of different groups was not different. The main bacterial strains were S. epidermidis, CoNS, E. coli, S. pneumoniae, and K. pneumoniae. | No corticosteroids in the previous two weeks |
| [113] | CS | To verify if topical administration of Lactobacillus lactis W136 to the nasal and sinus cavities would be safe for CRS patients refractory to medical and surgical treatment | 27 (24 completed the study) CRSwNP: 17 CRSsNP: 7 | USA | (54.9) | M: 11 F: 13 | AS: 18 AL: 5 | Nasal swab and brushing samples pre-ESS and post-ESS | SMT and MT (sequencing, OTUs) | Conventional culture: Oropharyngeal flora, CoNS, S. aureus, and P. aeruginosa were the most common bacteria. | No antibiotics and corticosteroids in the previous month |
| [87] | CSS | To investigate the prevalence, diversity, and abundance of archaea in the human sinuses and any associations with disease state | 60 CRSwNP: 16 CRSsNP: 15 DC: 9 HC: 20 | New Zealand (Most of the subjects were European (44/60, 73.3%)) | CRSwNP: (52.8) | M: 37 (14) F: 23 (2) | AS: 19 (6) | MMSS collected during ESS | MT (sequencing, ASVs; and digital PCR) | Phyla Euryarchaeota and Thaumarchaeota were detected. The most abundant bacteria were Corynebacterium, Staphylococcus, Moraxella, Lawsonella, and Haemophilus. CRSwNP subjects showed significantly decreased alpha diversity than HCs. | Most of the patients (13/16, 81.3%) had not taken antibiotics in the previous month |
| [114] | CSS | To establish associations among medication usage, the sinus microbiota, and patients’ clinical outcomes | 236 CRSwNP: 79 CRSsNP: 77 DC: 45 HC: 35 | New Zealand (Most of the subjects were European (191/236, 80.9%)) | 18–82 CRSwNP: 18–75 (46.0) | M: 100 (26) F: 136 (53) | AS: 78 (72) | MMSS and tissue samples intra-operatively collected | MT (sequencing, ASVs) | The most common bacteria were Corynebacterium and Staphylococcus The number of observed ASVs was significantly lower when compared to HCs. | Most of the patients (67/78, 85.9%) had not taken antibiotics in the previous month. |
| [115] | CSS | To analyze the bacterial flora of the nose and paranasal sinuses in CRS patients who underwent ESS over 65 years of age compared to a younger group of patients (<40 years) | 529 (269 completed the study) CRSwNP: 150 CRSsNP: 119 | Poland | >18 | M: 147 F: 122 | AS, AL, and AERD | MMSS collected during ESS | SMT | The most common bacteria was S. aureus. There were no statistically significant differences between the occurrence of a particular type of bacteria and the presence of NP in both age groups. | No antibiotics and systemic corticosteroids in the previous month |
| [59] | CSS | To investigate whether the sinus microbiota in CRSwNP is associated with eosinophilic inflammation | 37 CRSwNP: 31 (ECRSwNP: 21 NECRSwNP: 10) DC: 6 | Republic of Korea | ECRSwNP: (50.6) NECRSwNP: (37.3) | NA | AT: (13, ECRSwNP: 10, NECRSwNP: 3) AS: (8, ECRSwNP: 8) * Patients with AERD were excluded | MMSS | MT (sequencing, OTUs) | The most common bacteria were Firmicutes (mainly Staphylococci), Actinobacteria (mainly Corynebacterium, Bifidobacterium, and Propionibacterium species), and Proteobacteria (mainly Moraxella, Pseudomonas, Enterobacter and Aggregatibacter). ECRSwNP: RA of Anaerococcus, Tepidimonas, and Mesorhizobium were significantly decreased and Lachnoclostridium increased compared to those in DCs. ECRSwNP patients had higher asthma incidence and clinical severity scores. NECRSwNP: RA of Lachnospiraceae was increased compared with that in DCs. Deinococcus, Sphingomonas, and Lactobacillus were positively correlated with serum extracellular vesicles (EVs). | No antibiotics and systemic corticosteroids for at least the previous month |
| [55] | CSS | To characterize the normal microbiome, assess for any geographical or clinical influences, and identify any changes associated with CRS within and across geographical sites | 532 (410 reached the final stage of analysis) CRSwNP: 172 CRSsNP: 99 DC: 139 | Australia New Zealand Thailand India Brazil Chile The Netherlands Canada USA | 20–75 | NA | AS and AsS | MMSS collected during ESS | MT (sequencing, ASVs) | The most abundant bacteria were Corynebacterium, Staphylococcus, Streptococcus, Haemophilus, and Moraxella. Corynebacterium was significantly reduced and Streptococcus increased compared to DCs. | Clinicians were free to treat patients |
| [98] | CSS | To compare the bacterial flora in CRSwNP and CRSsNP patients and investigate a possible link between the type of bacterial flora and the coexistence | 470 (458 completed the study) CRSwNP: 245 CRSNsNP: 213 | Poland | M: (50.6) W: (49.8) | M: 248 F: 222 | AS: 104 (83) AL: 87 (52) AsS and other non-steroidal anti-inflammatory drugs: 53 (44) | MMSS collected during ESS | SMT | Gram-negative intestinal bacilli Enterobacteriaceae, CoNS and streptococci, and S. aureus were the most common bacteria. No statistically significant relationship was found between bacterial flora and the presence of asthma, hypersensitivity to drugs, or allergy. No statistical significance between the occurrence of a particular flora and the multiplicity of operations. | No antibiotics in the previous month |
| [88] | CSS | To characterize the microbiome dysbiosis in AERD patients | 37 AERD: 17 HC: 17 | USA | NA | NA | AERD: (17) | Inferior turbinate swab samples | MT (sequencing, OTUs) | AERD subjects showed reduced bacterial diversity (fewer species per sample and lower Shannon Diversity indexes). Moraxella, Corynebacterium, Pseudomonas, Staphylococcus, Sphingomonas, Streptococcus, Propionibacterium, and Eikenella showed the highest RA. Overabundance of Eikenella corrodens, M. catarrhalis, and Pseudomonas (Proteobacteria phylum) and underabundance of Corynebacterium in AERD patients compared to HCs. | No recent use of antibiotics and corticosteroids |
| [89] | CSS | To analyze the effects of antibiotics on the nasal microbiome and secreted proteome in CRS patients | 99 CRSwNP: 40 CRSsNP: 30 DC: 29 | Republic of Korea | CRSwNP: (48.8) | M: 67 (27) W 32 (13) | AS: 4 (2) AT 31 (13) AR: 33 (8) | Nasal secretion samples from MM | MT (sequencing, OTUs) | Corynebacterium (Actinobacteria), Staphylococcaceae (Firmicutes), Streptococcaceae (Firmicutes), Burkholderiaceae (Proteobacteria), Lachnospiraceae (Firmicutes), Veillonellaceae (Firmicutes), Propionibacteriaceae (Actinobacteria), and Moraxellacea (Proteobacteria) showed the highest level of RA. Shannon and Simpson indexes were significantly decreased across CRSwNP to DCs. The sinonasal microbiota of the CRSwNP showed significantly decreased bacterial diversity. Firmicutes and Bacteroidetes (Prevotellaceae) were significantly decreased compared to DCs. Cyanobacteria, Staphylococcaceae, Propionibacteriaceae, and Moraxellaceae were significantly increased compared to DCs. In the NABX (subjects who had not taken antibiotics three months before sampling) group, the Shannon and Simpson indexes were significantly decreased compared to DCs. Shannon and Simpson indexes were significantly lower in the ABX group than in the NABX group. Streptococcaceae, Lachnospiraceae, and Neisseriaceae were significantly decreased in the ABX group compared to the levels in the NABX group. | Some patients (24/40, 60%) had not taken antibiotics in the previous three months |
| [60] | CSS | To evaluate the bacterial community composition on distinct types of CRSwNP patients (ECRSwNP and NECRSwNP) | 73 CRSwNP: 34 (ECRSwNP: 16 NECRSwNP: 18) HC *: 39 | China | ECRsWNP: (48.3) NECRSwNP: (28.5) | M: 36 (18, ECRSwNP: 11, NECRSwNP: 7) F: 37 (16, ECRSwNP: 5, NECRSwNP: 11) | AS: (2, ECRSwNP: 2) * Patients with AERD were excluded | MMSS collected during FESS | MT (sequencing, OTUs) | The most common bacteria were Firmicutes, Actinobacteria, Proteobacteria, and Bacteroidetes. The most abundant genera were Staphylococcus, Corynebacterium, and Dolosigranulum. The diversity of sinus microbiota (Chao1 and Shannon indexes) was significantly lower in the CRSwNP than in DC. ECRSwNP: Firmicutes, Actinobacteria, and Proteobacteria. Staphylococcus, Corynebacterium, and Moraxella. NECRSwNP: Firmicutes, Actinobacteria and Bacteroidetes. Staphylococcus, Dolosigranulum, and Corynebacterium. Staphylococcus was significantly lower in the ECRSwNP compared to HC. The Shannon index significantly decreased only in the NECRSwNP, but not in the ECRSwNP, compared to HCs. Staphylococcus (S. aureus) abundance was the lowest in the ECRSwNP. The abundance of S. aureus was the highest in the NECRSwNP. The abundance of Moraxella was significantly decreased in the NECRSwNP compared with that in the ECRSwNP. The abundance of Haemophilus was significantly increased in the NECRSwNP compared to HC. | No antibiotics or corticosteroids in the previous month |
| [64] | CS | To explore nasal microbial diversity effects on the pathogenesis and prognosis of CRSwNP | 147 CRSwNP: 77 (NP recurrent: 12, NP non-recurrent: 65) CRSsNP: 36 DC: 34 | China | CRSwNP: (46.4) (NP recurrent: (48.6), NP non-recurrent: (49.7)) | M: 86 (43, NP recurrent: 5, NP non-recurrent: 36) F: 61 (34, NP recurrent: 7, NP non-recurrent: 29) | AS: 14 (11, NP recurrent: 3, NP non-recurrent: 8) AR: 22 (14, NP recurrent: 4, NP non-recurrent: 10) | MMSS collected during ESS and MM secretions after 1-year post-ESS | MT (sequencing, OTUs) | The most abundant bacteria were Lactobacillus, Corynebacterium, Staphylococcus, Streptococcus, Escherichia–Shigella, Enterobacter, Haemophilus, Moraxella, and Propionibacterium. The RA of Actinobacteria (Corynebacterium), Chlamydia, and Dolosigranulum was significantly lower than that in DCs. Lactobacillus, Escherichia–Shigella, Turicibacter, Clostridium, Enterococcus, and Romboutsia were positively correlated with the severity of CRSwNP (Lund–Mackay CT score). Smoking status, asthma, or allergic rhinitis did not change the microbiome distribution. Faecalibaculum had a significant negative correlation with the TNSS of patients with CRSwNP. The abundance of Actinobacteria after surgery was significantly higher than before in the NP non-recurrent group, while there was no significant change in nasal flora in the NP recurrent group. | No antibiotics or corticosteroids in the previous month |
| [94] | CSS | To assess the microbial composition in CRS patients, comparing different sampling methods and disease subtypes | 22 CRSwNP: 8 CRSsNP: 6 DC: 8 | Republic of Korea | 21–76 CRSwNP: (48) | M: 12 (3) F: 10 (5) | AS: 2 (2) AT: 6 (1) | MMSS, tissue biopsied from the uncinate process (UT) and NP tissue collected during ESS | MT (sequencing, 16S rRNA) | Bacteroidetes, Firmicutes, Proteobacteria, Actinobacteria, and Fusobacteria were the dominant phyla. The Shannon index was significantly decreased in NP compared to UT. Firmicutes was remarkably reduced in NP, whereas Proteobacteria was more abundant in NP than in UT. Sphingomonas and Sediminibacterium were enriched in the NP, while Ralstonia was reduced in NP. Prevotella was significantly and inversely correlated with disease severity. | No antibiotics and systemic corticosteroids in the previous month |
