Carbon Source and Substrate Surface Affect Biofilm Formation by the Plant-Associated Bacterium Pseudomonas donghuensis P482
Abstract
:1. Introduction
2. Results
2.1. EPS-Congo Red Binding Affected in P482 Mutants
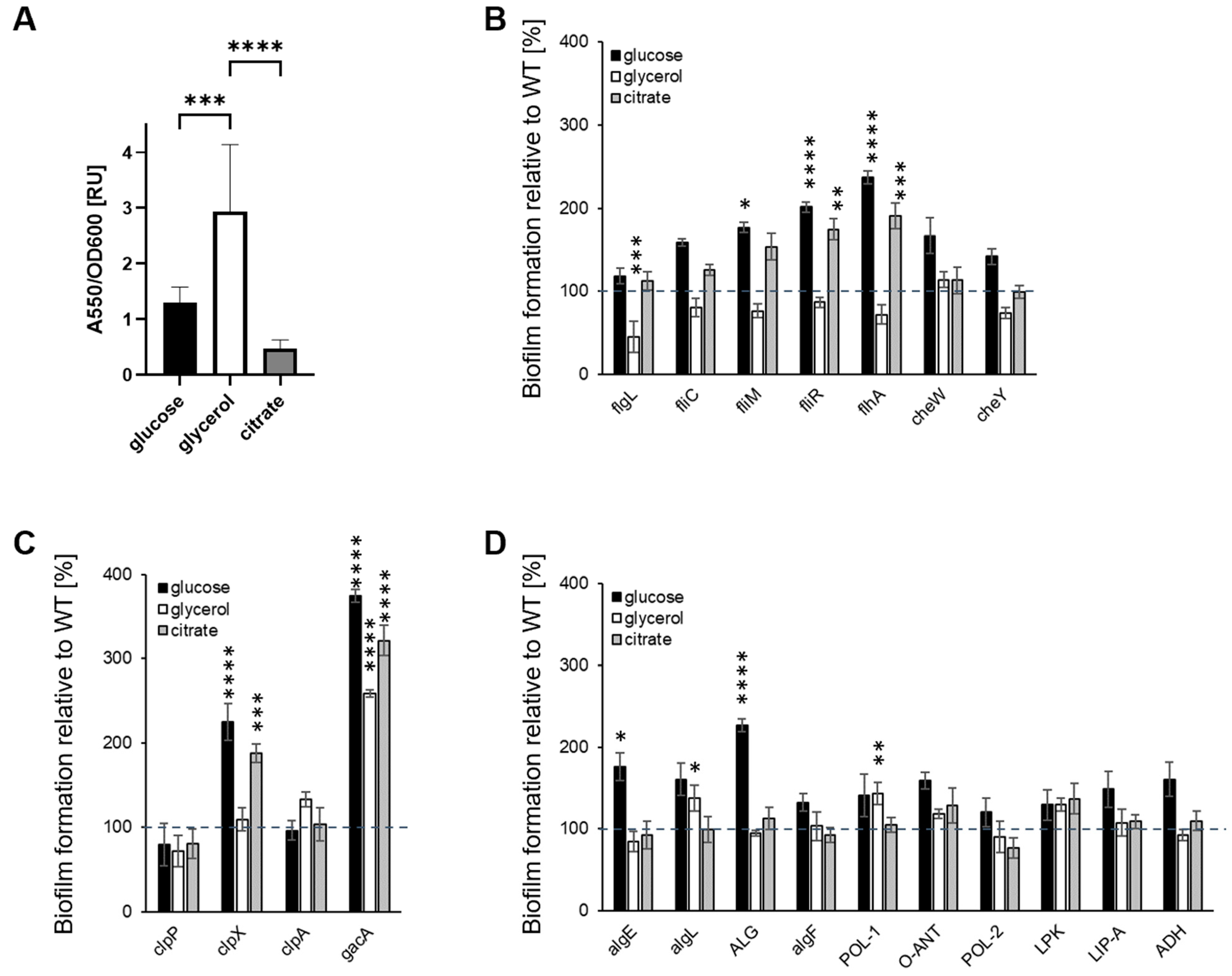
2.2. Carbon Source Influences Biofilm Formation on Polystyrene
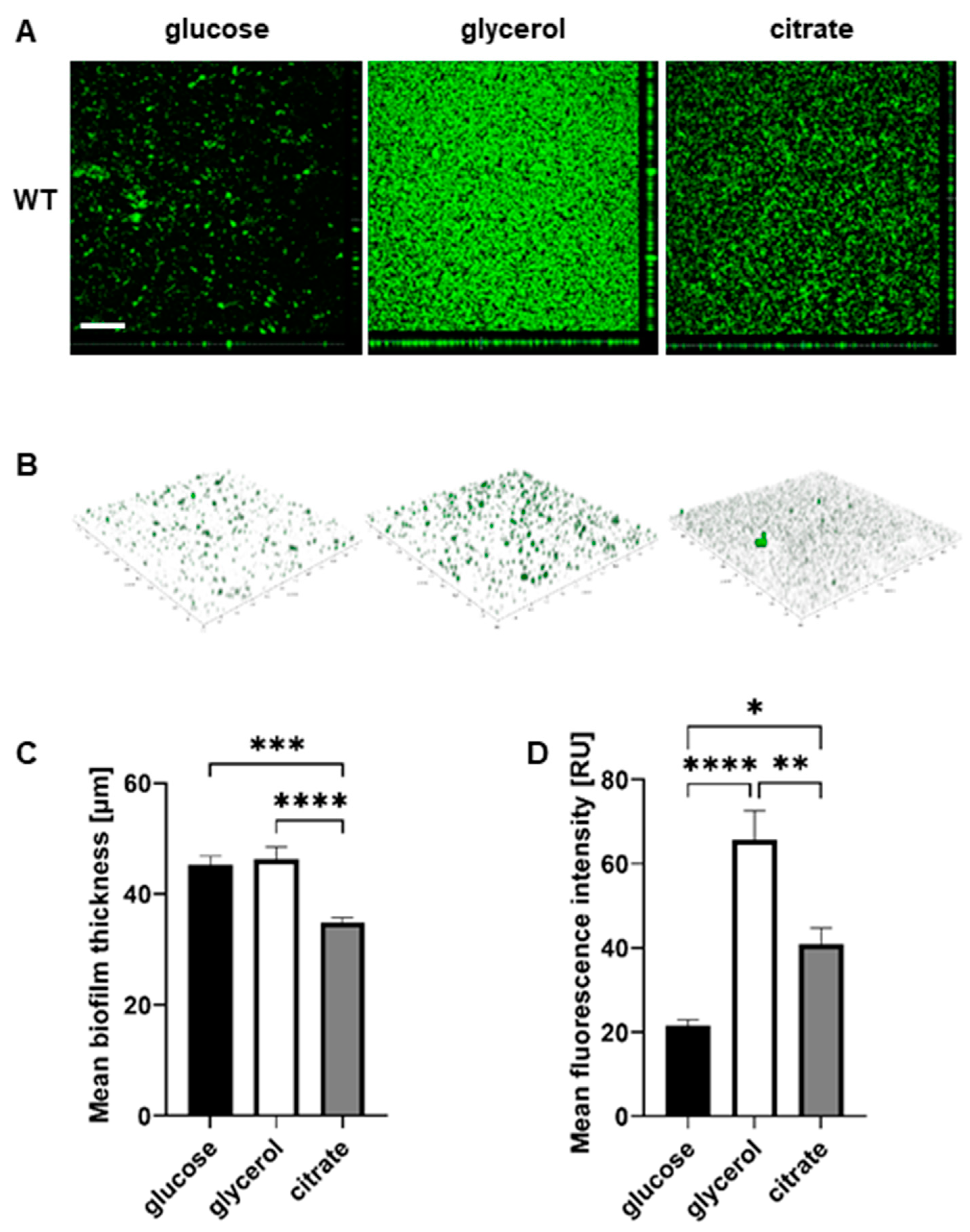
2.3. The Nature of the Abiotic Surface Impacts Biofilm Biomass and Architecture
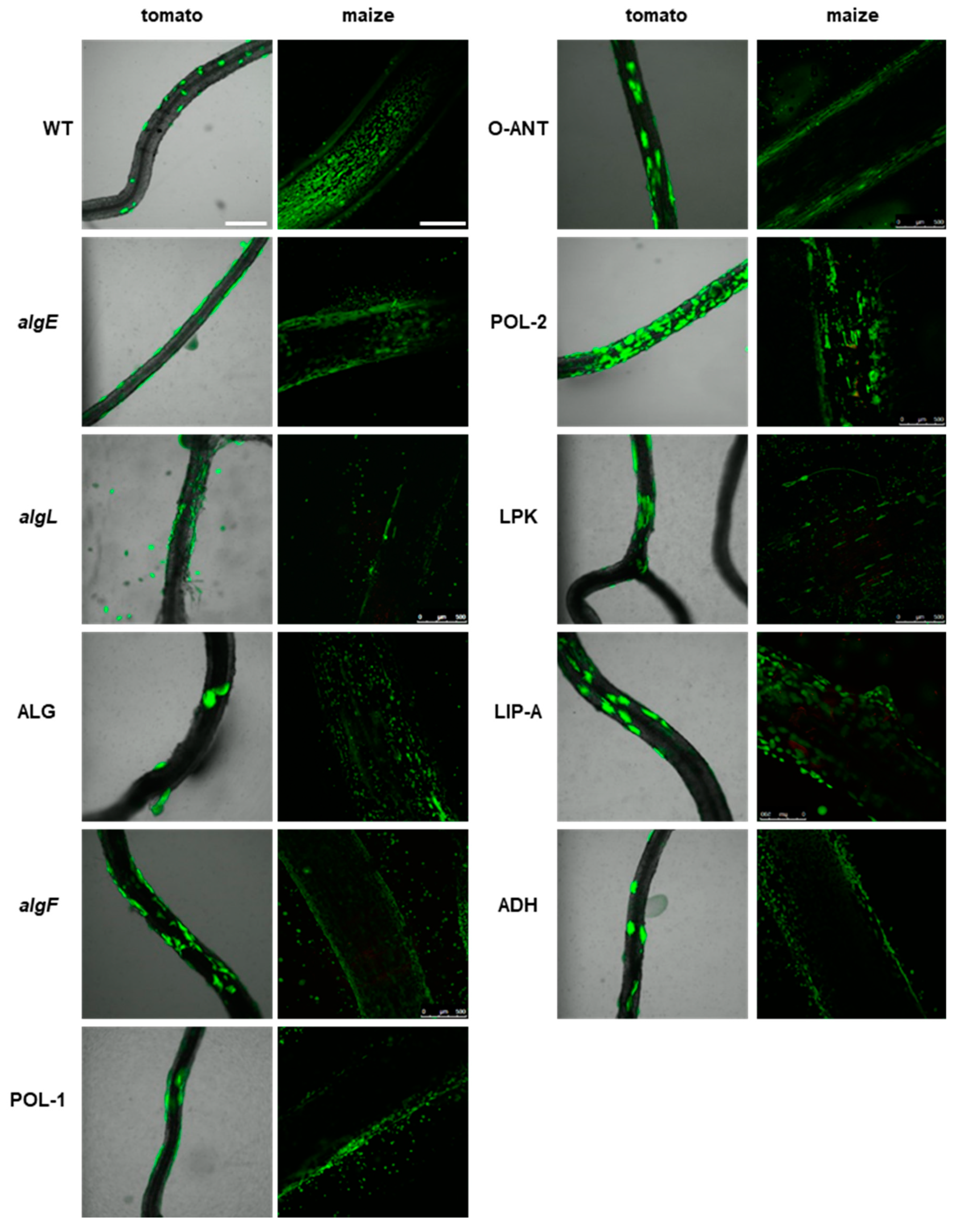
2.4. P482 Biofilm-Defective Mutants Remain Capable of Colonizing Tomato and Maize Roots
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Cell Micrography
4.3. Insertion Mutagenesis
4.4. Fluorescent Labelling of P482 Strains
4.5. Colony Morphology and Congo Red-Staining Assay
4.6. Biofilm Formation Assays
4.6.1. Crystal Violet Staining
4.6.2. Biofilm Formation on Glass
4.7. Tomato Rhizosphere Colonization
4.8. Maize Rhizosphere Colonization
4.9. Motility Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flemming, H.C.; Wuertz, S. Bacteria and Archaea on Earth and Their Abundance in Biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial Biofilms: From the Natural Environment to Infectious Diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as Complex Differentiated Communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial Life on Surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.E.; O’Toole, G.A. Microbial Biofilms: From Ecology to Molecular Genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [Google Scholar] [CrossRef]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The Biofilm Life Cycle: Expanding the Conceptual Model of Biofilm Formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, C.; Chen, Z.; Allan, E.; Van Der Mei, H.C.; Busscher, H.J. Emergent Heterogeneous Microenvironments in Biofilms: Substratum Surface Heterogeneity and Bacterial Adhesion Force-Sensing. FEMS Microbiol. Rev. 2018, 1, 259–272. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A.; Kolter, R. Flagellar and Twitching Motility Are Necessary for Pseudomonas aeruginosa Biofilm Development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef]
- Klausen, M.; Heydorn, A.; Ragas, P.; Lambertsen, L.; Aaes-Jørgensen, A.; Molin, S.; Tolker-Nielsen, T. Biofilm Formation by Pseudomonas aeruginosa Wild Type, Flagella and Type IV Pili Mutants. Mol. Microbiol. 2003, 48, 1511–1524. [Google Scholar] [CrossRef]
- De Weger, L.A.M.; Van Loosdrecht, M.C.; Klaassen, H.E.; Lugtenberg, B. Mutational Changes in Physicochemical Cell Surface Properties of Plant-Growth-Stimulating Pseudomonas spp. Do Not Influence the Attachment Properties of the Cells Downloaded From. J. Bacteriol. 1989, 171, 2756–2761. [Google Scholar] [CrossRef]
- Houry, A.; Briandet, R.; Aymerlch, S.; Gohar, M. Involvement of Motility and Flagella in Bacillus Cereus Biofilm Formation. Microbiology 2010, 156, 1009–1018. [Google Scholar] [CrossRef]
- Merritt, P.M.; Danhorn, T.; Fuqua, C. Motility and Chemotaxis in Agrobacterium tumefaciens Surface Attachment and Biofilm Formation. Proc. J. Bacteriol. 2007, 189, 8005–8014. [Google Scholar] [CrossRef]
- Schurr, M.J. Which Bacterial Biofilm Exopolysaccharide Is Preferred, Psl or Alginate? J. Bacteriol. 2013, 195, 1623–1626. [Google Scholar] [CrossRef] [PubMed]
- Nakao, R.; Ramstedt, M.; Wai, S.N.; Uhlin, B.E. Enhanced Biofilm Formation by Escherichia coli LPS Mutants Defective in Hep Biosynthesis. PLoS ONE 2012, 7, e51241. [Google Scholar] [CrossRef]
- Makin, S.A.; Beveridge, T.J. The Influence of A-Band and B-Band Lipopolysaccharide on the Surface Characteristics and Adhesion of Pseudomonas aeruginosa to Surfaces. Microbiology 1996, 142, 299–307. [Google Scholar] [CrossRef]
