Biomarker Landscape in RASopathies
Abstract
1. Introduction
2. Molecular Biomarkers of RASopathies
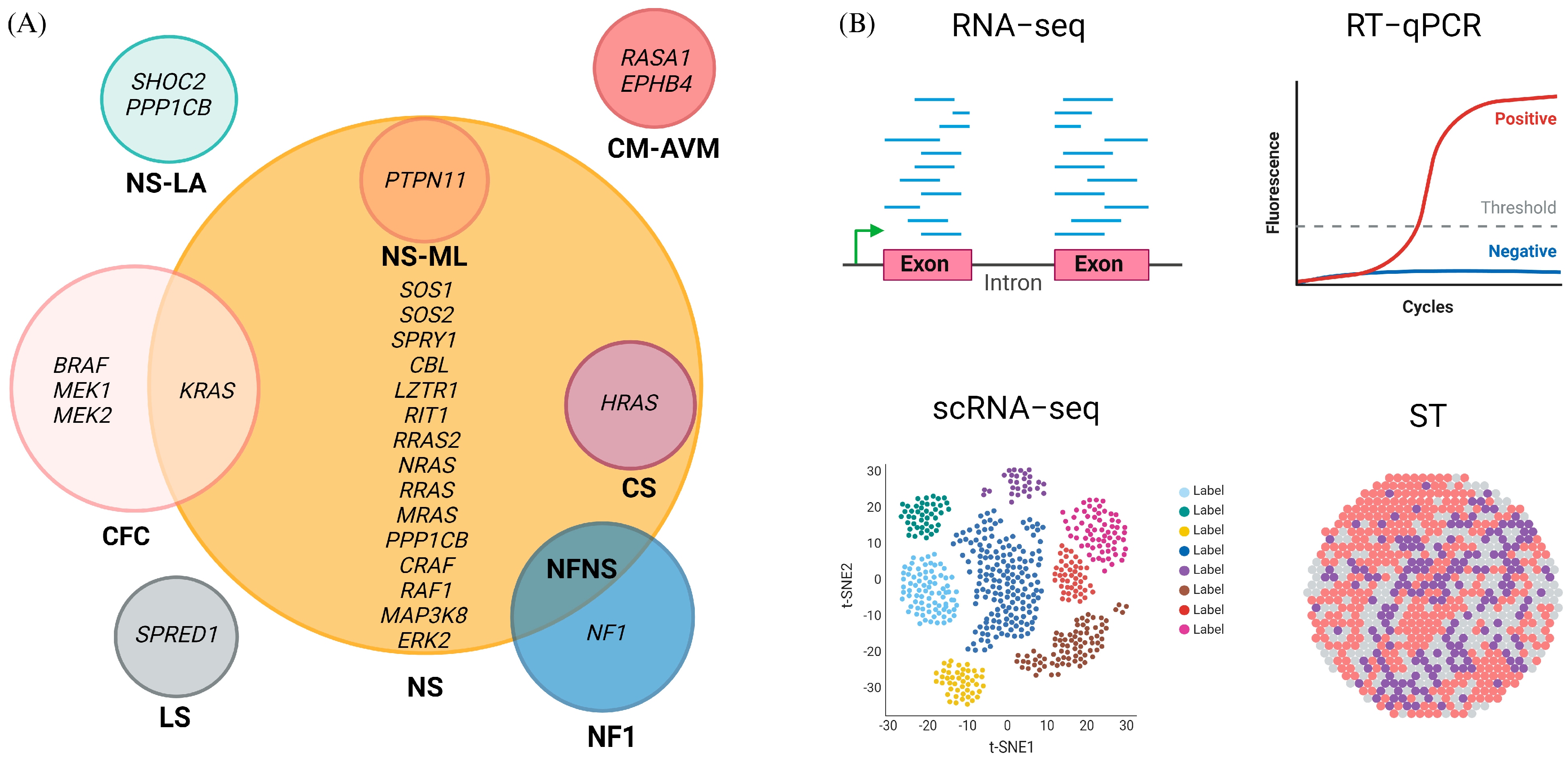
2.1. Genes as Biomarkers
2.2. mRNA Biomarkers
2.3. ncRNA Biomarkers
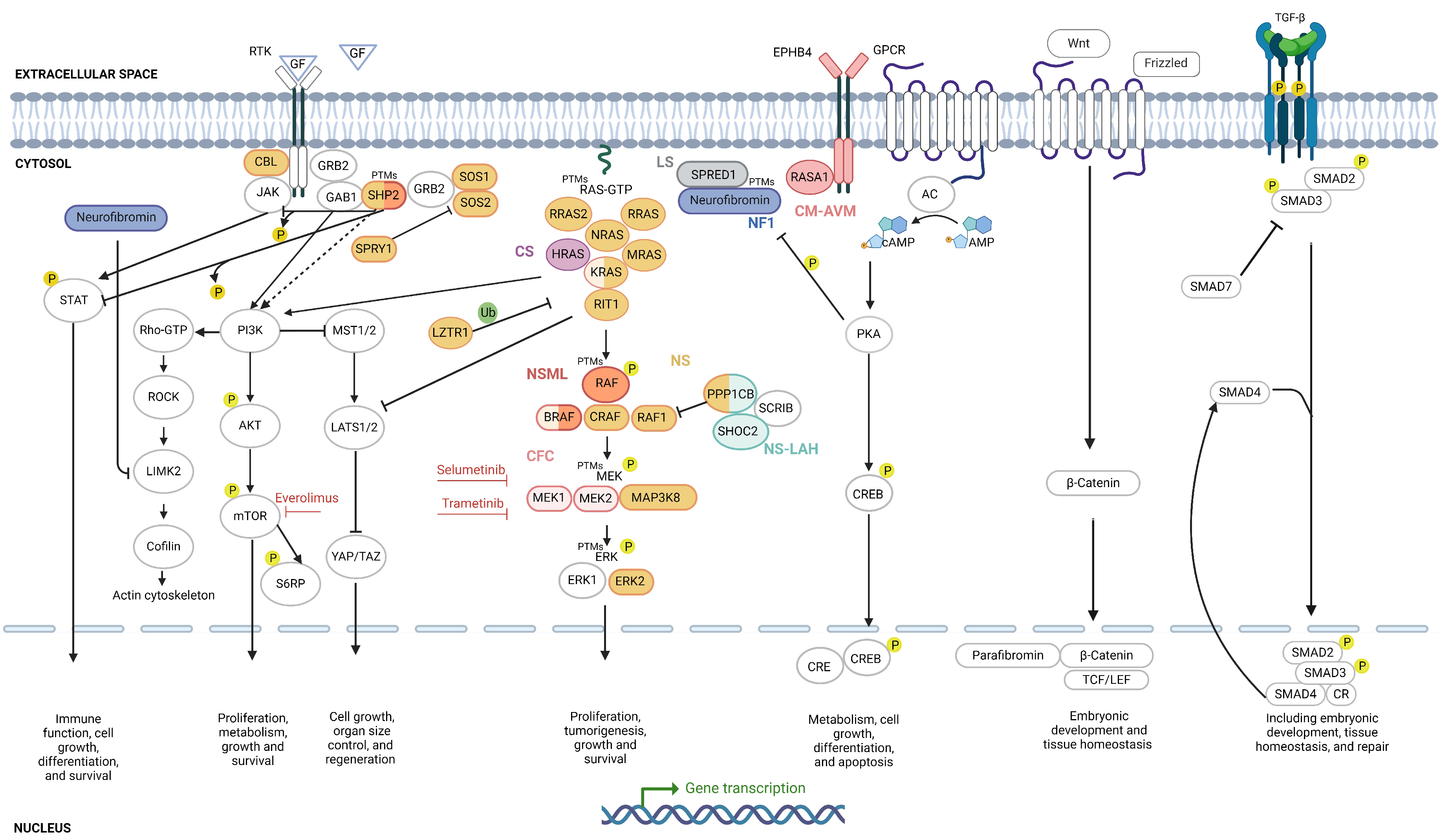
2.4. Protein Biomarkers
2.4.1. RAS/MAPK Biomarkers
2.4.2. PI3K/AKT/mTOR Biomarkers
2.4.3. Rho/ROCK/LIMK2/Cofilin Biomarkers
2.4.4. cAMP/PKA Biomarkers
2.4.5. JAK/STAT Biomarkers
2.4.6. Hippo Pathway Biomarkers
2.4.7. Wnt/β-Catenin Biomarkers
2.4.8. TGF-β Pathway Biomarkers
3. Metabolite Biomarkers
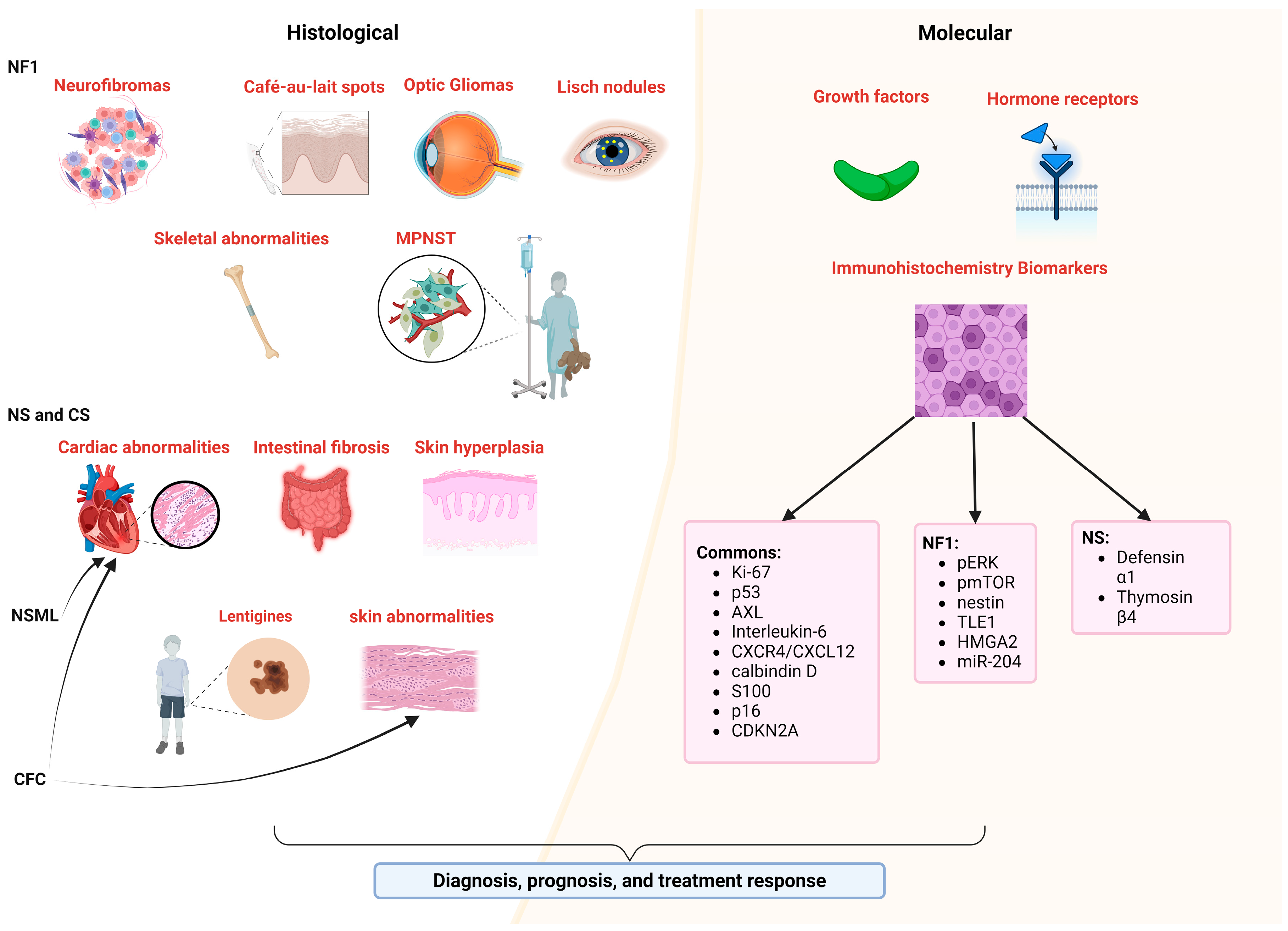
4. Biomarkers in Histology and Molecular Tissue Characterization
5. Physiologic Biomarkers
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rauen, K.A. The RASopathies. Annu. Rev. Genom. Hum. Genet. 2013, 14, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Tidyman, W.E.; Rauen, K.A. The RASopathies: Developmental Syndromes of Ras/MAPK Pathway Dysregulation. Curr. Opin. Genet. Dev. 2009, 19, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Rauen, K.A. Defining RASopathy. Dis. Models Mech. 2022, 15, dmm049344. [Google Scholar] [CrossRef]
- Montero-Bullón, J.F.; González-Velasco, Ó.; Isidoro-García, M.; Lacal, J. Integrated in Silico MS-Based Phosphoproteomics and Network Enrichment Analysis of RASopathy Proteins. Orphanet J. Rare Dis. 2021, 16, 303. [Google Scholar] [CrossRef] [PubMed]
- Saint-Laurent, C.; Mazeyrie, L.; Yart, A.; Edouard, T. Novel Therapeutic Perspectives in Noonan Syndrome and RASopathies. Eur. J. Pediatr. 2023, 183, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, H.; Ahrens-Nicklas, R.C.; Calderon-Anyosa, R.J.C.; Ritter, A.L.; Lin, K.Y.; Rossano, J.W.; Quartermain, M.D.; Banerjee, A. Genotype–Phenotype Association by Echocardiography Offers Incremental Value in Patients with Noonan Syndrome with Multiple Lentigines. Pediatr. Res. 2021, 90, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Huckstadt, V.; Chinton, J.; Gomez, A.; Obregon, M.G.; Gravina, L.P. Noonan Syndrome with Loose Anagen Hair with Variants in the PPP1CB Gene: First Familial Case Reported. Am. J. Med. Genet. Part A 2021, 185, 1256–1260. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cheng, S.; Fu, Y.; Yuan, H. Case Report: A de Novo RASopathy-Causing SHOC2 Variant in a Chinese Girl with Noonan Syndrome-like with Loose Anagen Hair. Front. Genet. 2022, 13, 1040124. [Google Scholar] [CrossRef] [PubMed]
- Bertola, D.R.; Castro, M.A.A.; Yamamoto, G.L.; Honjo, R.S.; Ceroni, J.R.; Buscarilli, M.M.; Freitas, A.B.; Malaquias, A.C.; Pereira, A.C.; Jorge, A.A.L.; et al. Phenotype–Genotype Analysis of 242 Individuals with RASopathies: 18-Year Experience of a Tertiary Center in Brazil. Am. J. Med. Genet. Part C Semin. Med. Genet. 2020, 184, 896–911. [Google Scholar] [CrossRef]
- Pabst, L.; Carroll, J.; Lo, W.; Truxal, K.V. Moyamoya Syndrome in a Child with Legius Syndrome: Introducing a Cerebral Vasculopathy to the SPRED1 Phenotype? Am. J. Med. Genet. Part A 2021, 185, 223–227. [Google Scholar] [CrossRef]
- Scorrano, G.; David, E.; Calì, E.; Chimenz, R.; La Bella, S.; Di Ludovico, A.; Di Rosa, G.; Gitto, E.; Mankad, K.; Nardello, R.; et al. The Cardiofaciocutaneous Syndrome: From Genetics to Prognostic–Therapeutic Implications. Genes 2023, 14, 2111. [Google Scholar] [CrossRef] [PubMed]
- Leoni, C.; Paradiso, F.V.; Foschi, N.; Tedesco, M.; Pierconti, F.; Silvaroli, S.; Diego, M.D.; Birritella, L.; Pantaleoni, F.; Rendeli, C.; et al. Prevalence of Bladder Cancer in Costello Syndrome: New Insights to Drive Clinical Decision-Making. Clin. Genet. 2022, 101, 454–458. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, C.L.; Flanagan, S.; Murati, M.; Boull, C.; McGough, E.; Ameduri, R.; Weigel, B.; Maguiness, S. Successful Management of an Arteriovenous Malformation with Trametinib in a Patient with Capillary-Malformation Arteriovenous Malformation Syndrome and Cardiac Compromise. Pediatr. Dermatol. 2022, 39, 316–319. [Google Scholar] [CrossRef] [PubMed]
- Báez-Flores, J.; Rodríguez-Martín, M.; Lacal, J. The Therapeutic Potential of Neurofibromin Signaling Pathways and Binding Partners. Commun. Biol. 2023, 6, 436. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Mo, J.; Brosseau, J.P.; Shipman, T.; Wang, Y.; Liao, C.P.; Cooper, J.M.; Allaway, R.J.; Gosline, S.J.C.; Guinney, J.; et al. Spatiotemporal Loss of NF1 in Schwann Cell Lineage Leads to Different Types of Cutaneous Neurofibroma Susceptible to Modification by the Hippo Pathway. Cancer Discov. 2019, 9, 114–129. [Google Scholar] [CrossRef] [PubMed]
- Lodi, M.; Boccuto, L.; Carai, A.; Cacchione, A.; Miele, E.; Colafati, G.S.; Camassei, F.D.; de Palma, L.; de Benedictis, A.; Ferretti, E.; et al. Low-Grade Gliomas in Patients with Noonan Syndrome: Case-Based Review of the Literature. Diagnostics 2020, 10, 582. [Google Scholar] [CrossRef] [PubMed]