| [61] | CSS | To investigate the changes in the clinical, histopathological, and hematological properties and the gut and airway microbiota in CRSwNP endotypes | 58 CRSwNP: 46 (ECRSwNP: 32 NECRSwNP: 14) HC: 12 | Sudan | CRSwNP: (34.7) (ECRSwNP: 34.8 NECRSwNP: (34.7)) | M: 26 (20, ECRSwNP: 13, NECRSwNP: 7) F: 32 (26, ECRSwNP: 19, NECRSwNP: 7) | AS: (4, ECRSwNP:4) AL: (1, ECRSwNP: 1) | MMSS (17) and fecal samples (10) | MT (sequencing, OTUs) | In the airway: Reduced alpha diversity in comparison to HCs. Moraxella, Parvimonas, and Porphyromonas increased more in the ECRSwNP than in the NECRSwNP. These bacteria were positively correlated with CT scores and severe disease. Prevotella 9, Succinivibrio, Lawsonella, and Exiguobacterium significantly decreased in ECRSwNP. These bacteria were negatively associated with CT scores and endoscopic score eosinophil percentage. In the gut microbiome: Actinobacteria phylum and its major genus (Bifidobacteria) were remarkably reduced in CRSwNP, mainly in ECRSwNP. Bifidobacterium and Barnesiell were negatively associated with CT score and endoscopic score. The abundance of Enterobacterales; Enterobacteriaceae; and several genera, such as Enterobacter, increased in NECRSwNP. | No antibiotics or systemic corticosteroids in the previous month |
| [78] | CS | To undertake a comprehensive multi-omics assessment of NP tissue transcriptome, proteome, and associated bacterial and fungal microbiome in CRSwNP patients | 3 CRSwNP: 3 | New Zealand (all patients were of European ancestry) | CRSwNP: 46–59 | M: (3) F: (0) | AS: (1) | NP tissue | MT (sequencing, ZOTUs) | The most abundant bacterial genera were Staphylococcus, Corynebacterium, Dolosigranulum, Anaerococcus, and Propionibacterium. The most abundant fungal genera were Malassezia; Candida; Rhodotorula; and unclassified members of Malasseziales, Dothideomycetes, Mycosphaerellaceae, and Phaeophaeriacea. | No antibiotics and corticosteroids in the previous month |
| [65] | CS | To explore the effects of nasal microbial diversity and inflammatory types on the prognosis of NPs | 77 and DC CRSwNP: 77 (NP recurrent: 12, NP non-recurrent: 62) | China | CRSwNP: (NP recurrent (48.6), NP non-recurrent (49.7)) | M: (34, NP recurrent: 5, NP non-recurrent: 29) F: (43, NP recurrent: 7, NP non-recurrent: 36) | AS: (11, NP recurrent: 3, NP non-recurrent: 8) AR: (14, NP recurrent: 4 and NP non-recurrent: 10) | MMSS and NP tissue and MM secretions after 1-year post-ESS | MT (sequencing, OTUs) | The most abundant bacteria were Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes, Fusobacteria, and Spirochaetae. Actinobacteria, Corynebacterium, and Dolosigranulum were significantly lower than in DCs. There was no significant difference in nasal microbiome richness between NP recurrent and non-recurrent groups. At the genus level, the dominant bacteria were Lactobacillus, Staphylococcus, Streptococcus, and Bacteroides. Faecalibaculum was negatively correlated with the overall nasal symptoms. The RA of Actinomycetes and Corynebacterium was significantly higher, and Staphylococci was significantly lower, in the NP non-recurrent group than in the NP recurrent group. | No antibiotics and corticosteroids in the previous month |
| [116] | CSS | To identify and characterize prophages present in S. aureus from patients suffering from CRS, concerning CRS disease phenotype and severity | 67 CRSwNP: 30 CRSsNP: 28 DC: 9 | Australia | NA | NA | NA | Samples collected during ESS | SMT * | S. aureus clinical isolates were obtained (in silico: intact prophages encoding human immune evasion cluster genes and more frequent in patients with more severe disease). | NA |
| [79] | CS | To analyze the alteration in the sinonasal microbiome in CRSwNP and CRSsNP patients after FESS | 35 CRSwNP: 20 CRSsNP: 15 | India | 12–76 (40.0) | M: 19 F: 16 | NA | MMSS pre-FESS and post-FESS | SMT | Pre-FESS cultures: MRSA predominantly, followed by S. aureus, Pseudomonas, E. coli, and Aspergillus. Post-FESS culture: S. aureus and E. coli. | Postoperatively patients were prescribed antibiotics |
| [117] | CSS | To investigate the potential role of Pantoea dispersa in rhinosinusitis | 390 (274 completed the study) | Taiwan | 20–99 (53.6) | M: 156 F: 118 | AS: 8 AR: 75 | Nasal swab samples | SMT | Seven CRSwNP patients had culture growth of P. dispersa. | NA |
| [40] | CSS | To examine the bacterial communities of the sphenoidal sinus in Iranian patients with and without CRS | 36 CRSwNP: 18 DC: 18 | Iran | CRSwNP: 30–63 (42.7) | M: 18 (9) F: 18 (8) | AS: 6 | Sphenoidal sinus surface mucosa swab samples collected during FESS | MT (qPCR) | The most common bacteria were Actinobacteria (Corynebacterium) and Staphylococcus spp. S. pneumoniae and H. influenza were not detected in any of the samples. S. haemolyticus and P. aeruginosa were significantly more prevalent than DCs. | No antibiotics in the previous month |
| [92] | CS | To determine whether altered nasal microbiota constituents could be used as biomarkers to predict CRSwNP recurrence | 85 (60 with complete clinical information for the establishment of a predictive model of CRSwNP recurrence) CRSwNP: 85 (NP recurrent: 39, NP non-recurrent: 46) | China | CRSwNP: 18–73 (46.4) (NP recurrent: (46.2), NP non-recurrent (46.5) | M: (64, recurrent: 28, non-recurrent: 36) F: (21, NP recurrent: 11, NP non-recurrent: 10) | AS: (17, NP recurrent: 15, NP non-recurrent: 2) | MMSS collected during ESS * | MT (sequencing, OTUs) | There was no significant difference in community diversity (OTUs, Shannon diversity index, and Chao richness), but both groups (recurrence and non-recurrence) showed distinct composition. Genera from the Proteobacteria and Firmicutes phyla were the major taxa that differed in abundance between both groups. Campylobacter, Bdellovibrio, and Aggregatibacter were more abundant than in the recurrence group. Actinobacillus, Gemella, and Moraxella were more abundant in non-recurrence. Shewanella and Preptostreptococcus exhibited a decrease in abundance, and Friedmanniella, Curvibacter, and Pseudoxanthomonas were more abundant in recurrence than non-recurrence. Porphyromonas, Bacteroides, Moryella, Aggregatibacter, Butyrivibrio, Shewanella, Pseudoxanthomonas, Friedmanniella, Limnobacter, and Curvibacter were the most important taxa discriminating recurrence from non-recurrence specimens. | No antibiotics or corticosteroids in the previous month |
| [118] | CSS | To explore the difference between sinus bacteriology in CRSwNP and CRSsNP patients and to analyze the differences in culture results from swabs taken from the MM versus the ethmoid sinus | 448 CRSwNP: 160 CRSsNP: 288 | Jordan | (40.0) CRSwNP: (39.7) | M: (96) F: (64) | NA | Ethmoid sinus and MMSS collected during FESS | SMT | The most common bacteria were MRSA, followed by CoNS and S. aureus. | No antibiotics or corticosteroids before surgery |
| [119] | CSS | To investigate the changes in microbiota and cytokines levels in CRSwNP and CRSsNP patients. | 36 CRSwNP: 12 CRSsNP: 10 DC: 15 | China | CRSwNP: (47.3) | M: 24 (9) F: (13/3) | NA | Secretions collected from the middle nasal canal, maxillary sinus, and ethmoid sinus intra-operatively collected | MT (sequencing, OTUs) | The most common bacteria were Staphylococcus, Corynebacterium, Porphyromonas, Serratia, Pseudomonas, Fusobacterium, Carnobacterium, Dolosigranulim, Cultibacterium (formerly Propionibacterium acnes), and Lawsonella. Beta diversity was significantly different between CRSwNP and DC. The abundance of C. propinquum and Carnobacterium maltaromaticum in CRSwNP differed from that in DC. Lawsonella, Moraxella, Corynebacterium, Carnobacterium, and Hafnia–Obesumbacterium were different at the genus level. | No corticosteroids in the previous month |
| [82] | CSS | To examine evidence of microbial exposure in subjects by probing serum samples of CRS patients and controls for seroreactivity to microbial protein-directed IgG and IgA | 118 CRS: 39 DC: 79 | USA | NA | NA | NA | Serum samples | MT (CRS-focused Nucleic Acid Programmable Protein Array (NAPPA)) | CRSwNP patients showed elevated sero-reactivity against S. aureus. Influenza A virus (H1N1 and H3N2) and rhinovirus B14 were identified. | No antibiotics and corticosteroids in the previous month |
| [91] | CSS | To demonstrate the role of bacteria in the pathogenesis of fungal ball (FB) versus CRSwNP and investigate the differences in microbiome profiles through a comparative analysis of microbial community diversity | 42 CRSwNP: 10 FB: 49 DC: 4 | China | CRSwNP: (41.3) | M: 15 (7) F: 28 (3)) | NA | MM and superior meatus swab samples | MT (sequencing, OTUs) | The major abundant phyla were Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria. The abundance of TM7 (Saccharimonadia), Chloroflexi, and Bacteroidete significantly differed between the CRSwNP and DC groups. The RA of Ruminococcacea from the phylum of clostridia and Comamonadaceae from the phylum of Burkholderiales was significantly higher, while that of Lactobacillus, Bacteroides S24-7, and Desulfovibrio was significantly lower than DC. The RA of Haemophilus was increased in CRSwNP. | No antibiotics in the previous month |
| [62] | CSS | To compare the nasal bacteriology between ECRSwNP and nECRSwNP patients | CRSwNP: 295 | Taiwan | CRSwNP: 20–84 (46.1) | M: (205) F: (90) | AS: (14, ECRSwNP: 9, NECRSwNP: 5) AR: (124, ECRSwNP: 70, NECRSwNP: 54) AsS: (1, NECRSwNP: 1) | MMSS pre-FESS | SMT | The most common bacteria were S. aureus and CoNS. Culture rates were similar between ECRSwNP and nECRSwNP. Gram-negative aerobes (mainly H. influenzae and C. koseri) were significantly more isolated from the NECRSwNP than the ECRSwNP. | No antibiotics in the previous week |
| [84] | CSS | To evaluate the microbial composition in the context of the inflammatory environment in patients suffering from CRSwNP, AERD, and CRSsNP | 80 CRSwNP: 20 CRSsNP: 20 AERD: 20 DC: 20 | Austria | CRSwNP: (49.4) AERD: (46.8)) | M: 50 (CRSwNP: 16, AERD: 12) F: 30 (CRSwNP: 4, AERD: 8) | AS: 42 (CRSwNP: 16, AERD: 20) AL: 32 (CRSwNP: 8, AERD: 8) | Anterior naris swab samples and MMSS | MT (sequencing, ASV) | Corynebacteria and staphylococci showed the highest RA in both CRSwNP and AERD. No alpha and beta diversity difference was observed between the DC, the CRSwNP, and the AERD groups. Dolosigranulum was less prevalent, and Lawsonella was more prevalent in patients with NP than in DCs. A higher RA of staphylococci in the MM in AERD patients compared to CRSwNP was observed, as well as of Lawsonella in patients suffering from CRSwNP in MM and anterior naris compared to DC. | No corticosteroids in the previous two weeks |