- Dekkers, L.C.; Van Der Bij, A.J.; Mulders, I.H.M.; Phoelich, C.C.; Wentwoord, R.A.R.; Glandorf, D.C.M.; Wijffelman, C.A.; Lugtenberg, B.J.J. Role of the O-Antigen of Lipopolysaccharide, and Possible Roles of Growth Rate and of NADH: Ubiquinone Oxidoreductase (Nuo) in Competitive Tomato Root-Tip Colonization by Pseudomonas fluorescens WCS365. Mol. Plant-Microbe Interact. 1998, 11, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Heydorn, A.; Ersbøll, B.; Kato, J.; Hentzer, M.; Parsek, M.R.; Tolker-Nielsen, T.; Givskov, M.; Molin, S. Statistical Analysis of Pseudomonas aeruginosa Biofilm Development: Impact of Mutations in Genes Involved in Twitching Motility, Cell-to-Cell Signaling, and Stationary-Phase Sigma Factor Expression. Appl. Environ. Microbiol. 2002, 68, 2008–2017. [Google Scholar] [CrossRef] [PubMed]
- Tolker-Nielsen, T.; Brinch, U.C.; Ragas, P.C.; Andersen, J.B.; Jacobsen, C.S.; Molin, S. Development and Dynamics of Pseudomonas sp. Biofilms. J. Bacteriol. 2000, 182, 6482–6489. [Google Scholar] [CrossRef]
- Brown, M.R.W.; Williams, P. The Influence of Environment on Envelope Properties Affecting Survival of Bacteria in Infections. Annu. Rev. Microbiol. 1985, 39, 527–556. [Google Scholar] [CrossRef]
- Petrova, O.E.; Sauer, K. Sticky Situations: Key Components That Control Bacterial Surface Attachment. J. Bacteriol. 2012, 194, 2413–2425. [Google Scholar] [CrossRef]
- Marti¬nez, L.C.; Vadyvaloo, V. Mechanisms of Post-Transcriptional Gene Regulation in Bacterial Biofilms. Front. Cell Infect. Microbiol. 2014, 4, 1–15. [Google Scholar] [CrossRef]
- Whitchurch, C.B.; Erova, T.E.; Emery, J.A.; Sargent, J.L.; Harris, J.M.; Semmler, A.B.T.; Young, M.D.; Mattick, J.S.; Wozniak, D.J. Phosphorylation of the Pseudomonas aeruginosa Response Regulator AlgR Is Essential for Type IV Fimbria-Mediated Twitching Motility. J. Bacteriol. 2002, 184, 4544–4554. [Google Scholar] [CrossRef]
- Friedman, L.; Kolter, R. Two Genetic Loci Produce Distinct Carbohydrate-Rich Structural Components of the Pseudomonas aeruginosa Biofilm Matrix. J. Bacteriol. 2004, 186, 4457–4465. [Google Scholar] [CrossRef]
- Ryder, C.; Byrd, M.; Wozniak, D.J. Role of Polysaccharides in Pseudomonas aeruginosa Biofilm Development. Curr. Opin. Microbiol. 2007, 10, 644–648. [Google Scholar] [CrossRef]
- Karatan, E.; Watnick, P. Signals, Regulatory Networks, and Materials That Build and Break Bacterial Biofilms. Microbiol. Mol. Biol. Rev. 2009, 73, 310–347. [Google Scholar] [CrossRef]
- Caiazza, N.C.; Merritt, J.H.; Brothers, K.M.; O’Toole, G.A. Inverse Regulation of Biofilm Formation and Swarming Motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 2007, 189, 3603–3612. [Google Scholar] [CrossRef]
- Haas, D.; Keel, C. Regulation of Antibiotic Production in Root-Colonizing Peudomonas spp. and Relevance for Biological Control of Plant Disease. Annu. Rev. Phytopathol. 2003, 41, 117–153. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; de Bruijn, I.; Nybroe, O.; Ongena, M. Natural Functions of Lipopeptides from Bacillus and Pseudomonas: More than Surfactants and Antibiotics. FEMS Microbiol. Rev. 2010, 34, 1037–1062. [Google Scholar] [CrossRef]
- Duque, E.; de la Torre, J.; Bernal, P.; Molina-Henares, M.A.; Alaminos, M.; Espinosa-Urgel, M.; Roca, A.; Fernández, M.; de Bentzmann, S.; Ramos, J.L. Identification of Reciprocal Adhesion Genes in Pathogenic and Non-Pathogenic Pseudomonas. Environ. Microbiol. 2013, 15, 36–48. [Google Scholar] [CrossRef]
- Cheng, X.; de Bruijn, I.; van der Voort, M.; Loper, J.E.; Raaijmakers, J.M. The Gac Regulon of Pseudomonas fluorescens SBW25. Environ. Microbiol. Rep. 2013, 5, 608–619. [Google Scholar] [CrossRef]
- Koch, M.D.; Black, M.E.; Han, E.; Shaevitz, J.W.; Gitai, Z. Pseudomonas aeruginosa Distinguishes Surfaces by Stiffness Using Retraction of Type IV Pili. Proc. Natl. Acad. Sci. USA 2022, 119, e2119434119. [Google Scholar] [CrossRef]
- Blanco-Romero, E.; Garrido-Sanz, D.; Durán, D.; Rybtke, M.; Tolker-Nielsen, T.; Redondo-Nieto, M.; Rivilla, R.; Martín, M. Role of Extracellular Matrix Components in Biofilm Formation and Adaptation of Pseudomonas ogarae F113 to the Rhizosphere Environment. Front. Microbiol. 2024, 15, 1728. [Google Scholar] [CrossRef]
- Bais, H.P.; Fall, R.; Vivanco, J.M. Biocontrol of Bacillus subtilis against Infection of Arabidopsis Roots by Pseudomonas syringae Is Facilitated by Biofilm Formation and Surfactin Production. Plant Physiol. 2004, 134, 307–319. [Google Scholar] [CrossRef]
