Plantaginis Semen Ameliorates Hyperuricemia Induced by Potassium Oxonate
Abstract
:1. Introduction
2. Results
2.1. Clarification of Constituents in PS by UPLC-QE-Orbitrap-MS
2.2. Identification of Blood-Migrating Constituents in PS by UPLC-QE-Orbitrap-MS
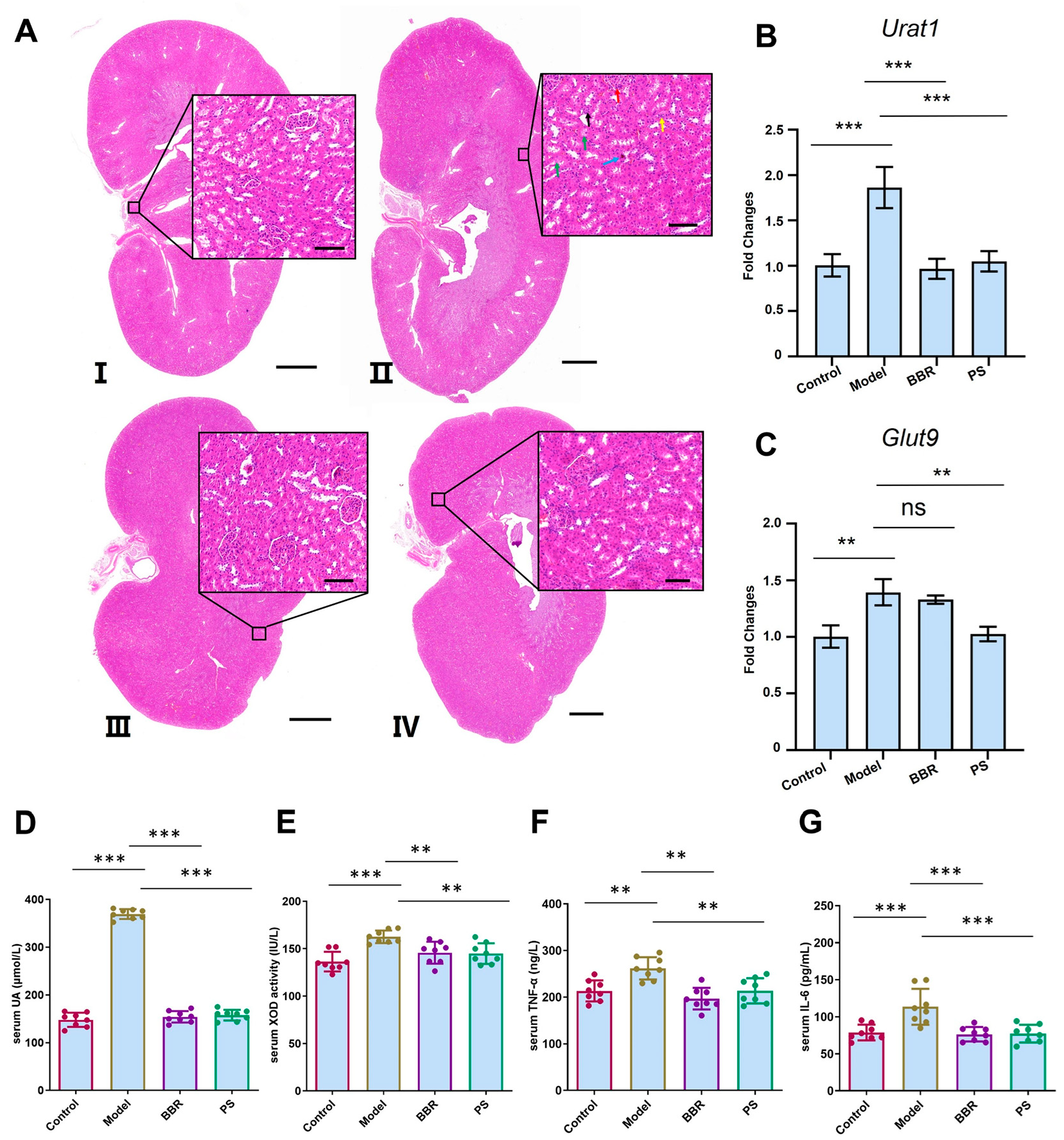
2.3. Plantaginis Semen Ethanol Extract Reversed Rat Urine Acid, Cytokines Levels, and mRNA Expression of Uric Acid Transporter Induced by Potassium Oxonate
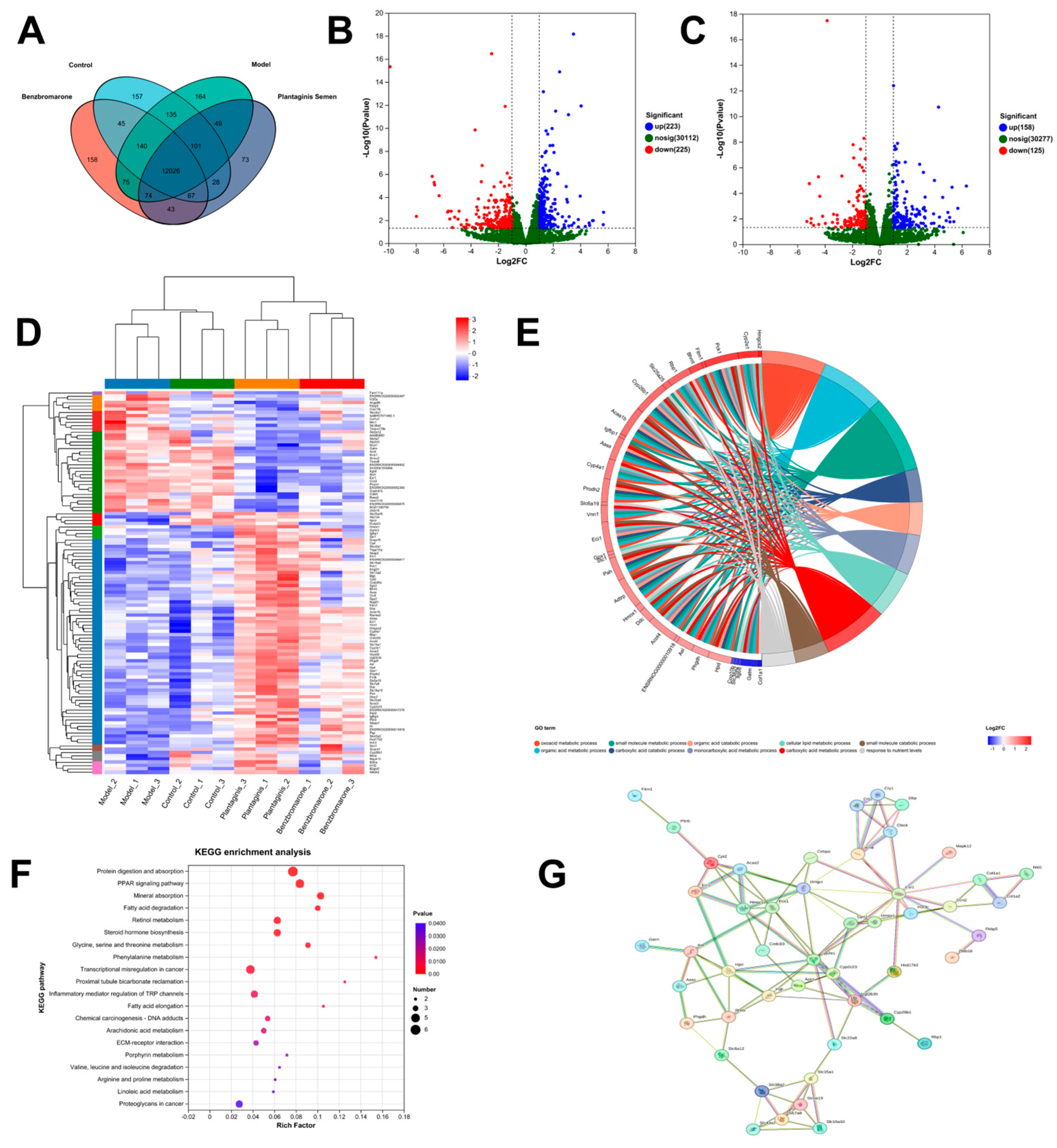
2.4. Differential Expression Genes and Related Pathways Analyzed by RNA Sequencing
2.5. Differential Expression Genes Were Verified by RT-qPCR
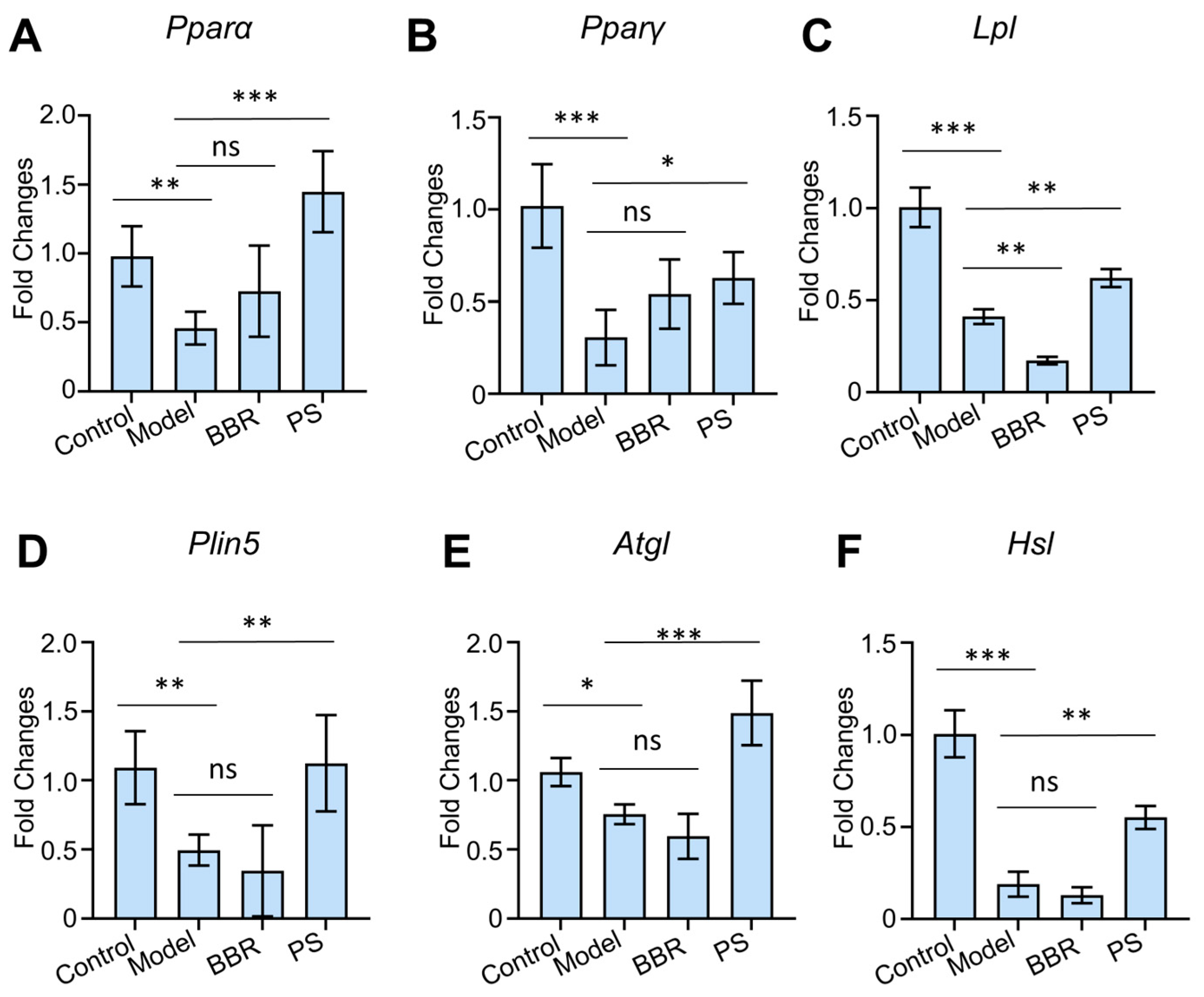
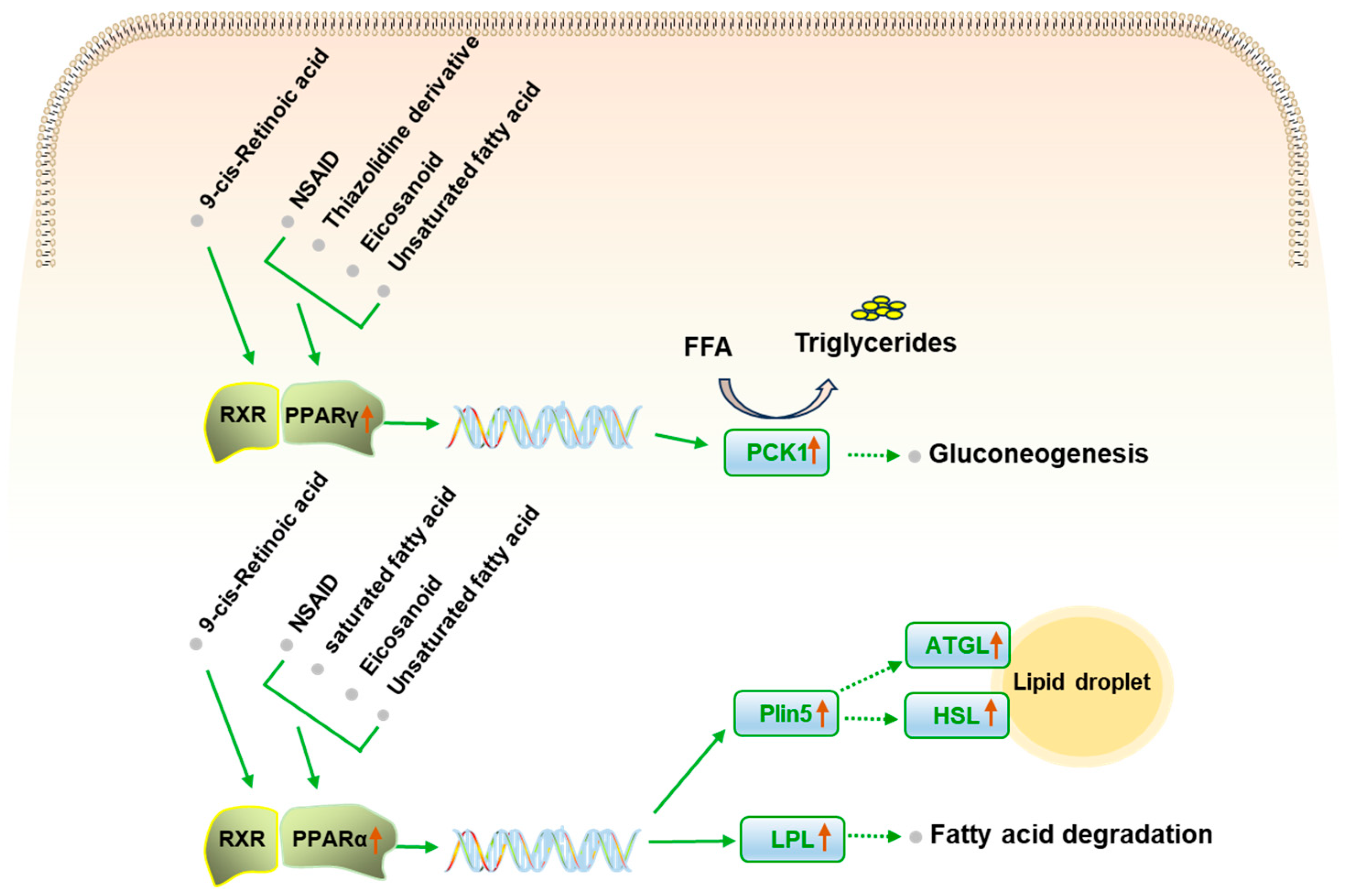
2.6. PS Regulates PPAR Signaling Pathway
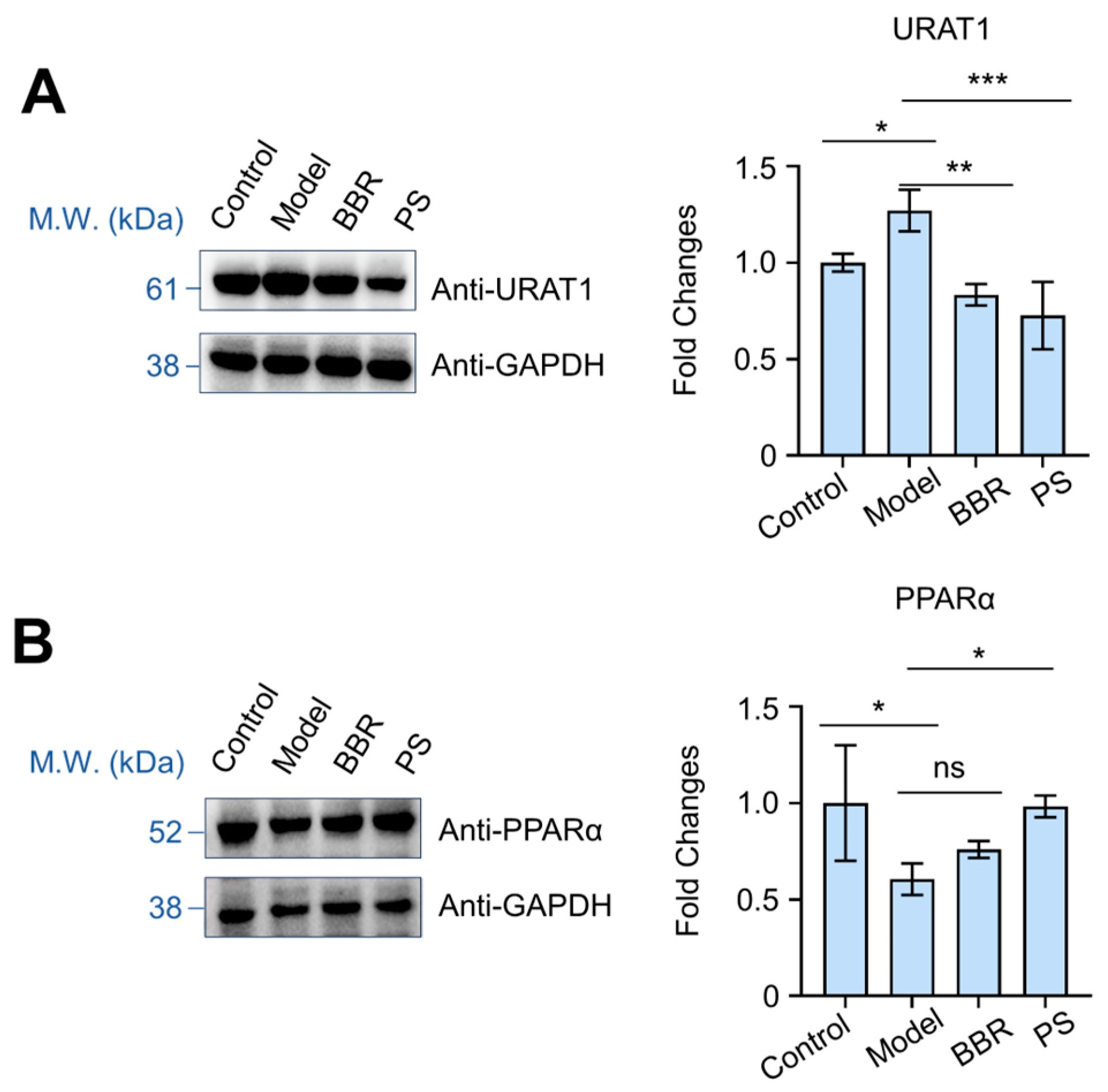
2.7. PS Regulates URAT1 and PPARα Proteins Expression
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Samples Preparation and Standard Solutions
4.3. Analysis of PS Components Migrating into the Blood
4.4. Qualitative Analysis by UPLC-QE-Orbitrap-MS
4.5. HUA Rat Model Establishment and Drugs Administration
4.6. Biochemical Index Detection
4.7. Hematoxylin–Eosin (H&E) Staining
4.8. RNA Sequencing
4.9. Real-Time Quantitative PCR
4.10. Western Blotting
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Acaa2 | Acetyl-CoA acyltransferase 2 |
| Acot4 | Acyl-CoA thioesterase 4 |
| Atgl | Adipose triglyceride lipase |
| Bhmt | Betaine-homocysteine S-Methyltransferase |
| Col1a1 | Collagen Type I Alpha 1 Chain |
| Cpt2 | Carnitine palmitoyltransferase 2 |
| DEGs | Differential expression genes |
| DEMs | Differentially expressed metabolites |
| ELISA | Enzyme-linked immunosorbent assay |
| Fkbp5 | FKBP prolyl isomerase 5 |
| H&E | Hematoxylin–eosin |
| Hsl | Hormone-sensitive lipase |
| HUA | Hyperuricemia |
| Gatm | Glycine amidinotransferase |
| Glut9 | Glucose transporter 9 |
| IL-1β | Interleukin-1β |
| IL-6 | Interleukin-6 |
| Lpl | Lipoprotein lipase |
| Mgll | Monoacylglycerol lipase |
| Pck1 | Phosphoenolpyruvate carboxykinase 1 |
| Phgdh | 3-Phosphoglycerate dehydrogenase |
| PI3K | Phosphatidylinositol 3-kinase |
| Plin5 | Perilipin-5 |
| PS | Plantaginis semen |
| PPAR | Peroxisome proliferators-activated receptors |
| RNA-seq | RNA sequencing |
| RT-qPCR | Real-time quantitative PCR |
| Slc7a8 | Solute carrier family 7 member 8 |
| TGF-β1 | Transforming growth factor beta 1 |
| TNF | Tumor necrosis factor |
| UA | Uric acid |
| UPLC-QE-Orbitrap-MS | Ultra-high-performance liquid chromatography–tandem quadrupole electrostatic field–orbital trap–mass spectrometry |
| Urat1 | Urate anion transporter 1 |
| XOD | Xanthine oxidase |
References
- Zhu, J.W.; Zeng, Y.; Zhang, H.Y.; Qu, Y.Y.; Ying, Z.Y.; Sun, Y.J.; Hu, Y.; Chen, W.W.; Yang, H.Z.; Yang, J.; et al. The Association of Hyperuricemia and Gout with the Risk of Cardiovascular Diseases: A Cohort and Mendelian Randomization Study in UK Biobank. Front. Med. 2022, 8, 817150. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zhang, M.; Huang, S.; Lan, X.Z.; Zheng, J.; Luo, H.; He, Y.; Lei, W. Hyperuricemia: A key contributor to endothelial dysfunction in cardiovascular diseases. FASEB J. 2023, 37, e23012. [Google Scholar] [CrossRef] [PubMed]
- Piani, F.; Agnoletti, D.; Borghi, C. Advances in pharmacotherapies for hyperuricemia. Expert Opin. Pharmacol. 2023, 24, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Thong, B.Y.H.; Vultaggio, A.; Rerkpattanapipat, T.; Schrijvers, R. Prevention of Drug Hypersensitivity Reactions: Prescreening and Premedication. J. Allergy Clin. Immunol. Pract. 2021, 9, 2958–2966. [Google Scholar] [CrossRef] [PubMed]
- Felser, A.; Lindinger, P.W.; Schnell, D.; Kratschmar, D.V.; Odermatt, A.; Mies, S.; Jenö, P.; Krähenbühl, S. Hepatocellular toxicity of benzbromarone: Effects on mitochondrial function and structure. Toxicology 2014, 324, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.L.; Gao, Y.Y.; Yang, Y.X.; Zhu, Q.F.; Guan, H.Y.; He, X.; Zhang, C.L.; Wang, Y.; Xu, G.B.; Zou, S.H.; et al. Amelioration effects of α-viniferin on hyperuricemia and hyperuricemia-induced kidney injury in mice. Phytomedicine 2023, 116, 154868. [Google Scholar] [CrossRef] [PubMed]
- Chinese Pharmacopoeia Commission. Chinese Pharmacopoeia; China Science and Technology Press: Beijing, China, 2020. [Google Scholar]
- Li, C.; Wen, R.; Liu, W.; Liu, Q.; Yan, L.P.; Wu, J.X.; Guo, Y.J.; Li, S.Y.; Gong, Q.F.; Yu, H. Diuretic Effect and Metabolomics Analysis of Crude and Salt-Processed Plantaginis Semen. Front. Pharmacol. 2020, 11, 563157. [Google Scholar] [CrossRef]