| [63] | CSS | To characterize nasal dysbiosis in a cohort of ECRSwNP patients and compare their nasal microbiomes with those of HCs | 88 ECRSwNP: 34 Patients without CRSwNP: 10 HC: 44 | China | 18–79 ECRSwNP: 18–67 (43.8) | M: 69 (30) F: 19 (4) | AS: 7 AT: 7 | MM brush samples | MT (sequencing, ASVs, and OTUs) | ECRSwNP had higher bacterial alpha diversity (Shannon and Chao1 indexes, intra-individual bacterial diversity). The most dominant phyla were Actinobacteria and Firmicutes. ECRSwNP was defined by increased RA of Sphingomonas, Moraxellaceae, Bacteroides, Bifidobacterium, Ruminococcus, and Faecalibacterium, as well as by decreased abundances of Ralstonia, Propionibacterium, and Propionibacter. Sphingomonas was more abundant in ECRSwNP than in HCs. Parabacteroides, Akkermansia, Devosia, Sutterella, and Desulfovibrio positively correlated with the SNOT-20 score. The abundances of Dyella, Gordonia, and Moraxella were positively correlated with LM (Lund–Mackay) CT scores, whereas the abundances of Gemmiger, Faecalibacterium, Anaeroplasma, Sutterella, Blautia, Geobacillus, Bifidobacterium, Sphingomonas, Dorea, Roseburia, and Ruminococcus negatively correlated with LM CT scores. | No antibiotics and corticosteroids in the previous week |
| [83] | RCT | To assess the longitudinal effect of corticosteroid therapy on sinus microbiota in CRSwNP patients | 44 CRSwNP: 29 DC: 15 | Canada | NA | M: 25 (14) W 18 (14) | AS: 18 (18) AsS: 11 (11) | MMSS | SMT and MT (microbiotyping using MALDI-TOF-MS) | The most prevalent organisms were Staphylococcus and Corynebacterium. The difference in the number of isolated organisms and the difference between Gram-positive and Gram-negative isolates were not statistically significant between HCs and CRSwNP patients. | No antibiotics and/or corticosteroids for at least the previous month |
| [120] | CSS | To evaluate the nasal microbiome, NP inflammation mediators, and inflammatory cell infiltration in CRSwNP patients | 77 CRSwNP: 77 | China | CRSwNP: 46.4 | M: (34) F: (43) | AS: (11) AL: (14) | MMSS collected during ESS | MT (sequencing, OTUs) | The most common genus was Enterobacter. | No antibiotics and corticosteroids for at least the previous month |
| [75] | CSS | To investigate the fungal and bacterial microbiome of sinus mucosa in CRSwNP and CRSsNP patients versus HC | 92 CRSwNP: 31 CRSsNP: 31 HC: 30 | USA | (50.0) | M: 47 F: 45 | AS: 32 (20) | Ethmoid tissue and skull base collected during ESS | MT (sequencing, ASVs) | Two patients (2/31, 6.5%) had positive fungal cultures. The mycobiome composition was not significantly different between HC and CRSwNP. Saccharomycetales and Cutaneotrichosporon were lower among CRSwNP, and Alternaria species were higher among CRSwNP. Beta diversity at the ASV and the genus level differed significantly between CRSwNP and HC. CRSwNP had a significantly greater abundance of Alternaria and Ramularia and a significantly lower abundance of Cutaneotrichosporon than HC. CRSwNP had significantly lower alpha diversity compared with HC. | NA |
3.2.2. Relationship between Microbial Dysbiosis and Inflammation in CRSwNP
| Ref. | Sample Type | Methodology | Results/Conclusion |
|---|---|---|---|
| [73] | MMSS + blood samples | SMT + NA | More Gram-negative aerobic and facultative anaerobic bacteria in the normal subgroup than in the subgroup with increased blood eosinophils. |
| [100] | MMSS + tissue samples from the uncinated processor posterior part of the inferior nasal turbinate | SMT + histological analysis (hematoxylin and eosin staining) | Differences in the distribution of neutrophils and macrophages in nasal mucosa were significant between smokers with pathogenic bacteria and non-smokers without pathogenic bacteria. |
| [97] | MMSS + blood samples | SMT + standard hospital protocol to determine serum biomarkers | CRSwNP patients colonized with Gram-negative bacteria have a similar inflammation pattern to those colonized with only Gram-positive bacteria. Higher serum IgE levels were associated with non-Pseudomonas Gram-negative bacteria. |
| [121] (Ex vivo) | NP tissues | ELISA and nasal polyp explant tissue stimulation model | Aspergillus niger stimulation increased pro-inflammatory cytokines tumor necrosis alpha (TNF-α), granulocyte-macrophage colony-stimulating factor (GM-CSF), and interleukin- (IL-6), while having little effect on the remodeling cytokines basic fibroblast growth factor (bFGF) and transforming growth factor beta 1 (TGF-b1). Cladosporium sphaerospermum, Alternaria alternata, and Penicillium notatum stimulation reduced pro-inflammatory cytokines TNF-α and IL-6 but induced a dose-dependent increase in remodeling cytokines bFGF and TGF-b1. |
| [102] (Ex vivo) | Nasal lavage and sino-nasal mucus samples | MT + transfected human embryonic kidney (HEK293) cells | Two patients only showed toll-like receptor 4 (TLR4) activation correlated positively with TNF and with a more elevated bacterial abundance but lower bacterial diversity than non-activated samples. The microbiota of samples with TLR activity was mainly composed of Gram-negative bacteria (Gammaproteobacteria, including the genera Haemophilus or Moraxella). Samples without TLR activity showed microorganisms commonly implicated in CRSwNP (Staphylococcus, Corynebacterium, or Moraxella). |
| [103] | Sinus brushing samples | MT + qRT-PCR | Most of the CRSwNP patients cluster into a subgroup typically dominated by Corynebacterium. Most of the CRSwNP patients cluster into a subgroup associated with peroxisome proliferator-activated receptor gamma (PPAR-y) and retinoic acid-inducible gene I (RIG-I) signaling pathways and a significant increase in IL-5. |
| [104] | Tissue biopsies collected from the bulla ethmoidalis | MT + Cytometric Bead Array (CBA) and transfected human embryonic kidney (HEK293) cells | CRSwNP patients were grouped in subject clusters associated with Staphylococcus, Corynebacterium, Streptococcus, and Propionibacterium. IL-8, cluster of differentiation (CD68)-positive macrophages, eosinophils, neutrophils, plasma cells, and IL-5 were significantly elevated in CRSwNP patients. |
| [72] | Sinonasal mucosa samples + saliva or venous blood | SMT and confocal laser scanning microscopy assay + qRT-PCR and genotyping | Bacterial biofilms were more frequently found in samples from subjects with nonfunctional taste receptor 2 member 38 (TAS2R38). |
| [24] | MMSS + nasal tissue samples | MT + UniCAP systems | IgE and IL-5 were negatively correlated with Geobacter anodireducens/sulfurreducens and Pelomonas puraquae. IL-5 was negatively correlated with Corynebacterium macginleyi and eosinophil cationic protein (ECP) with Corynebacterium accolens, Corynebacterium macginleyi, and S. pneumoniae. ECP and IL-5 were positively correlated with E. coli (CRSwNP+AS patients). Specific IgE to staphylococcal enterotoxins (SE-IgE) was significantly more frequent in CRSwNP with asthma and correlated with disease severity. |
| [69] | Nasal and sinus swab samples + tissue samples | SMT + qPCR and immunochemistry | Patients with normal flora (S. epidermidis) showed a significant seven-fold upregulation of the trefoil factor 3 (TFF3) gene in middle nasal turbinate and nasal samples compared to the same tissue specimen from patients with sterile swabs. |
| [110] | Ethmoidal sinus biopsy specimens | MT + histological analysis | CRSwNP were dominated by Moraxella and Stenotrophomonas but had less Streptococcus and Veillonella than DCs. T and B lymphocytes and macrophages were significantly elevated in the CRSwNP cohort compared to DCs. |
| [58] | Swab samples and specimens pre-ESS + blood samples | SMT + limulus amoebocyte lysate (LAL) assay, immunohistochemistry, and immunofluorescence | An abundant lipopolysaccharide (LPS) was found in the peripheral blood, especially for NECRSwNP patients. LPS levels were positively correlated with glucocorticoid receptor-beta (GR-β) expression in CRSwNP. Neutrophils and macrophages were the principal inflammatory cells containing LPS. |
| [59] | MMSS + NP tissues | MT + flow cytometry and ELISA | The RA of Lachnoclostridium showed a positive correlation with IL-5-producing innate lymphoid cells (ILC2s) and IL-5 levels. In contrast, the RA of Anaerococcus showed a negative correlation with IL-5-producing ILC2 and IL-5 levels in tissues. |
| [122] | Middle nasal passage swab samples + blood serum and nasal lavage fluid | SMT + enzyme immunoassay | The level of cytokines in the nasal lavage fluid (IL-8) and blood serum (IL-1β) was correlated with the total number of microorganisms and the concentration of Enterobacteriaceae and Staphylococci. |
| [64] | MMSS and MM secretions + NP tissues | MT + immunoassay | Enterobacter in patients with the IL-5-positive NP group was higher than that of the IL-5-negative NP group. |
| [78] | NP tissues | MT + transcriptome (RNA sequencing) and proteome analysis | Corynebacterium and Anaerococcus were negatively associated with tissue structural proteins (vimentin (VIM) and histoneHIST1H4A), actin beta (ACTB), and glyceraldehyde 3 phosphate dehydrogenase (GAPDH). Anaerococcus positively correlated with collagen proteins COL6A3 and COL1A2. Streptococcus positively correlated with immunoglobulin kappa constant (IGKC) and immunoglobulin heavy constant gamma 1 (IGHG1). |
| [65] | MMSS + NP tissues | MT + immunoassays and immunohistochemical staining | The RA of Actinomycetes and Corynebacterium was significantly higher, and Staphylococci was significantly lower in the NP non-recurrent group than in the NP recurrent group. The median expression levels of IFN-γ, IL-8, IL-17A, IL-17E, and IL-18 were significantly higher in the NP recurrent group than in the NP non-recurrent group. |
| [119] | Secretions collected from the middle nasal canal, maxillary sinus, and ethmoid sinus | MT + ELISA | The abundance of Cultibacterium acnes was positively related to IL-2 level, while Staphylococcus caprae negatively correlated with IFN-γ. |