- Timmusk, S.; Grantcharova, N.; Wagner, E.G.H. Paenibacillus Polymyxa Invades Plant Roots and Forms Biofilms. Appl. Environ. Microbiol. 2005, 71, 7292–7300. [Google Scholar] [CrossRef]
- Fujishige, N.A.; Kapadia, N.N.; De Hoff, P.L.; Hirsch, A.M. Investigations of Rhizobium Biofilm Formation. FEMS Microbiol. Ecol. 2006, 56, 195–206. [Google Scholar] [CrossRef]
- Ude, S.; Arnold, D.L.; Moon, C.D.; Timms-Wilson, T.; Spiers, A.J. Biofilm Formation and Cellulose Expression among Diverse Environmental Pseudomonas Isolates. Environ. Microbiol. 2006, 8, 1997–2011. [Google Scholar] [CrossRef]
- Rudrappa, T.; Splaine, R.E.; Biedrzycki, M.L.; Bais, H.P. Cyanogenic Pseudomonads Influence Multitrophic Interactions in the Rhizosphere. PLoS ONE 2008, 3, e2073. [Google Scholar] [CrossRef]
- Van de Broek, A.; Lambrecht, M.; Vanderleyden, J. Bacterial Chemotactic Motility Is Important for the Initiation of Wheat Root Colonization by Azospirillum brasilense. Microbiology 1998, 144, 2599–2606. [Google Scholar] [CrossRef]
- Ramos, C.; Mølbak, L.; Molin, S. Bacterial Activity in the Rhizosphere Analyzed at the Single-Cell Level by Monitoring Ribosome Contents and Synthesis Rates. Appl. Environ. Microbiol. 2000, 66, 801–809. [Google Scholar] [CrossRef]
- Yousef-Coronado, F.; Travieso, M.L.; Espinosa-Urgel, M. Different, Overlapping Mechanisms for Colonization of Abiotic and Plant Surfaces by Pseudomonas putida. FEMS Microbiol. Lett. 2008, 288, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Fuqua, C. Passing the Baton between Laps: Adhesion and Cohesion in Pseudomonas putida Biofilms: MicroCommentary. Mol. Microbiol. 2010, 77, 533–536. [Google Scholar] [CrossRef]
- Barahona, E.; Navazo, A.; Yousef-Coronado, F.; Aguirre De Cárcer, D.; Martínez-Granero, F.; Espinosa-Urgel, M.; Martín, M.; Rivilla, R. Efficient Rhizosphere Colonization by Pseudomonas fluorescens F113 Mutants Unable to Form Biofilms on Abiotic Surfaces. Environ. Microbiol. 2010, 12, 3185–3195. [Google Scholar] [CrossRef]
- Espinosa-Urgel, M.; Salido, A.; Ramos, J.-L. Genetic Analysis of Functions Involved in Adhesion of Pseudomonas putida to Seeds. J. Bacteriol. 2000, 182, 2363–2369. [Google Scholar] [CrossRef]
- Krzyzanowska, D.M.; Ossowicki, A.; Rajewska, M.; Maciag, T.; Jablonska, M.; Obuchowski, M.; Heeb, S.; Jafra, S. When Genome-Based Approach Meets the “Old but Good”: Revealing Genes Involved in the Antibacterial Activity of Pseudomonas sp. P482 against Soft Rot Pathogens. Front. Microbiol. 2016, 7, 782. [Google Scholar] [CrossRef]
- Ossowicki, A.; Jafra, S.; Garbeva, P. The Antimicrobial Volatile Power of the Rhizospheric Isolate Pseudomonas donghuensis P482. PLoS ONE 2017, 12, e0174362. [Google Scholar] [CrossRef]
- Matuszewska, M.; Maciąg, T.; Rajewska, M.; Wierzbicka, A.; Jafra, S. The Carbon Source-Dependent Pattern of Antimicrobial Activity and Gene Expression in Pseudomonas donghuensis P482. Sci. Rep. 2021, 11, 10994. [Google Scholar] [CrossRef]
- Matuszewska, M. Investigation of the Influence of Glucose, Glycerol and Iron Ions on the Synthesis of Antimicrobials and Formation of Biofilm by Pseudomonas donghuensis P482. Ph.D. Thesis, University of Gdańsk, Gdańsk, Poland, 2023. [Google Scholar]
- Krzyzanowska, D.; Obuchowski, M.; Bikowski, M.; Rychlowski, M.; Jafra, S. Colonization of Potato Rhizosphere by GFP-Tagged Bacillus subtilis MB73/2, Pseudomonas Sp. P482 and Ochrobactrum Sp. A44 Shown on Large Sections of Roots Using Enrichment Sample Preparation and Confocal Laser Scanning Microscopy. Sensors 2012, 12, 17608–17619. [Google Scholar] [CrossRef]
- Krzyżanowska, D.M.; Jabłońska, M.; Kaczyński, Z.; Czerwicka-Pach, M.; Macur, K.; Jafra, S. Host-Adaptive Traits in the Plant-Colonizing Pseudomonas donghuensis P482 Revealed by Transcriptomic Responses to Exudates of Tomato and Maize. Sci. Rep. 2023, 13, 9445. [Google Scholar] [CrossRef]
