Alterations in Tumor Aggression Following Androgen Receptor Signaling Restoration in Canine Prostate Cancer Cell Lines
Abstract
1. Introduction
2. Results
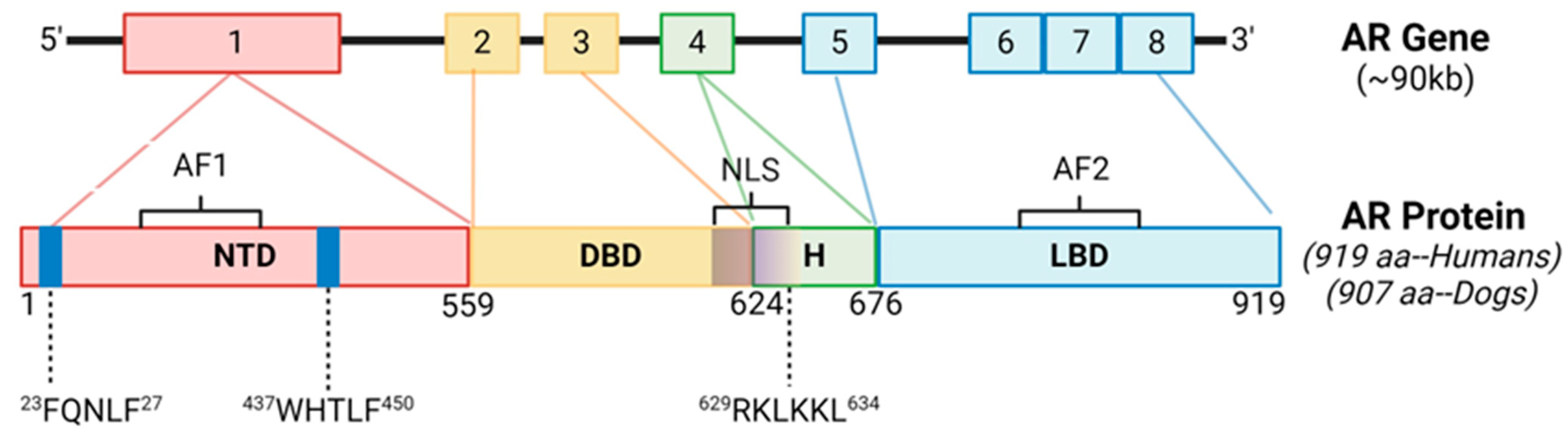
2.1. Androgen Receptor Gene Is Highly Conserved between Canines and Humans
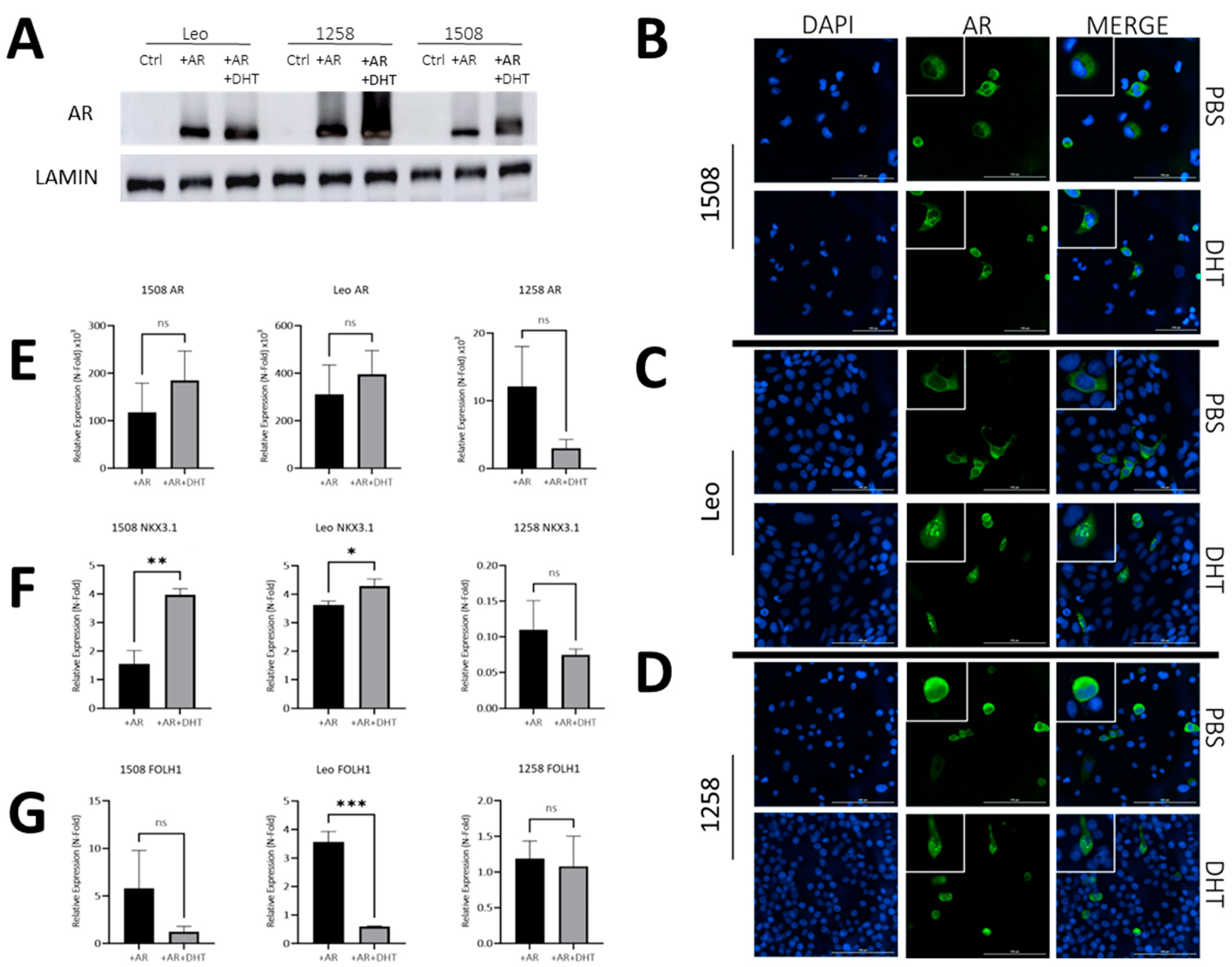
2.2. Androgen Receptor Transfection and Treatment with DHT Results in Nuclear Localization and Expression of Downstream Target Genes in Canines
