Unveiling the Secrets of Oil Palm Genetics: A Look into Omics Research
Abstract
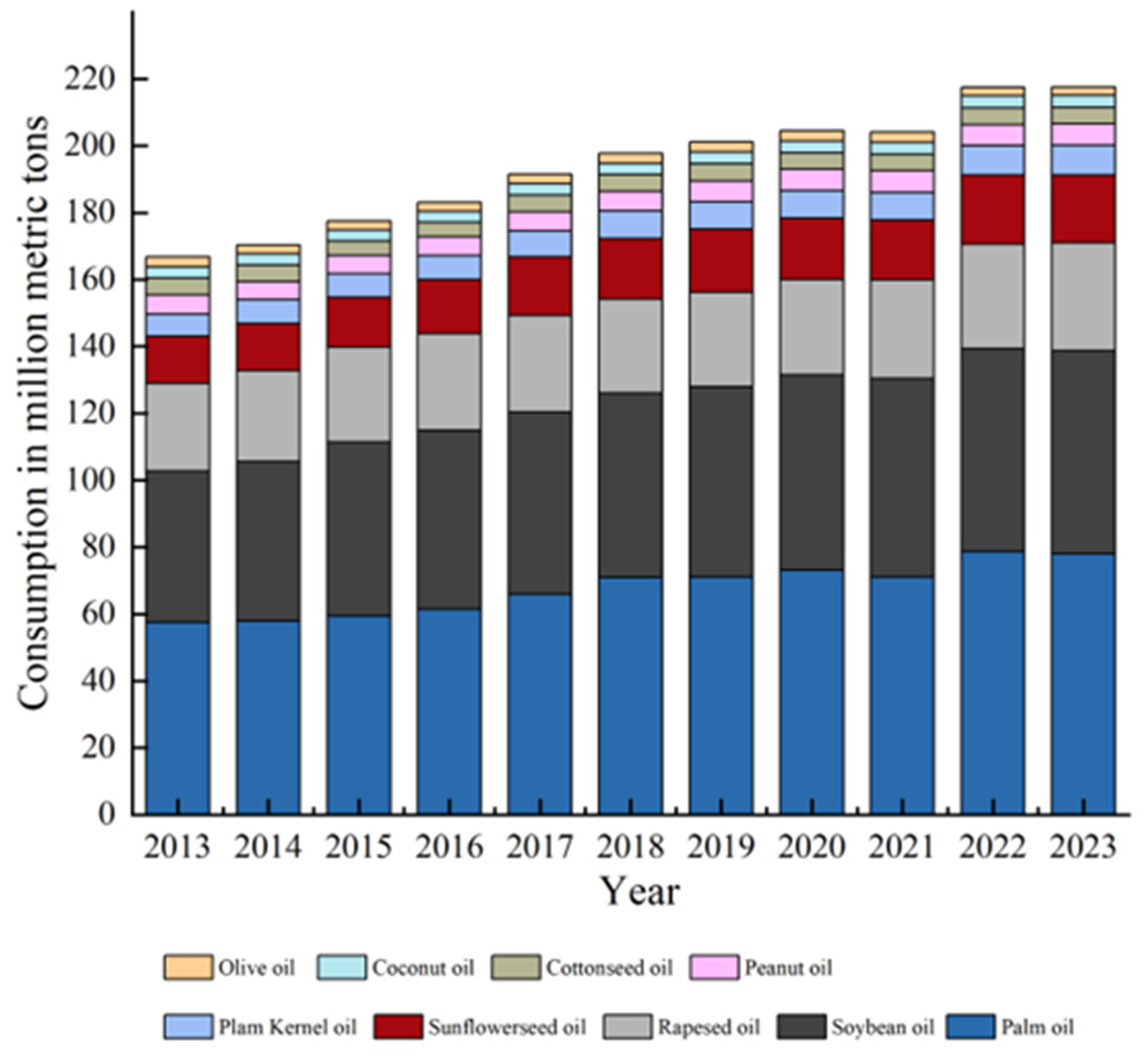
1. Introduction
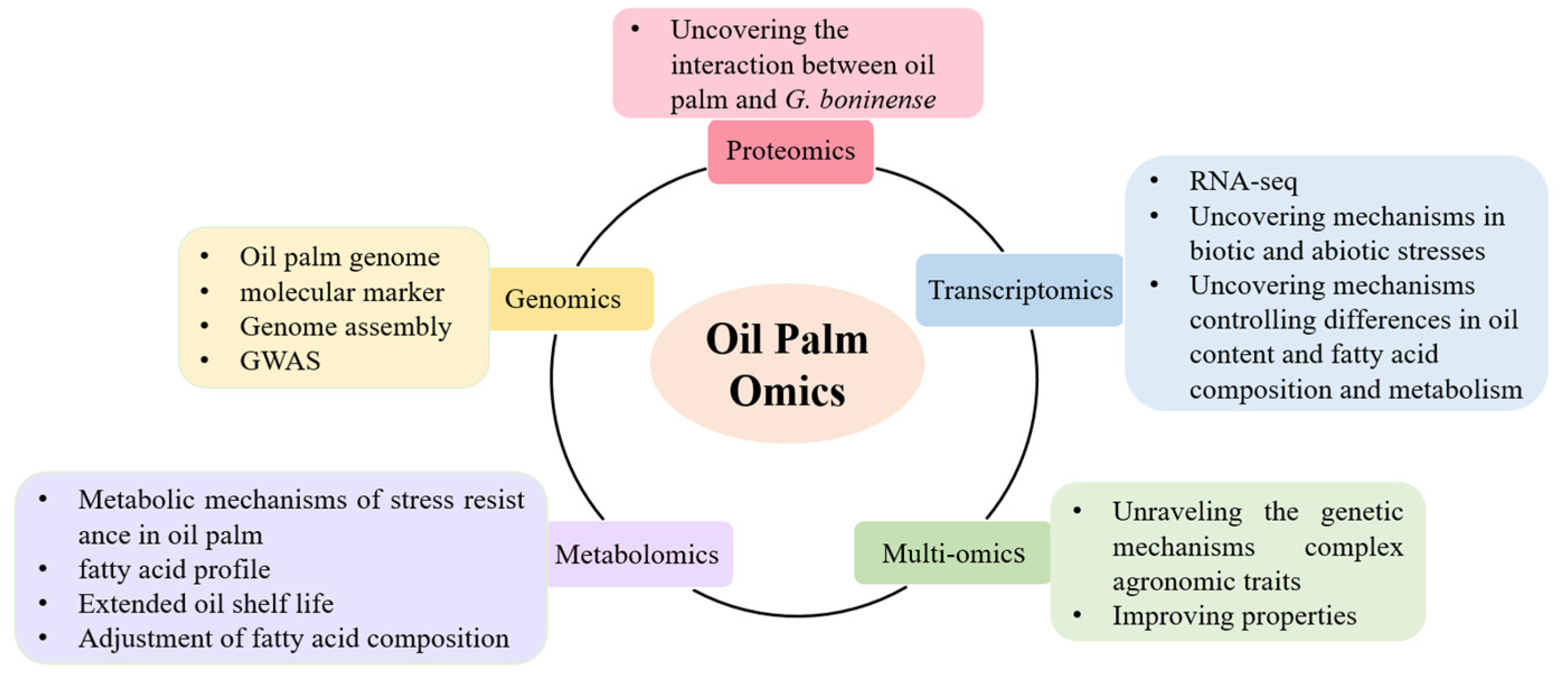
2. Oil Palm Genomics
2.1. Oil Palm Genome Sequencing
2.2. Research on Molecular Markers for Important Traits in Oil Palm
| Molecular Markers | Important Traits | References |
|---|---|---|
| SNP, SSR | oil content | [35] |
| SSR | hull thickness | [36] |
| SNP | stem height | [37] |
| SSR | drought-tolerant | [38] |
| SSR; | drought-tolerant | [38] |
| SNP | drought-tolerant | [39] |
| SSR | illegitimate progenies | [40] |
| SSR | identification of QTLs in oil palm | [41] |
| SSR | cold tolerance | [42] |
| SSR | identification of QTLs associated with callogenesis and embryogenesis in oil palm | [43] |
| EST-SSR | fruit exocarp colour | [30] |
| RFLP | genotypic discrimination | [44] |
| SNP | fatty acid content | [45] |
| SNP | height of oil palm | [46] |
| Intron polymorphisms (IP) | oil palm fatty acid composition | [47] |
| SSR | genetic diversity | [48,49,50] |
| SSR | sex ratio | [51] |
| SNP | vitamin E biosynthesis | [52] |
| SNP-CAPS | fruit traits | [53] |
2.3. Gene Resource Mining and Function Analysis of Important Traits in Oil Palm
| Gene Family | Function | Reference |
|---|---|---|
| Abiotic stress | ||
| CBF | Increased tolerance to low temperatures | [73] |
| ABA | Enhance resistance to drought | [74] |
| bZIP | Highly expressed under low temperature, salinity, and drought stress conditions | [75] |
| AP2/ERF | Increased tolerance to salt, low temperatures, and drought | [76] |
| HRE2(ERF-VII) | HRE2 responds to flood-induced hypoxia | [77] |
| MYB | Increased tolerance to stresses such as salinity, cold, and drought | [78] |
| WRKY | Involved in abiotic stress responses such as drought, salinity, and heat | [79] |
| EgSPCH(bHLH) | EgSPCH produces more pores in response to salt stress | [80] |
| ARF | EgARF responds to abiotic stresses (cold, drought, and salt stress) | [81] |
| CAT | EgCAT enhances tolerance to low temperature, drought, and salt stress | [82] |
| Biotic stress | ||
| CNL R gene | Expression in the early stages of oil palm defence mechanisms | [83] |
| AGLU1 RCH10 | Increasing oil palm resistance towards G. boninense | [84] |
| LCC | EgLCC24 gene detects oil palm seedlings resistant to G. boninense | [85] |
| Fatty acids | ||
| MADS-box | Fruit development and oil accumulation | [86] |
| MAD | Mediating medium-chain fatty acid (MCFA) anabolism in oil palm mesocarp | [87] |
| GDSL | Enhanced lipid content | [65] |
| ACBP | Lipid accumulation | [88] |
| Others | ||
| CAD | Lignin biosynthesis | [89] |
3. Oil Palm Transcriptomics
| Trait for Transcriptome Analysis | Transcription Factor | Summary | Reference |
|---|---|---|---|
| Somatic embryogenesis | FTIP, FRIGIDA-LIKE, and NF-YA, | The reproductive time of plants may be related to somatic embryogenesis potential | [100] |