- Luscan, A.; Shackleford, G.G.; Masliah-Planchon, J.; Laurendeau, I.; Ortonne, N.; Varin, J.; Lallemand, F.; Leroy, K.; Dumaine, V.; Hivelin, M.; et al. The Activation of the WNT Signaling Pathway Is a Hallmark in Neurofibromatosis Type 1 Tumorigenesis. Clin. Cancer Res. 2014, 20, 358–371. [Google Scholar] [CrossRef] [PubMed]
- Ou, F.S.; Michiels, S.; Shyr, Y.; Adjei, A.A.; Oberg, A.L. Biomarker Discovery and Validation: Statistical Considerations. J. Thorac. Oncol. 2021, 16, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Califf, R.M. Biomarker Definitions and Their Applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef]
- McCann, M.R.; De la Rosa, M.V.G.; Rosania, G.R.; Stringer, K.A. L-Carnitine and Acylcarnitines: Mitochondrial Biomarkers for Precision Medicine. Metabolites 2021, 11, 51. [Google Scholar] [CrossRef]
- Gurusamy, N.; Rajasingh, S.; Sigamani, V.; Rajasingh, R.; Isai, D.G.; Czirok, A.; Bittel, D.; Rajasingh, J. Noonan Syndrome Patient-Specific Induced Cardiomyocyte Model Carrying SOS1 Gene Variant c.1654A>G. Exp. Cell Res. 2021, 400, 112508. [Google Scholar] [CrossRef] [PubMed]
- Jafry, M.; Sidbury, R. RASopathies. Clin. Dermatol. 2020, 38, 455–461. [Google Scholar] [CrossRef]
- Motta, M.; Pannone, L.; Pantaleoni, F.; Bocchinfuso, G.; Radio, F.C.; Cecchetti, S.; Ciolfi, A.; Di Rocco, M.; Elting, M.W.; Brilstra, E.H.; et al. Enhanced MAPK1 Function Causes a Neurodevelopmental Disorder within the RASopathy Clinical Spectrum. Am. J. Hum. Genet. 2020, 107, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Tamburrino, F.; Mazzanti, L.; Scarano, E.; Gibertoni, D.; Sirolli, M.; Zioutas, M.; Schiavariello, C.; Perri, A.; Mantovani, A.; Rossi, C.; et al. Lipid Profile in Noonan Syndrome and Related Disorders: Trend by Age, Sex and Genotype. Front. Endocrinol. 2023, 14, 1209339. [Google Scholar] [CrossRef] [PubMed]
- Zenker, M. Clinical Overview on RASopathies. Am. J. Med. Genet. Part C Semin. Med. Genet. 2022, 190, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Linglart, L.; Gelb, B.D. Congenital Heart Defects in Noonan Syndrome: Diagnosis, Management, and Treatment. Am. J. Med. Genet. Part C Semin. Med. Genet. 2020, 184, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Yuan, K.; Chen, L. Molecular Biomarkers, Network Biomarkers, and Dynamic Network Biomarkers for Diagnosis and Prediction of Rare Diseases. Fundam. Res. 2022, 2, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Legius, E.; Messiaen, L.; Wolkenstein, P.; Pancza, P.; Avery, R.A.; Berman, Y.; Blakeley, J.; Babovic-Vuksanovic, D.; Cunha, K.S.; Ferner, R.; et al. Revised Diagnostic Criteria for Neurofibromatosis Type 1 and Legius Syndrome: An International Consensus Recommendation. Genet. Med. 2021, 23, 1506–1513. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Moog, U.; Felbor, U.; Has, C.; Zirn, B. Disorders Caused by Genetic Mosaicism. Dtsch. Arztebl. Int. 2020, 117, 119–125. [Google Scholar] [CrossRef]
- Chang, C.A.; Perrier, R.; Kurek, K.C.; Estrada-Veras, J.; Lehman, A.; Yip, S.; Hendson, G.; Diamond, C.; Pinchot, J.W.; Tran, J.M.; et al. Novel Findings and Expansion of Phenotype in a Mosaic RASopathy Caused by Somatic KRAS Variants. Am. J. Med. Genet. Part A 2021, 185, 2829–2845. [Google Scholar] [CrossRef] [PubMed]
- Amani, V.; Riemondy, K.A.; Fu, R.; Griesinger, A.M.; Grimaldo, E.; De Sousa, G.R.; Gilani, A.; Hemenway, M.; Foreman, N.K.; Donson, A.M.; et al. Integration of Single-Nuclei RNA-Sequencing, Spatial Transcriptomics and Histochemistry Defines the Complex Microenvironment of NF1-Associated Plexiform Neurofibromas. Acta Neuropathol. Commun. 2023, 11, 158. [Google Scholar] [CrossRef] [PubMed]
- Douben, H.; Hoogeveen-Westerveld, M.; Nellist, M.; Louwen, J.; Haan, M.K.D.; Punt, M.; Van Ommeren, B.; Van Unen, L.; Elfferich, P.; Kasteleijn, E.; et al. Functional Assays Combined with Pre-mRNA-Splicing Analysis Improve Variant Classification and Diagnostics for Individuals with Neurofibromatosis Type 1 and Legius Syndrome. Hum. Mutat. 2023, 2023, 9628049. [Google Scholar] [CrossRef]
- Koster, R.; Brandão, R.D.; Tserpelis, D.; van Roozendaal, C.E.P.; van Oosterhoud, C.N.; Claes, K.B.M.; Paulussen, A.D.C.; Sinnema, M.; Vreeburg, M.; van der Schoot, V.; et al. Pathogenic Neurofibromatosis Type 1 (NF1) RNA Splicing Resolved by Targeted RNAseq. npj Genom. Med. 2021, 6, 95. [Google Scholar] [CrossRef] [PubMed]
- Hartung, A.M.; Swensen, J.; Uriz, I.E.; Lapin, M.; Kristjansdottir, K.; Petersen, U.S.S.; Bang, J.M.V.; Guerra, B.; Andersen, H.S.; Dobrowolski, S.F.; et al. The Splicing Efficiency of Activating HRAS Mutations Can Determine Costello Syndrome Phenotype and Frequency in Cancer. PLoS Genet. 2016, 12, e1006039. [Google Scholar] [CrossRef] [PubMed]
- Devonshire, A.S.; Sanders, R.; Wilkes, T.M.; Taylor, M.S.; Foy, C.A.; Huggett, J.F. Application of next Generation qPCR and Sequencing Platforms to mRNA Biomarker Analysis. Methods 2013, 59, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Hanses, U.; Kleinsorge, M.; Roos, L.; Yigit, G.; Li, Y.; Barbarics, B.; El-Battrawy, I.; Lan, H.; Tiburcy, M.; Hindmarsh, R.; et al. Intronic CRISPR Repair in a Preclinical Model of Noonan Syndrome-Associated Cardiomyopathy. Circulation 2020, 142, 1059–1076. [Google Scholar] [CrossRef] [PubMed]