| [84] | Anterior naris and MMSS + nasal-secretion samples | MT + electrochemiluminescence technology | Corynebacteria and Staphylococci were the most abundant taxa in all patient groups. Dolosigranulum was also less prevalent, and Lawsonella was more prevalent in patients with NP than in DCs. AERD patients showed an increased RA of Staphylococcus and a decreased RA of Corynebacterium compared to other patient groups. Elevated levels of type 2 response-associated cytokines IL-5, IL-9, and Eotaxin-3 in patients with NP. Eotaxin, chemokine CCL17, and IL-6 levels were significantly elevated in CRSwNP patients compared to DCs. Staphylococcus and IL-5 showed a moderately strong correlation, while Corynebacterium accolens and Eotaxin-3 were negatively correlated. |
| [63] | MM brush samples + NP | MT + histological analysis | Sphingomonas, Akkermansia, Blautia, Devosia, Desulfovibrio, Parabacteroides, and Bacteroides were positively correlated with absolute tissue eosinophil count. The abundances of Bifidobacterium, Blautia, Desulfovibrio, Gemmiger, and Bacteroides were positively correlated with the percentage of tissue eosinophils. |
| [120] | MMSS + NP tissue samples + blood samples | MT + immunoassays and hematoxylin-and-eosin staining, immunohistochemical staining, and flow cytometry | Enterobacter was significantly higher in the IL-5-positive NP group than in the IL-5-negative NP group. IL-17 and IL-27 were negatively correlated with Enterobacter and Anaerococcus. IL-8 was negatively correlated with Enterobacter and Staphylococcus. IL-18 was positively correlated with Candidatus–Arthromitus and Arthrofilaria and negatively correlated with Haemophilus. IL-27 was positively correlated with Faecalibaculum. Arthrofilaria was positively correlated, and Moraxella and Peptostreptococcus were negatively correlated with blood neutrophil counts. Lactobacillus and Enterobacter were positively correlated with the degree of neutrophil infiltration in NP tissue. Porphyromonas was negatively correlated with tissue neutrophil infiltration. |
3.2.3. Effect of the Modulation of the Microbiota on the CRSwNP Treatment
4. Discussion
4.1. Microbiota Composition in CRSwNP
4.2. Relationship between Microbial Dysbiosis and Inflammation in CRSwNP
4.3. Effect of the Modulation of the Microbiota on the CRSwNP Treatment
5. Conclusions
6. Future Directions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AERD | aspirin-exacerbated respiratory disease |
| ASV | amplicon sequence variant |
| bFGF | casic fibroblast growth factor |
| CMV | cytomegalovirus |
| CoNS | coagulase-negative staphylococci |
| COPD | chronic obstructive pulmonary disease |
| CRD | chronic respiratory disease |
| CRS | chronic rhinosinusitis |
| CRSsNP | chronic respiratory disease without nasal polyps |
| CRSwNP | chronic respiratory disease with nasal polyps |
| CT | computed tomography |
| EBV | Epstein–Barr virus |
| ECP | eosinophilic cationic protein |
| ECRSwNP | eosinophilic chronic respiratory disease with nasal polyps |
| ELISA | enzyme-linked immunosorbent assay |
| EPOS | European Position Paper on chronic rhinosinusitis |
| ESS | endoscopic sinus surgery |
| FESS | functional endoscopic sinus surgery |
| GM-CSF | granulocyte-macrophage colony-stimulating factor |
| GR | glucocorticoid receptor |
| GRADE | Grading of Recommendations, Assessment, Development, and Evaluations |
| HHV-6 | human herpesvirus 6 |
| HSV-1 | herpes simplex virus type 1 |
| IgE | immunoglobulin E |
| IGHG1 | immunoglobulin heavy constant gamma 1 |
| IGKC | ommunoglobulin kappa constant |
| IL | interleukin |
| ILC | innate lymphoid cells |
| ITS | internal transcribed spacer |
| LPS | lipopolysaccharide |
| MRSA | methicillin-resistant Staphylococcus aureus |
| NECRSwNP | non-eosinophilic chronic respiratory disease with nasal polyps |
| NOS | Newcastle–Ottawa Scale |
| NP | nasal polyp |
| NSAID-ERD | nonsteroidal anti-inflammatory drug-exacerbated respiratory disease |
| OTU | operational taxonomic unit |
| PCR | polymerase chain reaction |
| PICO | Population, Intervention, Control, and Outcome |
| PPAR | peroxisome proliferator-activated receptor |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| PROSPERO | International Prospective Register of Systematic Reviews |
| QOL | quality of life |
| RIG-I | retinoic acid-inducible gene I |
| RoB | risk of bias |
| RoB 2 | version 2 of the Cochrane risk-of-bias tool |
| rRNA | ribosomal ribonucleic acid |
| TAS2R38 | taste receptor 2 member 38 |
| TFF3 | trefoil family factor peptide 3 |
| Th2 | T-helper type 2 |
| TLR | Toll-like receptors |
| TNF | tumor necrosis factor |
References
- GBD 2019 Chronic Respiratory Diseases Collaborators. Global Burden of Chronic Respiratory Diseases and Risk Factors, 1990–2019: An Update from the Global Burden of Disease Study 2019. EClinicalMedicine 2023, 59, 101936. [Google Scholar] [CrossRef]
- Bachert, C.; Marple, B.; Schlosser, R.J.; Hopkins, C.; Schleimer, R.P.; Lambrecht, B.N.; Bröker, B.M.; Laidlaw, T.; Song, W.-J. Adult Chronic Rhinosinusitis. Nat. Rev. Dis. Prim. 2020, 6, 86. [Google Scholar] [CrossRef] [PubMed]
- de Loos, D.D.; Lourijsen, E.S.; Wildeman, M.A.M.; Freling, N.J.M.; Wolvers, M.D.J.; Reitsma, S.; Fokkens, W.J. Prevalence of Chronic Rhinosinusitis in the General Population Based on Sinus Radiology and Symptomatology. J. Allergy Clin. Immunol. 2019, 143, 1207–1214. [Google Scholar] [CrossRef] [PubMed]
- Bachert, C.; Zhang, N.; Holtappels, G.; De Lobel, L.; van Cauwenberge, P.; Liu, S.; Lin, P.; Bousquet, J.; Van Steen, K. Presence of IL-5 Protein and IgE Antibodies to Staphylococcal Enterotoxins in Nasal Polyps Is Associated with Comorbid Asthma. J. Allergy Clin. Immunol. 2010, 126, 962–968.e6. [Google Scholar] [CrossRef] [PubMed]
- Farina, D.; Ravanelli, M.; Borghesi, A.; Maroldi, R. Flying through Congested Airspaces: Imaging of Chronic Rhinosinusitis. Insights Imaging 2010, 1, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Ting, F.; Hopkins, C. Outcome Measures in Chronic Rhinosinusitis. Curr. Otorhinolaryngol. Rep. 2018, 6, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Abdulrashid, N.A.; Ali, O.I.; Elsharkawy, M.A. Effect of Photobiomodulation Therapy on Headache, and Fatigue in Patients with Chronic Rhinosinusitis: A Randomized Controlled Study. Lasers Med. Sci. 2024, 39, 62. [Google Scholar] [CrossRef] [PubMed]
- Wahid, N.W.; Smith, R.; Clark, A.; Salam, M.; Philpott, C.M. The Socioeconomic Cost of Chronic Rhinosinusitis Study. Rhinology 2020, 58, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Fokkens, W.J.; Lund, V.J.; Hopkins, C.; Hellings, P.W.; Kern, R.; Reitsma, S.; Toppila-Salmi, S.; Bernal-Sprekelsen, M.; Mullol, J.; Alobid, I.; et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2020. Rhinology 2020, 58, 1–464. [Google Scholar] [CrossRef]
- Grayson, J.W.; Cavada, M.; Harvey, R.J. Clinically Relevant Phenotypes in Chronic Rhinosinusitis. J. Otolaryngol. Head Neck Surg. 2019, 48, 23. [Google Scholar] [CrossRef]
- Cho, S.H.; Kim, D.W.; Gevaert, P. Chronic Rhinosinusitis without Nasal Polyps. J. Allergy Clin. Immunol. Pract. 2016, 4, 575. [Google Scholar] [CrossRef] [PubMed]
- Snidvongs, K.; Sangubol, M.; Poachanukoon, O. Pediatric Versus Adult Chronic Rhinosinusitis. Curr. Allergy Asthma Rep. 2020, 20, 29. [Google Scholar] [CrossRef]
- Bartosik, T.J.; Liu, D.T.; Campion, N.J.; Villazala-Merino, S.; Janik, S.; Dahm, V.; Mueller, C.A.; Vyskocil, E.; Stanek, V.; Quint, T.; et al. Differences in Men and Women Suffering from CRSwNP and AERD in Quality of Life. Eur. Arch. Oto-Rhino-Laryngol. 2021, 278, 1419–1427. [Google Scholar] [CrossRef] [PubMed]
- Ahern, S.; Cervin, A. Inflammation and Endotyping in Chronic Rhinosinusitis—A Paradigm Shift. Medicina 2019, 55, 95. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, K.; Shi, J.; Sun, Y. Endotyping Difficult-to-Treat Chronic Rhinosinusitis with Nasal Polyps by Structured Histopathology. Int. Arch. Allergy Immunol. 2023, 184, 1036–1046. [Google Scholar] [CrossRef]
- Vizuete, J.A.C.; Sastre, J.; Bernal, A.D.C.; Picado, C.; Moragón, E.M.; García, J.M.I.; Serrano, C.C.; Gutiérrez, F.J.Á.; Miret, J.M. Asthma, Rhinitis, and Nasal Polyp Multimorbidities. Arch. Bronconeumol. 2019, 55, 146–155. [Google Scholar] [CrossRef]
- Khan, A.; Vandeplas, G.; Huynh, T.M.T.; Joish, V.N.; Mannent, L.; Tomassen, P.; Van Zele, T.; Cardell, L.O.; Arebro, J.; Olze, H.; et al. The Global Allergy and Asthma European Network (GALEN) Rhinosinusitis Cohort: A Large European Cross-Sectional Study of Chronic Rhinosinusitis Patients with and without Nasal Polyps. Rhinology 2019, 57, 32–42. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, A.; Emmanuel, B.; Garcia, D.; Rosta, E. Systematic Literature Review of Humanistic and Economic Burdens of Chronic Rhinosinusitis with Nasal Polyposis. Curr. Med. Res. Opin. 2020, 36, 1913–1926. [Google Scholar] [CrossRef]
- Stevens, W.W.; Schleimer, R.P.; Kern, R.C. Chronic Rhinosinusitis with Nasal Polyps. J. Allergy Clin. Immunol. Pract. 2016, 4, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Alobid, I.; Antón, E.; Armengot, M.; Chao, J.; Colás, C.; del Cuvillo, A.; Dávila, I.; Dordal, M.T.; Escobar, C.; Fernández-Parra, B.; et al. SEAIC-SEORL. Consensus Document on Nasal Polyposis. POLINA Project. J. Investig. Allergol. Clin. Immunol. 2011, 21 (Suppl. 1), 1–58. [Google Scholar]
- Chen, S.; Zhou, A.; Emmanuel, B.; Thomas, K.; Guiang, H. Systematic Literature Review of the Epidemiology and Clinical Burden of Chronic Rhinosinusitis with Nasal Polyposis. Curr. Med. Res. Opin. 2020, 36, 1897–1911. [Google Scholar] [CrossRef] [PubMed]
- Al-Ahmad, M.; Alsaleh, S.; Al-Reefy, H.; Al Abduwani, J.; Nasr, I.; Al Abri, R.; Alamadi, A.M.H.; Fraihat, A.A.; Alterki, A.; Abuzakouk, M.; et al. Expert Opinion on Biological Treatment of Chronic Rhinosinusitis with Nasal Polyps in the Gulf Region. J. Asthma Allergy 2022, 15, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sudiro, M.; Kuntara, A.; Waldi, D. Correlation of Lund-Mackay Score on Computed Tomography Scan and Nasoendoscopic Score in Chronic Rhinosinusitis. Acta Inform. Medica AIM J. Soc. Med. Inform. Bosnia Herzeg. Cas. Drus. Za Med. Inform. BiH 2023, 31, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Chalermwatanachai, T.; Vilchez-Vargas, R.; Holtappels, G.; Lacoere, T.; Jáuregui, R.; Kerckhof, F.M.; Pieper, D.H.; Van De Wiele, T.; Vaneechoutte, M.; Van Zele, T.; et al. Chronic Rhinosinusitis with Nasal Polyps Is Characterized by Dysbacteriosis of the Nasal Microbiota. Sci. Rep. 2018, 8, 7926. [Google Scholar] [CrossRef] [PubMed]
- Lal, D.; Brar, T.; Ramkumar, S.P.; Li, J.; Kato, A.; Zhang, L. Genetics and Epigenetics of Chronic Rhinosinusitis. J. Allergy Clin. Immunol. 2023, 151, 848–868. [Google Scholar] [CrossRef] [PubMed]