- Gao, J.; Yu, X.; Xie, Z. Draft Genome Sequence of High-Siderophore-Yielding Pseudomonas sp. Strain HYS. J. Bacteriol. 2012, 194, 4121. [Google Scholar] [CrossRef]
- Agaras, B.C.; Iriarte, A.; Valverde, C.F. Genomic Insights into the Broad Antifungal Activity, Plant-Probiotic Properties, and Their Regulation, in Pseudomonas donghuensis Strain SVBP6. PLoS ONE 2018, 13, e0194088. [Google Scholar] [CrossRef]
- Marin-Bruzos, M.; Grayston, S.J.; Forge, T.; Nelson, L.M. Isolation and Characterization of Streptomycetes and Pseudomonad Strains with Antagonistic Activity against the Plant Parasitic Nematode Pratylenchus penetrans and Fungi Associated with Replant Disease. Biol. Control. 2021, 158, 104599. [Google Scholar] [CrossRef]
- Ghafoor, A.; Hay, I.D.; Rehm, B.H.A. Role of Exopolysaccharides in Pseudomonas aeruginosa Biofilm Formation and Architecture. Appl. Environ. Microbiol. 2011, 77, 5238–5246. [Google Scholar] [CrossRef]
- Bogino, P.C.; de las Oliva, M.M.; Sorroche, F.G.; Giordano, W. The Role of Bacterial Biofilms and Surface Components in Plant-Bacterial Associations. Int. J. Mol. Sci. 2013, 14, 15838–15859. [Google Scholar] [CrossRef]
- Maunders, E.; Welch, M. Matrix Exopolysaccharides; the Sticky Side of Biofilm Formation. FEMS Microbiol. Lett. 2017, 364, fnx120. [Google Scholar] [CrossRef]
- Wang, B.X.; Cady, K.C.; Oyarce, G.C.; Ribbeck, K.; Lauba, M.T. Two-Component Signaling Systems Regulate Diverse Virulence-Associated Traits in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2021, 87, 1–18. [Google Scholar] [CrossRef]
- Wei, Q.; Ma, L.Z. Biofilm Matrix and Its Regulation in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2013, 14, 20983–21005. [Google Scholar] [CrossRef]
- Li, J.; Yang, Y.; Dubern, J.F.; Li, H.; Halliday, N.; Chernin, L.; Gao, K.; Cámara, M.; Liu, X. Regulation of GacA in Pseudomonas chlororaphis Strains Shows a Niche Specificity. PLoS ONE 2015, 10, e0137553. [Google Scholar] [CrossRef]
- Ramey, B.E.; Koutsoudis, M.; Bodman, S.B.V.; Fuqua, C. Biofilm Formation in Plant-Microbe Associations. Curr. Opin. Microbiol. 2004, 7, 602–609. [Google Scholar] [CrossRef]
- Albareda, M.; Dardanelli, M.S.; Sousa, C.; Megías, M.; Temprano, F.; Rodríguez-Navarro, D.N. Factors Affecting the Attachment of Rhizospheric Bacteria to Bean and Soybean Roots. FEMS Microbiol. Lett. 2006, 259, 67–73. [Google Scholar] [CrossRef]
- Danhorn, T.; Fuqua, C. Biofilm Formation by Plant-Associated Bacteria. Annu. Rev. Microbiol. 2007, 61, 401–422. [Google Scholar] [CrossRef]
- Espinosa-Urgel, M.; Ramos-González, M.I. Becoming Settlers: Elements and Mechanisms for Surface Colonization by Pseudomonas putida. Environ. Microbiol. 2023, 25, 1575–1593. [Google Scholar] [CrossRef]
- Blanco-Romero, E.; Redondo-Nieto, M.; Martínez-Granero, F.; Garrido-Sanz, D.; Ramos-González, M.I.; Martín, M.; Rivilla, R. Genome-Wide Analysis of the FleQ Direct Regulon in Pseudomonas fluorescens F113 and Pseudomonas putida KT2440. Sci. Rep. 2018, 8, 13145. [Google Scholar] [CrossRef]
- Zboralski, A.; Filion, M. Genetic Factors Involved in Rhizosphere Colonization by Phytobeneficial Pseudomonas spp. Comput. Struct. Biotechnol. J. 2020, 18, 3539–3554. [Google Scholar] [CrossRef]
- Ajijah, N.; Fiodor, A.; Pandey, A.K.; Rana, A.; Pranaw, K. Plant Growth-Promoting Bacteria (PGPB) with Biofilm-Forming Ability: A Multifaceted Agent for Sustainable Agriculture. Diversity 2023, 15, 112. [Google Scholar] [CrossRef]
- Costa-Gutierrez, S.B.; Adler, C.; Espinosa-Urgel, M.; de Cristóbal, R.E. Pseudomonas putida and Its Close Relatives: Mixing and Mastering the Perfect Tune for Plants. Appl. Microbiol. Biotechnol. 2022, 106, 3351–3367. [Google Scholar] [CrossRef]
- Rossi, C.S.; Barrientos-Moreno, L.; Paone, A.; Cutruzzolà, F.; Paiardini, A.; Espinosa-Urgel, M.; Rinaldo, S. Nutrient Sensing and Biofilm Modulation: The Example of L-Arginine in Pseudomonas. Int. J. Mol. Sci. 2022, 23, 4386. [Google Scholar] [CrossRef]
- Sauer, K.; Cullen, M.C.; Rickard, A.H.; Zeef, L.A.H.; Davies, D.G.; Gilbert, P. Characterization of Nutrient-Induced Dispersion in Pseudomonas aeruginosa PAO1 Biofilm. J. Bacteriol. 2004, 186, 7312–7326. [Google Scholar] [CrossRef]