- Drenckhahn, J.D.; Nicin, L.; Akhouaji, S.; Krück, S.; Blank, A.E.; Schänzer, A.; Yörüker, U.; Jux, C.; Tombor, L.; Abplanalp, W.; et al. Cardiomyocyte Hyperplasia and Immaturity but Not Hypertrophy Are Characteristic Features of Patients with RASopathies. J. Mol. Cell. Cardiol. 2023, 178, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Coccia, E.; Valeri, L.; Zuntini, R.; Caraffi, S.G.; Peluso, F.; Pagliai, L.; Vezzani, A.; Pietrangiolillo, Z.; Leo, F.; Melli, N.; et al. Prenatal Clinical Findings in RASA1-Related Capillary Malformation-Arteriovenous Malformation Syndrome. Genes 2023, 14, 549. [Google Scholar] [CrossRef]
- Brosseau, J.P.; Sathe, A.A.; Wang, Y.; Nguyen, T.; Glass, D.A.; Xing, C.; Le, L.Q. Human Cutaneous Neurofibroma Matrisome Revealed by Single-Cell RNA Sequencing. Acta Neuropathol. Commun. 2021, 9, 11. [Google Scholar] [CrossRef]
- Imada, E.L.; Strianese, D.; Edward, D.P.; alThaqib, R.; Price, A.; Arnold, A.; Al-Hussain, H.; Marchionni, L.; Rodriguez, F.J. RNA-Sequencing Highlights Differential Regulated Pathways Involved in Cell Cycle and Inflammation in Orbitofacial Neurofibromas. Brain Pathol. 2022, 32, e13007. [Google Scholar] [CrossRef]
- Yan, H.; Bu, P. Non-Coding RNA in Cancer. Essays Biochem. 2021, 65, 625–639. [Google Scholar] [CrossRef]
- de Carvalho, J.B.; de Morais, G.L.; Vieira, T.C.d.S.; Rabelo, N.C.; Llerena, J.C.; de Carvalho Gonzalez, S.M.; de Vasconcelos, A.T.R. miRNA Genetic Variants Alter Their Secondary Structure and Expression in Patients with RASopathies Syndromes. Front. Genet. 2019, 10, 1144. [Google Scholar] [CrossRef]
- Yu, T.T.; Xu, Q.F.; Li, S.Y.; Huang, H.J.; Dugan, S.; Shao, L.; Roggenbuck, J.A.; Liu, X.T.; Liu, H.Z.; Hirsch, B.A.; et al. Deletion at an 1q24 Locus Reveals a Critical Role of Long Noncoding RNA DNM3OS in Skeletal Development. Cell Biosci. 2021, 11, 47. [Google Scholar] [CrossRef]
- Amirnasr, A.; Verdijk, R.M.; van Kuijk, P.F.; Kartal, P.; Vriends, A.L.M.; French, P.J.; van Royen, M.E.; Taal, W.; Sleijfer, S.; Wiemer, E.A.C. Deregulated microRNAs in Neurofibromatosis Type 1 Derived Malignant Peripheral Nerve Sheath Tumors. Sci. Rep. 2020, 10, 2927. [Google Scholar] [CrossRef]
- Khosravi, T.; Oladnabi, M. The Role of miRNAs and lncRNAs in Neurofibromatosis Type 1. J. Cell. Biochem. 2023, 124, 17–30. [Google Scholar] [CrossRef]
- Nix, J.S.; Yuan, M.; Imada, E.L.; Ames, H.; Marchionni, L.; Gutmann, D.H.; Rodriguez, F.J. Global microRNA Profiling Identified miR-10b-5p as a Regulator of Neurofibromatosis 1 (NF1)-Glioma Migration. Neuropathol. Appl. Neurobiol. 2021, 47, 96–107. [Google Scholar] [CrossRef]
- Mulero-Navarro, S.; Sevilla, A.; Roman, A.C.; Lee, D.F.; D’Souza, S.L.; Pardo, S.; Riess, I.; Su, J.; Cohen, N.; Schaniel, C.; et al. Myeloid Dysregulation in a Human Induced Pluripotent Stem Cell Model of PTPN11-Associated Juvenile Myelomonocytic Leukemia. Cell Rep. 2015, 13, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Herman, A.B.; Tsitsipatis, D.; Gorospe, M. Integrated lncRNA Function upon Genomic and Epigenomic Regulation. Mol. Cell 2022, 82, 2252–2266. [Google Scholar] [CrossRef] [PubMed]
- Tritto, V.; Ferrari, L.; Esposito, S.; Zuccotti, P.; Bianchessi, D.; Natacci, F.; Saletti, V.; Eoli, M.; Riva, P. Non-Coding RNA and Tumor Development in Neurofibromatosis Type 1: ANRIL Rs2151280 Is Associated with Optic Glioma Development and a Mild Phenotype in Neurofibromatosis Type 1 Patients. Genes 2019, 10, 892. [Google Scholar] [CrossRef] [PubMed]
- Gyorffy, B.; Schafer, R. Biomarkers Downstream of RAS: A Search for Robust Transcriptional Targets. Curr. Cancer Drug Targets 2010, 10, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.L.; Philips, M.R. Post-Translational Modification of RAS Proteins. Curr. Opin. Struct. Biol. 2021, 71, 180–192. [Google Scholar] [CrossRef]
- Jang, H.H. Regulation of Protein Degradation by Proteasomes in Cancer. J. Cancer Prev. 2018, 23, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Dohlman, H.G.; Campbell, S.L. Regulation of Large and Small G Proteins by Ubiquitination. J. Biol. Chem. 2019, 294, 18613–18623. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.E.; Yoon, J.Y.; Jeong, W.J.; Jeon, S.H.; Park, Y.; Yoon, J.B.; Park, Y.N.; Kim, H.; Choi, K.Y. H-Ras Is Degraded by Wnt/β-Catenin Signaling via β-TrCP-Mediated Polyubiquitylation. J. Cell Sci. 2009, 122, 842–848. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, H.; Yin, G.; Kawaguchi, R.; Popov, K.I.; Temple, B.; Sasaki, M.; Kofuji, S.; Wolfe, K.; Kofuji, K.; Okumura, K.; et al. Identification of Lysine Methylation in the Core GTPase Domain by GoMADScan. PLoS ONE 2019, 14, e0219436. [Google Scholar] [CrossRef] [PubMed]
- Castellano, E.; Santos, E. Functional Specificity of Ras Isoforms: So Similar but so Different. Genes Cancer 2011, 2, 216–231. [Google Scholar] [CrossRef] [PubMed]