- Zhao, H.; Gao, Q.; Kong, L.Z.; Tang, W.W.; Jiao, Y.Y.; Wang, Y.L.; Yu, Z.; Feng, Y. Study on Network Pharmacological Analysis and Preliminary Validation to Understand the Mechanisms of Plantaginis Semen in Treatment of Gouty Nephropathy. Evid.-Based Complement. Altern. Med. 2020, 2020, 8861110. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Xu, J.; Wang, R.; Tang, W.; Kong, L.; Wang, W.; Wang, L.; Zhang, Y.; Ma, W. Plantaginis Semen polysaccharides ameliorate renal damage through regulating NLRP3 inflammasome in gouty nephropathy rats. Food Funct. 2021, 12, 2543–2553. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Qi, M.; Tong, R.; Wang, D.; Ding, L.; Li, Z.; Huang, C.; Wang, Z.; Yang, L. Plantago asiatica L. Seed Extract Improves Lipid Accumulation and Hyperglycemia in High-Fat Diet-Induced Obese Mice. Int. J. Mol. Sci 2017, 18, 1393. [Google Scholar] [CrossRef]
- Qin, N.K.; Qin, M.; Shi, W.J.; Kong, L.B.; Wang, L.T.; Xu, G.; Guo, Y.Y.; Zhang, J.Y.; Ma, Q. Investigation of pathogenesis of hyperuricemia based on untargeted and targeted metabolomics. Sci. Rep. 2022, 12, 13980. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Shi, W.J.; Wang, L.T.; Qin, N.K.; Wang, C.X.; Guo, Y.Y.; Xu, G.; Fang, J.; Yu, X.; Ma, Q. Lipidomics study of the therapeutic mechanism of Plantaginis Semen in potassium oxonate-induced hyperuricemia rat. BMC Complement. Med. Ther. 2021, 21, 175. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Cao, Y.; Qu, Y.; Li, T.; Wang, J.; Yang, Y.; Zhang, C.; Sun, Y. Integrating UPLC-QE-Orbitrap-MS technology and network pharmacological method to reveal the mechanism of Bailemian capsule to relieve insomnia. Nat. Prod. Res. 2022, 36, 2554–2558. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Tian, P.; Zhang, D.W.; Zhou, H.Y.; Sun, W.; Wang, Y.Y.; Han, D.E. Quantitative analysis of 16 components in Cyclocarya paliurus leaf by UPLC-QE-Orbitrap-MS. China J. Chin. Mater. Medica 2018, 43, 4077–4083. [Google Scholar]
- Zhou, Z.; Chen, B.; Chen, S.; Lin, M.; Chen, Y.; Jin, S.; Chen, W.; Zhang, Y. Applications of Network Pharmacology in Traditional Chinese Medicine Research. Evid. -Based Complement. Altern. Med. 2020, 2020, 1646905. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Jin, X.; Ma, Y.; Yang, Y.; Li, J.; Liang, L.; Liu, R.; Li, Z. A comprehensive application: Molecular docking and network pharmacology for the prediction of bioactive constituents and elucidation of mechanisms of action in component-based Chinese medicine. Comput. Biol. Chem. 2021, 90, 107402. [Google Scholar] [CrossRef] [PubMed]
- Fura, A. Role of pharmacologically active metabolites in drug discovery and development. Drug Discov. Today 2006, 11, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yuan, S.; Deng, Y.; Wang, X.; Wu, S.; Chen, X.; Li, Y.; Ouyang, J.; Lin, D.; Quan, H.; et al. The dysregulation of immune cells induced by uric acid: Mechanisms of inflammation associated with hyperuricemia and its complications. Front. Immunol. 2023, 14, 1282890. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, A.; Ishaq, M.; Zhao, L.; Safdar, B.; Rehman, A.U.; Munir, M.; Raza, A.; Nadeem, M.; Iqbal, W.; Wang, C. Natural compounds with xanthine oxidase inhibitory activity: A review. Chem. Biol. Drug Des. 2019, 93, 387–418. [Google Scholar] [CrossRef]
- Ren, Q.; Tao, S.; Guo, F.; Wang, B.; Yang, L.; Ma, L.; Fu, P. Natural flavonol fisetin attenuated hyperuricemic nephropathy via inhibiting IL-6/JAK2/STAT3 and TGF-beta/SMAD3 signaling. Phytomedicine 2021, 87, 153552. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Zhao, X.; Wang, F.; Wang, G. A brief review of urate transporter 1 (URAT1) inhibitors for the treatment of hyperuricemia and gout: Current therapeutic options and potential applications. Eur. J. Pharmacol. 2021, 907, 174291. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.J.; Takeda, M.; Enomoto, A.; Fujimura, M.; Miyazaki, H.; Anzai, N.; Endou, H. Interactions of urate transporter URAT1 in human kidney with uricosuric drugs. Nephrology 2011, 16, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Novikov, A.; Fu, Y.; Huang, W.; Freeman, B.; Patel, R.; van Ginkel, C.; Koepsell, H.; Busslinger, M.; Onishi, A.; Nespoux, J.; et al. SGLT2 inhibition and renal urate excretion: Role of luminal glucose, GLUT9, and URAT1. Am. J. Physiol. Renal. Physiol. 2019, 316, F173–F185. [Google Scholar] [CrossRef] [PubMed]
- Pitere, R.R.; van Heerden, M.B.; Pepper, M.S.; Ambele, M.A. Slc7a8 Deletion Is Protective against Diet-Induced Obesity and Attenuates Lipid Accumulation in Multiple Organs. Biology 2022, 11, 311. [Google Scholar] [CrossRef]