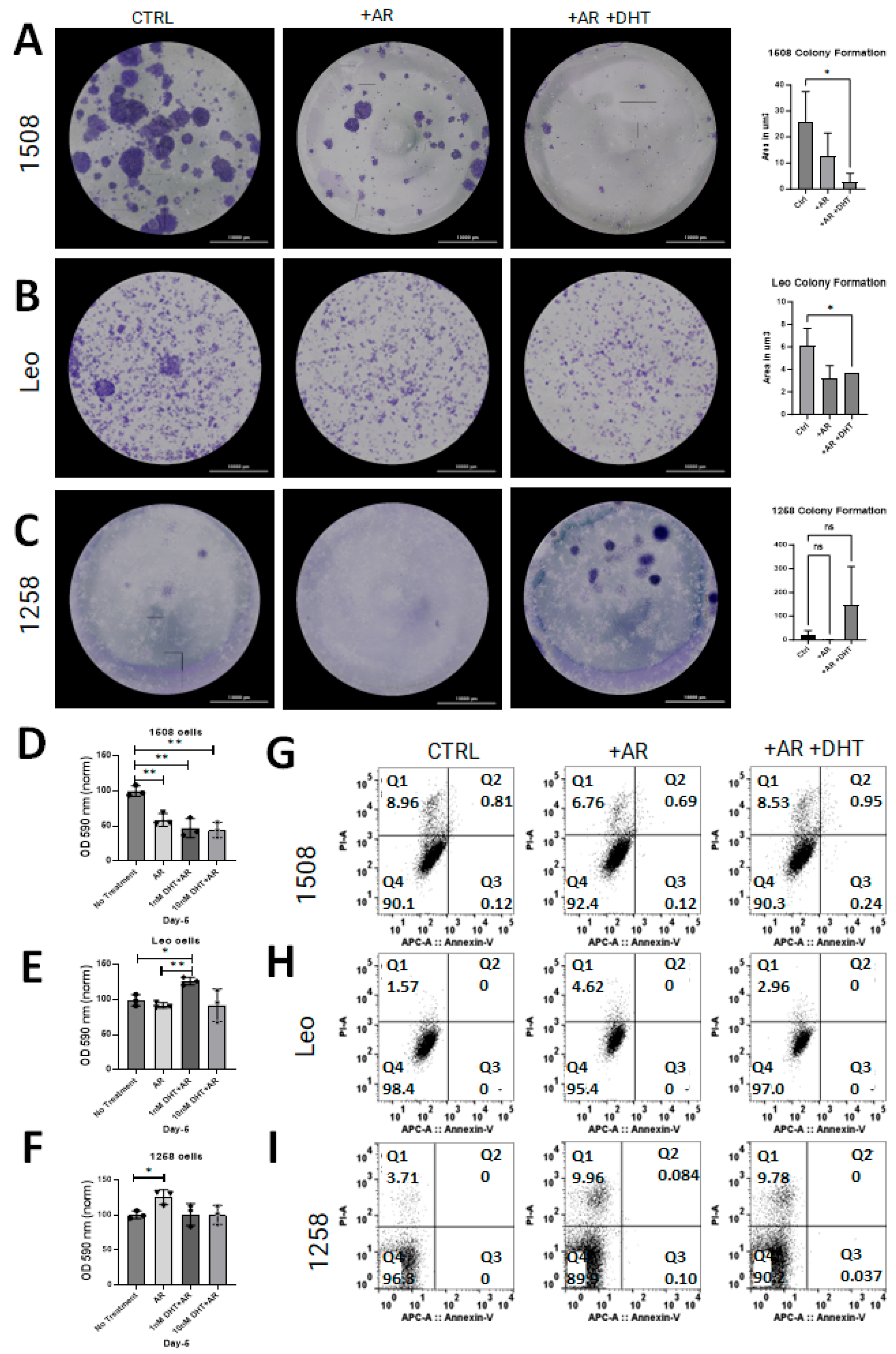
2.3. AR Signaling Restoration Decreases Proliferation in Canine PCa Cell Line 1508 and Leo but Decreases Metabolic Activity in 1258
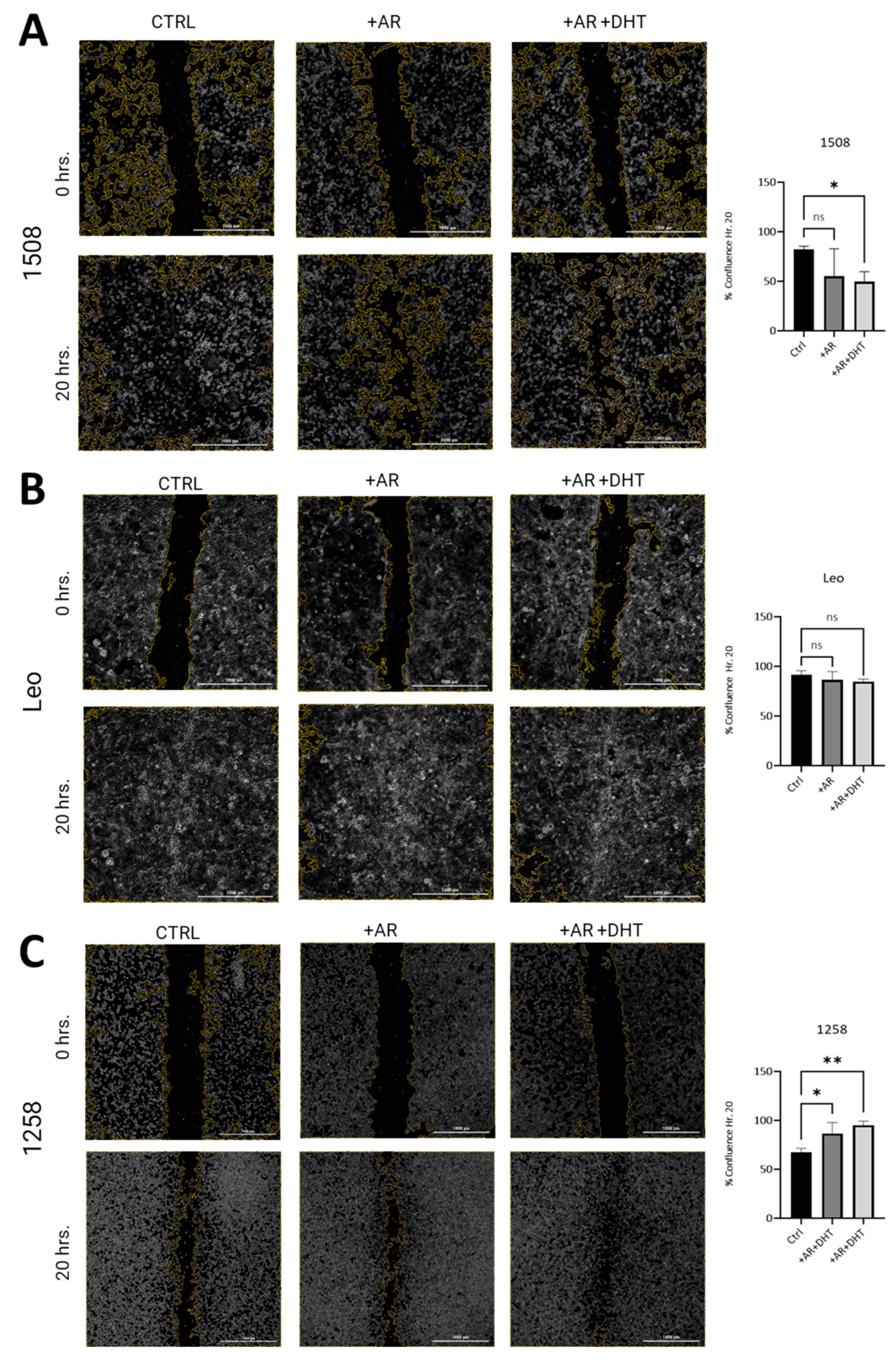
2.4. AR Signaling Restoration Decreases Migration in Canine PCa Cell Line 1508 and Leo but Increases It in 1258
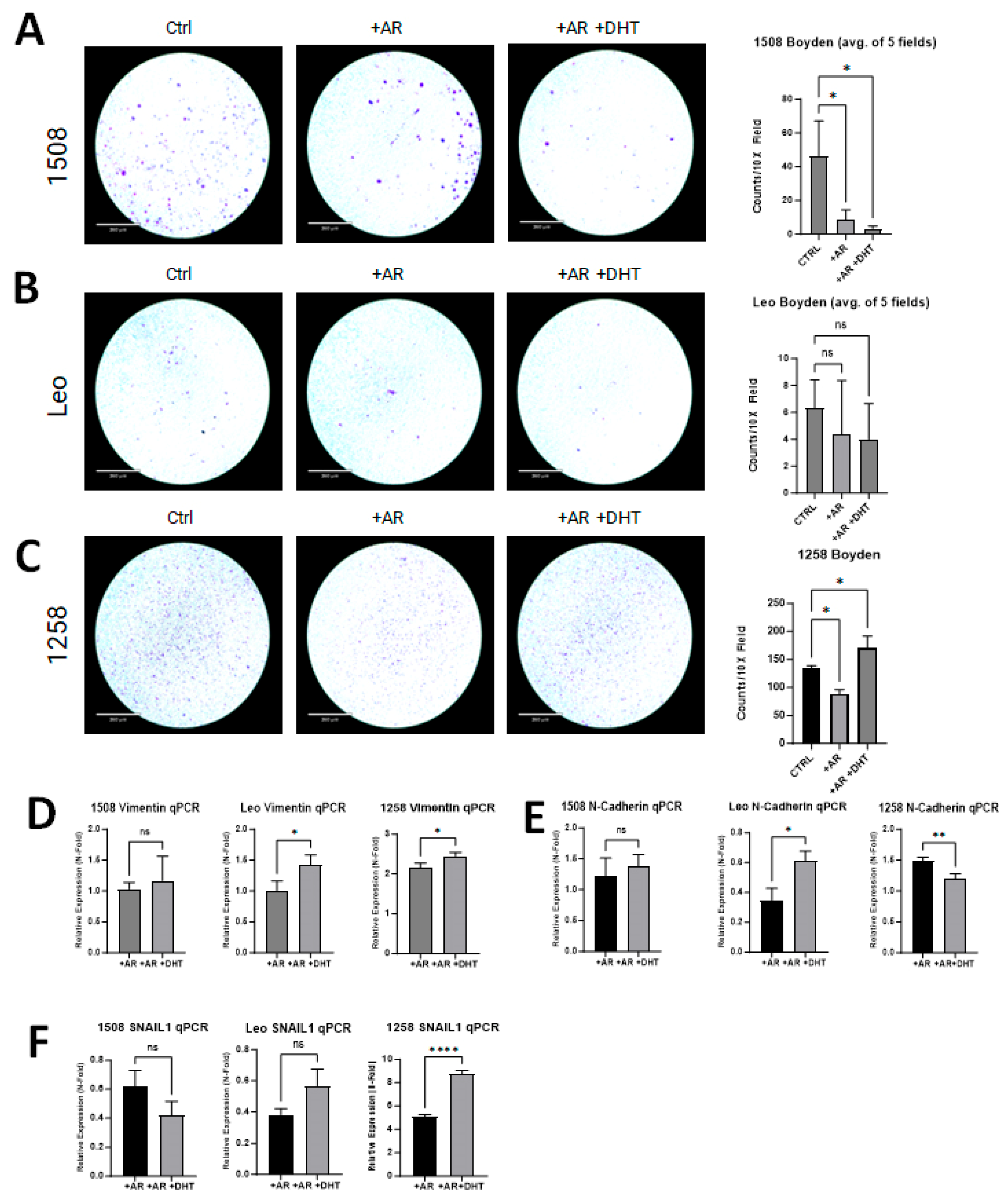