- Harmsen, M.; Yang, L.; Pamp, S.J.; Tolker-Nielsen, T. An Update on Pseudomonas aeruginosa Biofilm Formation, Tolerance, and Dispersal. FEMS Immunol. Med. Microbiol. 2010, 59, 253–268. [Google Scholar] [CrossRef]
- Huynh, T.T.; McDougald, D.; Klebensberger, J.; Al Qarni, B.; Barraud, N.; Rice, S.A.; Kjelleberg, S.; Schleheck, D. Glucose Starvation-Induced Dispersal of Pseudomonas aeruginosa Biofilms Is Camp and Energy Dependent. PLoS ONE 2012, 7, e42874. [Google Scholar] [CrossRef]
- Scoffield, J.; Silo-Suh, L. Glycerol Metabolism Promotes Biofilm Formation by Pseudomonas aeruginosa. Can. J. Microbiol. 2016, 62, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Deutscher, J. The Mechanisms of Carbon Catabolite Repression in Bacteria. Curr. Opin. Microbiol. 2008, 11, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Görke, B.; Stülke, J. Carbon Catabolite Repression in Bacteria: Many Ways to Make the Most out of Nutrients. Nat. Rev. Microbiol. 2008, 6, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Rojo, F. Carbon Catabolite Repression in Pseudomonas: Optimizing Metabolic Versatility and Interactions with the Environment. FEMS Microbiol. Rev. 2010, 34, 658–684. [Google Scholar] [CrossRef] [PubMed]
- Bruckner, R.; Titgemeyer, F. Carbon Catabolite Repression in Bacteria: Choice of the Carbon Source and Autoregulatory Limitation of Sugar Utilization. FEMS Microbiol. Lett. 2002, 209, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Zeng, M.; You, J.; Xie, Z. Pseudomonas Donghuensis HYS Virulence towards Caenorhabditis elegans Is Regulated by the Cbr/Crc System. Sci. Rep. 2019, 9, 8772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Gao, Q.; Chen, W.; Qin, H.; Hengzhuang, W.; Chen, Y.; Yang, L.; Zhang, G. Regulation of PQS Quorum Sensing via Catabolite Repression Control in Pseudomonas aeruginosa. Microbiology 2013, 159, 1931–1936. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The Involvement of Cell-to-Cell Signals in the Development of a Bacterial Biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Nivens, D.E.; Ohman, D.E.; Williams, J.; Franklin, M.J. Role of Alginate and Its O Acetylation in Formation of Pseudomonas aeruginosa Microcolonies and Biofilms. J. Bacteriol. 2001, 183, 1047–1057. [Google Scholar] [CrossRef]
- Dumitrache, A.; Klingeman, D.M.; Natzke, J.; Rodriguez, M.; Giannone, R.J.; Hettich, R.L.; Davison, B.H.; Brown, S.D. Specialized Activities and Expression Differences for Clostridium thermocellum Biofilm and Planktonic Cells. Sci. Rep. 2017, 7, 43583. [Google Scholar] [CrossRef]
- Park, S.; Park, Y.H.; Lee, C.R.; Kim, Y.R.; Seok, Y.J. Glucose Induces Delocalization of a Flagellar Biosynthesis Protein from the Flagellated Pole. Mol. Microbiol. 2016, 101, 795–808. [Google Scholar] [CrossRef]
- Ozer, E.; Yaniv, K.; Chetrit, E.; Boyarski, A.; Meijler, M.M.; Berkovich, R.; Kushmaro, A.; Alfonta, L. An inside Look at a Biofilm: Pseudomonas aeruginosa Flagella Biotracking. Sci. Adv. 2021, 7, 8581–8592. [Google Scholar] [CrossRef] [PubMed]
- Conrad, J.C. Physics of Bacterial Near-Surface Motility Using Flagella and Type IV Pili: Implications for Biofilm Formation. Res. Microbiol. 2012, 163, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Burrows, L.L. Pseudomonas aeruginosa Twitching Motility: Type IV Pili in Action. Annu. Rev. Microbiol. 2012, 66, 493–520. [Google Scholar] [CrossRef] [PubMed]
- Fazli, M.; Almblad, H.; Rybtke, M.L.; Givskov, M.; Eberl, L.; Tolker-Nielsen, T. Regulation of Biofilm Formation in Pseudomonas and Burkholderia Species. Environ. Microbiol. 2014, 16, 1961–1981. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.; Silistre, H.; Lovelock, L.; Wright, V.J.; Chan, K.G.; Hong, K.W.; Williams, P.; Cámara, M.; Heeb, S. Genome-Wide Mapping of the RNA Targets of the Pseudomonas aeruginosa Riboregulatory Protein RsmN. Nucleic Acids Res. 2018, 46, 6823–6840. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Contreras, M.; Martín, M.; Villacieros, M.; O’Gara, F.; Bonilla, I.; Rivilla, R. Phenotypic Selection and Phase Variation Occur during Alfalfa Root Colonization by Pseudomonas fluorescens F113. J. Bacteriol. 2002, 184, 1587–1596. [Google Scholar] [CrossRef] [PubMed]