| Drought | MYBs, HOXs, NF-Ys | A potential miRNA target gene for oil palm under water deficit conditions | [101] |
| Pi-deficiency | PHL7, NIP6–1 and 14–3-3 | Involved in the lack of regulation and domestication of oil palm Pi | [102] |
| Waterlogging | LBD41, HRA1, HRE2 | Respond to waterlogging | [77] |
| High somatic embryogenesis rate | LE, DDX28, HD-ZIP, NPF | The physiological state related to somatic embryogenesis potential was revealed | [103] |
| Bud Rot Disease | Clon34 and Clon57 | Control pathogen | [104] |
| Defoliation stress | / | Elucidating the mechanism of oil palm sex determination. | [105] |
| Bagworm stress | P450, GST and CCE AChE, nAChR, ACCase, GABA, RyR | Insecticide targets for detoxification and modification potential | [106] |
| Aluminium stress | DREB1F, NAC | Induces the expression of internal detoxification enzymes | [107] |
| Water deprivation | NF-YA3, HOX32, GRF1 | It plays a certain role in the response of oil palm seedlings to drought and salt stress. | [108] |
| Salt stress | MYC, G-Box, ABRE, TATA-box | Using identified genes to try to mitigate salt stress | [109] |
| G. boninense infection | target gene #5, #13, #16, #6 and #15, #1, #9, #12, and #14 | It may be related to early infection reaction to G. boninense | [110] |
4. Oil Palm Proteomics
5. Oil Palm Metabolomics
6. Conclusions and Outlook
- (1)
- Constructing more high-quality oil palm transcriptome databases, further studying the biological functions of non-coding RNAs, and conducting research on the molecular mechanisms of sexual reproduction in oil palm.
- (2)
- Strengthening the research on oil palm growth, development, and defence metabolism. Targeted metabolomics is crucial for an understanding of specific metabolites or changes in metabolic pathways under different environmental conditions. By combining targeted and non-targeted metabolomics, we can strengthen the research on the identification and interactions between primary and secondary metabolites, the regulation mechanisms of metabolites in oil palm growth and development, and the role of secondary metabolites in the defence mechanism.
- (3)
- Developing and applying low-cost and rapid techniques for oil palm genome determination, and strengthening research on genetic diversity and epigenetic modification in response to stress in oil palm species.
- (4)
- Exploring the responses of oil palm to multiple stresses and the genetic mechanism of complex agronomic traits by combining multi-omics technologies and integrating genomics, transcriptomics, lipidomics, metabolomics, and proteomics, so that we can identify regulatory genes of complex agronomic traits and to clarify the mechanism of action and metabolic pathways, so as to provide a theoretical basis and genetic resources for modern molecular breeding.