- Ratner, N.; Miller, S.J. A RASopathy Gene Commonly Mutated in Cancer: The Neurofibromatosis Type 1 Tumour Suppressor. Nat. Rev. Cancer 2015, 15, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, B.; Yi, Y.; Chen, D.Y.; Weber, J.D.; Gutmann, D.H. Proteomic Analysis Reveals Hyperactivation of the Mammalian Target of Rapamycin Pathway in Neurofibromatosis 1-Associated Human and Mouse Brain Tumors. Cancer Res. 2005, 65, 2755–2760. [Google Scholar] [CrossRef]
- Bergoug, M.; Doudeau, M.; Godin, F.; Mosrin, C.; Vallée, B.; Bénédetti, H. Neurofibromin Structure, Functions and Regulation. Cells 2020, 9, 2365. [Google Scholar] [CrossRef]
- McFall, T.; Stites, E.C. Identification of RAS Mutant Biomarkers for EGFR Inhibitor Sensitivity Using a Systems Biochemical Approach. Cell Rep. 2021, 37, 110096. [Google Scholar] [CrossRef]
- Koliou, X.; Fedonidis, C.; Kalpachidou, T.; Mangoura, D. Nuclear Import Mechanism of Neurofibromin for Localization on the Spindle and Function in Chromosome Congression. J. Neurochem. 2016, 136, 78–91. [Google Scholar] [CrossRef]
- Feng, L.; Yunoue, S.; Tokuo, H.; Ozawa, T.; Zhang, D.; Patrakitkomjorn, S.; Ichimura, T.; Saya, H.; Araki, N. PKA Phosphorylation and 14-3-3 Interaction Regulate the Function of Neurofibromatosis Type I Tumor Suppressor, Neurofibromin. FEBS Lett. 2004, 557, 275–282. [Google Scholar] [CrossRef]
- Harder, A.; Rosche, M.; Reuß, D.E.; Holtkamp, N.; Uhlmann, K.; Friedrich, R.; Mautner, V.F.; Von Deimling, A. Methylation Analysis of the Neurofibromatosis Type 1 (NF1) Promoter in Peripheral Nerve Sheath Tumours. Eur. J. Cancer 2004, 40, 2820–2828. [Google Scholar] [CrossRef]
- Beauclair, G.; Bridier-Nahmias, A.; Zagury, J.F.; Säb, A.; Zamborlini, A. JASSA: A Comprehensive Tool for Prediction of SUMOylation Sites and SIMs. Bioinformatics 2015, 31, 3483–3491. [Google Scholar] [CrossRef]
- Yan, W.; Markegard, E.; Dharmaiah, S.; Urisman, A.; Drew, M.; Esposito, D.; Scheffzek, K.; Nissley, D.V.; McCormick, F.; Simanshu, D.K. Structural Insights into the SPRED1-Neurofibromin-KRAS Complex and Disruption of SPRED1-Neurofibromin Interaction by Oncogenic EGFR. Cell Rep. 2020, 32, 107909. [Google Scholar] [CrossRef]
- Pandit, B.; Sarkozy, A.; Pennacchio, L.A.; Carta, C.; Oishi, K.; Martinelli, S.; Pogna, E.A.; Schackwitz, W.; Ustaszewska, A.; Landstrom, A.; et al. Gain-of-Function RAF1 Mutations Cause Noonan and LEOPARD Syndromes with Hypertrophic Cardiomyopathy. Nat. Genet. 2007, 39, 1007–1012. [Google Scholar] [CrossRef]
- Razzaque, M.A.; Nishizawa, T.; Komoike, Y.; Yagi, H.; Furutani, M.; Amo, R.; Kamisago, M.; Momma, K.; Katayama, H.; Nakagawa, M.; et al. Germline Gain-of-Function Mutations in RAF1 Cause Noonan Syndrome. Nat. Genet. 2007, 39, 1013–1017. [Google Scholar] [CrossRef]
- Heidorn, S.J.; Milagre, C.; Whittaker, S.; Nourry, A.; Niculescu-Duvas, I.; Dhomen, N.; Hussain, J.; Reis-Filho, J.S.; Springer, C.J.; Pritchard, C.; et al. Kinase-Dead BRAF and Oncogenic RAS Cooperate to Drive Tumor Progression through CRAF. Cell 2010, 140, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.E.; Allanson, J.E.; Tartaglia, M.; Gelb, B.D. Noonan Syndrome. Lancet 2013, 381, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Galperin, E.; Wilson, P.; Abdelmoti, L.; Norcross, R.; Palayam, M. Proteins of the Ubiquitin System in the Shoc2—ERK1/2 Signaling Axis and Noonan-like Syndrome with Loose Anagen Hair (NSLAH) RASopathy. FASEB J. 2022, 36, r1999. [Google Scholar] [CrossRef]
- Bivona, T.G.; Quatela, S.E.; Bodemann, B.O.; Ahearn, I.M.; Soskis, M.J.; Mor, A.; Miura, J.; Wiener, H.H.; Wright, L.; Saba, S.G.; et al. PKC Regulates a Farnesyl-Electrostatic Switch on K-Ras That Promotes Its Association with Bcl-XL on Mitochondria and Induces Apoptosis. Mol. Cell 2006, 21, 481–493. [Google Scholar] [CrossRef]
- Choi, B.H.; Philips, M.R.; Chen, Y.; Lu, L.; Dai, W. K-Ras Lys-42 Is Crucial for Its Signaling, Cell Migration, and Invasion. J. Biol. Chem. 2018, 293, 17574–17581. [Google Scholar] [CrossRef] [PubMed]
- Ottaiano, A.; Normanno, N.; Facchini, S.; Cassata, A.; Nappi, A.; Romano, C.; Silvestro, L.; Stefano, A.D.; Rachiglio, A.M.; Roma, C.; et al. Study of Ras Mutations’ Prognostic Value in Metastatic Colorectal Cancer: Storia Analysis. Cancers 2020, 12, 1919. [Google Scholar] [CrossRef] [PubMed]
- Simão, S.; Agostinho, R.R.; Martínez-Ruiz, A.; Araújo, I.M. Regulation of Ras Signaling by S-Nitrosylation. Antioxidants 2023, 12, 1562. [Google Scholar] [CrossRef] [PubMed]
- Martin-Vega, A.; Cobb, M.H. Navigating the ERK1/2 MAPK Cascade. Biomolecules 2023, 13, 1555. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; You, J.; Luo, W.; Yue, J.; Ma, L.; Xiao, W.; Zhu, D.; Wu, Z.; Wang, D.; Nadiminty, N.; et al. The N-Terminal Kinase Suppressor of Ras Complex Has a Weak Nucleoside Diphosphate Kinase Activity. Thorac. Cancer 2010, 1, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Lauriol, J.; Thul, J.; Behnke-Hall, K.; Logeswaran, T.; Schänzer, A.; Böğürcü, N.; Garvalov, B.K.; Zenker, M.; Gelb, B.D.; et al. Rapidly Progressive Hypertrophic Cardiomyopathy in an Infant with Noonan Syndrome with Multiple Lentigines: Palliative Treatment with a Rapamycin Analog. Am. J. Med. Genet. Part A 2015, 167, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Ranza, E.; Guimier, A.; Verloes, A.; Capri, Y.; Marques, C.; Auclair, M.; Mathieu-Dramard, M.; Morin, G.; Thevenon, J.; Faivre, L.; et al. Overlapping Phenotypes between SHORT and Noonan Syndromes in Patients with PTPN11 Pathogenic Variants. Clin. Genet. 2020, 98, 10–18. [Google Scholar] [CrossRef]