- Kay, E.; Humair, B.; Dénervaud, V.; Riedel, K.; Spahr, S.; Eberl, L.; Valverde, C.; Haas, D. Two GacA-Dependent Small RNAs Modulate the Quorum-Sensing Response in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 6026–6033. [Google Scholar] [CrossRef] [PubMed]
- Mawla, G.D. Functions of Alternative ClpP Subunits in Pseudomonas aeruginosa. Ph.D. Thesis, Massachusetts Institute of Technology, Boston, MA, USA, 2020. [Google Scholar]
- Hall, B.M.; Breidenstein, E.B.M.; de la Fuente-Núñez, C.; Reffuveille, F.; Mawla, G.D.; Hancock, R.E.W.; Baker, T.A. Two Isoforms of Clp Peptidase in Pseudomonas aeruginosa Control Distinct Aspects of Cellular Physiology. J. Bacteriol. 2017, 199, 10–1128. [Google Scholar] [CrossRef]
- Mawla, G.D.; Hall, B.M.; Cárcamo-Oyarce, G.; Grant, R.A.; Zhang, J.J.; Kardon, J.R.; Ribbeck, K.; Sauer, R.T.; Baker, T.A. ClpP1P2 Peptidase Activity Promotes Biofilm Formation in Pseudomonas aeruginosa. Mol. Microbiol. 2021, 115, 1094–1109. [Google Scholar] [CrossRef]
- Marr, A.K.; Overhage, J.; Bains, M.; Hancock, R.E.W. The Lon Protease of Pseudomonas aeruginosa Is Induced by Aminoglycosides and Is Involved in Biofilm Formation and Motility. Microbiology 2007, 153, 474–482. [Google Scholar] [CrossRef]
- Miller, C.G. Protein Degradation and Proteolytic Modification. In Escherichia coli and Salmonella: Cellular and Molecular Biology; Neidhardt, F.C., Ed.; American Society for Microbiology: Washington, DC, USA, 1996; Volume 1, pp. 938–954. [Google Scholar]
- Gerth, U.; Kock, H.; Kusters, I.; Michalik, S.; Switzer, R.L.; Hecker, M. Clp-Dependent Proteolysis down-Regulates Central Metabolic Pathways in Glucose-Starved Bacillus subtilis. J. Bacteriol. 2008, 190, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Molière, N.; Hoßmann, J.; Schäfer, H.; Turgay, K. Role of Hsp100/Clp Protease Complexes in Controlling the Regulation of Motility in Bacillus subtilis. Front. Microbiol. 2016, 7, 315. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; van Hullebusch, E.D.; Neu, T.R.; Nielsen, P.H.; Seviour, T.; Stoodley, P.; Wingender, J.; Wuertz, S. The Biofilm Matrix: Multitasking in a Shared Space. Nat. Rev. Microbiol. 2023, 21, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J. The Biofilm Matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Ma, L.; Jackson, K.D.; Landry, R.M.; Parsek, M.R.; Wozniak, D.J. Analysis of Pseudomonas aeruginosa Conditional Psl Variants Reveals Roles for the Psl Polysaccharide in Adhesion and Maintaining Biofilm Structure Postattachment. J. Bacteriol. 2006, 188, 8213–8221. [Google Scholar] [CrossRef]
- Vassoler Serrato, R. Bacterial Alginate Biosynthesis and Metabolism. In Alginate—Applications and Future Perspectives; Severo, I.A., Mariano, A.B., Vargas, J.V.C., Eds.; IntechOpen: London, UK, 2022. [Google Scholar]
- Bertani, B.; Ruiz, N. Function and Biogenesis of Lipopolysaccharides. Ecosal Plus 2018, 8. [Google Scholar] [CrossRef]
- Figaj, D.; Ambroziak, P.; Przepiora, T.; Skorko-Glonek, J. The Role of Proteases in the Virulence of Plant Pathogenic Bacteria. Int. J. Mol. Sci. 2019, 20, 672. [Google Scholar] [CrossRef] [PubMed]
- She, P.; Wang, Y.; Liu, Y.; Tan, F.; Chen, L.; Luo, Z.; Wu, Y. Effects of Exogenous Glucose on Pseudomonas aeruginosa Biofilm Formation and Antibiotic Resistance. Microbiol. Open 2019, 8, e933. [Google Scholar] [CrossRef]
- Colvin, K.M.; Irie, Y.; Tart, C.S.; Urbano, R.; Whitney, J.C.; Ryder, C.; Howell, P.L.; Wozniak, D.J.; Parsek, M.R. The Pel and Psl Polysaccharides Provide Pseudomonas aeruginosa Structural Redundancy within the Biofilm Matrix. Environ. Microbiol. 2012, 14, 1913–1928. [Google Scholar] [CrossRef]
- Palmer, J.; Flint, S.; Brooks, J. Bacterial Cell Attachment, the Beginning of a Biofilm. J. Ind. Microbiol. Biotechnol. 2007, 34, 577–588. [Google Scholar] [CrossRef]