2.5. AR Signaling Restoration Decreases Invasion and Markers of EMT in Some Canine PCa Cell Lines but Increases in Others
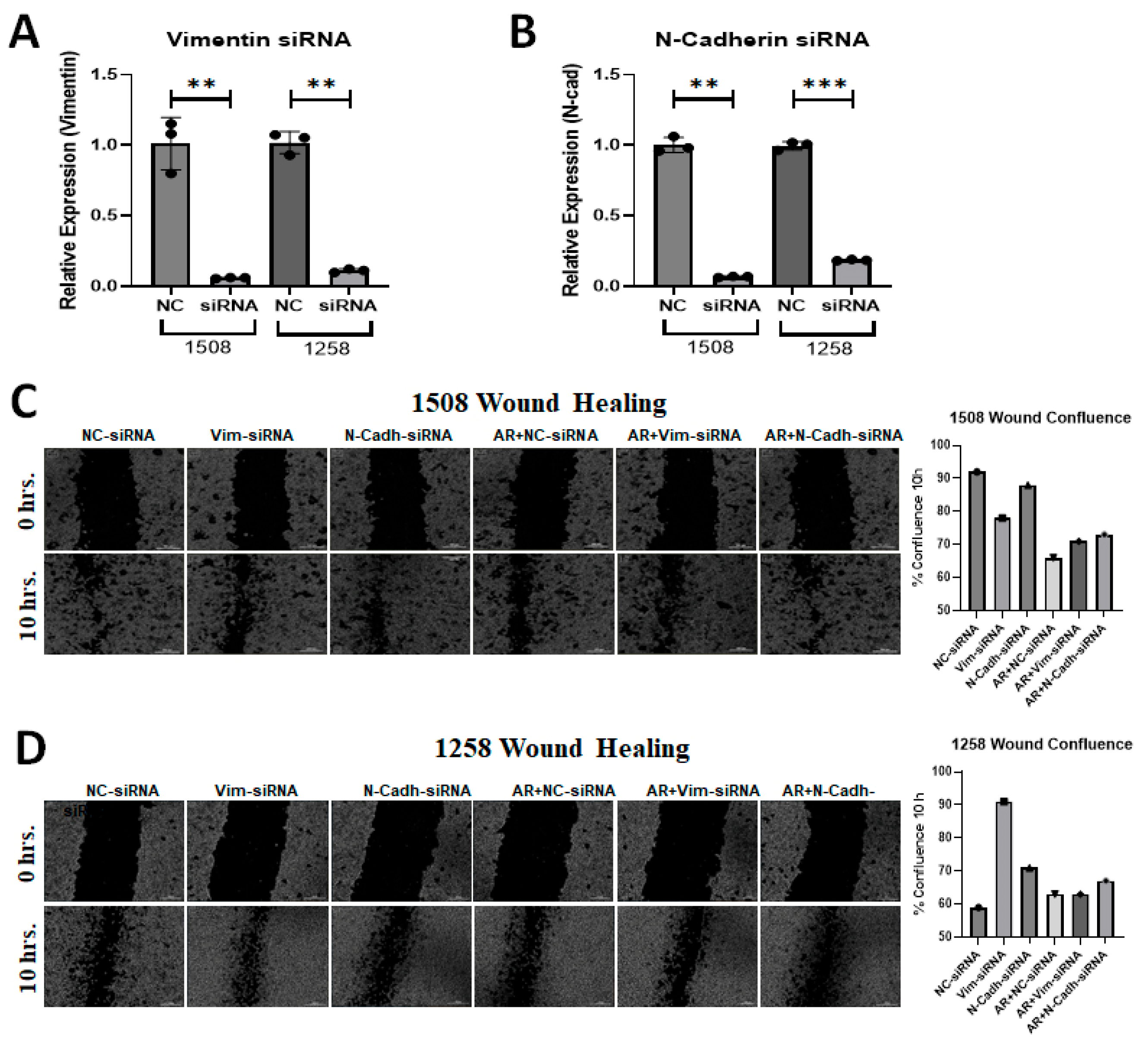
2.6. Vimentin Regulates Migration of 1258 but Not 1508
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cell Lines and Cell Culture Materials
5.2. Androgen Receptor Plasmid Construction, Transfection, and Sequence Alignment
5.3. siRNA Construction and Transfection