- Endo, M.; Yamamoto, H.; Setsu, N.; Kohashi, K.; Takahashi, Y.; Ishii, T.; Iida, K.I.; Matsumoto, Y.; Hakozaki, M.; Aoki, M.; et al. Prognostic Significance of AKT/mTOR and MAPK Pathways and Antitumor Effect of mTOR Inhibitor in NF1-Related and Sporadic Malignant Peripheral Nerve Sheath Tumors. Clin. Cancer Res. 2013, 19, 450–461. [Google Scholar] [CrossRef]
- Solares, I.; Viñal, D.; Morales-Conejo, M.; Rodriguez-Salas, N.; Feliu, J. Novel Molecular Targeted Therapies for Patients with Neurofibromatosis Type 1 with Inoperable Plexiform Neurofibromas: A Comprehensive Review. ESMO Open 2021, 6, 100223. [Google Scholar] [CrossRef] [PubMed]
- Ugwu, N.; Cheraghlou, S.; Ko, C.J.; Cohen, J.M. Incidence, Survival, and Prognostic Factors Associated with Malignant Nodular Hidradenoma in the United States. J. Am. Acad. Dermatol. 2023, 88, 875–877. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, N.J.; Prabhu, S.P.; Reddy, A.T.; Fisher, M.J.; Packer, R.; Goldman, S.; Robison, N.J.; Gutmann, D.H.; Viskochil, D.H.; Allen, J.C.; et al. A Phase II Study of Continuous Oral mTOR Inhibitor Everolimus for Recurrent, Radiographic-Progressive Neurofibromatosis Type 1-Associated Pediatric Low-Grade Glioma: A Neurofibromatosis Clinical Trials Consortium Study. Neuro Oncol. 2020, 22, 1527–1535. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, K.; Wakioka, T.; Nishinakamura, H.; Kamio, M.; Yang, L.; Inoue, M.; Hasegawa, M.; Yonemitsu, Y.; Komiya, S.; Yoshimura, A. The Sprouty-Related Protein, Spred, Inhibits Cell Motility, Metastasis, and Rho-Mediated Actin Reorganization. Oncogene 2004, 23, 5567–5576. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Diggs-Andrews, K.A.; Gianino, S.M.; Gutmann, D.H. Neurofibromatosis-1 Heterozygosity Impairs CNS Neuronal Morphology in a cAMP/PKA/ROCK-Dependent Manner. Mol. Cell. Neurosci. 2012, 49, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, T.; Araki, N.; Yunoue, S.; Tokuo, H.; Feng, L.; Patrakitkomjorn, S.; Hara, T.; Ichikawa, Y.; Matsumoto, K.; Fuji, K.; et al. The Neurofibromatosis Type 1 Gene Product Neurofibromin Enhances Cell Motility by Regulating Actin Filament Dynamics via the Rho-ROCK-LIMK2-Cofilin Pathway. J. Biol. Chem. 2005, 280, 39524–39533. [Google Scholar] [CrossRef] [PubMed]
- Langdon, Y.; Tandon, P.; Paden, E.; Duddy, J.; Taylor, J.M.; Conlon, F.L. SHP-2 Acts via ROCK to Regulate the Cardiac Actin Cytoskeleton. Development 2012, 139, 948–957. [Google Scholar] [CrossRef]
- Chen, M.; Lu, L.; Cheng, D.; Zhang, J.; Liu, X.; Zhang, J.; Zhang, T. Icariin Promotes Osteogenic Differentiation in a Cell Model with NF1 Gene Knockout by Activating the cAMP/PKA/CREB Pathway. Molecules 2023, 28, 5128. [Google Scholar] [CrossRef] [PubMed]
- Mazuelas, H.; Magallón-Lorenz, M.; Uriarte-Arrazola, I.; Negro, A.; Rosas, I.; Blanco, I.; Castellanos, E.; Lázaro, C.; Gel, B.; Carrió, M.; et al. Unbalancing cAMP and Ras/MAPK Pathways as a Therapeutic Strategy for Cutaneous Neurofibromas. JCI Insight 2024, 9, 168826. [Google Scholar] [CrossRef]
- Biayna, J.; Mazuelas, H.; Gel, B.; Terribas, E.; Dumbovic, G.; Rosas, I.; Fernández-Rodriguez, J.; Blanco, I.; Castellanos, E.; Carrió, M.; et al. Using Antisense Oligonucleotides for the Physiological Modulation of the Alternative Splicing of NF1 Exon 23a during PC12 Neuronal Differentiation. Sci. Rep. 2021, 11, 3661. [Google Scholar] [CrossRef]
- Machado Almeida, P.; Lago Solis, B.; Stickley, L.; Feidler, A.; Nagoshi, E. Neurofibromin 1 in Mushroom Body Neurons Mediates Circadian Wake Drive through Activating cAMP–PKA Signaling. Nat. Commun. 2021, 12, 5758. [Google Scholar] [CrossRef]
- Xue, C.; Yao, Q.; Gu, X.; Shi, Q.; Yuan, X.; Chu, Q.; Bao, Z.; Lu, J.; Li, L. Evolving Cognition of the JAK-STAT Signaling Pathway: Autoimmune Disorders and Cancer. Signal Transduct. Target. Ther. 2023, 8, 204. [Google Scholar] [CrossRef] [PubMed]
- Miot, H.A.; Criado, P.R.; de Castro, C.C.S.; Ianhez, M.; Talhari, C.; Ramos, P.M. JAK-STAT Pathway Inhibitors in Dermatology. An. Bras. Dermatol. 2023, 98, 656–677. [Google Scholar] [CrossRef] [PubMed]
- Erdogan, F.; Radu, T.B.; Orlova, A.; Qadree, A.K.; de Araujo, E.D.; Israelian, J.; Valent, P.; Mustjoki, S.M.; Herling, M.; Moriggl, R.; et al. JAK-STAT Core Cancer Pathway: An Integrative Cancer Interactome Analysis. J. Cell. Mol. Med. 2022, 26, 2049–2062. [Google Scholar] [CrossRef] [PubMed]