- Hook, A.L.; Chang, C.Y.; Yang, J.; Luckett, J.; Cockayne, A.; Atkinson, S.; Mei, Y.; Bayston, R.; Irvine, D.J.; Langer, R.; et al. Combinatorial Discovery of Polymers Resistant to Bacterial Attachment. Nat. Biotechnol. 2012, 30, 868–875. [Google Scholar] [CrossRef]
- Romero, M.; Luckett, J.; Figueredo, G.P.; Carabelli, A.M.; Scurr, D.; Hook, A.L.; Dubern, J.-F.; Ison, E.; Kammerling, L.; da Silva, A.C.; et al. Micro Topographical Instruction of Bacterial Attachment, Biofilm Formation and in Vivo Host Response. Prepr. BioRxiv 2022. [Google Scholar] [CrossRef]
- Kimkes, T.E.P.; Heinemann, M. How Bacteria Recognise and Respond to Surface Contact. FEMS Microbiol. Rev. 2019, 44, 106–122. [Google Scholar] [CrossRef]
- De-la-Pinta, I.; Cobos, M.; Ibarretxe, J.; Montoya, E.; Eraso, E.; Guraya, T.; Quindós, G. Effect of Biomaterials Hydrophobicity and Roughness on Biofilm Development. J. Mater. Sci. Mater. Med. 2019, 30, 1–11. [Google Scholar] [CrossRef]
- Loo, C.Y.; Young, P.M.; Lee, W.H.; Cavaliere, R.; Whitchurch, C.B.; Rohanizadeh, R. Superhydrophobic, Nanotextured Polyvinyl Chloride Films for Delaying Pseudomonas aeruginosa Attachment to Intubation Tubes and Medical Plastics. Acta Biomater. 2012, 8, 1881–1890. [Google Scholar] [CrossRef]
- Ozkan, E.; Mondal, A.; Singha, P.; Douglass, M.; Hopkins, S.P.; Devine, R.; Garren, M.; Manuel, J.; Warnock, J.; Handa, H. Fabrication of Bacteria- and Blood-Repellent Superhydrophobic Polyurethane Sponge Materials. ACS Appl. Mater. Interfaces 2020, 12, 51160–51173. [Google Scholar] [CrossRef]
- Montgomerie, Z.; Popat, K.C. Improved Hemocompatibility and Reduced Bacterial Adhesion on Superhydrophobic Titania Nanoflower Surfaces. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 119, 111503. [Google Scholar] [CrossRef]
- Zheng, S.; Bawazir, M.; Dhall, A.; Kim, H.E.; He, L.; Heo, J.; Hwang, G. Implication of Surface Properties, Bacterial Motility, and Hydrodynamic Conditions on Bacterial Surface Sensing and Their Initial Adhesion. Front. Bioeng. Biotechnol. 2021, 9, 643722. [Google Scholar] [CrossRef]
- Alexander, M.R.; Williams, P. Water Contact Angle Is Not a Good Predictor of Biological Responses to Materials. Biointerphases 2017, 12, 9843. [Google Scholar] [CrossRef]
- Capdevila, S.; Martínez-Granero, F.M.; Sánchez-Contreras, M.; Rivilla, R.; Martín, M. Analysis of Pseudomonas fluorescens F113 Genes Implicated in Flagellar Filament Synthesis and Their Role in Competitive Root Colonization. Microbiology 2004, 150, 3889–3897. [Google Scholar] [CrossRef]
- Rudrappa, T.; Biedrzycki, M.L.; Bais, H.P. Causes and Consequences of Plant-Associated Biofilms. FEMS Microbiol. Ecol. 2008, 64, 153–166. [Google Scholar] [CrossRef]
- Lugtenberg, B.J.J.; Kravchenko, L.V.; Simons, M. Tomato Seed and Root Exudate Sugars: Composition, Utilization by Pseudomonas Biocontrol Strains and Role in Rhizosphere Colonization. Environ. Microbiol. 1999, 1, 439–446. [Google Scholar] [CrossRef]
- Walker, T.S.; Bais, H.P.; Déziel, E.; Schweizer, H.P.; Rahme, L.G.; Fall, R.; Vivanco, J.M. Pseudomonas aeruginosa-Plant Root Interactions. Pathogenicity, Biofilm Formation, and Root Exudation. Plant Physiol. 2004, 134, 320–331. [Google Scholar] [CrossRef]
- De Weert, S.; Vermeiren, H.; Mulders, I.H.M.; Kuiper, I.; Hendrickx, N.; Bloemberg, G.V.; Vanderleyden, J.; De Mot, R.; Lugtenberg, B.J.J. Flagella-Driven Chemotaxis Towards Exudate Components Is an Important Trait for Tomato Root Colonization by Pseudomonas fluorescens. Mol. Plant Microbe Interact. 2002, 15, 1173–1180. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; Volume 1. [Google Scholar]
- Thoma, S.; Schobert, M. An Improved Escherichia Coli Donor Strain for Diparental Mating. FEMS Microbiol. Lett. 2009, 294, 127–132. [Google Scholar] [CrossRef]
- Krzyżanowska, D.M.; Maciąg, T.; Ossowicki, A.; Rajewska, M.; Kaczyński, Z.; Czerwicka, M.; Rąbalski, Ł.; Czaplewska, P.; Jafra, S. Ochrobactrum quorumnocens Sp. Nov., a Quorum Quenching Bacterium from the Potato Rhizosphere, and Comparative Genome Analysis with Related Type Strains. PLoS ONE 2019, 14, e0210874. [Google Scholar] [CrossRef]