- Heredero-Jung, D.H.; Elena-Pérez, S.; García-Sánchez, A.; Estravís, M.; Isidoro-García, M.; Sanz, C.; Dávila, I. Interleukin 5 Receptor Subunit Alpha Expression as a Potential Biomarker in Patients with Nasal Polyposis. Biomedicines 2023, 11, 1966. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.J.; Garcia-Sanchez, A.; Estravis, M.; Gil-Melcón, M.; Isidoro-Garcia, M.; Sanz, C.; Davila, I. Genetics and Epigenetics of Nasal Polyposis: A Systematic Review. J. Investig. Allergol. Clin. Immunol. 2021, 31, 196–211. [Google Scholar] [CrossRef]
- Pérez-Pazos, J.; García-Sánchez, A.; Estravís, M.; Moreno-Jimenez, E.; Morgado, N.; Gómez-García, M.; Ramos-González, J.; Gil-Melcón, M.; Martín-García, C.; Muñoz-Bellido, F.; et al. Beyond T2-Asthma Biomarkers: Risk Stratification for NSAID-Exacerbated Respiratory Disease. ERJ Open Res. 2024, 10, 00909–02023. [Google Scholar] [CrossRef]
- Huntley, K.S.; Raber, J.; Fine, L.; Bernstein, J.A. Influence of the Microbiome on Chronic Rhinosinusitis With and Without Polyps: An Evolving Discussion. Front. Allergy 2021, 2, 737086. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in Health and Diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Khaledi, M.; Poureslamfar, B.; Alsaab, H.O.; Tafaghodi, S.; Hjazi, A.; Singh, R.; Alawadi, A.H.; Alsaalamy, A.; Qasim, Q.A.; Sameni, F. The Role of Gut Microbiota in Human Metabolism and Inflammatory Diseases: A Focus on Elderly Individuals. Ann. Microbiol. 2024, 74, 1. [Google Scholar] [CrossRef]
- Riccio, P.; Rossano, R. The Human Gut Microbiota Is Neither an Organ nor a Commensal. FEBS Lett. 2020, 594, 3262–3271. [Google Scholar] [CrossRef] [PubMed]
- Hillman, E.T.; Lu, H.; Yao, T.; Nakatsu, C.H. Microbial Ecology along the Gastrointestinal Tract. Microbes Environ. 2017, 32, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Malard, F.; Dore, J.; Gaugler, B.; Mohty, M. Introduction to Host Microbiome Symbiosis in Health and Disease. Mucosal Immunol. 2021, 14, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Gelardi, M.; Giancaspro, R.; Cassano, M. Biofilm in Sino-Nasal Infectious Diseases: The Role Nasal Cytology in the Diagnostic Work up and Therapeutic Implications. Eur. Arch. Oto-Rhino-Laryngol. 2023, 280, 1523–1528. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.; Schleimer, R.; Kern, R.C. The Etiology and Pathogenesis of Chronic Rhinosinusitis: A Review of Current Hypotheses. Curr. Allergy Asthma Rep. 2015, 15, 41. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.Z.; Li, Y.C.; Wang, X.D.; Lu, X.X.; Hu, C.H.; He, S.; Liu, X. The Microbiology of Chronic Rhinosinusitis with and without Nasal Polyps. Eur. Arch. Oto-Rhino-Laryngol. 2018, 275, 1439–1447. [Google Scholar] [CrossRef]
- Huang, J.; Xu, Y. Autoimmunity: A New Focus on Nasal Polyps. Int. J. Mol. Sci. 2023, 24, 8444. [Google Scholar] [CrossRef]
- Chegini, Z.; Shariati, A.; Asghari, A.; Rajaeih, S.; Ghorbani, M.; Jalessi, M.; Mirshekar, M.; Razavi, S. Molecular Analysis of Dominant Paranasal Sinus Bacteria in Patients with and without Chronic Rhinosinusitis. Arch. Microbiol. 2022, 204, 327. [Google Scholar] [CrossRef] [PubMed]
- Soyka, M.B.; Wawrzyniak, P.; Eiwegger, T.; Holzmann, D.; Treis, A.; Wanke, K.; Kast, J.I.; Akdis, C.A. Defective Epithelial Barrier in Chronic Rhinosinusitis: The Regulation of Tight Junctions by IFN-γ and IL-4. J. Allergy Clin. Immunol. 2012, 130, 1087–1096.e10. [Google Scholar] [CrossRef] [PubMed]
- Pothoven, K.L.; Norton, J.E.; Hulse, K.E.; Suh, L.A.; Carter, R.G.; Rocci, E.; Harris, K.E.; Shintani-Smith, S.; Conley, D.B.; Chandra, R.K.; et al. Oncostatin M Promotes Mucosal Epithelial Barrier Dysfunction, and Its Expression Is Increased in Patients with Eosinophilic Mucosal Disease. J. Allergy Clin. Immunol. 2015, 136, 737–746.e4. [Google Scholar] [CrossRef] [PubMed]
- Brescia, G.; Alessandrini, L.; Marioni, G. Structured Histopathology for Endotyping and Planning Rational Treatment in Chronic Rhinosinusitis. Am. J. Otolaryngol. Head Neck Med. Surg. 2021, 42, 102795. [Google Scholar] [CrossRef] [PubMed]
- Goulioumis, A.K.; Kourelis, K.; Gkorpa, M.; Danielides, V. Pathogenesis of Nasal Polyposis: Current Trends. Indian J. Otolaryngol. Head Neck Surg. 2023, 75, 733–741. [Google Scholar] [CrossRef]
- Estravís, M.; Pérez-Pazos, J.; Moreno-Jimenez, E.; Triviño, J.C.; García-Sánchez, A.; Gómez-García, M.; Morgado, N.; Ramos-González, J.; Gil-Melcón, M.; Martín-García, C.; et al. Transcriptomics Reveals New Regulatory Mechanisms Involved in Benralizumab Response. Allergy Eur. J. Allergy Clin. Immunol. 2023, 78, 3023–3026. [Google Scholar] [CrossRef]
- Estravís, M.; Pérez-Pazos, J.; Martin, M.J.; Ramos-González, J.; Gil-Melcón, M.; Martín-García, C.; García-Sánchez, A.; Sanz, C.; Dávila, I. Quantitative and Qualitative Methods of Evaluating Response to Biologics in Severe Asthma Patients: Results from a Real-World Study. J. Allergy Clin. Immunol. Pract. 2023, 11, 949–951.e2. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome Definition Re-Visited: Old Concepts and New Challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Guyatt, G.H.; Oxman, A.D.; Vist, G.E.; Kunz, R.; Falck-Ytter, Y.; Alonso-Coello, P.; Schünemann, H.J. GRADE: An Emerging Consensus on Rating Quality of Evidence and Strength of Recommendations. BMJ 2008, 336, 924–926. [Google Scholar] [CrossRef]
- Methley, A.M.; Campbell, S.; Chew-Graham, C.; McNally, R.; Cheraghi-Sohi, S. PICO, PICOS and SPIDER: A Comparison Study of Specificity and Sensitivity in Three Search Tools for Qualitative Systematic Reviews. BMC Health Serv. Res. 2014, 14, 579. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A Web and Mobile App for Systematic Reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef] [PubMed]
- Sterne, J.A.C.; Savović, J.; Page, M.J.; Elbers, R.G.; Blencowe, N.S.; Boutron, I.; Cates, C.J.; Cheng, H.Y.; Corbett, M.S.; Eldridge, S.M.; et al. RoB 2: A Revised Tool for Assessing Risk of Bias in Randomised Trials. BMJ 2019, 366, l4898. [Google Scholar] [CrossRef] [PubMed]
- Stang, A. Critical Evaluation of the Newcastle-Ottawa Scale for the Assessment of the Quality of Nonrandomized Studies in Meta-Analyses. Eur. J. Epidemiol. 2010, 25, 603–605. [Google Scholar] [CrossRef] [PubMed]
- Modesti, P.A.; Reboldi, G.; Cappuccio, F.P.; Agyemang, C.; Remuzzi, G.; Rapi, S.; Perruolo, E.; Parati, G.; Agostoni, P.; Barros, H.; et al. Panethnic Differences in Blood Pressure in Europe: A Systematic Review and Meta-Analysis. PLoS ONE 2016, 11, e0147601. [Google Scholar] [CrossRef]
- Paramasivan, S.; Bassiouni, A.; Shiffer, A.; Dillon, M.R.; Cope, E.K.; Cooksley, C.; Ramezanpour, M.; Moraitis, S.; Ali, M.J.; Bleier, B.; et al. The International Sinonasal Microbiome Study: A Multicentre, Multinational Characterization of Sinonasal Bacterial Ecology. Allergy Eur. J. Allergy Clin. Immunol. 2020, 75, 2033–2045. [Google Scholar] [CrossRef]
- Cao, P.P.; Bin Li, H.; Wang, B.F.; Bin Wang, S.; You, X.J.; Cui, Y.H.; Wang, D.Y.; Desrosiers, M.; Liu, Z. Distinct Immunopathologic Characteristics of Various Types of Chronic Rhinosinusitis in Adult Chinese. J. Allergy Clin. Immunol. 2009, 124, 478–484.e2. [Google Scholar] [CrossRef]
- Kim, D.; Assiri, A.M.; Kim, J.H. Recent Trends in Bacteriology of Adult Patients with Chronic Rhinosinusitis. J. Clin. Med. 2019, 8, 1889. [Google Scholar] [CrossRef]
- Bin Wang, S.; Chen, S.M.; Zhu, K.S.; Zhou, B.; Chen, L.; Zou, X.Y. Increased Lipopolysaccharide Content Is Positively Correlated with Glucocorticoid Receptor-Beta Expression in Chronic Rhinosinusitis with Nasal Polyps. Immunity, Inflamm. Dis. 2020, 8, 605–614. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, S.H.; Lim, J.Y.; Kim, D.; Jeong, I.S.; Lee, D.K.; Jang, Y.J. Association between the Sinus Microbiota with Eosinophilic Inflammation and Prognosis in Chronic Rhinosinusitis with Nasal Polyps. Exp. Mol. Med. 2020, 52, 978–987. [Google Scholar] [CrossRef]
- Feng, T.; Miao, P.; Liu, B.; Liu, Y.; Bao, X.; Xu, J.; Ren, N.; Li, Y.; Shi, J.; Cao, W.; et al. Sinus Microbiota in Patients With Eosinophilic and Non-Eosinophilic Chronic Rhinosinusitis With Nasal Polyps. Front. Cell. Infect. Microbiol. 2021, 11, 672355. [Google Scholar] [CrossRef]
- Abbas, E.E.; Li, C.; Xie, A.; Lu, S.; Tang, L.; Liu, Y.; Elfadil, A.; Wen, S. Distinct Clinical Pathology and Microbiota in Chronic Rhinosinusitis With Nasal Polyps Endotypes. Laryngoscope 2021, 131, E34–E44. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.S.; Liang, K.L. Comparison of Bacteriology Between Eosinophilic and Noneosinophilic Chronic Rhinosinusitis With Nasal Polyps. Otolaryngol. Head Neck Surg. 2023, 168, 1545–1550. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Xie, R.; Xiong, X.; Hu, Z.; Mao, X.; Wang, X.; Zhang, J.; Sun, P.; Yue, Z.; Wang, W.; et al. Alterations of Nasal Microbiome in Eosinophilic Chronic Rhinosinusitis. J. Allergy Clin. Immunol. 2023, 151, 1286–1295.e2. [Google Scholar] [CrossRef]
- Gan, W.; Zhang, H.; Yang, F.; Liu, S.; Liu, F.; Meng, J. The Influence of Nasal Bacterial Microbiome Diversity on the Pathogenesis and Prognosis of Chronic Rhinosinusitis Patients with Polyps. Eur. Arch. Oto-Rhino-Laryngol. 2021, 278, 1075–1088. [Google Scholar] [CrossRef]
- Gan, W.; Zhang, H.; Yang, F.; Liu, S.; Liu, F.; Meng, J. The Influence of Nasal Microbiome Diversity and Inflammatory Patterns on the Prognosis of Nasal Polyps. Sci. Rep. 2021, 11, 6364. [Google Scholar] [CrossRef]
- Lee, C.W.; Lee, B.J.; Yoo, S.H.; Yi, J.S. Relationship between Positive Bacterial Culture in Maxillary Sinus and Surgical Outcomes in Chronic Rhinosinusitis with Nasal Polyps. Auris Nasus Larynx 2014, 41, 446–449. [Google Scholar] [CrossRef] [PubMed]
- Stern, S.; Hadar, T.; Nachalon, Y.; Ben-Zvi, H.; Soudry, E.; Yaniv, E. Bacteriology of the Middle Meatus in Chronic Rhinosinusitis with and without Polyposis. ORL 2016, 78, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Saibene, A.M.; Vassena, C.; Pipolo, C.; Trimboli, M.; De Vecchi, E.; Felisati, G.; Drago, L. Odontogenic and Rhinogenic Chronic Sinusitis: A Modern Microbiological Comparison. Int. Forum Allergy Rhinol. 2016, 6, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Mihalj, M.; Bujak, M.; Butković, J.; Zubčić, Ž.; Levak, M.T.; Čes, J.; Kopić, V.; Lončar, M.B.; Mihalj, H. Differential Expression of TFF1 and TFF3 in Patients Suffering from Chronic Rhinosinusitis with Nasal Polyposis. Int. J. Mol. Sci. 2019, 20, 5461. [Google Scholar] [CrossRef]
- Korkmaz, H.; Ocal, B.; Tatar, E.C.; Tatar, I.; Ozdek, A.; Saylam, G.; Celik, H.H. Biofilms in Chronic Rhinosinusitis with Polyps: Is Eradication Possible? Eur. Arch. Otorhinolaryngol. 2014, 271, 2695–2702. [Google Scholar] [CrossRef]