- Chan, G.; Kalaitzidis, D.; Neel, B.G. The Tyrosine Phosphatase Shp2 (PTPN11) in Cancer. Cancer Metastasis Rev. 2008, 27, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Seif, F.; Khoshmirsafa, M.; Aazami, H.; Mohsenzadegan, M.; Sedighi, G.; Bahar, M. The Role of JAK-STAT Signaling Pathway and Its Regulators in the Fate of T Helper Cells. Cell Commun. Signal. 2017, 15, 23. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Keng, V.W.; Patmore, D.M.; Kendall, J.J.; Patel, A.V.; Jousma, E.; Jessen, W.J.; Choi, K.; Tschida, B.R.; Silverstein, K.A.T.; et al. Insertional Mutagenesis Identifies a STAT3/Arid1b/β-Catenin Pathway Driving Neurofibroma Initiation. Cell Rep. 2016, 14, 1979–1990. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Meng, Z.; Chen, R.; Guan, K.-L. The Hippo Pathway: Biology and Pathophysiology. Annu. Rev. Biochem. 2019, 88, 577–604. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, E. Ras and the Hippo Pathway in Cancer. In Conquering RAS: From Biology to Cancer Therapy; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 25–39. ISBN 978-0-12-803541-2. [Google Scholar]
- Faden, D.L.; Asthana, S.; Tihan, T.; De Risi, J.; Kliot, M. Whole Exome Sequencing of Growing and Non-Growing Cutaneous Neurofibromas from a Single Patient with Neurofibromatosis Type 1. PLoS ONE 2017, 12, e0170348. [Google Scholar] [CrossRef]
- Vélez-Reyes, G.L.; Koes, N.; Ryu, J.H.; Kaufmann, G.; Berner, M.; Weg, M.T.; Wolf, N.K.; Rathe, S.K.; Ratner, N.; Moriarity, B.S.; et al. Transposon Mutagenesis-Guided Crispr/Cas9 Screening Strongly Implicates Dysregulation of Hippo/Yap Signaling in Malignant Peripheral Nerve Sheath Tumor Development. Cancers 2021, 13, 1584. [Google Scholar] [CrossRef]
- Zhao, H.; Ming, T.; Tang, S.; Ren, S.; Yang, H.; Liu, M.; Tao, Q.; Xu, H. Wnt Signaling in Colorectal Cancer: Pathogenic Role and Therapeutic Target. Mol. Cancer 2022, 21, 144. [Google Scholar] [CrossRef] [PubMed]
- Jeong, W.J.; Ro, E.J.; Choi, K.Y. Interaction between Wnt/β-Catenin and RAS-ERK Pathways and an Anti-Cancer Strategy via Degradations of β-Catenin and RAS by Targeting the Wnt/β-Catenin Pathway. npj Precis. Oncol. 2018, 2, 5. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Gan, W.J. Wnt/β-Catenin Signaling Pathway in the Development and Progression of Colorectal Cancer. Cancer Manag. Res. 2023, 15, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Tompa, M.; Nagy, A.; Komoly, S.; Kalman, B. Wnt Pathway Markers in Molecular Subgroups of Glioblastoma. Brain Res. 2019, 1718, 114–125. [Google Scholar] [CrossRef]
- Noda, S.; Takahashi, A.; Hayashi, T.; Tanuma, S.I.; Hatakeyama, M. Determination of the Catalytic Activity of LEOPARD Syndrome-Associated SHP2 Mutants toward Parafibromin, a Bona Fide SHP2 Substrate Involved in Wnt Signaling. Biochem. Biophys. Res. Commun. 2016, 469, 1133–1139. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Budi, E.H. Specificity, Versatility, and Control of TGF-b Family Signaling. Sci. Signal. 2019, 12, aav5183. [Google Scholar] [CrossRef] [PubMed]
- Tzavlaki, K.; Moustakas, A. TGF-Β Signaling. Biomolecules 2020, 10, 487. [Google Scholar] [CrossRef] [PubMed]
- Torres, K.C.L.; Lima, G.; Simões e Silva, A.C.; Lubambo, I.; Rodrigues, L.O.; Rodrigues, L.; Silveira, K.D.; Vieira, É.L.M.; Romano-Silva, M.A.; Miranda, D.M. Immune Markers in the RASopathy Neurofibromatosis Type 1. J. Neuroimmunol. 2016, 295–296, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Park, J.S.; Kim, D.; Kim, B.; Lee, J.H.; Nam, Y.; Yoo, H.W.; Lee, B.H.; Han, Y.M. SHP2 Mutations Induce Precocious Gliogenesis of Noonan Syndrome-Derived iPSCs during Neural Development in Vitro. Stem Cell Res. Ther. 2020, 11, 209. [Google Scholar] [CrossRef]
- Jin, Q.; Ma, R.C.W. Metabolomics in Diabetes and Diabetic Complications: Insights from Epidemiological Studies. Cells 2021, 10, 2832. [Google Scholar] [CrossRef]
- Occelli, C.; Levraut, J.; Pourcher, T. Metabolomics, the Future of Biomarkers? Eur. J. Emerg. Med. 2024, 31, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.S.; Jansen, A.K.; Rodrigues, L.O.C.; Matos, C.M.; Souza, M.L.R.; Miranda, D.M.; de Rezende, N.A. Increased Insulin Sensitivity in Individuals with Neurofibromatosis Type 1. Arch. Endocrinol. Metab. 2018, 62, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.S.; Jansen, A.K.; Rodrigues, L.O.C.; Matos, C.M.; Souza, M.L.R.; de Souza, J.F.; de Fátima Haueisen Sander Diniz, M.; Barreto, S.M.; Diniz, L.M.; de Rezende, N.A.; et al. Lower Fasting Blood Glucose in Neurofibromatosis Type 1. Endocr. Connect. 2016, 5, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Tritz, R.; Benson, T.; Harris, V.; Hudson, F.Z.; Mintz, J.; Zhang, H.; Kennard, S.; Chen, W.; Stepp, D.W.; Csanyi, G.; et al. Nf1 Heterozygous Mice Recapitulate the Anthropometric and Metabolic Features of Human Neurofibromatosis Type 1. Transl. Res. 2021, 228, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Noronha, R.M.; Villares, S.M.F.; Torres, N.; Quedas, E.P.S.; Homma, T.K.; Albuquerque, E.V.A.; Moraes, M.B.; Funari, M.F.A.; Bertola, D.R.; Jorge, A.A.L.; et al. Noonan Syndrome Patients beyond the Obvious Phenotype: A Potential Unfavorable Metabolic Profile. Am. J. Med. Genet. Part A 2021, 185, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Fahrner, J.A.; Frazier, A.; Bachir, S.; Walsh, M.F.; Applegate, C.D.; Thompson, R.; Halushka, M.K.; Murphy, A.M.; Gunay-Aygun, M. A Rasopathy Phenotype with Severe Congenital Hypertrophic Obstructive Cardiomyopathy Associated with a PTPN11 Mutation and a Novel Variant in SOS1. Am. J. Med. Genet. A 2012, 158A, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Gripp, K.W.; Kawame, H.; Viskochil, D.H.; Nicholson, L. Elevated Catecholamine Metabolites in Patients with Costello Syndrome. Am. J. Med. Genet. 2004, 128A, 48–51. [Google Scholar] [CrossRef] [PubMed]