Author Contributions
Funding
Conflicts of Interest
References
- Damayant, S.; Andry, S.; Khairurrijal; Kartasasmita, R.E. Isolation of β-carotene from Palm (Elaeis guineensis Jacq.) Oil Using Transesterification-adsorption-desorption Method and its Characterization. J. Appl. Sci. 2014, 14, 2615–2621. [Google Scholar] [CrossRef]
- Babu, B.K.; Mathur, R.K.; Anitha, P.; Ravichandran, G.; Bhagya, H.P. Phenomics, genomics of oil palm (Elaeis guineensis Jacq.): Way forward for making sustainable and high yielding quality oil palm. Physiol. Mol. Biol. Plants 2021, 27, 587–604. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Ong-Abdullah, M.; Low, E.-T.L.; Manaf, M.A.A.; Rosli, R.; Nookiah, R.; Ooi, L.C.-L.; Ooi, S.E.; Chan, K.-L.; Halim, M.A.; et al. Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 2013, 500, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Sundram, K.; Sambanthamurthi, R.; Tan, Y.A. Palm fruit chemistry and nutrition. Asia Pac. J. Clin. Nutr. 2003, 12, 355–362. [Google Scholar]
- Mba, O.I.; Dumont, M.-J.; Ngadi, M. Palm Oil: Processing, Characterization and Utilization in the Food Industry—A Review. Food Biosci. 2015, 10, 26–41. [Google Scholar] [CrossRef]
- Nainggolan, M.; Sinaga, A.G. Characteristics of fatty acid composition and minor constituents of red palm olein and palm kernel oil combination. J. Adv. Pharm. Technol. Res. 2021, 12, 22–26. [Google Scholar] [CrossRef]
- Barcelos, E.; Rios Sde, A.; Cunha, R.N.; Lopes, R.; Motoike, S.Y.; Babiychuk, E.; Skirycz, A.; Kushnir, S. Oil palm natural diversity and the potential for yield improvement. Front. Plant Sci. 2015, 6, 190. [Google Scholar] [CrossRef]
- Rival, A.; Beule, T.; Barre, P.; Hamon, S.; Duval, Y.; Noirot, M. Comparative flow cytometric estimation of nuclear DNA content in oil palm (Elaeis guineensis Jacq) tissue cultures and seed-derived plants. Plant Cell Rep. 1997, 16, 884–887. [Google Scholar] [CrossRef] [PubMed]
- Pellicer, J.; Leitch, I.J. The Plant DNA C-values database (release 7.1): An updated online repository of plant genome size data for comparative studies. New Phytol. 2020, 226, 301–305. [Google Scholar] [CrossRef]
- Jin, J.; Lee, M.; Bai, B.; Sun, Y.; Qu, J.; Rahmadsyah; Alfiko, Y.; Lim, C.H.; Suwanto, A.; Sugiharti, M.; et al. Draft genome sequence of an elite Dura palm and whole-genome patterns of DNA variation in oil palm. DNA Res. 2016, 23, 527–533. [Google Scholar] [CrossRef]
- Utomo, C.; Tanjung, Z.A.; Aditama, R.; Buana, R.F.N.; Pratomo, A.D.M.; Tryono, R.; Liwang, T. Draft Genome Sequence of the Phytopathogenic Fungus Ganoderma boninense, the Causal Agent of Basal Stem Rot Disease on Oil Palm. Am. Soc. Microbiol. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Ong, A.-L.; Teh, C.-K.; Mayes, S.; Massawe, F.; Appleton, D.R.; Kulaveerasingam, H. An Improved Oil Palm Genome Assembly as a Valuable Resource for Crop Improvement and Comparative Genomics in the Arecoideae Subfamily. Plants 2020, 9, 1476. [Google Scholar] [CrossRef] [PubMed]
- Mohd Sanusi, N.S.N.; Rosli, R.; Chan, K.-L.; Halim, M.A.A.; Ting, N.-C.; Singh, R.; Low, E.-T.L. Integrated consensus genetic map and genomic scaffold re-ordering of oil palm (Elaeis guineensis) genome. Comput. Biol. Chem. 2023, 102, 107801. [Google Scholar] [CrossRef] [PubMed]
- Sanusi, N.S.N.M.; Rosli, R.; Halim, M.A.A.; Chan, K.-L.; Nagappan, J.; Azizi, N.; Amiruddin, N.; Tatarinova, T.V.; Low, E.-T.L. PalmXplore: Oil palm gene database. Database 2018, 2018, bay095. [Google Scholar] [CrossRef] [PubMed]
- Uthaipaisanwong, P.; Chanprasert, J.; Shearman, J.R.; Sangsrakru, D.; Yoocha, T.; Jomchai, N.; Jantasuriyarat, C.; Tragoonrung, S.; Tangphatsornruang, S. Characterization of the chloroplast genome sequence of oil palm (Elaeis guineensis Jacq.). Gene 2012, 500, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Jaligot, E.; Rival, A.; Beulé, T.; Dussert, S.; Verdeil, J.L. Somaclonal variation in oil palm (Elaeis guineensis Jacq.): The DNA methylation hypothesis. Plant Cell Rep. 2000, 19, 684–690. [Google Scholar] [CrossRef]
- Low, E.T.; Rosli, R.; Jayanthi, N.; Mohd-Amin, A.H.; Azizi, N.; Chan, K.L.; Maqbool, N.J.; Maclean, P.; Brauning, R.; McCulloch, A.; et al. Analyses of hypomethylated oil palm gene space. PLoS ONE 2014, 9, e86728. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ong-Abdullah, M.; Ordway, J.M.; Jiang, N.; Ooi, S.-E.; Kok, S.-Y.; Sarpan, N.; Azimi, N.; Hashim, A.T.; Ishak, Z.; Rosli, S.K.; et al. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 2015, 525, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Sarpan, N.; Taranenko, E.; Ooi, S.-E.; Low, E.-T.L.; Espinoza, A.; Tatarinova, T.V.; Ong-Abdullah, M. DNA methylation changes in clonally propagated oil palm. Plant Cell Rep. 2020, 39, 1219–1233. [Google Scholar] [CrossRef]
- Ooi, S.-E.; Choo, C.-N.; Sarpan, N.; Wong, C.-K.; Wong, W.-C. Karma-EgDEF1 methylation in Elaeis guineensis clonal mother palms that produced high mantling rates in the second clonal generation. Vitr. Cell. Dev. Biol. Plant 2024, 60, 176–182. [Google Scholar] [CrossRef]
- Billotte, N.; Marseillac, N.; Risterucci, A.M.; Adon, B.; Brottier, P.; Baurens, F.C.; Singh, R.; Herrán, A.; Asmady, H.; Billot, C.; et al. Microsatellite-based high density linkage map in oil palm (Elaeis guineensis Jacq.). Theor. Appl. Genet. 2005, 110, 754–765. [Google Scholar] [CrossRef] [PubMed]
- Jaligot, E.; Beulé, T.; Rival, A. Methylation-sensitive RFLPs: Characterisation of two oil palm markers showing somaclonal variation-associated polymorphism. Theor. Appl. Genet. 2002, 104, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Cheah, S.C. Analysis of the inheritance of AFLP markers in an interspecific cross of oil palm using the pseudo-testcross strategy. J. Oil Palm Res. 1999, 11, 64–73. [Google Scholar]
- Mayes, S.; Jack, P.L.; Corley, R.H.; Marshall, D.F. Construction of a RFLP genetic linkage map for oil palm (Elaeis guineensis Jacq.). Genome 1997, 40, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Mohan, M.; Nair, S.; Bhagwat, A.; Krishna, T.G.; Yano, M.; Bhatia, C.R.; Sasaki, T. Genome mapping, molecular markers and marker-assisted selection in crop plants. Mol. Breed. 1997, 3, 87–103. [Google Scholar] [CrossRef]