- Singh, P.; Mehta, R.; Agarwal, S.; Mishra, P. Bacterial Biofilm on the Sinus Mucosa of Healthy Subjects and Patients with Chronic Rhinosinusitis (with or without Nasal Polyposis). J. Laryngol. Otol. 2015, 129, 46–49. [Google Scholar] [CrossRef][Green Version]
- Cantone, E.; Negri, R.; Roscetto, E.; Grassia, R.; Catania, M.R.; Capasso, P.; Maffei, M.; Soriano, A.A.; Leone, C.A.; Iengo, M.; et al. In Vivo Biofilm Formation, Gram-Negative Infections and TAS2R38 Polymorphisms in CRSw NP Patients. Laryngoscope 2018, 128, E339–E345. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Lu, X.; Bo, M.; Qing, H.; Wang, X.; Zhang, L. The Microbiology of Chronic Rhinosinusitis with and without Nasal Polyps. Acta Otolaryngol. 2014, 134, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.C.; Bassiouni, A.; Tanjararak, K.; Vreugde, S.; Wormald, P.J.; Psaltis, A.J. Role of Fungi in Chronic Rhinosinusitis through ITS Sequencing. Laryngoscope 2018, 128, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Simpson, C.A.; Yang, H.H.; Suh, J.D.; Wang, M.B.; Lagishetty, V.; Liang, F.; Jacobs, J.P. Fungal and Bacterial Microbiome in Sinus Mucosa of Patients with and without Chronic Rhinosinusitis. Laryngoscope 2024, 134, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Gelber, J.T.; Cope, E.K.; Goldberg, A.N.; Pletcher, S.D. Evaluation of Malassezia and Common Fungal Pathogens in Subtypes of Chronic Rhinosinusitis. Int. Forum Allergy Rhinol. 2016, 6, 950–955. [Google Scholar] [CrossRef] [PubMed]
- Hoggard, M.; Zoing, M.; Biswas, K.; Taylor, M.W.; Douglas, R.G. The Sinonasal Mycobiota in Chronic Rhinosinusitis and Control Patients. Rhinology 2019, 57, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Hoggard, M.; Jacob, B.; Wheeler, D.; Zoing, M.; Chang, K.; Biswas, K.; Middleditch, M.; Douglas, R.G.; Taylor, M.W. Multiomic Analysis Identifies Natural Intrapatient Temporal Variability and Changes in Response to Systemic Corticosteroid Therapy in Chronic Rhinosinusitis. Immunity, Inflamm. Dis. 2021, 9, 90–107. [Google Scholar] [CrossRef]
- Babu, H.; Sharma, R.; Sharma, V.K.; Goyal, R.; Rana, A.K. Does FESS Alter the Sinonasal Microbiome. Egypt. J. Ear Nose Throat Allied Sci. 2022, 23, 1–6. [Google Scholar] [CrossRef]
- Costa, C.; Garzaro, M.; Boggio, V.; Sidoti, F.; Simeone, S.; Raimondo, L.; Cavallo, G.P.; Pecorari, G.; Cavallo, R. Detection of Herpesviruses 1–6 and Community-Acquired Respiratory Viruses in Patients with Chronic Rhinosinusitis with Nasal Polyposis. Intervirology 2014, 57, 101–105. [Google Scholar] [CrossRef]
- Rowan, N.R.; Lee, S.; Sahu, N.; Kanaan, A.; Cox, S.; Phillips, C.D.; Wang, E.W. The Role of Viruses in the Clinical Presentation of Chronic Rhinosinusitis. Am. J. Rhinol. Allergy 2015, 29, e197–e200. [Google Scholar] [CrossRef] [PubMed]
- Lal, D.; Song, L.; Brar, T.; Cope, E.K.; Keim, P.; Williams, S.; Chung, Y.; Murugan, V.; LaBaer, J.; Magee, D.M. Antibody Responses to the Host Microbiome in Chronic Rhinosinusitis. Int. Forum Allergy Rhinol. 2023, 13, 1503–1510. [Google Scholar] [CrossRef]
- Alammar, Y.; Rousseau, S.; Desrosiers, M.; Tewfik, M.A. The Effect of Corticosteroids on Sinus Microbiota in Chronic Rhinosinusitis Patients with Nasal Polyposis. Am. J. Rhinol. Allergy 2023, 37, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Bartosik, T.J.; Campion, N.J.; Freisl, K.; Liu, D.T.; Gangl, K.; Stanek, V.; Tu, A.; Pjevac, P.; Hausmann, B.; Eckl-Dorna, J.; et al. The Nasal Microbiome in Patients Suffering from Non-Steroidal Anti-Inflammatory Drugs-Exacerbated Respiratory Disease in Absence of Corticosteroids. Front. Immunol. 2023, 14, 1112345. [Google Scholar] [CrossRef] [PubMed]
- Koeller, K.; Herlemann, D.P.R.; Schuldt, T.; Ovari, A.; Guder, E.; Podbielski, A.; Kreikemeyer, B.; Olzowy, B. Microbiome and Culture Based Analysis of Chronic Rhinosinusitis Compared to Healthy Sinus Mucosa. Front. Microbiol. 2018, 9, 643. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Yang, F.; Tang, Y.; Zhou, D.; Qing, D.; Hu, J.; Liu, S.; Liu, F.; Meng, J. The Difference in Nasal Bacterial Microbiome Diversity between Chronic Rhinosinusitis Patients with Polyps and a Control Population. Int. Forum Allergy Rhinol. 2019, 9, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Wagner Mackenzie, B.; West, A.G.; Waite, D.W.; Lux, C.A.; Douglas, R.G.; Taylor, M.W.; Biswas, K. A Novel Description of the Human Sinus Archaeome During Health and Chronic Rhinosinusitis. Front. Cell. Infect. Microbiol. 2020, 10, 398. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.A.; Domissy, A.; Simon, R.A.; White, A.A.; Modena, B.D. Dysbiosis in Aspirin-Exacerbated Respiratory Disease. Int. Forum Allergy Rhinol. 2021, 11, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Han, D.; Mo, J.H.; Kim, Y.M.; Kim, D.W.; Choi, H.G.; Park, J.W.; Shin, H.W. Antibiotic-Dependent Relationships between the Nasal Microbiome and Secreted Proteome in Nasal Polyps. Allergy Asthma Immunol. Res. 2021, 13, 589–608. [Google Scholar] [CrossRef]
- Lal, D.; Keim, P.; Delisle, J.; Barker, B.; Rank, M.A.; Chia, N.; Schupp, J.M.; Gillece, J.D.; Cope, E.K. Mapping and Comparing Bacterial Microbiota in the Sinonasal Cavity of Healthy, Allergic Rhinitis, and Chronic Rhinosinusitis Subjects. Int. Forum Allergy Rhinol. 2017, 7, 561–569. [Google Scholar] [CrossRef]
- Shi, P.; Wei, H.; Liu, X.; Dong, S.; He, S.; Zeng, Y.; Yang, T.; Liu, C.; Li, Y. The Nasal Bacteria Microbiome Comparison Among Fungal Ball Sinusitis, Chronic Sinusitis with Polyps. Indian J. Microbiol. 2023, 63, 120–128. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, J.; Hao, Y.; Wang, B.; Wang, Y.; Liu, Q.; Zhao, J.; Li, Y.; Wang, P.; Wang, X.; et al. Predicting the Recurrence of Chronic Rhinosinusitis with Nasal Polyps Using Nasal Microbiota. Allergy Eur. J. Allergy Clin. Immunol. 2022, 77, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Wos-Oxley, M.L.; Chaves-Moreno, D.; Jáuregui, R.; Oxley, A.P.A.; Kaspar, U.; Plumeier, I.; Kahl, S.; Rudack, C.; Becker, K.; Pieper, D.H. Exploring the Bacterial Assemblages along the Human Nasal Passage. Environ. Microbiol. 2016, 18, 2259–2271. [Google Scholar] [CrossRef]
- Cho, S.W.; Kim, D.Y.; Choi, S.; Won, S.; Kang, H.R.; Yi, H. Microbiome Profiling of Uncinate Tissue and Nasal Polyps in Patients with Chronic Rhinosinusitis Using Swab and Tissue Biopsy. PLoS ONE 2021, 16, e0249688. [Google Scholar] [CrossRef]
- Liu, C.M.; Kohanski, M.A.; Mendiola, M.; Soldanova, K.; Dwan, M.G.; Lester, R.; Nordstrom, L.; Price, L.B.; Lane, A.P. Impact of Saline Irrigation and Topical Corticosteroids on the Postsurgical Sinonasal Microbiota. Int. Forum Allergy Rhinol. 2015, 5, 185–190. [Google Scholar] [CrossRef]
- Varvyanskaya, A.; Lopatin, A. Efficacy of Long-Term Low-Dose Macrolide Therapy in Preventing Early Recurrence of Nasal Polyps after Endoscopic Sinus Surgery. Int. Forum Allergy Rhinol. 2014, 4, 533–541. [Google Scholar] [CrossRef]
- Tabet, P.; Endam, L.M.; Boisvert, P.; Boulet, L.P.; Desrosiers, M. Gram-Negative Bacterial Carriage in Chronic Rhinosinusitis with Nasal Polyposis Is Not Associated with More Severe Inflammation. Int. Forum Allergy Rhinol. 2015, 5, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Stryjewska-Makuch, G.; Janik, M.A.; Klamińska-Cebula, H.; Kolebacz, B.; Ścierski, W.; Lisowska, G. Bacteriological Analysis of Selected Phenotypes of Chronic Rhinosinusitis with Co-Existing Asthma, Allergy and Hypersensitivity to Non-Steroidal Anti-Inflammatory Drugs. Postep. Dermatologii i Alergol. 2021, 38, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.B.; Hong, S.W.; Kim, D.K.; Jeon, S.G.; Kim, K.R.; Cho, S.H.; Gho, Y.S.; Jee, Y.K.; Kim, Y.K. Decreased Diversity of Nasal Microbiota and Their Secreted Extracellular Vesicles in Patients with Chronic Rhinosinusitis Based on a Metagenomic Analysis. Allergy Eur. J. Allergy Clin. Immunol. 2014, 69, 517–526. [Google Scholar] [CrossRef]
- Uhliarova, B.; Adamkov, M.; Svec, M.; Calkovska, A. The Effect of Smoking on CT Score, Bacterial Colonization and Distribution of Inflammatory Cells in the Upper Airways of Patients with Chronic Rhinosinusitis. Inhal. Toxicol. 2014, 26, 419–425. [Google Scholar] [CrossRef]
- Ramakrishnan, V.R.; Frank, D.N. Impact of Cigarette Smoking on the Middle Meatus Microbiome in Health and Chronic Rhinosinusitis. Int. Forum Allergy Rhinol. 2015, 5, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Biswas, K.; Chang, A.; Hoggard, M.; Radcliff, F.J.; Jiang, Y.; Taylor, M.W.; Darveau, R.; Douglas, R.G. Toll-like Receptor Activation by Sino-Nasal Mucus in Chronic Rhinosinusitis. Rhinology 2017, 55, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Cope, E.K.; Goldberg, A.N.; Pletcher, S.D.; Lynch, S.V. Compositionally and Functionally Distinct Sinus Microbiota in Chronic Rhinosinusitis Patients Have Immunological and Clinically Divergent Consequences. Microbiome 2017, 5, 53. [Google Scholar] [CrossRef] [PubMed]
- Hoggard, M.; Waldvogel-Thurlow, S.; Zoing, M.; Chang, K.; Radcliff, F.J.; Mackenzie, B.W.; Biswas, K.; Douglas, R.G.; Taylor, M.W. Inflammatory Endotypes and Microbial Associations in Chronic Rhinosinusitis. Front. Immunol. 2018, 9, 2065. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Hoggard, M.; Zoing, M.; Jiang, Y.; Biswas, K.; Taylor, M.W.; Douglas, R.G. The Effect of Medical Treatments on the Bacterial Microbiome in Patients with Chronic Rhinosinusitis: A Pilot Study. Int. Forum Allergy Rhinol. 2018, 8, 890–899. [Google Scholar] [CrossRef] [PubMed]
- Karunasagar, A.; Jalastagi, R.; Naik, A.; Rai, P. Detection of Bacteria by 16S RRNA PCR and Sequencing in Culture-Negative Chronic Rhinosinusitis. Laryngoscope 2018, 128, 2223–2225. [Google Scholar] [CrossRef] [PubMed]
- Ooi, M.L.; Jothin, A.; Bennett, C.; Ooi, E.H.; Vreugde, S.; Psaltis, A.J.; Wormald, P.J. Manuka Honey Sinus Irrigations in Recalcitrant Chronic Rhinosinusitis: Phase 1 Randomized, Single-Blinded, Placebo-Controlled Trial. Int. Forum Allergy Rhinol. 2019, 9, 1470–1477. [Google Scholar] [CrossRef] [PubMed]
- Vandelaar, L.J.; Hanson, B.; Marino, M.; Yao, W.C.; Luong, A.U.; Arias, C.A.; Ramakrishnan, V.; Citardi, M.J. Analysis of Sinonasal Microbiota in Exacerbations of Chronic Rhinosinusitis Subgroups. OTO Open 2019, 3, 2473974X19875100. [Google Scholar] [CrossRef] [PubMed]
- Zakiah, A.M.; Perkasa, M.F.; Utami, A.D.; Djamin, R.; Bahar, B.; Hamid, F.; Punagi, A.Q. Microbiome Profile in Chronic Rhinosinusitis with and without Polyps of Makassar, Indonesia. Otorhinolaryngol. Clin. 2019, 11, 55–63. [Google Scholar] [CrossRef]