- Zhang, J.; Song, Y.; Shi, Q.; Fu, L. Research progress on FASN and MGLL in the regulation of abnormal lipid metabolism and the relationship between tumor invasion and metastasis. Front. Med. 2021, 15, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhang, Y.; Zhang, X.; Yao, Y.; Zheng, Y.; Cui, X.; Liu, C.; Wang, Q.; Li, J.Z. Long non-coding RNA Bhmt-AS attenuates hepatic gluconeogenesis via modulation of Bhmt expression. Biochem. Biophys. Res. Commun. 2019, 516, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Blom, H.J.; De Vriese, A.S. Why are homocysteine levels increased in kidney failure? A metabolic approach. J. Lab. Clin. Med. 2002, 139, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Clar, J.; Gri, B.; Calderaro, J.; Birling, M.C.; Herault, Y.; Smit, G.P.; Mithieux, G.; Rajas, F. Targeted deletion of kidney glucose-6 phosphatase leads to nephropathy. Kidney Int. 2014, 86, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Misawa, K.; Hasegawa, T.; Mishima, E.; Jutabha, P.; Ouchi, M.; Kojima, K.; Kawai, Y.; Matsuo, M.; Anzai, N.; Nagasaki, M. Contribution of Rare Variants of the SLC22A12 Gene to the Missing Heritability of Serum Urate Levels. Genetics 2020, 214, 1079–1090. [Google Scholar] [CrossRef] [PubMed]
- Lamichane, S.; Dahal Lamichane, B.; Kwon, S.M. Pivotal Roles of Peroxisome Proliferator-Activated Receptors (PPARs) and Their Signal Cascade for Cellular and Whole-Body Energy Homeostasis. Int. J. Mol. Sci. 2018, 19, 949. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.G.; Shang, Y.J.; Zhang, J.; Zhang, J.R.; Li, W.J.; Jiao, B.H. Hypouricemic effects of phenylpropanoid glycosides acteoside of Scrophularia ningpoensis on serum uric acid levels in potassium oxonate-pretreated Mice. Am. J. Chin. Med. 2008, 36, 149–157. [Google Scholar] [CrossRef]
- Wan, Y.; Zou, B.; Zeng, H.; Zhang, L.; Chen, M.; Fu, G. Inhibitory effect of verbascoside on xanthine oxidase activity. Int. J. Biol. Macromol. 2016, 93, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.X.; Wang, J.; Zhang, S.W.; Zhu, J.X.; Li, M.; Huang, W.H.; Wan, J.Y.; Yao, H.Q.; Wang, C.Z.; Yuan, C.S. Antigout Effects of: Xanthine Oxidase Inhibitory Activities Assessed by Electrochemical Biosensing Method. Evid.-Based Complement. Altern. Med. 2018, 2018, 1364617. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.X.; Yang, H.Y.; Hu, W.Q.; Cheng, J.; Liu, Y.; Yi, L.T.; Cheng, H.Y. Active components from Lagotis brachystachya maintain uric acid homeostasis by inhibiting renal TLR4-NLRP3 signaling in hyperuricemic mice. Inflammopharmacology 2021, 29, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Kim, O.K.; Yun, J.M.; Lee, M.; Kim, D.; Lee, J. Hypouricemic Effects of Chrysanthemum indicum L. and Cornus officinalis on Hyperuricemia-Induced HepG2 Cells, Renal Cells, and Mice. Plants 2021, 10, 1668. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, Z.; Luo, J.; Jiang, Y.; Li, L.; Chen, Y.; Zhang, L.; Huang, Q.; Cao, Y.; Zhou, P.; et al. Apigenin ameliorates hyperuricemic nephropathy by inhibiting URAT1 and GLUT9 and relieving renal fibrosis via the Wnt/beta-catenin pathway. Phytomedicine 2021, 87, 153585. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Gao, H.; Zhang, Y.; Wang, S.; Lu, M.; Dai, X.; Liu, Y.; Shi, H.; Xu, T.; Yin, J.; et al. Apigenin Ameliorates Hyperuricemia and Renal Injury through Regulation of Uric Acid Metabolism and JAK2/STAT3 Signaling Pathway. Pharmaceuticals 2022, 15, 1442. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, Y.; Takada, T.; Saito, H.; Hirata, H.; Ota-Kontani, A.; Tsuchiya, Y.; Suzuki, H. Identification of Inhibitory Activities of Dietary Flavonoids against URAT1, a Renal Urate Re-Absorber: In Vitro Screening and Fractional Approach Focused on Rooibos Leaves. Nutrients 2022, 14, 575. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Gu, Q.; Nie, S.M.; Wang, J.D.; Zhao, H.; Zhai, B.W.; Zhang, M.Y.; Fu, Y.J. Untargeted metabolomics reveals the regulatory effect of geniposidic acid on lipid accumulation in HepG2 cells and Caenorhabditis elegans and validation in hyperlipidemic hamsters. Phytomedicine 2023, 125, 155295. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Zhao, C.; Wang, Y.; Peng, Y.; Cheng, J.; Li, Z.; Wu, L.; Jin, M.; Feng, H. Aucubin inhibited lipid accumulation and oxidative stress via Nrf2/HO-1 and AMPK signalling pathways. J. Cell. Mol. Med. 2019, 23, 4063–4075. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, S.; Dai, X.; Liu, T.; Liu, Y.; Shi, H.; Yin, J.; Xu, T.; Zhang, Y.; Zhao, D.; et al. Simiao San alleviates hyperuricemia and kidney inflammation by inhibiting NLRP3 inflammasome and JAK2/STAT3 signaling in hyperuricemia mice. J. Ethnopharmacol. 2023, 312, 116530. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Cheng, J.; Zhao, P.; Chen, K.L.; Li, J. Screening the Best Compatibility of Selaginella moellendorffii Prescription on Hyperuricemia and Gouty Arthritis and Its Mechanism. Evid.