- Ho, C.-L.; Kwan, Y.-Y.; Choi, M.-C.; Tee, S.-S.; Ng, W.-H.; Lim, K.-A.; Lee, Y.-P.; Ooi, S.-E.; Lee, W.-W.; Tee, J.-M.; et al. Analysis and functional annotation of expressed sequence tags (ESTs) from multiple tissues of oil palm (Elaeis guineensis Jacq.). BMC Genom. 2007, 8, 381. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, T.; Meng, X.; Song, J.; Zhang, C.; Gao, P. Genetic Mapping and QTL Analysis of Fruit Traits in Melon (Cucumis melo L.). Curr. Issues Mol. Biol. 2023, 45, 3419–3433. [Google Scholar] [CrossRef]
- Cardona, C.C.C.; Coronado, Y.M.; Conronado, A.C.M.; Ochoa, I.J.B. Genetic diversity in oil palm (Elaeis guineensis Jacq) using RAM (Random Amplified Microsatellites). Bragantia 2018, 77, 546–556. [Google Scholar] [CrossRef]
- Sunilkumar, K.; Murugesan, P.; Mathur, R.K.; Rajesh, M.K. Genetic diversity in oil palm (Elaeis guineensis and Elaeis oleifera) germplasm as revealed by microsatellite (SSR) markers. Indian J. Agric. Sci. 2020, 90, 741–745. [Google Scholar] [CrossRef]
- Suraninpong, P.; Nuanlaong, S. Comparative transcriptome profiling and molecular marker development for oil palm fruit color. Sci. Rep. 2022, 12, 15507. [Google Scholar] [CrossRef]
- Mohd Shaha, F.R.; Liew, P.L.; Qamaruz Zaman, F.; Nulit, R.; Barin, J.; Rolland, J.; Yong, H.Y.; Boon, S.H. Genotyping by sequencing for the construction of oil palm (Elaeis guineensis Jacq.) genetic linkage map and mapping of yield related quantitative trait loci. PeerJ 2024, 12, e16570. [Google Scholar] [CrossRef] [PubMed]
- Kwong, Q.B.; Teh, C.K.; Ong, A.L.; Heng, H.Y.; Lee, H.L.; Mohamed, M.; Low, J.Z.-B.; Apparow, S.; Chew, F.T.; Mayes, S.; et al. Development and Validation of a High-Density SNP Genotyping Array for African Oil Palm. Mol. Plant 2016, 9, 1132–1141. [Google Scholar] [CrossRef] [PubMed]
- Babu, B.K.; Mathur, R.K.; Ravichandran, G.; Anita, P.; Venu, M.V.B. Genome wide association study (GWAS) and identification of candidate genes for yield and oil yield related traits in oil palm (Eleaeis guineensis) using SNPs by genotyping-based sequencing. Genomics 2020, 112, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Wibowo, C.S.; Apriyanto, A.; Ernawan, R.; Neing, D.; Susilo, R.; Cordell, H.J.; Gatehouse, A.M.R.; Edwards, M.G. Genetic variants associated with leaf spot disease resistance in oil palm (Elaeis guineensis): A genome-wide association study. Plant Pathol. 2023, 72, 1626–1636. [Google Scholar] [CrossRef]
- Yaakub, Z.; Kamaruddin, K.; Singh, R.; Mustafa, S.; Marjuni, M.; Ting, N.C.; Amiruddin, M.D.; Leslie, L.E.; Cheng-Li, O.L.; Sritharan, K.; et al. An integrated linkage map of interspecific backcross 2 (BC(2)) populations reveals QTLs associated with fatty acid composition and vegetative parameters influencing compactness in oil palm. BMC Plant Biol. 2020, 20, 356. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yarra, R.; Zhao, Z.; Jin, L.; Cao, H. Development of SSR markers based on transcriptome data and association mapping analysis for fruit shell thickness associated traits in oil palm (Elaeis guineensis Jacq.). 3 Biotech 2020, 10, 280. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Xia, J.H.; Zou, Z.; Ye, J.; Rahmadsyah; Alfiko, Y.; Jin, J.; Lieando, J.V.; Purnamasari, M.I.; Lim, C.H.; et al. A consensus linkage map of oil palm and a major QTL for stem height. Sci. Rep. 2015, 5, 8232. [Google Scholar] [CrossRef] [PubMed]
- Nuanlaong, S.; Wuthisuthimethavee, S.; Azzeme, A.M.; Suraninpong, P. Optimized Method for the Identification of Candidate Genes and Molecular Maker Development Related to Drought Tolerance in Oil Palm (Elaeis guineensis Jacq.). Plants 2022, 11, 2317. [Google Scholar] [CrossRef]
- Suraninpong, P.; Thongkhao, K.; Azzeme, A.M.; Suksa-Ard, P. Monitoring Drought Tolerance in Oil Palm: Choline Monooxygenase as a Novel Molecular Marker. Plants 2023, 12, 3089. [Google Scholar] [CrossRef]
- Zolkafli, S.H.; Ithnin, M.; Chan, K.-L.; Zainol Abidin, M.I.; Ismail, I.; Ting, N.C.; Ooi, L.C.-L.; Singh, R. Optimal set of microsatellite markers required to detect illegitimate progenies in selected oil palm (Elaeis guineensis Jacq.) breeding crosses. Breed. Sci. 2021, 71, 253–260. [Google Scholar] [CrossRef]
- Bhagya, H.P.; Kalyana Babu, B.; Gangadharappa, P.M.; Naika, M.B.N.; Satish, D.; Mathur, R.K. Identification of QTLs in oil palm (Elaeis guineensis Jacq.) using SSR markers through association mapping. J. Genet. 2020, 99, 19. [Google Scholar] [CrossRef] [PubMed]
- Zulfugarov, I.S.; Tovuu, A.; Eu, Y.J.; Dogsom, B.; Poudyal, R.S.; Nath, K.; Hall, M.; Banerjee, M.; Yoon, U.C.; Moon, Y.H.; et al. Production of superoxide from Photosystem II in a rice (Oryza sativa L.) mutant lacking PsbS. BMC Plant Biol. 2014, 14, 242. [Google Scholar] [CrossRef] [PubMed]
- Ting, N.C.; Jansen, J.; Nagappan, J.; Ishak, Z.; Chin, C.W.; Tan, S.G.; Cheah, S.C.; Singh, R. Identification of QTLs associated with callogenesis and embryogenesis in oil palm using genetic linkage maps improved with SSR markers. PLoS ONE 2013, 8, e53076. [Google Scholar] [CrossRef] [PubMed]
- Jack, P.L.; Dimitrijevic, T.A.F.; Mayes, S. Assessment of nuclear, mitochondrial and chloroplast RFLP markers in oil palm (Elaeis guineensis Jacq.). Theor. Appl. Genet. 1995, 90, 643–649. [Google Scholar] [CrossRef]
- Xia, W.; Luo, T.; Dou, Y.; Zhang, W.; Mason, A.S.; Huang, D.; Huang, X.; Tang, W.; Wang, J.; Zhang, C.; et al. Identification and Validation of Candidate Genes Involved in Fatty Acid Content in Oil Palm by Genome-Wide Association Analysis. Front. Plant Sci. 2019, 10, 1263. [Google Scholar] [CrossRef]
- Somyong, S.; Phetchawang, P.; Bihi, A.K.; Sonthirod, C.; Kongkachana, W.; Sangsrakru, D.; Jomchai, N.; Pootakham, W.; Tangphatsornruang, S. A SNP variation in an expansin (EgExp4) gene affects height in oil palm. PeerJ 2022, 10, e13046. [Google Scholar] [CrossRef]