- Biswas, K.; Cavubati, R.; Gunaratna, S.; Hoggard, M.; Waldvogel-Thurlow, S.; Hong, J.; Chang, K.; Wagner Mackenzie, B.; Taylor, M.W.; Douglas, R.G. Comparison of Subtyping Approaches and the Underlying Drivers of Microbial Signatures for Chronic Rhinosinusitis. mSphere 2019, 4, e00679-18. [Google Scholar] [CrossRef]
- Takeda, K.; Sakakibara, S.; Yamashita, K.; Motooka, D.; Nakamura, S.; El Hussien, M.A.; Katayama, J.; Maeda, Y.; Nakata, M.; Hamada, S.; et al. Allergic Conversion of Protective Mucosal Immunity against Nasal Bacteria in Patients with Chronic Rhinosinusitis with Nasal Polyposis. J. Allergy Clin. Immunol. 2019, 143, 1163–1175.e15. [Google Scholar] [CrossRef] [PubMed]
- Wagner Mackenzie, B.; Baker, J.; Douglas, R.G.; Taylor, M.W.; Biswas, K. Detection and Quantification of Staphylococcus in Chronic Rhinosinusitis. Int. Forum Allergy Rhinol. 2019, 9, 1462–1469. [Google Scholar] [CrossRef]
- Endam, L.M.; Alromaih, S.; Gonzalez, E.; Madrenas, J.; Cousineau, B.; Renteria, A.E.; Desrosiers, M. Intranasal Application of Lactococcus Lactis W136 Is Safe in Chronic Rhinosinusitis Patients With Previous Sinus Surgery. Front. Cell. Infect. Microbiol. 2020, 10, 440. [Google Scholar] [CrossRef]
- Lux, C.A.; Wagner Mackenzie, B.; Johnston, J.; Zoing, M.; Biswas, K.; Taylor, M.W.; Douglas, R.G. Antibiotic Treatment for Chronic Rhinosinusitis: Prescription Patterns and Associations With Patient Outcome and the Sinus Microbiota. Front. Microbiol. 2020, 11, 595555. [Google Scholar] [CrossRef] [PubMed]
- Leszczyńska, J.; Stryjewska-Makuch, G.; Ścierski, W.; Lisowska, G. Bacterial Flora of the Nose and Paranasal Sinuses among Patients over 65 Years Old with Chronic Rhinosinusitis Who Underwent Endoscopic Sinus Surgery. Clin. Interv. Aging 2020, 15, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Nepal, R.; Houtak, G.; Shaghayegh, G.; Bouras, G.; Shearwin, K.; Psaltis, A.J.; Wormald, P.; Vreugde, S. Prophages Encoding Human Immune Evasion Cluster Genes Are Enriched in Staphylococcus Aureus Isolated from Chronic Rhinosinusitis Patients with Nasal Polyps. Microb. Genom. 2021, 7, 000726. [Google Scholar] [CrossRef]
- Su, Y.W.; Huang, W.H.; Yeh, C.F. Pantoea Dispersa Rhinosinusitis: Clinical Aspects of a Rare Sinonasal Pathogen. Eur. Arch. Oto-Rhino-Laryngol. 2022, 279, 4389–4395. [Google Scholar] [CrossRef]
- Odat, H.; Al-Ashqar, R.; Al-Ali, R.A.; Al-Qudah, M. The Bacteriology of Middle Meatus and Ethmoid Sinuses in Chronic Rhinosinusitis: Epidemiological Analysis and Surgical Impact. Int. Tinnitus J. 2022, 26, 110–114. [Google Scholar] [CrossRef]
- Zhu, Z.S.; Lan, J.; Wei, R.; Xu, Y.; Hong, Y.; Bao, W.; He, G. Microbiome and Th Cytokines Association in Chronic Rhinosinusitis with or without Nasal Polyp. Laryngoscope Investig. Otolaryngol. 2023, 8, 335–345. [Google Scholar] [CrossRef]
- Gan, W.; Xiang, Y.; Wei, B.; Liu, S.; Liu, F. The Inflammatory Microenvironment of Nasal Polyps in Patients with Chronic Rhinosinusitis and the Relationship of This Microenvironment with the Nasal Microbiome. Asian J. Surg. 2024, 47, 124–133. [Google Scholar] [CrossRef]
- Sproson, E.L.; Thomas, K.M.; Lau, L.C.; Harries, P.G.; Howarth, P.H.; Salib, R.J. Common Airborne Fungi Induce Species-Specific Effects on Upper Airway Inflammatory and Remodelling Responses. Rhinology 2016, 54, 51–55. [Google Scholar] [CrossRef]
- Dobretsov, K.G.; Kolenchukova, O.; Sipkin, A.; Bellussi, L.M.; Ciprandi, G.; Passali, D. A Randomized, Double-Blind, Placebo- -Controlled Study to Investigate the Use of Bacteriophages in Patients with Chronic Rhinosinusitis with Nasal Polyps. Otolaryngol. Pol. 2021, 75, 33–37. [Google Scholar] [CrossRef]
- Ancona, G.; Alagna, L.; Alteri, C.; Palomba, E.; Tonizzo, A.; Pastena, A.; Muscatello, A.; Gori, A.; Bandera, A. Gut and Airway Microbiota Dysbiosis and Their Role in COVID-19 and Long-COVID. Front. Immunol. 2023, 14, 1080043. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.S.B.; Raes, J.; Bork, P. The Human Gut Microbiome: From Association to Modulation. Cell 2018, 172, 1198–1215. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, T.; Jiang, L.; Dinu, I.; Groizeleau, J.; Kozyrskyj, A.L.; Greenwood, C.M.T.; Arrieta, M.C. The Rise to Power of the Microbiome: Power and Sample Size Calculation for Microbiome Studies. Mucosal Immunol. 2022, 15, 1060–1070. [Google Scholar] [CrossRef] [PubMed]
- Kers, J.G.; Saccenti, E. The Power of Microbiome Studies: Some Considerations on Which Alpha and Beta Metrics to Use and How to Report Results. Front. Microbiol. 2022, 12, 796025. [Google Scholar] [CrossRef] [PubMed]
- Kumpitsch, C.; Koskinen, K.; Schöpf, V.; Moissl-Eichinger, C. The Microbiome of the Upper Respiratory Tract in Health and Disease. BMC Biol. 2019, 17, 87. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liang, Q.; Ren, Y.; Guo, C.; Ge, X.; Wang, L.; Cheng, Q.; Luo, P.; Zhang, Y.; Han, X. Immunosenescence: Molecular Mechanisms and Diseases. Signal Transduct. Target. Ther. 2023, 8, 200. [Google Scholar] [CrossRef] [PubMed]
- Whelan, F.J.; Verschoor, C.P.; Stearns, J.C.; Rossi, L.; Luinstra, K.; Loeb, M.; Smieja, M.; Johnstone, J.; Surette, M.G.; Bowdish, D.M.E. The Loss of Topography in the Microbial Communities of the Upper Respiratory Tract in the Elderly. Ann. Am. Thorac. Soc. 2014, 11, 513–521. [Google Scholar] [CrossRef]
- Jain, R.; Hoggard, M.; Biswas, K.; Zoing, M.; Jiang, Y.; Douglas, R. Changes in the Bacterial Microbiome of Patients with Chronic Rhinosinusitis after Endoscopic Sinus Surgery. Int. Forum Allergy Rhinol. 2017, 7, 7–15. [Google Scholar] [CrossRef]
- Shah, S.J.; Hawn, V.S.; Zhu, N.; Fang, C.H.; Gao, Q.; Akbar, N.A.; Abuzeid, W.M. Postoperative Infection Rate and Associated Factors Following Endoscopic Sinus Surgery. Ann. Otol. Rhinol. Laryngol. 2022, 131, 5–11. [Google Scholar] [CrossRef]
- Szaleniec, J.; Bezshapkin, V.; Krawczyk, A.; Kopera, K.; Zapała, B.; Gosiewski, T.; Kosciolek, T. Determinants of the Microbiome Spatial Variability in Chronic Rhinosinusitis. Rhinology 2024, 62, 119–126. [Google Scholar] [CrossRef]
- Antonopoulos, D.A.; Huse, S.M.; Morrison, H.G.; Schmidt, T.M.; Sogin, M.L.; Young, V.B. Reproducible Community Dynamics of the Gastrointestinal Microbiota Following Antibiotic Perturbation. Infect. Immun. 2009, 77, 2367–2375. [Google Scholar] [CrossRef] [PubMed]
- Gerardi, D.; Santostasi, N.; Torge, D.; Rinaldi, F.; Bernardi, S.; Bianchi, S.; Piattelli, M.; Varvara, G. Regenerative Potential of Platelet—Rich Fibrin in Maxillary Sinus Floor Lift Techniques: A Systematic Review. J. Biol. Regul. Homeost. Agents 2023, 37, 2357–2369. [Google Scholar] [CrossRef]
- Szaleniec, J.; Gibała, A.; Hartwich, P.; Hydzik-Sobocińska, K.; Konior, M.; Gosiewski, T.; Szaleniec, M. Challenging the Gold Standard: Methods of Sampling for Microbial Culture in Patients with Chronic Rhinosinusitis. Eur. Arch. Oto-Rhino-Laryngol. 2021, 278, 4795–4803. [Google Scholar] [CrossRef]
- Lee, J.T.; Frank, D.N.; Ramakrishnan, V. Microbiome of the Paranasal Sinuses: Update and Literature Review. Am. J. Rhinol. Allergy 2016, 30, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact Sequence Variants Should Replace Operational Taxonomic Units in Marker-Gene Data Analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Walters, K.E.; Martiny, J.B.H. Alpha-, Beta-, and Gamma-Diversity of Bacteria Varies across Habitats. PLoS ONE 2020, 15, e0233872. [Google Scholar] [CrossRef]
- Copeland, E.; Leonard, K.; Carney, R.; Kong, J.; Forer, M.; Naidoo, Y.; Oliver, B.G.G.; Seymour, J.R.; Woodcock, S.; Burke, C.M.; et al. Chronic Rhinosinusitis: Potential Role of Microbial Dysbiosis and Recommendations for Sampling Sites. Front. Cell. Infect. Microbiol. 2018, 8, 57. [Google Scholar] [CrossRef]
- Hufnagl, K.; Pali-Schöll, I.; Roth-Walter, F.; Jensen-Jarolim, E. Dysbiosis of the Gut and Lung Microbiome Has a Role in Asthma. Semin. Immunopathol. 2020, 42, 75–93. [Google Scholar] [CrossRef]
- Garcia-Nuñez, M.; Millares, L.; Pomares, X.; Ferrari, R.; Pérez-Brocal, V.; Gallego, M.; Espasa, M.; Moya, A.; Monsó, E. Severity-Related Changes of Bronchial Microbiome in Chronic Obstructive Pulmonary Disease. J. Clin. Microbiol. 2014, 52, 4217–4223. [Google Scholar] [CrossRef] [PubMed]
- Ege, M.J.; Mayer, M.; Normand, A.-C.; Genuneit, J.; Cookson, W.O.; Phil, D.; Braun-Fahrländer, C.; Heederik, D.; Piarrroux, R.; von Mutius, E. Exposure to Environmental Microorganisms and Childhood Asthma. N. Engl. J. Med. 2011, 364, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.J.; Jithpratuck, W.; Wasylik, K.; Sriaroon, P.; Dishaw, L.J. Associations of Microbial Diversity with Age and Other Clinical Variables among Pediatric Chronic Rhinosinusitis (CRS) Patients. Microorganisms 2023, 11, 422. [Google Scholar] [CrossRef] [PubMed]
- Abrahamsson, T.R.; Jakobsson, H.E.; Andersson, A.F.; Björkstén, B.; Engstrand, L.; Jenmalm, M.C. Low Gut Microbiota Diversity in Early Infancy Precedes Asthma at School Age. Clin. Exp. Allergy 2014, 44, 842–850. [Google Scholar] [CrossRef]
- Man, W.H.; De Steenhuijsen Piters, W.A.A.; Bogaert, D. The Microbiota of the Respiratory Tract: Gatekeeper to Respiratory Health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef]
- Slattery, J.; Macfabe, D.F.; Frye, R.E. The Significance of the Enteric Microbiome on the Development of Childhood Disease: A Review of Prebiotic and Probiotic Therapies in Disorders of Childhood. Clin. Med. Insights Pediatr. 2016, 10, CMPed.S38338. [Google Scholar] [CrossRef]
- Mulder, B.; Pouwels, K.B.; Schuiling-Veninga, C.C.M.; Bos, H.J.; de Vries, T.W.; Jick, S.S.; Hak, E. Antibiotic Use during Pregnancy and Asthma in Preschool Children: The Influence of Confounding. Clin. Exp. Allergy 2016, 46, 1214–1226. [Google Scholar] [CrossRef] [PubMed]
- Maxfield, A.Z.; Korkmaz, H.; Gregorio, L.L.; Busaba, N.Y.; Gray, S.T.; Holbrook, E.H.; Guo, R.; Bleier, B.S. General Antibiotic Exposure Is Associated with Increased Risk of Developing Chronic Rhinosinusitis. Laryngoscope 2017, 127, 296–302. [Google Scholar] [CrossRef]
- Konovalovas, A.; Armalytė, J.; Klimkaitė, L.; Liveikis, T.; Jonaitytė, B.; Danila, E.; Bironaitė, D.; Mieliauskaitė, D.; Bagdonas, E.; Aldonytė, R. Human Nasal Microbiota Shifts in Healthy and Chronic Respiratory Disease Conditions. BMC Microbiol. 2024, 24, 150. [Google Scholar] [CrossRef]
- Mccauley, K.; Durack, J.; Valladares, R.; Douglas, W.; Lin, D.L.; Calatroni, A.; Lebeau, P.K.; Hoang, T. Distinct nasal airway bacterial microbiotas differentially relate to exacerbation in pediatric patients with asthma. J. Allergy Clin. Immunol. 2019, 144, 1187–1197. [Google Scholar] [CrossRef]
- Wang, Z.; Maschera, B.; Lea, S.; Kolsum, U.; Michalovich, D.; Van Horn, S.; Traini, C.; Brown, J.R.; Hessel, E.M.; Singh, D. Airway Host-Microbiome Interactions in Chronic Obstructive Pulmonary Disease. Respir. Res. 2019, 20, 113. [Google Scholar] [CrossRef] [PubMed]