-Based Complement. Altern. Med. 2019, 2019, 7263034. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, R.; Pasalar, P.; Shokri, H.; Shabani, M.; Emamgholipour, S. Evidence for the effect of soluble uric acid in augmenting endoplasmic reticulum stress markers in human peripheral blood mononuclear cells. J. Physiol. Biochem. 2022, 78, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Al Shanableh, Y.; Hussein, Y.Y.; Saidwali, A.H.; Al-Mohannadi, M.; Aljalham, B.; Nurulhoque, H.; Robelah, F.; Al-Mansoori, A.; Zughaier, S.M. Prevalence of asymptomatic hyperuricemia and its association with prediabetes, dyslipidemia and subclinical inflammation markers among young healthy adults in Qatar. BMC Endocr. Disord. 2022, 22, 21. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Wan, L.; Chen, X.; Li, X.; Hao, X.; Li, X.; Li, J.; Ding, H. Conjugated Linoleic Acid Ameliorates High Fructose-Induced Hyperuricemia and Renal Inflammation in Rats via NLRP3 Inflammasome and TLR4 Signaling Pathway. Mol. Nutr. Food Res. 2019, 63, e1801402. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cui, T.; Ci, X.; Zhao, F.; Sun, Y.; Li, Y.; Liu, R.; Wu, W.; Yi, X.; Liu, C. The effect of polymorphism of uric acid transporters on uric acid transport. J. Nephrol. 2019, 32, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Lutkewitte, A.J.; Finck, B.N. Regulation of Signaling and Metabolism by Lipin-mediated Phosphatidic Acid Phosphohydrolase Activity. Biomolecules 2020, 10, 1386. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Cui, T.; Liu, Y.; Wu, S.; Han, C.; Li, J. Astragalus membranaceus and Salvia miltiorrhiza ameliorate diabetic kidney disease via the ”gut-kidney axis”. Phytomedicine 2023, 121, 155129. [Google Scholar] [CrossRef]
- Chen, M.; Dong, J.; Zhao, X.; Yin, X.; Wu, K.; Wang, Q.; Liu, X.; Wu, Y.; Gong, Z. Cadmium influence on lipid metabolism in Sprague-Dawley rats through linoleic acid and glycerophospholipid metabolism pathways. J. Biochem. Mol. Toxicol. 2023, 37, e23412. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Ridgway, N.D. Substrate channeling in the glycerol-3-phosphate pathway regulates the synthesis, storage and secretion of glycerolipids. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158438. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Baro, M.R.; Coleman, R.A. Mitochondrial acyltransferases and glycerophospholipid metabolism. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, L.; Siersbaek, M.; Mandrup, S. PPARs: Fatty acid sensors controlling metabolism. Semin. Cell Dev. Biol. 2012, 23, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Su, M.; Liu, M.; Sheng, Y.; Zhu, L.; Yang, L.; Mu, R.; Zou, J.; Liu, X.; Liu, L. Hepatic retinaldehyde deficiency is involved in diabetes deterioration by enhancing PCK1- and G6PC-mediated gluconeogenesis. Acta Pharm. Sin. B 2023, 13, 3728–3743. [Google Scholar] [CrossRef] [PubMed]
- Bougarne, N.; Weyers, B.; Desmet, S.J.; Deckers, J.; Ray, D.W.; Staels, B.; De Bosscher, K. Molecular Actions of PPARalpha in Lipid Metabolism and Inflammation. Endocr. Rev. 2018, 39, 760–802. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.Y.; Lim, C.M.; Park, H.M.; Kim, J.; Pham, T.H.; Yang, Y.; Lee, H.P.; Hong, J.T.; Yoon, D.Y. MMPP promotes adipogenesis and glucose uptake via binding to the PPARgamma ligand binding domain in 3T3-L1 MBX cells. Front. Pharmacol. 2022, 13, 994584. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.A.; Shen, C.; Wei, X.; Zhang, X.; Wang, S.; Chen, X.; Torres, M.; Lu, Y.; Lin, L.L.; Wang, H.H.; et al. The mechanisms to dispose of misfolded proteins in the endoplasmic reticulum of adipocytes. Nat. Commun. 2023, 14, 3132. [Google Scholar] [CrossRef] [PubMed]
- Krizanac, M.; Mass Sanchez, P.B.; Schroder, S.K.; Weiskirchen, R.; Asimakopoulos, A. Lipid-Independent Regulation of PLIN5 via IL-6 through the JAK/STAT3 Axis in Hep3B Cells. Int. J. Mol. Sci. 2023, 24, 7219. [Google Scholar] [CrossRef] [PubMed]
- Klyuyeva, A.V.; Belyaeva, O.V.; Goggans, K.R.; Krezel, W.; Popov, K.M.; Kedishvili, N.Y. Changes in retinoid metabolism and signaling associated with metabolic remodeling during fasting and in type I diabetes. J. Biol. Chem. 2021, 296, 100323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, Y.; Lin, S.; Zhang, S.; Qiu, M. Hyperuricemia as a possible risk factor for abnormal lipid metabolism in the Chinese population: A cross-sectional study. Ann. Palliat. Med. 2021, 10, 11454–11463. [Google Scholar] [CrossRef] [PubMed]
- Trommelen, J.; Tomé, D.; van Loon, L.J.C. Gut amino acid absorption in humans: Concepts and relevance for postprandial metabolism. Clin. Nutr. Open Sci. 2021, 36, 43–55. [Google Scholar] [CrossRef]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef] [PubMed]