- Li, J.; Yang, Y.; Sun, X.; Liu, R.; Xia, W.; Shi, P.; Zhou, L.; Wang, Y.; Wu, Y.; Lei, X.; et al. Development of Intron Polymorphism Markers and Their Association with Fatty Acid Component Variation in Oil Palm. Front. Plant Sci. 2022, 13, 885418. [Google Scholar] [CrossRef]
- Myint, K.A.; Yaakub, Z.; Rafii, M.Y.; Oladosu, Y.; Samad, M.Y.A.; Ramlee, S.I.; Mustaffa, S.; Arolu, F.; Abdullah, N.; Marjuni, M.; et al. Genetic Diversity Assessment of MPOB-Senegal Oil Palm Germplasm Using Microsatellite Markers. BioMed Res. Int. 2021, 2021, 6620645. [Google Scholar] [CrossRef] [PubMed]
- Zaki, N.M.; Singh, R.; Rosli, R.; Ismail, I. Elaeis oleifera Genomic-SSR Markers: Exploitation in Oil Palm Germplasm Diversity and Cross-Amplification in Arecaceae. Int. J. Mol. Sci. 2012, 13, 4069–4088. [Google Scholar] [CrossRef]
- Ting, N.C.; Zaki, N.M.; Rosli, R.; Low, E.T.; Ithnin, M.; Cheah, S.C.; Tan, S.G.; Singh, R. SSR mining in oil palm EST database: Application in oil palm germplasm diversity studies. J. Genet. 2010, 89, 135–145. [Google Scholar] [CrossRef]
- Somyong, S.; Poopear, S.; Sunner, S.K.; Wanlayaporn, K.; Jomchai, N.; Yoocha, T.; Ukoskit, K.; Tangphatsornruang, S.; Tragoonrung, S. ACC oxidase and miRNA 159a, and their involvement in fresh fruit bunch yield (FFB) via sex ratio determination in oil palm. Mol. Genet. Genom. 2016, 291, 1243–1257. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Xia, W.; Mason, A.S.; Huang, D.; Sun, X.; Fan, H.; Xiao, Y. Developing functional markers for vitamin E biosynthesis in oil palm. PLoS ONE 2021, 16, e0259684. [Google Scholar] [CrossRef]
- Venu, M.V.B.; Babu, B.K. Identification of fruit forms using CAPS marker among the oil palm indigenous germplasm. J. Oilseeds Res. 2023, 37. [Google Scholar] [CrossRef]
- Ithnin, M.; Xu, Y.; Marjuni, M.; Serdari, N.M.; Amiruddin, M.D.; Low, E.-T.L.; Tan, Y.-C.; Yap, S.-J.; Ooi, L.C.L.; Nookiah, R.; et al. Multiple locus genome-wide association studies for important economic traits of oil palm. Tree Genet. Genomes 2017, 13, 103. [Google Scholar] [CrossRef]
- Ithnin, M.; Vu, W.T.; Shin, M.-G.; Suryawanshi, V.; Sherbina, K.; Zolkafli, S.H.; Serdari, N.M.; Amiruddin, M.D.; Abdullah, N.; Mustaffa, S.; et al. Genomic diversity and genome-wide association analysis related to yield and fatty acid composition of wild American oil palm. Plant Sci. 2021, 304, 110731. [Google Scholar] [CrossRef]
- Osorio-Guarín, J.A.; Garzón-Martínez, G.A.; Delgadillo-Duran, P.; Bastidas, S.; Moreno, L.P.; Enciso-Rodríguez, F.E.; Cornejo, O.E.; Barrero, L.S. Genome-wide association study (GWAS) for morphological and yield-related traits in an oil palm hybrid (Elaeis oleifera × Elaeis guineensis) population. BMC Plant Biol. 2019, 19, 533. [Google Scholar] [CrossRef] [PubMed]
- Babu, B.K.; Mathur, R.K.; Ravichandran, G.; Venu, M.V.B. Genome-wide association study (GWAS) for stem height increment in oil palm (Elaeis guineensis) germplasm using SNP markers. Tree Genet. Genomes 2019, 15, 40. [Google Scholar] [CrossRef]
- Bhagya, H.P.; Kalyana Babu, B.; Mathur, R.K.; Ramajayam, D.; Ravichandran, G.; Anitha, P. Oil palm (Elaeis guineensis Jacq.) germplasm genome-wide association analysis for the oil yield traits utilizing microsatellite markers. Ind. Crops Prod. 2024, 218, 118934. [Google Scholar] [CrossRef]
- Teh, C.-K.; Ong, A.-L.; Kwong, Q.-B.; Apparow, S.; Chew, F.-T.; Mayes, S.; Mohamed, M.; Appleton, D.; Kulaveerasingam, H. Genome-wide association study identifies three key loci for high mesocarp oil content in perennial crop oil palm. Sci. Rep. 2016, 6, 19075. [Google Scholar] [CrossRef]
- Babu, B.K.; Mathur, R.K.; Ravichandran, G.; Anitha, P.; Venu, M.V.B. Genome-wide association study for leaf area, rachis length and total dry weight in oil palm (Eleaeis guineensis) using genotyping by sequencing. PLoS ONE 2019, 14, e0220626. [Google Scholar] [CrossRef]
- Babu, K.; Mathur, R.K.; Venu, M.V.; Shil, S.; Ravichandran, G.; Anita, P.; Bhagya, H.P. Genome-wide association study (GWAS) of major QTLs for bunch and oil yield related traits in Elaeis guineensis L. Plant Sci. 2021, 305, 110810. [Google Scholar] [CrossRef]
- Teh, C.-K.; Muaz, S.D.; Tangaya, P.; Fong, P.-Y.; Ong, A.-L.; Mayes, S.; Chew, F.-T.; Kulaveerasingam, H.; Appleton, D. Characterizing haploinsufficiency of SHELL gene to improve fruit form prediction in introgressive hybrids of oil palm. Sci. Rep. 2017, 7, 3118. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Low, E.-T.L.; Ooi, L.C.-L.; Ong-Abdullah, M.; Nookiah, R.; Ting, N.-C.; Marjuni, M.; Chan, P.-L.; Ithnin, M.; Manaf, M.A.A.; et al. The oil palm VIRESCENS gene controls fruit colour and encodes a R2R3-MYB. Nat. Commun. 2014, 5, 4106. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.-L.; Tatarinova, T.V.; Rosli, R.; Amiruddin, N.; Azizi, N.; Halim, M.A.A.; Sanusi, N.S.N.M.; Jayanthi, N.; Ponomarenko, P.; Triska, M.; et al. Evidence-based gene models for structural and functional annotations of the oil palm genome. Biol. Direct 2017, 12, 21. [Google Scholar] [CrossRef]
- Zhang, Y.; Bai, B.; Lee, M.; Alfiko, Y.; Suwanto, A.; Yue, G.H. Cloning and characterization of EgGDSL, a gene associated with oil content in oil palm. Sci. Rep. 2018, 8, 11406. [Google Scholar] [CrossRef]
- Jin, Y.; Yuan, Y.; Gao, L.; Sun, R.; Chen, L.; Li, D.; Zheng, Y. Characterization and Functional Analysis of a Type 2 Diacylglycerol Acyltransferase (DGAT2) Gene from Oil Palm (Elaeis guineensis Jacq.) Mesocarp in Saccharomyces cerevisiae and Transgenic Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1791. [Google Scholar] [CrossRef] [PubMed]
- Rosli, R.; Chan, P.-L.; Chan, K.-L.; Amiruddin, N.; Low, E.-T.L.; Singh, R.; Harwood, J.L.; Murphy, D.J. In silico characterization and expression profiling of the diacylglycerol acyltransferase gene family (DGAT1, DGAT2, DGAT3 and WS/DGAT) from oil palm, Elaeis guineensis. Plant Sci. 2018, 275, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Y.; Zhang, Q.; Jin, Y.H.; Zou, J.X.; Zheng, Y.S.; Li, D.D. A MADS-box gene, EgMADS21, negatively regulates EgDGAT2 expression and decreases polyunsaturated fatty acid accumulation in oil palm (Elaeis guineensis Jacq.). Plant Cell Rep. 2020, 39, 1505–1516. [Google Scholar] [CrossRef]