- Alexeyev, M.F. The PKNOCK Series of Broad-Host-Range Mobilizable Suicide Vectors for Gene Knockout and Targeted DNA Insertion into the Chromosome of Gram-Negative Bacteria. Biotechniques 1999, 26, 824–828. [Google Scholar] [CrossRef]
- Belas, R. Biofilms, Flagella, and Mechanosensing of Surfaces by Bacteria. Trends Microbiol. 2014, 22, 517–527. [Google Scholar] [CrossRef]
- Sampedro, I.; Parales, R.E.; Krell, T.; Hill, J.E. Pseudomonas Chemotaxis. FEMS Microbiol. Rev. 2015, 39, 17–46. [Google Scholar] [CrossRef]
- Bouteiller, M.; Dupont, C.; Bourigault, Y.; Latour, X.; Barbey, C.; Konto-ghiorghi, Y.; Merieau, A. Pseudomonas Flagella: Generalities and Specificities. Int. J. Mol. Sci. 2021, 22, 3337. [Google Scholar] [CrossRef] [PubMed]
- Heeb, S.; Haas, D. Regulatory Roles of the GacS/GacA Two-Component System in Plant-Associated and Other Gram-Negative Bacteria. Mol. Plant Microbe Interact. 2001, 14, 1351–1363. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.G.; Leveau, J.H.J.; Lindow, S.E. Improved Gfp and InaZ Broad-Host-Range Promoter-Probe Vectors. Mol. Plant Microbe Interact. 2000, 13, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2010, 47, e2437. [Google Scholar] [CrossRef]
- Ha, D.-G.; Kuchma, S.L.; O’Toole, G.A. Plate-Based Assay for Swimming Motility in Pseudomonas aeruginosa. Methods Mol. Biol. 2014, 1149, 59–65. [Google Scholar] [CrossRef]
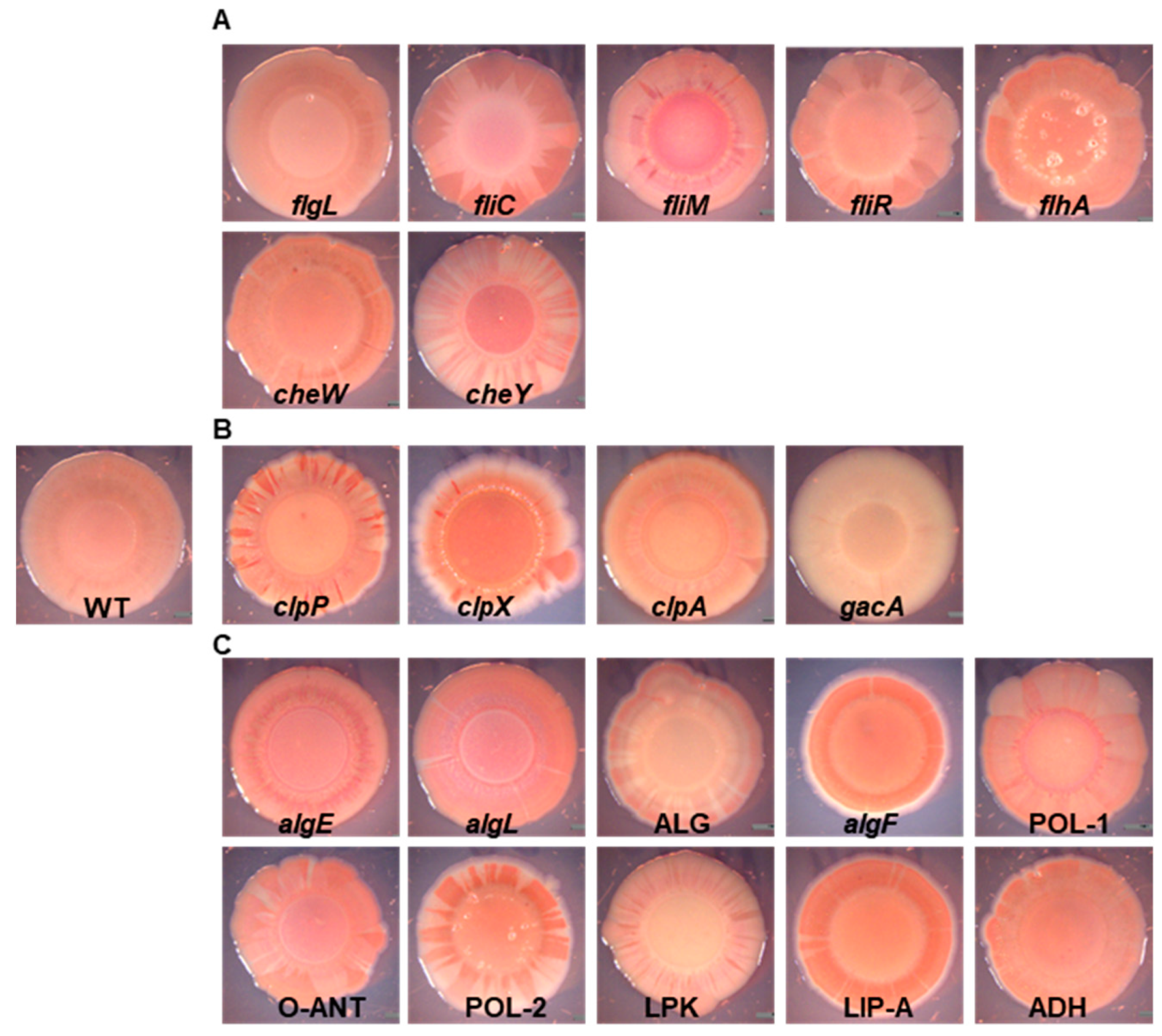



| Mutant’s Name | Locus | Annotated Gene | Product | Function in Biofilm Formation |
|---|---|---|---|---|
| flgL | BV82_0864 | flgL | flagellar hook-associated protein 3 | Motility/Chemotaxis |
| fliC | BV82_0868 | fliC | bacterial flagellin N-terminal helical region family protein | |
| fliM | BV82_0887 | fliM | flagellar motor switch protein FliM | |
| fliR | BV82_0892 | fliR | flagellar biosynthetic protein FliR | |
| flhA | BV82_0894 | flhA | flagellar biosynthesis protein FlhA | |
| cheW | BV82_0850 | cheW | chemotaxis protein CheW | |
| cheY | BV82_0883 | cheY | chemotaxis protein CheY | |
| clpP | BV82_1101 | clpP | ATP-dependent Clp protease proteolytic subunit ClpP | Proteases/Regulatory |
| clpX | BV82_1102 | clpX | ATP-dependent Clp protease ATP-binding subunit ClpX | |
| clpA | BV82_1251 | clpA | ATP-dependent Clp protease ATP-binding subunit ClpA | |
| gacA | BV82_3318 | gacA | GacA, a response regulator of the two-component GacS/GacA system | |
| algE | BV82_4694 | algE | alginate production protein AlgE | Biofilm matrix/Adhesion |
| algL | BV82_4697 | algL | alginate lyase | |
| ALG | BV82_4698 | - | alginate O-acetyltransferase | |
| algF | BV82_4700 | algF | alginate O-acetyltransferase AlgF | |
| POL-1 | BV82_5088 | - | polysaccharide biosynthesis family protein | |
| O-ANT | BV82_5092 | - | O-antigen ligase family protein | |
| POL-2 | BV82_5095 | - | polysaccharide biosynthesis/export family protein | |
| LPS | BV82_1850 | - | lipopolysaccharide kinase family protein | |
| LIP-A | BV82_0543 | - | lipid-A-disaccharide synthase | |
| ADH | BV82_3824 | - | adhesin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajewska, M.; Maciąg, T.; Narajczyk, M.; Jafra, S. Carbon Source and Substrate Surface Affect Biofilm Formation by the Plant-Associated Bacterium Pseudomonas donghuensis P482. Int. J. Mol. Sci. 2024, 25, 8351. https://doi.org/10.3390/ijms25158351
Rajewska M, Maciąg T, Narajczyk M, Jafra S. Carbon Source and Substrate Surface Affect Biofilm Formation by the Plant-Associated Bacterium Pseudomonas donghuensis P482. International Journal of Molecular Sciences. 2024; 25(15):8351. https://doi.org/10.3390/ijms25158351
Chicago/Turabian StyleRajewska, Magdalena, Tomasz Maciąg, Magdalena Narajczyk, and Sylwia Jafra. 2024. "Carbon Source and Substrate Surface Affect Biofilm Formation by the Plant-Associated Bacterium Pseudomonas donghuensis P482" International Journal of Molecular Sciences 25, no. 15: 8351. https://doi.org/10.3390/ijms25158351