- Marciano, D.P.; Snyder, M.P. Personalized Metabolomics. In High-Throughput Metabolomics: Methods and Protocols; D’Alessandro, A., Ed.; Springer: New York, NY, USA, 2019; pp. 447–456. ISBN 978-1-4939-9236-2. [Google Scholar]
- Wallis, D.; Stemmer-Rachamimov, A.; Adsit, S.; Korf, B.; Pichard, D.; Blakeley, J.; Sarin, K.Y. Status and Recommendations for Incorporating Biomarkers for Cutaneous Neurofibromas Into Clinical Research. Neurology 2021, 97, S42–S49. [Google Scholar] [CrossRef] [PubMed]
- Hilal, N.; Chen, Z.; Chen, M.H.; Choudhury, S. RASopathies and Cardiac Manifestations. Front. Cardiovasc. Med. 2023, 10, 1176828. [Google Scholar] [CrossRef]
- Meier, A.B.; Raj Murthi, S.; Rawat, H.; Toepfer, C.N.; Santamaria, G.; Schmid, M.; Mastantuono, E.; Schwarzmayr, T.; Berutti, R.; Cleuziou, J.; et al. Cell Cycle Defects Underlie Childhood-Onset Cardiomyopathy Associated with Noonan Syndrome. iScience 2022, 25, 103596. [Google Scholar] [CrossRef]
- Siegel, D.H.; Mann, J.A.; Krol, A.L.; Rauen, K.A. Dermatological Phenotype in Costello Syndrome: Consequences of Ras Dysregulation in Development. Br. J. Dermatol. 2012, 166, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Magaki, S.; Hojat, S.A.; Wei, B.; So, A.; Yong, W.H. An Introduction to the Performance of Immunohistochemistry. In Methods in Molecular Biology; NIH Public Access: Rockville Pike Bethesda, MD, USA, 2019; Volume 1897, pp. 289–298. [Google Scholar]
- Peltonen, S.; Kallionpää, R.A.; Peltonen, J. Neurofibromatosis Type 1 (NF1) Gene: Beyond Café Au Lait Spots and Dermal Neurofibromas. Exp. Dermatol. 2017, 26, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Helfferich, J.; Nijmeijer, R.; Brouwer, O.F.; Boon, M.; Fock, A.; Hoving, E.W.; Meijer, L.; den Dunnen, W.F.A.; de Bont, E.S.J.M. Neurofibromatosis Type 1 Associated Low Grade Gliomas: A Comparison with Sporadic Low Grade Gliomas. Crit. Rev. Oncol. Hematol. 2016, 104, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Ozarslan, B.; Russo, T.; Argenziano, G.; Santoro, C.; Piccolo, V. Cutaneous Findings in Neurofibromatosis Type 1. Cancers 2021, 13, 463. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, M.M.; Antonescu, C.R.; Fletcher, C.D.M.; Kim, A.; Lazar, A.J.; Quezado, M.M.; Reilly, K.M.; Stemmer-Rachamimov, A.; Stewart, D.R.; Viskochil, D.; et al. Histopathologic Evaluation of Atypical Neurofibromatous Tumors and Their Transformation into Malignant Peripheral Nerve Sheath Tumor in Patients with Neurofibromatosis 1—A Consensus Overview. Hum. Pathol. 2017, 67, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Dombi, E.; Tepas, K.; Fox, E.; Martin, S.; Wolters, P.; Balis, F.M.; Jayaprakash, N.; Turkbey, B.; Muradyan, N.; et al. Phase I Trial and Pharmacokinetic Study of Sorafenib in Children with Neurofibromatosis Type I and Plexiform Neurofibromas. Pediatr. Blood Cancer 2013, 60, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Meyerholz, D.K.; Ofori-Amanfo, G.K.; Leidinger, M.R.; Goeken, J.A.; Khanna, R.; Sieren, J.C.; Darbro, B.W.; Quelle, D.E.; Weimer, J.M. Immunohistochemical Markers for Prospective Studies in Neurofibromatosis-1 Porcine Models. J. Histochem. Cytochem. 2017, 65, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Guedes-Corrêa, J.; Cardoso, R. Immunohistochemical Markers for Schwannomas, Neurofibromas and Malignant Peripheral Nerve Sheath Tumors—What Can the Recent Literature Tell Us? Arq. Bras. Neurocir. Braz. Neurosurg. 2018, 37, 105–112. [Google Scholar] [CrossRef]
- Martin, E.; Acem, I.; Grünhagen, D.J.; Bovée, J.V.M.G.; Verhoef, C. Prognostic Significance of Immunohistochemical Markers and Genetic Alterations in Malignant Peripheral Nerve Sheath Tumors: A Systematic Review. Front. Oncol. 2020, 10, 594069. [Google Scholar] [CrossRef]
- Johansson, G.; Peng, P.C.; Huang, P.Y.; Chien, H.F.; Hua, K.T.; Kuo, M.L.; Chen, C.T.; Lee, M.J. Soluble AXL: A Possible Circulating Biomarker for Neurofibromatosis Type 1 Related Tumor Burden. PLoS ONE 2014, 9, e115916. [Google Scholar] [CrossRef]