- Chan, P.L.; Rose, R.J.; Abdul Murad, A.M.; Zainal, Z.; Ong, P.W.; Ooi, L.C.; Low, E.L.; Ishak, Z.; Yahya, S.; Song, Y.; et al. Early nodulin 93 protein gene: Essential for induction of somatic embryogenesis in oil palm. Plant Cell Rep. 2020, 39, 1395–1413. [Google Scholar] [CrossRef]
- Nizan, I.E.F.; Kamaruddin, K.; Ong, P.W.; Ramli, Z.; Singh, R.; Rose, R.J.; Chan, P.L. Overexpression of Oil Palm Early Nodulin 93 Protein Gene (EgENOD93) Enhances In Vitro Shoot Regeneration in Arabidopsis thaliana. Mol. Biotechnol. 2022, 64, 743–757. [Google Scholar] [CrossRef]
- Tregear, J.W.; Richaud, F.; Collin, M.; Esbelin, J.; Parrinello, H.; Cochard, B.; Nodichao, L.; Morcillo, F.; Adam, H.; Jouannic, S. Micro-RNA-Regulated SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) Gene Expression and Cytokinin Accumulation Distinguish Early-Developing Male and Female Inflorescences in Oil Palm (Elaeis guineensis). Plants 2022, 11, 685. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yarra, R. Genome-Wide Analysis of SPL/miR156 Module and Its Expression Analysis in Vegetative and Reproductive Organs of Oil Palm (Elaeis guineensis). Int. J. Mol. Sci. 2023, 24, 13658. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, Y.; Iqbal, A.; Qadri, R.; Shi, P.; Wang, Y.; Wu, Y.; Fan, H.; Wu, G. Correlation analysis of cold-related gene expression with physiological and biochemical indicators under cold stress in oil palm. PLoS ONE 2019, 14, e0225768. [Google Scholar] [CrossRef]
- Wang, L.; Lee, M.; Ye, B.; Yue, G.H. Genes, pathways and networks responding to drought stress in oil palm roots. Sci. Rep. 2020, 10, 21303. [Google Scholar] [CrossRef]
- Zhou, L.; Yarra, R. Genome-wide identification and expression analysis of bZIP transcription factors in oil palm (Elaeis guineensis Jacq.) under abiotic stress. Protoplasma 2022, 259, 469–483. [Google Scholar] [CrossRef]
- Zhou, L.; Yarra, R. Genome-Wide Identification and Characterization of AP2/ERF Transcription Factor Family Genes in Oil Palm under Abiotic Stress Conditions. Int. J. Mol. Sci. 2021, 22, 2821. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Kobayashi, M.J.; Marsoem, S.N.; Irawati, D.; Kosugi, A.; Kondo, T.; Tani, N. Transcriptomic responses of oil palm (Elaeis guineensis) stem to waterlogging at plantation in relation to precipitation seasonality. Front. Plant Sci. 2023, 14, 1213496. [Google Scholar] [CrossRef]
- Zhou, L.; Yarra, R.; Yang, Y.; Liu, Y.; Yang, M.; Cao, H. The oil palm R2R3-MYB subfamily genes EgMYB111 and EgMYB157 improve multiple abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Rep. 2022, 41, 377–393. [Google Scholar] [CrossRef]
- Lee, F.C.; Yeap, W.C.; Appleton, D.R.; Ho, C.-L.; Kulaveerasingam, H. Identification of drought responsive Elaeis guineensis WRKY transcription factors with sensitivity to other abiotic stresses and hormone treatments. BMC Genom. 2022, 23, 164. [Google Scholar] [CrossRef]
- Song, Z.; Wang, L.; Lai, C.; Lee, M.; Yang, Z.; Yue, G. EgSPEECHLESS Responses to Salt Stress by Regulating Stomatal Development in Oil Palm. Int. J. Mol. Sci. 2022, 23, 4659. [Google Scholar] [CrossRef]
- Jin, L.; Yarra, R.; Zhou, L.; Cao, H. The auxin response factor (ARF) gene family in Oil palm (Elaeis guineensis Jacq.): Genome-wide identification and their expression profiling under abiotic stresses. Protoplasma 2022, 259, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; John Martin, J.J.; Li, R.; Zeng, X.; Wu, Q.; Li, Q.; Fu, D.; Li, X.; Liu, X.; Ye, J.; et al. Catalase (CAT) Gene Family in Oil Palm (Elaeis guineensis Jacq.): Genome-Wide Identification, Analysis, and Expression Profile in Response to Abiotic Stress. Int. J. Mol. Sci. 2024, 25, 1480. [Google Scholar] [CrossRef] [PubMed]
- Rosli, R.; Amiruddin, N.; Ab Halim, M.A.; Chan, P.-L.; Chan, K.-L.; Azizi, N.; Morris, P.E.; Leslie Low, E.-T.; Ong-Abdullah, M.; Sambanthamurthi, R.; et al. Comparative genomic and transcriptomic analysis of selected fatty acid biosynthesis genes and CNL disease resistance genes in oil palm. PLoS ONE 2018, 13, e0194792. [Google Scholar] [CrossRef] [PubMed]
- Hanin, A.N.; Parveez, G.K.A.; Rasid, O.A.; Masani, M.Y.A. Biolistic-mediated oil palm transformation with alfalfa glucanase (AGLU1) and rice chitinase (RCH10) genes for increasing oil palm resistance towards Ganoderma boninense. Ind. Crops Prod. 2020, 144, 112008. [Google Scholar] [CrossRef]
- Faizah, R.; Putranto, R.A.; Raharti, V.R.; Supena, N.; Sukma, D.; Budiani, A.; Wening, S.; Sudarsono, S. Defense response changes in roots of oil palm (Elaeis guineensis Jacq.) seedlings after internal symptoms of Ganoderma boninense Pat. infection. BMC Plant Biol. 2022, 22, 139. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Wang, W.; Xu, X.; Li, S.-Y.; Zheng, Y.; Li, D.-D. Identification and Analysis of MADS-Box Genes Expressed in the Mesocarp of Oil Palm Fruit (Elaeis guineensis Jacq.). Biochem. Genet. 2023, 61, 2382–2400. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yan, J.; Yang, M.; Zou, J.; Zheng, Y.; Li, D. EgMADS3 directly regulates EgLPAAT to mediate medium-chain fatty acids (MCFA) anabolism in the mesocarp of oil palm. Plant Cell Rep. 2024, 43, 107. [Google Scholar] [CrossRef]
- Amiruddin, N.; Chan, P.-L.; Azizi, N.; Morris, P.E.; Chan, K.-L.; Ong, P.W.; Rosli, R.; Masura, S.S.; Murphy, D.J.; Sambanthamurthi, R.; et al. Characterization of Oil Palm Acyl-CoA-Binding Proteins and Correlation of Their Gene Expression with Oil Synthesis. Plant Cell Physiol. 2019, 61, 735–747. [Google Scholar] [CrossRef]