- El Ridi, R.; Tallima, H. Physiological functions and pathogenic potential of uric acid: A review. J. Adv. Res. 2017, 8, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Knopfel, E.B.; Vilches, C.; Camargo, S.M.R.; Errasti-Murugarren, E.; Staubli, A.; Mayayo, C.; Munier, F.L.; Miroshnikova, N.; Poncet, N.; Junza, A.; et al. Dysfunctional LAT2 Amino Acid Transporter Is Associated with Cataract in Mouse and Humans. Front. Physiol. 2019, 10, 688. [Google Scholar] [CrossRef] [PubMed]
- Morio, H.; Reien, Y.; Hirayama, Y.; Hashimoto, H.; Anzai, N. Protein kinase C activation upregulates human L-type amino acid transporter 2 function. J. Physiol. Sci. 2021, 71, 11. [Google Scholar] [CrossRef] [PubMed]
- Beale, E.G.; Harvey, B.J.; Forest, C. PCK1 and PCK2 as candidate diabetes and obesity genes. Cell Biochem. Biophys. 2007, 48, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Seo, J.W.; Kim, M.; Tae, D.; Seok, J.; Kim, Y.G.; Lee, S.H.; Kim, J.S.; Hwang, H.S.; Jeong, K.H.; et al. Urinary mRNA Signatures as Predictors of Renal Function Decline in Patients with Biopsy-Proven Diabetic Kidney Disease. Front. Endocrinol. 2021, 12, 774436. [Google Scholar] [CrossRef] [PubMed]
- Glorieux, G.; Mullen, W.; Duranton, F.; Filip, S.; Gayrard, N.; Husi, H.; Schepers, E.; Neirynck, N.; Schanstra, J.P.; Jankowski, J.; et al. New insights in molecular mechanisms involved in chronic kidney disease using high-resolution plasma proteome analysis. Nephrol. Dial. Transpl. 2015, 30, 1842–1852. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Feng, M.; Li, Z.; Zhou, M.; Xu, L.; Pan, K.; Wang, S.; Su, W.; Zhang, W. ETV5 Regulates Hepatic Fatty Acid Metabolism Through PPAR Signaling Pathway. Diabetes 2021, 70, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wang, J.; Ma, L.; Ge, Y.; Wang, X.M. The effect of lipid metabolism disorder on patients with hyperuricemia using Multi-Omics analysis. Sci. Rep. 2023, 13, 18211. [Google Scholar] [CrossRef]
- Pang, S.; Jiang, Q.; Sun, P.; Li, Y.; Zhu, Y.; Liu, J.; Ye, X.; Chen, T.; Zhao, F.; Yang, W. Hyperuricemia prevalence and its association with metabolic disorders: A multicenter retrospective real-world study in China. Ann. Transl. Med. 2021, 9, 1550. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.H.; Tsai, C.C.; Shyong, Y.J.; Yang, C.T.; Li, K.Y.; Lin, Y.W.; Chang, K.C.; Wang, M.H.; Wu, T.M.; Lin, F.H. Effects of Highly Oxygenated Water in a Hyperuricemia Rat Model. J. Healthc. Eng. 2020, 2020, 1323270. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Liu, X.J.; Lee, M.H.; Li, H.T. Vitamin C alleviates hyperuricemia nephropathy by reducing inflammation and fibrosis. J. Food Sci. 2021, 86, 3265–3276. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Liu, B.Y.; Wang, S.; Sui, M.H. Effects of acupuncture at Shu, Yuan, and Mu acupoints on blood serum uric acid and xanthine oxidase levels in a rat model of gout and hyperuricemia. J. Tradit. Chin. Med. 2017, 37, 841–845. [Google Scholar] [PubMed]
- Qian, X.W.; Jiang, Y.; Luo, Y.Y.; Jiang, Y.C. The Anti-hyperuricemia and Anti-inflammatory Effects of Atractylodes Macrocephala in Hyperuricemia and Gouty Arthritis Rat Models. Comb. Chem. High T. Scr. 2023, 26, 950–964. [Google Scholar]
- Li, R.; Tan, Y.; Li, Y.; Zhu, X.; Tang, X.; Zhang, L.; Chen, J. Effects of Tart Cherry Powder on Serum Uric Acid in Hyperuricemia Rat Model. Evid.-Based Complement. Altern. Med. 2020, 2020, 1454305. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, A.P.; Nikitina, V.A.; Krytskaya, D.U.; Shcherbakova, K.P.; Trofimov, A.N. Reference gene expression stability within the rat brain under mild intermittent ketosis induced by supplementation with medium-chain triglycerides. PLoS ONE 2023, 18, e0273224. [Google Scholar] [CrossRef] [PubMed]
- Ravingerova, T.; Carnicka, S.; Nemcekova, M.; Ledvenyiova, V.; Adameova, A.; Kelly, T.; Barlaka, E.; Galatou, E.; Khandelwal, V.K.; Lazou, A. PPAR-alpha activation as a preconditioning-like intervention in rats in vivo confers myocardial protection against acute ischaemia-reperfusion injury: Involvement of PI3K-Akt. Can. J. Physiol. Pharmacol. 2012, 90, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, M.; Kruijt, J.K.; Van Eck, M.; Van Berkel, T.J. Specific gene expression of ATP-binding cassette transporters and nuclear hormone receptors in rat liver parenchymal, endothelial, and Kupffer cells. J. Biol. Chem. 2003, 278, 25448–25453. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.F.; Du, C.K.; Su, X.Y. Forskolin induced remodeling of lipid droplets in rat adipocytes. Sheng Li Xue Bao Acta Physiol. Sin. 2019, 71, 379–387. [Google Scholar]