- Brems, H.; Pasmant, E.; Van Minkelen, R.; Wimmer, K.; Upadhyaya, M.; Legius, E.; Messiaen, L. Review and Update of SPRED1 Mutations Causing Legius Syndrome. Hum. Mutat. 2012, 33, 1538–1546. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, M.; Yao, Z. Molecular Screening Strategies for NF1-like Syndromes with Café-Au-Lait Macules (Review). Mol. Med. Rep. 2016, 14, 4023–4029. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guglielmi, F.; Kirschner, F.; Staderini, E.; Iavarone, F.; Fiorino, A.; Gallenzi, P. Proteomic Analysis of Salivary Inflammatory Biomarkers of Developmental Gingival Enlargements in Patients with West and Noonan Syndromes: A Preliminary Pilot Single-Center Retrospective Study. Eur. Rev. Med. Pharmacol. Sci. 2023, 27, 11093–11102. [Google Scholar] [CrossRef] [PubMed]
- Moniez, S.; Pienkowski, C.; Lepage, B.; Hamdi, S.; Daudin, M.; Oliver, I.; Jouret, B.; Cartault, A.; Diene, G.; Verloes, A.; et al. Noonan Syndrome Males Display Sertoli Cell-Specific Primary Testicular Insufficiency. Eur. J. Endocrinol. 2018, 179, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Xie, Y.M.; Wang, S.S.; Zhang, Z.W. Cardiovascular Abnormalities and Gene Mutations in Children with Noonan Syndrome. Front. Genet. 2022, 13, 915129. [Google Scholar] [CrossRef] [PubMed]
- Pierpont, E.I.; Hudock, R.L.; Foy, A.M.; Semrud-Clikeman, M.; Pierpont, M.E.; Berry, S.A.; Shanley, R.; Rubin, N.; Sommer, K.; Moertel, C.L. Social Skills in Children with RASopathies: A Comparison of Noonan Syndrome and Neurofibromatosis Type 1. J. Neurodev. Disord. 2018, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Pierpont, M.E.M.; Magoulas, P.L.; Adi, S.; Kavamura, M.I.; Neri, G.; Noonan, J.; Pierpont, E.I.; Reinker, K.; Roberts, A.E.; Shankar, S.; et al. Cardio-Facio-Cutaneous Syndrome: Clinical Features, Diagnosis, and Management Guidelines. Pediatrics 2014, 134, e1149. [Google Scholar] [CrossRef] [PubMed]
- Hebron, K.E.; Hernandez, E.R.; Yohe, M.E. The RASopathies: From Pathogenetics to Therapeutics. DMM Dis. Models Mech. 2022, 15, dmm049107. [Google Scholar] [CrossRef] [PubMed]
- Palit, A.; Inamadar, A.C. RASopathies: Dermatologists’ Viewpoints. Indian J. Dermatol. Venereol. Leprol. 2022, 88, 452–463. [Google Scholar] [CrossRef]
- Fowlkes, J.L.; Thrailkill, K.M.; Bunn, R.C. RASopathies: The Musculoskeletal Consequences and Their Etiology and Pathogenesis. Bone 2021, 152, 116060. [Google Scholar] [CrossRef]
- Lioncino, M.; Monda, E.; Verrillo, F.; Moscarella, E.; Calcagni, G.; Drago, F.; Marino, B.; Digilio, M.C.; Putotto, C.; Calabrò, P.; et al. Hypertrophic Cardiomyopathy in RASopathies: Diagnosis, Clinical Characteristics, Prognostic Implications, and Management. Heart Fail. Clin. 2022, 18, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Delogu, A.B.; Blandino, R.; Leoni, C.; Tartaglia, M.; Zampino, G. RASopathies and Sigmoid-Shaped Ventricular Septum Morphology: Evidence of a Previously Unappreciated Cardiac Phenotype. Pediatr. Res. 2023, 93, 752–754. [Google Scholar] [CrossRef] [PubMed]
- Mangels, R.; Blumenfeld, Y.J.; Homeyer, M.; Mrazek-Pugh, B.; Hintz, S.R.; Hudgins, L. RASopathies: A Significant Cause of Polyhydramnios? Prenat. Diagn. 2021, 41, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.E.; Baek, S.T. Neurodevelopmental Aspects of RASopathies. Mol. Cells 2019, 42, 441–447. [Google Scholar] [CrossRef] [PubMed]
- McGhee, C.A.; Honari, H.; Siqueiros-Sanchez, M.; Serur, Y.; van Staalduinen, E.K.; Stevenson, D.; Bruno, J.L.; Raman, M.M.; Green, T. Influences of RASopathies on Neuroanatomical Variation in Children. Biol. Psychiatry Cogn. Neurosci. Neuroimaging, 2024; in press. [Google Scholar] [CrossRef] [PubMed]
- Siano, M.A.; Pivonello, R.; Salerno, M.; Falco, M.; Mauro, C.; De Brasi, D.; Klain, A.; Sestito, S.; De Luca, A.; Pinna, V.; et al. Endocrine System Involvement in Patients with RASopathies: A Case Series. Front. Endocrinol. 2022, 13, 1030398. [Google Scholar] [CrossRef]
- Bizzarri, C.; Bottaro, G. Endocrine Implications of Neurofibromatosis 1 in Childhood. Horm. Res. Paediatr. 2015, 83, 232–241. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrito, N.; Báez-Flores, J.; Rodríguez-Martín, M.; Sastre-Rodríguez, J.; Coppola, A.; Isidoro-García, M.; Prieto-Matos, P.; Lacal, J. Biomarker Landscape in RASopathies. Int. J. Mol. Sci. 2024, 25, 8563. https://doi.org/10.3390/ijms25168563
Ferrito N, Báez-Flores J, Rodríguez-Martín M, Sastre-Rodríguez J, Coppola A, Isidoro-García M, Prieto-Matos P, Lacal J. Biomarker Landscape in RASopathies. International Journal of Molecular Sciences. 2024; 25(16):8563. https://doi.org/10.3390/ijms25168563
Chicago/Turabian StyleFerrito, Noemi, Juan Báez-Flores, Mario Rodríguez-Martín, Julián Sastre-Rodríguez, Alessio Coppola, María Isidoro-García, Pablo Prieto-Matos, and Jesus Lacal. 2024. "Biomarker Landscape in RASopathies" International Journal of Molecular Sciences 25, no. 16: 8563. https://doi.org/10.3390/ijms25168563
APA StyleFerrito, N., Báez-Flores, J., Rodríguez-Martín, M., Sastre-Rodríguez, J., Coppola, A., Isidoro-García, M., Prieto-Matos, P., & Lacal, J. (2024). Biomarker Landscape in RASopathies. International Journal of Molecular Sciences, 25(16), 8563. https://doi.org/10.3390/ijms25168563