- Yusuf, C.Y.L.; Nabilah, N.S.; Taufik, N.; Seman, I.A.; Abdullah, M.P. Genome-wide analysis of the CAD gene family reveals two bona fide CAD genes in oil palm. 3 Biotech 2022, 12, 149. [Google Scholar] [CrossRef]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 2016, 19, 286–302. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, Z.; Xu, H.; Li, Y.; Huang, S.; Cao, G.; Shi, M.; Zhu, J.; Zhou, J.; Li, R.; et al. ArecaceaeMDB: A comprehensive multi-omics database for Arecaceae breeding and functional genomics studies. Plant Biotechnol. J. 2023, 21, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Xia, W.; Mason, A.S.; Cao, Z.; Fan, H.; Zhang, B.; Zhang, J.; Ma, Z.; Peng, M.; Huang, D. Genetic control of fatty acid composition in coconut (Cocos nucifera), African oil palm (Elaeis guineensis), and date palm (Phoenix dactylifera). Planta 2019, 249, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Jin, L.; Yarra, R.; Cao, H.; Chen, P.; John Martin, J.J. Transcriptome analysis reveals key developmental and metabolic regulatory aspects of oil palm (Elaeis guineensis Jacq.) during zygotic embryo development. BMC Plant Biol. 2022, 22, 112. [Google Scholar] [CrossRef] [PubMed]
- Apriyanto, A.; Compart, J.; Fettke, J. Transcriptomic analysis of mesocarp tissue during fruit development of the oil palm revealed specific isozymes related to starch metabolism that control oil yield. Front. Plant Sci. 2023, 14, 1220237. [Google Scholar] [CrossRef] [PubMed]
- Chu, K.L.; Koley, S.; Jenkins, L.M.; Bailey, S.R.; Kambhampati, S.; Foley, K.; Arp, J.J.; Morley, S.A.; Czymmek, K.J.; Bates, P.D.; et al. Metabolic flux analysis of the non-transitory starch tradeoff for lipid production in mature tobacco leaves. Metab. Eng. 2022, 69, 231–248. [Google Scholar] [CrossRef]
- Apriyanto, A.; Ernawan, R.; Putra, M.K.A.; Susilo, R.; Ajambang, W.; Pancoro, A.; Wibowo, C.S. Gene expression profiles of oil palm leaves from different oil yields and genetic population: Transcriptomic dataset. Data Brief 2023, 48, 109043. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yarra, R.; Zhang, R.; Zhou, L.; Jin, L.; Martin, J.J.J.; Cao, H. Transcriptome analysis of oil palm pistil during pollination and fertilization to unravel the role of phytohormone biosynthesis and signaling genes. Funct. Integr. Genom. 2022, 22, 261–278. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, C.; Liang, Y.; Sun, R.; Gao, L.; Liu, T.; Li, D. Genome-wide association analysis of the lipid and fatty acid metabolism regulatory network in the mesocarp of oil palm (Elaeis guineensis Jacq.) based on small noncoding RNA sequencing. Tree Physiol. 2018, 39, 356–371. [Google Scholar] [CrossRef]
- Xia, W.; Dou, Y.; Liu, R.; Gong, S.; Huang, D.; Fan, H.; Xiao, Y. Genome-wide discovery and characterization of long noncoding RNAs in African oil palm (Elaeis guineensis Jacq.). PeerJ 2020, 8, e9585. [Google Scholar] [CrossRef]
- Ooi, S.-E.; Feshah, I.; Nuraziyan, A.; Sarpan, N.; Ata, N.; Lim, C.-C.; Choo, C.-N.; Wong, W.-C.; Wong, F.-H.; Wong, C.-K.; et al. Leaf transcriptomic signatures for somatic embryogenesis potential of Elaeis guineensis. Plant Cell Rep. 2021, 40, 1141–1154. [Google Scholar] [CrossRef]
- Salgado, F.F.; da Silva, T.L.C.; Vieira, L.R.; Silva, V.N.B.; Leão, A.P.; Costa, M.; Togawa, R.C.; de Sousa, C.A.F.; Grynberg, P.; Souza, M.T., Jr. The early response of oil palm (Elaeis guineensis Jacq.) plants to water deprivation: Expression analysis of miRNAs and their putative target genes, and similarities with the response to salinity stress. Front. Plant Sci. 2022, 13, 970113. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.-L.; Abdullah, S.N.A.; Ho, C.-L.; Musa, M.H.b.; Yeap, W.-C. Comparative transcriptome analysis reveals novel insights into transcriptional responses to phosphorus starvation in oil palm (Elaeis guineensis) root. BMC Genom. Data 2021, 22, 6. [Google Scholar] [CrossRef] [PubMed]
- Sahara, A.; Roberdi, R.; Wiendi, N.M.A.; Liwang, T. Transcriptome profiling of high and low somatic embryogenesis rate of oil palm (Elaeis guineensis Jacq. var. Tenera). Front. Plant Sci. 2023, 14, 1142868. [Google Scholar] [CrossRef] [PubMed]
- García-Gaona, M.; Botero-Rozo, D.; Araque, L.; Romero, H.M. The Dynamic Interaction between Oil Palm and Phytophthora palmivora in Bud Rot Disease: Insights from Transcriptomic Analysis and Network Modelling. J. Fungi 2024, 10, 164. [Google Scholar] [CrossRef] [PubMed]
- Apriyanto, A.; Ajambang, W. Transcriptomic dataset for early inflorescence stages of oil palm in response to defoliation stress. Data Brief 2022, 41, 107914. [Google Scholar] [CrossRef] [PubMed]
- Zainuddin, N.; Maidin, M.S.T.; Kamarudin, N.; Napiah, N.R.A.M.A.; Keni, M.F.; Masri, M.M.M. De novo transcriptome analysis of bagworm Metisa plana from highly infested oil palm estate in Perak revealed detoxification genes and potential insecticide targets. J. Asia Pac. Entomol. 2023, 26, 102039. [Google Scholar] [CrossRef]
- Mejia-Alvarado, F.S.; Botero-Rozo, D.; Araque, L.; Bayona, C.; Herrera-Corzo, M.; Montoya, C.; Ayala-Díaz, I.; Romero, H.M. Molecular network of the oil palm root response to aluminum stress. BMC Plant Biol. 2023, 23, 346. [Google Scholar] [CrossRef] [PubMed]
- Salgado, F.F.; Vieira, L.R.; Silva, V.N.B.; Leão, A.P.; Grynberg, P.; do Carmo Costa, M.M.; Togawa, R.C.; de Sousa, C.A.F.; Júnior, M.T.S. Expression analysis of miRNAs and their putative target genes confirm a preponderant role of transcription factors in the early response of oil palm plants to salinity stress. BMC Plant Biol. 2021, 21, 518. [Google Scholar] [CrossRef]
- Ferreira, T.M.M.; Ferreira Filho, J.A.; Leão, A.P.; de Sousa, C.A.F.; Souza, M.T., Jr. Structural and functional analysis of stress-inducible genes and their promoters selected from young oil palm (Elaeis guineensis) under salt stress. BMC Genom. 2022, 23, 735. [Google Scholar] [CrossRef]
- Faizah, R.; Putranto, R.A.; Wening, S.; Sukma, D.; Raharti, V.R.; Budiani, A.; Sudarsono, S.J.I.C.S.E.; Science, E. Differential expression of root specific genes of oil palm seedlings at early stage of Ganoderma boninense infection. IOP Conf. Ser. Earth Environ. Sci. 2020, 418, 012044. [Google Scholar] [CrossRef]