- Ma, D.; Wu, T.; Qu, Y.; Yang, J.; Cai, L.; Li, X.; Wang, Y. Astragalus polysaccharide prevents heart failure-induced cachexia by alleviating excessive adipose expenditure in white and brown adipose tissue. Lipids Health Dis. 2023, 22, 9. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, B.; Miklosz, A.; Zabielski, P.; Chabowski, A.; Gorski, J. Effect of tachycardia on mRNA and protein expression of the principal components of the lipolytic system in the rat’s heart ventricles. J. Physiol. Pharmacol. 2017, 68, 731–736. [Google Scholar] [PubMed]
- Slow, S.; Garrow, T.A. Liver choline dehydrogenase and kidney betaine-homocysteine methyltransferase expression are not affected by methionine or choline intake in growing rats. J. Nutr. 2006, 136, 2279–2283. [Google Scholar] [CrossRef] [PubMed]
- Caton, P.W.; Nayuni, N.K.; Murch, O.; Corder, R. Endotoxin induced hyperlactatemia and hypoglycemia is linked to decreased mitochondrial phosphoenolpyruvate carboxykinase. Life Sci. 2009, 84, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, P.; Dave, M.H.; Remy, C.; Kosiek, O.; Busque, S.M.; Dufner, M.; Geibel, J.P.; Verrey, F.; Wagner, C.A. An amino acid transporter involved in gastric acid secretion. Pflüg. Arch. 2006, 451, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Tyler, R.E.; Weinberg, B.Z.S.; Lovelock, D.F.; Ornelas, L.C.; Besheer, J. Exposure to the predator odor TMT induces early and late differential gene expression related to stress and excitatory synaptic function throughout the brain in male rats. Genes Brain Behav. 2020, 19, e12684. [Google Scholar] [CrossRef]
- Yamakawa, J.; Ishigaki, Y.; Takano, F.; Takahashi, T.; Yoshida, J.; Moriya, J.; Takata, T.; Tatsuno, T.; Sasaki, K.; Ohta, T.; et al. The Kampo medicines Orengedokuto, Bofutsushosan and Boiogito have different activities to regulate gene expressions in differentiated rat white adipocytes: Comprehensive analysis of genetic profiles. Biol. Pharm. Bull. 2008, 31, 2083–2089. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Chen, L.; He, M.; Luo, Y.; Zhou, M.; Zhang, N.; Yuan, J.; Wang, H.; Xie, Y. Molecular mechanistic insight into the anti-hyperuricemic effect of Eucommia ulmoides in mice and rats. Pharm. Biol. 2019, 57, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Zhang, X.; Tao, H.; Wu, Y.; Yan, J.; Liao, L.; Meng, J.; Lin, F. Ameliorative effect and mechanism of Yi-Suan-Cha against hyperuricemia in rats. J. Clin. Lab. Anal. 2021, 35, e23859. [Google Scholar] [CrossRef]



| NO. | tR/min | Formula | Adducts | Theoretical Value | Measured Value | Mass Error (ppm) | MS/MS | Identification | Label |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1.02 | C6H14O6 | +H | 183.0863 | 183.0864 | 0.55 | 147.1855, 98.9845 | Sorbitol | h |
| 2 | 2.81 | C6H8O7 | -H | 191.0186 | 191.0201 | 7.85 | 191.0199, 173.0089, 111.0089, 87.0088 | Citric acid | f |
| 3 * | 4.74 | C15H22O9 | -H | 345.1180 | 345.1195 | 4.35 | 299.8194, 139.0407, 89.3671 | Aucubin | c |
| 4 | 5.60 | C16H22O10 | +H | 375.1286 | 375.1279 | −1.87 | 287.0074, 195.0659, 177.0552, 167.0707, 149.0602, 121.0652 | Gardoside | c |
| 5 | 5.60 | C16H20O9 | +H | 357.1180 | 357.1175 | −1.40 | 195.0659, 177.0553, 149.0603, 121.0652 | Feruloylsucrose | i |
| 6 * | 5.61 | C16H22O10 | -H | 373.1129 | 373.1135 | 1.61 | 300.3118, 211.0611, 193.0512, 167.0711, 149.0609, 123.0453, 121.0660 | Geniposidic acid | c |
| 7 | 5.81 | C20H30O12 | -H | 461.1654 | 461.1672 | 3.90 | 461.1658, 315.1101, 161.0455, 135.0452 | Verbasoside | b |
| 8 | 6.30 | C9H12O4 | +H | 185.0808 | 185.0809 | 0.54 | 185.1539, 139.0766, 129.9802, 111.0751 | Pedicularis-lactone | c |
| 9 | 7.46 | C10H10O4 | -H | 193.0495 | 193.0500 | 2.59 | 193.0509, 116.9284, 96.9601 | Ferulic acid | f |
| 10 | 9.50 | C29H36O16 | -H | 639.1920 | 639.1956 | 5.63 | 141.5413 | Plantagoamidinic acid | e |
| 11 | 11.15 | C11H19N3O2 | -H | 224.1394 | 224.1405 | 4.91 | 224.1392, 141.0924 | Plantagoamidinic acid A | e |
| 12 | 11.72 | C21H22O12 | -H | 465.1028 | 465.1042 | 3.01 | 465.1032, 303.0506, 151.0393 | Plantagoside | a |
| 13 | 14.23 | C29H36O16 | -H | 639.1920 | 639.1954 | 5.32 | 477.1617, 179.0362, 161.0246, 135.0457 | Plantamajoside | b |
| 14 | 15.12 | C15H12O7 | -H | 303.0499 | 303.0514 | 4.95 | 303.0510, 151.0402, 107.0139 | Taxifolin | a |
| 15* | 15.41 | C29H36O15 | -H | 623.1970 | 623.2000 | 4.81 | 623.1987, 461.1670, 315.1082, 179.0352, 161.0246, 153.0559, 135.0453 | Acteoside | b |
| 16 | 15.41 | C29H34O15 | -H | 621.1814 | 621.1828 | 2.25 | 179.0350, 161.0246, 135.0453 | Orobanchoside | b |
| 17 | 15.94 | C23H26O11 | -H | 477.1391 | 477.1410 | 3.98 | 477.1409, 315.1057, 161.0234, 135.0448 | Calceolarioside B | b |