- Brook, I. Microbiology of Chronic Rhinosinusitis. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Zheng, H.; Han, S.; Tang, L.; Yang, X.; Chu, P.; Wang, P.; Lu, J.; Ge, W.; Ni, X. Detection of Nasal Microbiota in Pediatric Patients with Antrochoanal Polyps by TLDA. Int. J. Pediatr. Otorhinolaryngol. 2020, 130, 109811. [Google Scholar] [CrossRef] [PubMed]
- Moyano, R.O.; Tonetti, F.R.; Tomokiyo, M.; Kanmani, P.; Vizoso-Pinto, M.G.; Kim, H.; Quilodrán-Vega, S.; Melnikov, V.; Alvarez, S.; Takahashi, H.; et al. The Ability of Respiratory Commensal Bacteria to Beneficially Modulate the Lung Innate Immune Response Is a Strain Dependent Characteristic. Microorganisms 2020, 8, 727. [Google Scholar] [CrossRef]
- Fujieda, S.; Imoto, Y.; Kato, Y.; Ninomiya, T.; Tokunaga, T.; Tsutsumiuchi, T.; Yoshida, K.; Kidoguchi, M.; Takabayashi, T. Eosinophilic Chronic Rhinosinusitis. Allergol. Int. 2019, 68, 403–412. [Google Scholar] [CrossRef]
- Stryjewska-Makuch, G.; Janik, M.A.; Lisowska, G.; Kolebacz, B. Bacteriological Analysis of Isolated Chronic Sinusitis without Polyps. Postep. Dermatol. Alergol. 2018, 35, 375–380. [Google Scholar] [CrossRef]
- Yaniv, D.; Stern, D.; Vainer, I.; Ben Zvi, H.; Yahav, D.; Soudry, E. The Bacteriology of Recurrent Acute Exacerbations of Chronic Rhinosinusitis: A Longitudinal Analysis. Eur. Arch. Oto-Rhino-Laryngol. 2020, 277, 3051–3057. [Google Scholar] [CrossRef]
- Jeican, I.I.; Gheban, D.; Barbu-Tudoran, L.; Inișca, P.; Albu, C.; Ilieș, M.; Albu, S.; Vică, M.L.; Matei, H.V.; Tripon, S.; et al. Respiratory Nasal Mucosa in Chronic Rhinosinusitis with Nasal Polyps versus COVID-19: Histopathology, Electron Microscopy Analysis and Assessing of Tissue Interleukin-33. J. Clin. Med. 2021, 10, 4110. [Google Scholar] [CrossRef]
- Ver Heul, A.; Planer, J.; Kau, A.L. The Human Microbiota and Asthma. Clin. Rev. Allergy Immunol. 2019, 57, 350–363. [Google Scholar] [CrossRef]
- Brescia, G.; Contro, G.; Giacomelli, L.; Barion, U.; Frigo, A.C.; Marioni, G. Blood Eosinophilic and Basophilic Trends in Recurring and Non-Recurring Eosinophilic Rhinosinusitis with Nasal Polyps. Am. J. Rhinol. Allergy 2021, 35, 296–301. [Google Scholar] [CrossRef]
- Tosun, F.; Arslan, H.H.; Karslioglu, Y.; Deveci, M.S.; Durmaz, A. Relationship between Postoperative Recurrence Rate and Eosinophil Density of Nasal Polyps. Ann. Otol. Rhinol. Laryngol. 2010, 119, 455–459. [Google Scholar] [CrossRef]
- Cavaliere, C.; Masieri, S.; Begvarfaj, E.; Loperfido, A.; Baroncelli, S.; Cascone, F.; Ciofalo, A. Long-Term Perspectives on Chronic Rhinosinusitis with Nasal Polyps: Evaluating Recurrence Rates after Functional Endoscopic Sinus Surgery in the Biologics Era—A 5-Year Follow-Up Study. J. Pers. Med. 2024, 14, 297. [Google Scholar] [CrossRef] [PubMed]
- Bomar, L.; Brugger, S.D.; Yost, B.H.; Davies, S.S.; Lemon, K.P. Corynebacterium Accolens Releases Antipneumococcal Free Fatty Acids from Human Nostril and Skin Surface Triacylglycerols. MBio 2016, 7, e01725-15. [Google Scholar] [CrossRef] [PubMed]
- Menberu, M.A.; Liu, S.; Cooksley, C.; Hayes, A.J.; Psaltis, A.J.; Wormald, P.J.; Vreugde, S. Corynebacterium Accolens Has Antimicrobial Activity against Staphylococcus Aureus and Methicillin-Resistant s. Aureus Pathogens Isolated from the Sinonasal Niche of Chronic Rhinosinusitis Patients. Pathogens 2021, 10, 207. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Peña, S.; Rodríguez-Martínez, S.; Cancino-Diaz, M.E.; Cancino-Diaz, J.C. Staphylococcus Epidermidis Controls Opportunistic Pathogens in the Nose, Could It Help to Regulate SARS-CoV-2 (COVID-19) Infection? Life 2022, 12, 341. [Google Scholar] [CrossRef] [PubMed]
- Loperfido, A.; Cavaliere, C.; Begvarfaj, E.; Ciofalo, A.; D’Erme, G.; De Vincentiis, M.; Greco, A.; Millarelli, S.; Bellocchi, G.; Masieri, S. The Impact of Antibiotics and Steroids on the Nasal Microbiome in Patients with Chronic Rhinosinusitis: A Systematic Review According to PICO Criteria. J. Pers. Med. 2023, 13, 1583. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.C.; Adappa, N.D.; Cohen, N.A. Use of Topical Nasal Therapies in the Management of Chronic Rhinosinusitis. Laryngoscope 2013, 123, 2347–2359. [Google Scholar] [CrossRef]
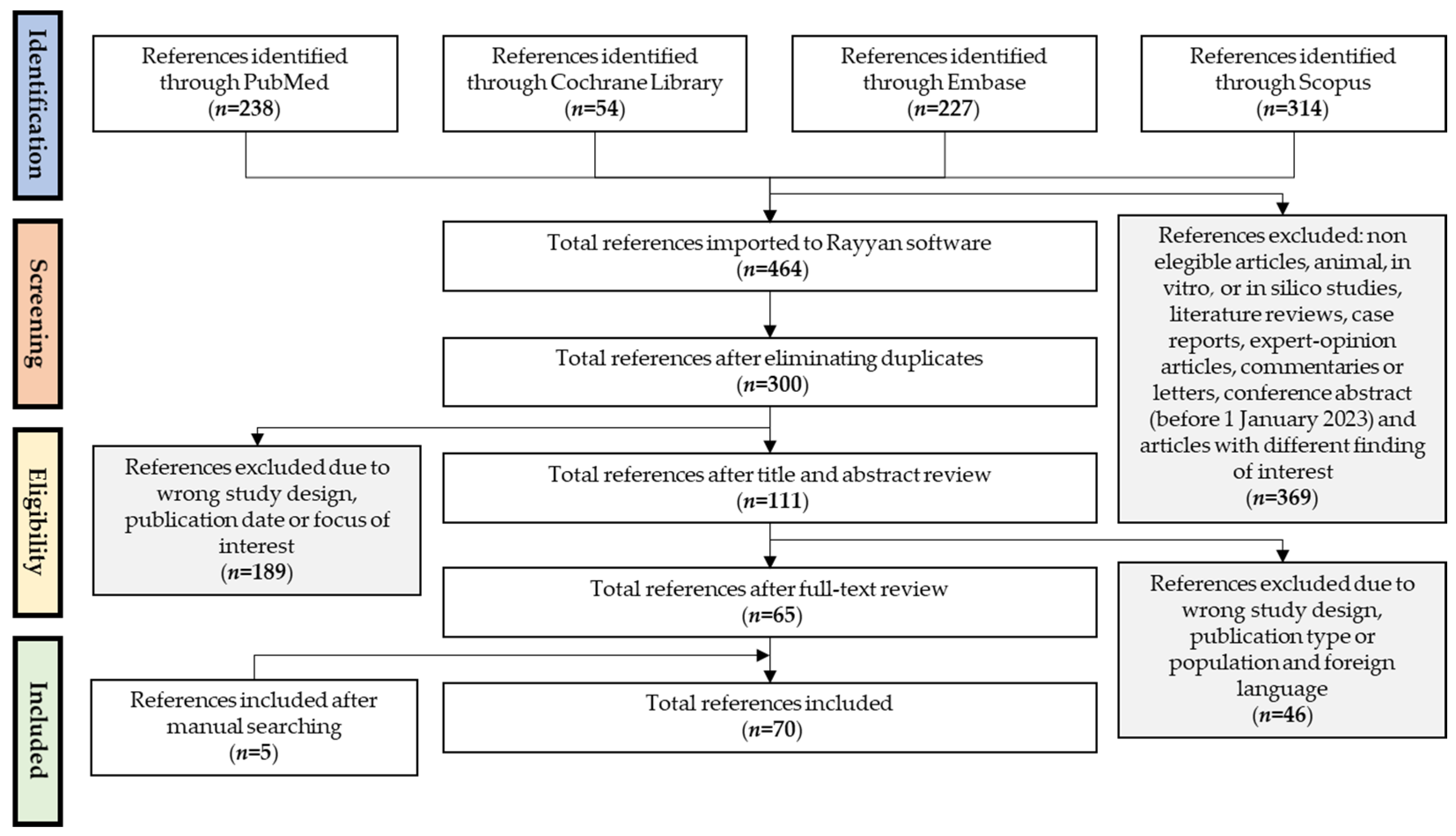
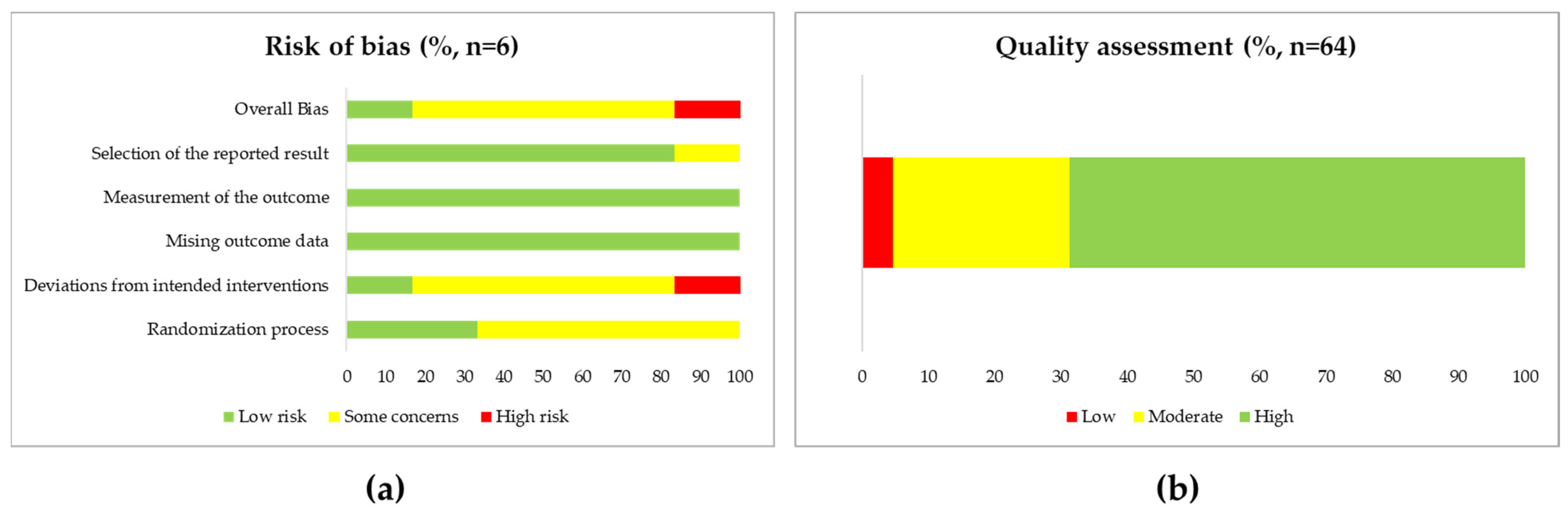
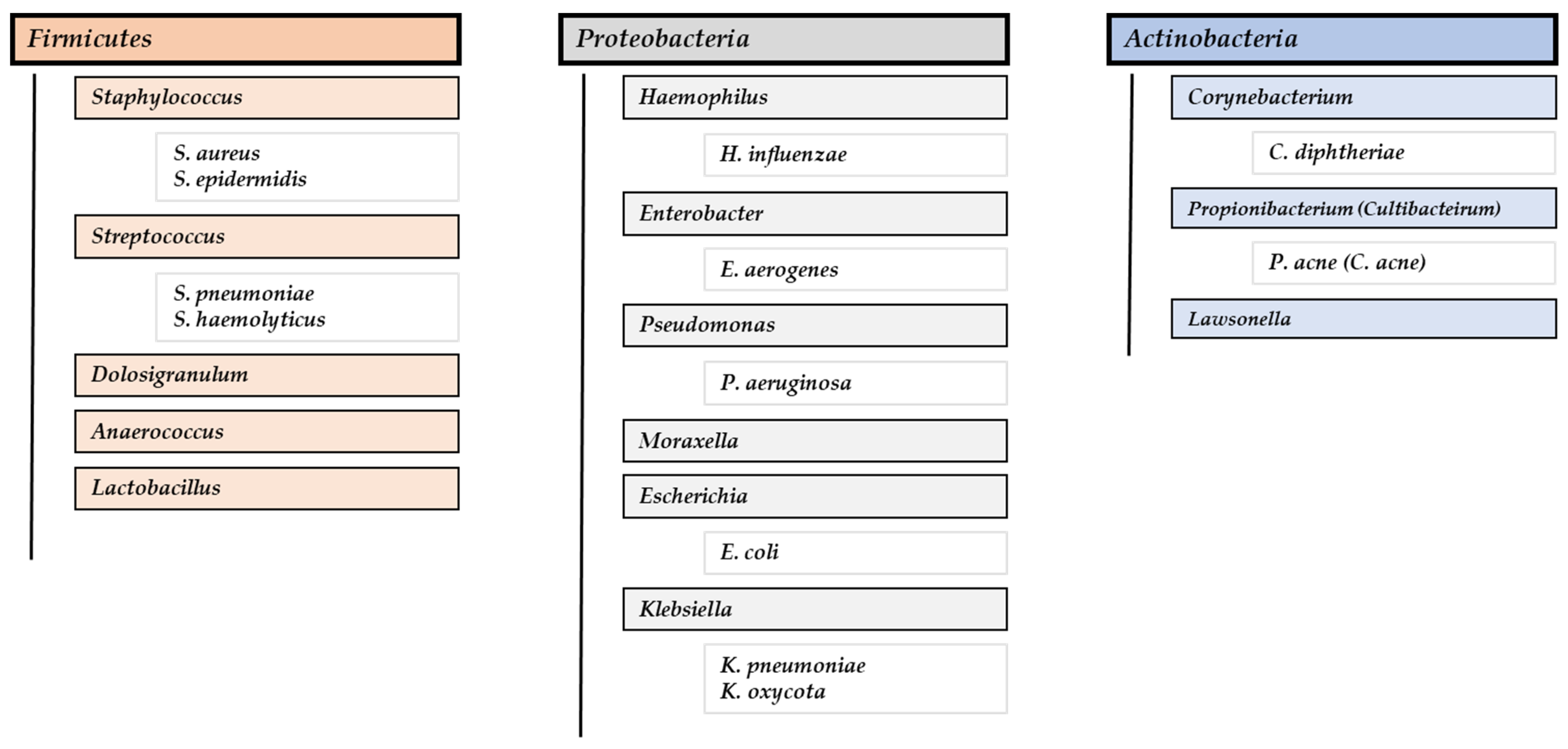
| Ref. | Population (P) | Intervention (I) | Comparison (C) | Outcome (O) |
|---|---|---|---|---|
| [96] | CRSwNP patients underwent bilateral FESS | Clarithromycin + MFNS Duration: Three or six months | MFNS | No significant changes in the nasal microbiota: The proportion of clarithromycin-resistant strains remained constant throughout the antibiotic treatment. Some clinical benefits were observed in the antibiotic-treated groups: lower CT scores and eosinophil cationic protein (ECP) levels than the control group. |
| [70] | CRSwNP patients underwent ESS | Clarithromycin + MFNS Duration: Eight weeks | MFNS | Adding clarithromycin to MFNS was associated with a significant eradication in biofilm formation (six of 12 biofilm-positive samples before surgery turned negative) when compared to MFNS alone (one of 11 biofilm-positive samples). It was not demonstrated that combined therapy was superior in improving tomography (CT) and symptom (SNOT-20) scores. |
| [95] | Post-surgical CRSwNP patients with patent maxillary antrostomy | Treatment of nasal saline irrigation or intranasal corticosteroid spray of any type * CRSwNP patients who used saline irrigation also used topical corticosteroids (spray or irrigation) | DCs | No significant changes in the nasal microbiota associated with saline irrigation or intranasal corticosteroid spray. |
| [105] | CRSwNP and CRSsNP patients for whom ESS was indicated | Doxycycline or prednisone Duration: Seven days * All patients continued with daily topical corticosteroids nasal sprays and regular sinonasal saline lavage | Placebo | In individual patients, doxycycline was associated with an increase in RA of Corynebacterium and Haemophilus compared to prednisone and untreated controls. |
| [107] | CRSwNP and CRSsNP patients underwent ESS | Manuka honey (MH) with augmented methylglyoxal (MXO) sinonasal rinses Duration: Two weeks and ten days of placebo tablets or culture-directed oral antibiotic tablets (control group) | Placebo (saline sinonasal rinses) | 50% of patients had a reduction in standard semiquantitative bacterial culture rate (mainly about S. aureus) after the MH + MXO treatment. A statistically significant reduction in LKS (Lund–Kennedy score) outcome was also observed after the treatment. |
| [113] | CRSwNP and CRSsNP patients with previous ESS | Topical administration of live Lactococcus lactis W126 Duration: Two weeks * All patients continued with saline irrigation | No placebo control was included | Treatment was associated with a reduced abundance of multiple species of Staphylococcus, Peptostreptococcae, and Enterobacialles, as well as an increased abundance of Dolosigranulum pigrum. The response pattern showed progressive improvement during the treatment and post-treatment (SNOT-22 scores, nasal congestion, post-nasal drip, and need to blow nose). |
| [89] | CRSwNP, CRSnNP patients, and DCs | Oral antibiotics treatment within three months before sampling | No antibiotics treatment | Alpha diversity (Shannon and Simpson indexes) was significantly lower in patients who had taken antibiotics than in patients who had not. Streptococcaceae, Lachnospiraceae, and Neisseriaceae were significantly decreased in patients who had taken antibiotics compared to patients who had not. |
| [122] * | CRSwNP patients underwent FESS | Intranasal gel with bacteriophage mixture (Otofag) Duration: Ten weeks | Placebo | The total number of bacteria significantly decreased in the experimental group compared to the placebo group. In detail, a significant decrease in the number of Streptococci and Enterobacteriaceae was observed in the experimental group. The decrease in cytokines in the nasal lavage fluid (IL-8) and blood serum (IL-1B) was proved by the total number of microorganisms and by the concentration of Enterobacteriaceae and Staphylococci. |
| [83] | CRSwNP patients | Prednisone + amoxicillin–clavulanate or prednisone alone Duration: Prednisone for three weeks and amoxicillin–clavulanate for two weeks * All patients were concurrent to their baseline intranasal saline irrigation and topical corticosteroid | DCs | Non-significant changes in the nasal microbiota. Micrococcus was isolated before both treatments but not post-treatment. After three months from treatment, an increase in the prevalence of Corynebacterium was observed in the group treated with prednisone and amoxicillin–clavulanate, as well as in Gram-negative (Pseudomonas) and Staphylococcus genera in the group treated only with prednisone. Both treatments showed significant short-term (after one month from treatment) improvement (SNOT-22), but not in the long term (after three months). |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-García, M.; Moreno-Jimenez, E.; Morgado, N.; García-Sánchez, A.; Gil-Melcón, M.; Pérez-Pazos, J.; Estravís, M.; Isidoro-García, M.; Dávila, I.; Sanz, C. The Role of the Gut and Airway Microbiota in Chronic Rhinosinusitis with Nasal Polyps: A Systematic Review. Int. J. Mol. Sci. 2024, 25, 8223. https://doi.org/10.3390/ijms25158223
Gómez-García M, Moreno-Jimenez E, Morgado N, García-Sánchez A, Gil-Melcón M, Pérez-Pazos J, Estravís M, Isidoro-García M, Dávila I, Sanz C. The Role of the Gut and Airway Microbiota in Chronic Rhinosinusitis with Nasal Polyps: A Systematic Review. International Journal of Molecular Sciences. 2024; 25(15):8223. https://doi.org/10.3390/ijms25158223
Chicago/Turabian StyleGómez-García, Manuel, Emma Moreno-Jimenez, Natalia Morgado, Asunción García-Sánchez, María Gil-Melcón, Jacqueline Pérez-Pazos, Miguel Estravís, María Isidoro-García, Ignacio Dávila, and Catalina Sanz. 2024. "The Role of the Gut and Airway Microbiota in Chronic Rhinosinusitis with Nasal Polyps: A Systematic Review" International Journal of Molecular Sciences 25, no. 15: 8223. https://doi.org/10.3390/ijms25158223
APA StyleGómez-García, M., Moreno-Jimenez, E., Morgado, N., García-Sánchez, A., Gil-Melcón, M., Pérez-Pazos, J., Estravís, M., Isidoro-García, M., Dávila, I., & Sanz, C. (2024). The Role of the Gut and Airway Microbiota in Chronic Rhinosinusitis with Nasal Polyps: A Systematic Review. International Journal of Molecular Sciences, 25(15), 8223. https://doi.org/10.3390/ijms25158223