- Saand, M.A.; Li, J.; Wu, Y.; Zhou, L.; Cao, H.; Yang, Y. Integrative Omics Analysis of Three Oil Palm Varieties Reveals (Tanzania × Ekona) TE as a Cold-Resistant Variety in Response to Low-Temperature Stress. Int. J. Mol. Sci 2022, 23, 14926. [Google Scholar] [CrossRef] [PubMed]
- Silva Rde, C.; Carmo, L.S.; Luis, Z.G.; Silva, L.P.; Scherwinski-Pereira, J.E.; Mehta, A. Proteomic identification of differentially expressed proteins during the acquisition of somatic embryogenesis in oil palm (Elaeis guineensis Jacq.). J. Proteom. 2014, 104, 112–127. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.S.; Liddell, S.; Ong Abdullah, M.; Wong, W.C.; Chin, C.F. Differential proteomic analysis of embryogenic lines in oil palm (Elaeis guineensis Jacq). J. Proteom. 2016, 143, 334–345. [Google Scholar] [CrossRef]
- Ribeiro, D.G.; de Almeida, R.F.; Fontes, W.; de Souza Castro, M.; de Sousa, M.V.; Ricart, C.A.O.; da Cunha, R.N.V.; Lopes, R.; Scherwinski-Pereira, J.E.; Mehta, A. Stress and cell cycle regulation during somatic embryogenesis plays a key role in oil palm callus development. J. Proteom. 2019, 192, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Aroonluk, S.; Roytrakul, S.; Jantasuriyarat, C. Identification and Characterization of Phosphoproteins in Somatic Embryogenesis Acquisition during Oil Palm Tissue Culture. Plants 2020, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Balotf, S.; Wilson, R.; Tegg, R.S.; Nichols, D.S.; Wilson, C.R. Shotgun Proteomics as a Powerful Tool for the Study of the Proteomes of Plants, Their Pathogens, and Plant-Pathogen Interactions. Proteomes 2022, 10, 5. [Google Scholar] [CrossRef]
- Loei, H.; Lim, J.; Tan, M.; Lim, T.K.; Lin, Q.S.; Chew, F.T.; Kulaveerasingam, H.; Chung, M.C. Proteomic analysis of the oil palm fruit mesocarp reveals elevated oxidative phosphorylation activity is critical for increased storage oil production. J. Proteome Res. 2013, 12, 5096–5109. [Google Scholar] [CrossRef]
- Kok, S.-Y.; Namasivayam, P.; Ee, G.C.-L.; Ong-Abdullah, M. Comparative proteomic analysis of oil palm (Elaeis guineensis Jacq.) during early fruit development. J. Proteom. 2021, 232, 104052. [Google Scholar] [CrossRef]
- Hassan, H.; Amiruddin, M.D.; Weckwerth, W.; Ramli, U.S. Deciphering key proteins of oil palm (Elaeis guineensis Jacq.) fruit mesocarp development by proteomics and chemometrics. Electrophoresis 2019, 40, 254–265. [Google Scholar] [CrossRef]
- Rodrigues-Neto, J.C.; Correia, M.V.; Souto, A.L.; Ribeiro, J.A.d.A.; Vieira, L.R.; Souza, M.T.; Rodrigues, C.M.; Abdelnur, P.V. Metabolic fingerprinting analysis of oil palm reveals a set of differentially expressed metabolites in fatal yellowing symptomatic and non-symptomatic plants. Metabolomics 2018, 14, 142. [Google Scholar] [CrossRef]
- Neto, J.C.R.; Vieira, L.R.; de Aquino Ribeiro, J.A.; de Sousa, C.A.F.; Júnior, M.T.S.; Abdelnur, P.V. Metabolic effect of drought stress on the leaves of young oil palm (Elaeis guineensis) plants using UHPLC–MS and multivariate analysis. Sci. Rep. 2021, 11, 18271. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, I.I.; Abdullah, S.N.A.; Saud, H.M.; Shaharuddin, N.A.; Isa, N.M. The Dynamic Responses of Oil Palm Leaf and Root Metabolome to Phosphorus Deficiency. Metabolites 2021, 11, 217. [Google Scholar] [CrossRef] [PubMed]
- Santiago, K.A.A.; Wong, W.C.; Goh, Y.K.; Tey, S.H.; Ting, A.S.Y. Pathogenicity of monokaryotic and dikaryotic mycelia of Ganoderma boninense revealed via LC–MS-based metabolomics. Sci. Rep. 2024, 14, 5330. [Google Scholar] [CrossRef] [PubMed]
- Zakaria, L. Basal Stem Rot of Oil Palm: The Pathogen, Disease Incidence, and Control Methods. Plant Dis. 2023, 107, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Neoh, B.K.; Teh, H.F.; Ng, T.L.; Tiong, S.H.; Thang, Y.M.; Ersad, M.A.; Mohamed, M.; Chew, F.T.; Kulaveerasingam, H.; Appleton, D.R. Profiling of metabolites in oil palm mesocarp at different stages of oil biosynthesis. J. Agric. Food Chem. 2013, 61, 1920–1927. [Google Scholar] [CrossRef]
- Rozali, N.L.; Tahir, N.I.; Hassan, H.; Othman, A.; Ramli, U.S. Identification of amines, amino and organic acids in oil palm (Elaeis guineensis Jacq.) spear leaf using GC- and LC/Q-TOF MS metabolomics platforms. Biocatal. Agric. Biotechnol. 2021, 37, 102165. [Google Scholar] [CrossRef]
| Traits | Number of Accessions | Chromosomes | Number of QTLs | Number of Candidate Genes | Reference |
|---|---|---|---|---|---|
| compact fruit bunch | 422 | 4, 8, 11 | 4 | / | [54] |
| oil content | 196 | / | 33 | 55 | [55] |
| fatty acid composition; nutrient content | 210 | 2, 13 | 6 | 3 | [34] |
| oil content | 471 | 3, 5, 10, 13, 15 | / | / | [56] |
| disease resistance | 96 | 6, 7, 9 | 5 | / | [57] |
| leaf area size; high yield; | 115 | / | / | 5 | [58] |
| trunk height | 132 | 5, 6 | / | / | [59] |
| stem height | 96 | 1, 2, 4, 14, 16 | 7 | / | [60] |
| high oil production | 310 | 1, 4, 7, 10, 11, 12, 15 | 43 | / | [61] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, W.; John Martin, J.J.; Li, X.; Liu, X.; Zhang, R.; Hou, M.; Cao, H.; Cheng, S. Unveiling the Secrets of Oil Palm Genetics: A Look into Omics Research. Int. J. Mol. Sci. 2024, 25, 8625. https://doi.org/10.3390/ijms25168625
Xu W, John Martin JJ, Li X, Liu X, Zhang R, Hou M, Cao H, Cheng S. Unveiling the Secrets of Oil Palm Genetics: A Look into Omics Research. International Journal of Molecular Sciences. 2024; 25(16):8625. https://doi.org/10.3390/ijms25168625
Chicago/Turabian StyleXu, Wen, Jerome Jeyakumar John Martin, Xinyu Li, Xiaoyu Liu, Ruimin Zhang, Mingming Hou, Hongxing Cao, and Shuanghong Cheng. 2024. "Unveiling the Secrets of Oil Palm Genetics: A Look into Omics Research" International Journal of Molecular Sciences 25, no. 16: 8625. https://doi.org/10.3390/ijms25168625
APA StyleXu, W., John Martin, J. J., Li, X., Liu, X., Zhang, R., Hou, M., Cao, H., & Cheng, S. (2024). Unveiling the Secrets of Oil Palm Genetics: A Look into Omics Research. International Journal of Molecular Sciences, 25(16), 8625. https://doi.org/10.3390/ijms25168625