| 18 | 16.34 | C21H20O11 | -H | 447.0922 | 447.0941 | 4.25 | 447.0935, 285.0405, 284.0324, 256.0373, 199.0396, 151.0041 | Cynaroside | a |
| 19 | 16.45 | C29H36O16 | -H | 639.1920 | 639.1954 | 5.32 | 639.1927, 477.1605, 315.1107, 179.0350, 161.0246, 153.0549, 135.0454, 84.7942 | Plantainoside D | b |
| 20 | 17.10 | C27H30O15 | -H | 593.1501 | 593.1522 | 3.54 | 285.0384, 284.0322 | Kaempferol-3-rutinoside | a |
| 21 * | 17.15 | C29H36O15 | -H | 623.1970 | 623.2000 | 4.81 | 623.1990, 461.1681, 179.0350, 161.0245, 135.0453 | Isoacteoside | b |
| 22 | 17.58 | C27H32O14 | -H | 579.1708 | 579.1725 | 2.94 | 271.0618, 151.0029 | Naringin | a |
| 23 * | 17.62 | C15H12O6 | +H | 289.0707 | 289.0704 | −1.04 | 271.0609, 179.0344, 163.0395, 153.0188 | Eriodictyol | a |
| 24 | 17.87 | C21H20O12 | +H | 465.1028 | 465.1031 | 0.65 | 302.0745, 300.3060 | Hyperoside | a |
| 25 | 17.96 | C21H20O10 | -H | 431.0973 | 431.0994 | 4.87 | 431.0989, 269.0454, 268.0375, 151.0038 | Apigenin 7-glucoside | a |
| 26 | 18.11 | C27H30O14 | -H | 577.1552 | 577.1575 | 3.99 | 413.0989, 269.0454, 268.0375, 183.0457 | Rhoifolin | a |
| 27 | 18.26 | C31H38O16 | -H | 665.2076 | 665.2112 | 5.41 | 503.1779, 181.5070, 161.0246 | 2-Acetylacteoside | b |
| 28 | 18.80 | C21H20O11 | +H | 449.1078 | 449.1080 | 0.45 | 449.0978, 287.0555 | Plantaginin | b |
| 29 * | 18.91 | C15H10O7 | -H | 301.0343 | 301.0358 | 4.98 | 301.0358, 254.8575, 121.6543 | Quercetin | a |
| 30 | 19.27 | C22H22O11 | -H | 461.1078 | 461.1094 | 3.47 | 461.1952, 181.7875 | Homoplantaginin | a |
| 31 * | 19.90 | C15H10O6 | -H | 285.0394 | 285.0408 | 4.91 | 285.0408, 199.0405, 175.0404, 151.0036, 133.0297, 107.0138, 83.0142 | Luteolin | a |
| 32 * | 21.05 | C15H10O5 | -H | 269.0444 | 269.0456 | 4.46 | 269.0456, 200.9979, 117.0348, 65.0031 | Apigenin | a |
| 33 | 24.74 | C15H10O5 | -H | 269.0444 | 269.0456 | 4.46 | 269.0456, 241.0506, 225.0557, 197.0611 | Baicalein | a |
| 34 | 29.15 | C9H8O4 | +H | 181.0495 | 181.0493 | −1.10 | 181.0718, 163.0395, 135.0446, 116.9860, 79.0545, 65.0390 | Caffeic acid | f |
| 35 | 29.77 | C30H48O4 | -H | 471.3469 | 471.3484 | 3.18 | 471.3484, 453.3378, 427.3596, 407.3329 | Maslinic acid | d |
| 36 | 31.03 | C18H32O2 | +H | 181.0495 | 181.0493 | −1.10 | 181.0718, 163.0395, 135.0446, 116.9860, 79.0545, 65.0390 | Caffeic acid | f |
| 37 | 31.39 | C30H48O3 | -H | 455.3520 | 455.3535 | 3.29 | 456.3588, 455.3534, 408.2341 | Oleanic acid | d |
| 38 | 31.58 | C18H30O2 | -H | 277.2162 | 277.2173 | 3.97 | 277.2173, 259.2071, 233.1541, 121.0296, 81.9533, 59.0138 | α-Linolenic acid | g |
| 39 * | 31.95 | C30H48O3 | -H | 455.3520 | 455.3533 | 2.85 | 455.3536, 300.0453, 181.7717, 147.1723, 115.9207, 99.9257 | Ursolic acid | d |
| NO. | tR/min | Formula | Adducts | Theoretical Value | Measured Value | Mass Error (ppm) | MS/MS | Identification |
|---|---|---|---|---|---|---|---|---|
| 1 | 2.53 | C6H8O7 | -H | 191.0186 | 191.0199 | 6.81 | 191.0200, 173.0090, 111.0088, 87.0088 | Citric acid a |
| 2 | 3.51 | C9H13NO9S | -H | 310.0227 | 310.0247 | 6.45 | 310.0974, 230.0188, 87.1158 | Citric acid j |
| 3 | 4.72 | C15H22O9 | -H | 345.1180 | 345.1193 | 3.77 | 299.9364, 139.0401, 89.3645 | Aucubin a |
| 4 | 4.79 | C22H30O16 | -H | 549.1450 | 549.1460 | 1.82 | 211.0613, 167.0714, 149.0607, 123.0452, 121.0660 | Geniposidic acid i |
| 5 | 4.96 | C16H22O13S | -H | 453.0697 | 453.0713 | 3.53 | 453.0710, 300.4093, 193.0514, 149.0610, 123.0451, 121.4576 | Geniposidic acid h |
| 6 | 5.60 | C16H22O10 | -H | 373.1129 | 373.1135 | 1.61 | 211.0614, 193.0512, 167.0714, 149.0609, 123.0452, 121.0660 | Geniposidic acid a |
| 7 | 6.09 | C15H20O9 | -H | 343.1024 | 343.1037 | 3.79 | 343.1034, 181.6917, 162.8395, 160.8422, 159.8602, 139.0766, 101.0247, 99.0089, 89.0247 | Aucubin d |
| 8 | 6.42 | C15H20O8 | -H | 327.1074 | 327.1089 | 4.59 | 165.0558, 119.1114, 99.0086, 89.0245 | Aucubin b |
| 9 | 6.92 | C16H20O9 | -H | 355.1024 | 355.1038 | 3.94 | 300.2832, 121.0297 | Geniposidic acid b |
| 10 | 7.11 | C10H10O4 | -H | 193.0495 | 193.0507 | 6.22 | 193.8154, 149.0606, 133.0296 | Ferulic acid a |
| 11 | 11.59 | C11H19N3O2 | -H | 224.1394 | 224.1406 | 5.35 | 224.1406, 141.0923 | Plantagoamidinic acid A a |
| 12 | 12.33 | C21H20O12 | -H | 463.0871 | 463.0890 | 4.10 | 151.0039, 107.0139 | Plantagoside d |
| 13 | 15.33 | C29H36O15 | -H | 623.1970 | 623.2006 | 5.78 | 623.2032, 461.1672, 179.0350, 161.0245, 135.0452 | Acteoside a |
| 14 | 15.77 | C21H20O11 | -H | 447.0922 | 447.0939 | 3.80 | 447.1942, 151.0037, 107.0140 | Plantagoside b |
| 15 | 15.77 | C21H20O11 | -H | 447.0922 | 447.0939 | 3.80 | 447.1942, 151.0037 | Cynaroside a |
| 16 | 17.04 | C29H36O15 | -H | 623.1970 | 623.2005 | 5.62 | 623.2021, 461.1671, 161.0244, 135.0457 | Isoacteoside a |
| 17 | 17.90 | C21H18O11 | -H | 445.0765 | 445.0783 | 4.04 | 445.1906, 269.0458 | Cynaroside d |
| 18 | 17.90 | C21H18O11 | -H | 445.0765 | 445.0783 | 4.04 | 445.1906, 269.0458, 117.0347, 65.3859 | Apigenin i |
| 19 | 17.94 | C21H22O11 | -H | 449.1078 | 449.1094 | 3.56 | 258.0529, 273.0766, 151.0039 | Cynaroside e |
| 20 | 18.76 | C24H40O8 | +H | 457.2796 | 457.2792 | −0.87 | 457.2742, 281.0787, 133.0863 | Linoleic acid i |
| 21 | 18.83 | C21H18O12 | -H | 461.0715 | 461.0732 | 3.69 | 461.1859, 285.0403, 199.0399, 175.0396, 151.0038, 133.0296, 107.0138 | Luteolin i |
| 22 | 19.13 | C15H10O9S | -H | 364.9962 | 364.9978 | 4.38 | 365.1043, 213.0549, 175.0405, 151.0033 | Luteolin h |
| 23 | 19.27 | C22H22O11 | -H | 461.1078 | 461.1094 | 3.47 | 461.1850, 285.0768, 165.1093, 151.0033 | Cynaroside g |
| 24 | 19.27 | C22H22O11 | -H | 461.1078 | 461.1094 | 3.47 | 461.1850, 181.7875 | Homoplantaginin a |
| 25 | 19.31 | C15H12O5 | -H | 271.0601 | 271.0613 | 4.43 | 271.1891, 227.1645, 151.0042, 119.0504, 116.9956, 65.6944 | Apigenin e |
| 26 | 19.77 | C15H10O6 | -H | 285.0394 | 285.0406 | 4.21 | 285.0402, 194.9880, 166.9930, 133.0293, 116.9960 | Apigenin f |
| 27 | 20.89 | C15H10O5 | -H | 269.0444 | 269.0452 | 2.97 | 269.1378, 225.1501, 200.9967, 182.9875, 116.9956 | Apigenin a |
| 28 | 23.19 | C20H34O3 | +H | 323.2581 | 323.2575 | −1.86 | 323.2375, 149.1331, 133.1017, 107.0858, 97.0651, 69.0703, 55.0547 | Linoleic acid k |
| 29 | 29.09 | C9H8O4 | +H | 181.0495 | 181.0494 | −0.55 | 181.1596, 163.0395, 135.0445, 116.9861, 79.0545, 65.0390 | Caffeic acid a |
| 30 | 29.91 | C18H30O3 | -H | 293.2111 | 293.2122 | 3.75 | 293.2117, 275.2018, 249.2219, 121.1017, 59.0137 | α-Linolenic acid f |
| 31 | 29.95 | C18H32O3 | -H | 295.2268 | 295.2278 | 3.39 | 295.2278, 277.2172, 121.5115, 59.0137 | α-Linolenic acid c |
| 32 | 29.96 | C18H30O2 | +H | 279.2319 | 279.2313 | −2.15 | 279.2327, 261.2224, 173.1333, 165.1279, 163.1124, 133.1024, 131.0859, 107.0857, 105.0704, 67.0546, 65.0388, 55.0548, 53.0390 | Linoleic acid d |
| 33 | 30.27 | C18H32O3 | +H | 297.2424 | 297.2420 | −1.35 | 279.2326, 191.1441, 181.1586, 165.1280, 149.1332, 133.1015, 123.1173, 107.0859, 69.0702, 55.0548 | Linoleic acid f |
| 34 | 30.42 | C18H30O | +H | 263.2369 | 263.2365 | −1.52 | 263.2372, 245.2271, 175.1492, 165.1640, 133.1016, 107.0859, 69.0702, 55.0548 | Linoleic acid b |
| 35 | 30.45 | C36H56O9 | -H | 631.3841 | 631.3865 | 3.80 | 456.3566, 455.3532, 408.2441 | Oleanolic acid i |
| 36 | 31.07 | C18H28O2 | -H | 275.2006 | 275.2015 | 3.27 | 275.2012, 231.2118, 121.7366, 79.9572, 59.0139 | α-Linolenic acid d |
| 37 | 31.52 | C18H30O2 | -H | 277.2162 | 277.2171 | 3.25 | 277.2171, 259.2079, 233.0141, 59.0137 | α-Linolenic acid a |
| 38 | 31.95 | C30H48O3 | -H | 455.3520 | 455.3537 | 3.73 | 455.3168, 300.1568, 181.5451, 115.9202, 99.9258 | Ursolic acid a |
| 39 | 32.01 | C18H32O2 | -H | 279.2319 | 279.2328 | 3.22 | 279.2328, 261.2221, 121.6766, 59.0138 | α-Linolenic acid e |
| 40 | 32.50 | C18H34O2 | +H | 283.2632 | 283.2625 | −2.47 | 177.1648, 135.1137, 109.1016, 107.0860, 71.0859, 69.0702, 57.0704, 55.0548 | Linoleic acid e |
| Genes | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|
| Urat1 | CTCTGCCTTTCTCCTGTTGA | CCCCTTGATGATGACCTTG |
| Glut9 | TTCGGGTCCTCCTTCCTCTA | GGACACAGTCACAGACCAGA |
| Slc7a8 | TCCACATTTGGTGGAGTCAA | TGGATCATGGCTAACACGCT |
| Bhmt | CGGGCAGACCGTACAATCC | CTTTCGTCACTCCCCAAGCA |
| Fkbp5 | GAACCCAATGCTGAGCTTATG | ATGTACTTGCCTCCCTTGAAG |
| Mgll | GCGGTGCCATCTCCATCC | GCTCCTGCCACTGCTATCC |
| Pparα | TCACACAATGCAATCCGTTT | GGCCTTGACCTTGTTCATGT |
| Pparγ | CATGCTTGTGAAGGATGCAAG | TTCTGAAACCGACAGTACTGACAT |
| Lpl | AGACTCGCTCTCAGATGCCCTAC | TCACTTTCAGCCACTGTGCCATAC |
| Plin5 | AAATCAGAGGAGCTGGTGGA | ACGCACAAAGTAGCCCTGTT |
| Atgl | CCCTGACTCGAGTTTCGGAT | CACATAGCGCACCCCTTGAA |
| Hsl | GCCAGCCACAACCTAGCAGAAC | CATCGCCCTCAAAGAAGAGCACTC |
| Gapdh | TCTCTGCTCCTCCCTGTTC | ACACCGACCTTCACCATCT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Wang, L.; Ji, L.; Mu, L.; Wang, K.; Xu, G.; Wang, S.; Ma, Q. Plantaginis Semen Ameliorates Hyperuricemia Induced by Potassium Oxonate. Int. J. Mol. Sci. 2024, 25, 8548. https://doi.org/10.3390/ijms25158548
Liu T, Wang L, Ji L, Mu L, Wang K, Xu G, Wang S, Ma Q. Plantaginis Semen Ameliorates Hyperuricemia Induced by Potassium Oxonate. International Journal of Molecular Sciences. 2024; 25(15):8548. https://doi.org/10.3390/ijms25158548
Chicago/Turabian StyleLiu, Tian, Liting Wang, Li Ji, Leixin Mu, Kaihe Wang, Guang Xu, Shifeng Wang, and Qun Ma. 2024. "Plantaginis Semen Ameliorates Hyperuricemia Induced by Potassium Oxonate" International Journal of Molecular Sciences 25, no. 15: 8548. https://doi.org/10.3390/ijms25158548