5.4. Cell Lysate and Protein Immunoblotting
5.5. Differential Gene Expression (RT-qPCR)
5.6. Immunofluorescence
5.7. MTT Assay
5.8. Clonogenic Assay
5.9. Flow Cytometry for Apoptosis
5.10. Migration Assay
5.11. Invasion Assay
5.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Schreiber-Agus, N. Mouse models of prostate cancer. Oncogene 1999, 18, 5349–5355. [Google Scholar] [CrossRef] [PubMed]
- Ryman-Tubb, T.; Lothion-Roy, J.H.; Metzler, V.M.; Harris, A.E.; Robinson, B.D.; Rizvanov, A.A.; Jeyapalan, J.N.; James, V.H.; England, G.; Rutland, C.S.; et al. Comparative pathology of dog and human prostate cancer. Vet. Med. Sci. 2022, 8, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Vasilatis, D.M.; Lucchesi, C.A.; Ghosh, P.M. Molecular Similarities and Differences between Canine Prostate Cancer and Human Prostate Cancer Variants. Biomedicines 2023, 11, 1100. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.L.; L’Eplattenier, H.; van den Ham, R.; Verseijden, F.; Jagtenberg, A.; Mol, J.A.; Teske, E. Androgen receptor CAG repeat polymorphisms in canine prostate cancer. J. Vet. Intern. Med. 2008, 22, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.L.; van den Ham, R.; Mol, J.; Teske, E. Immunostaining of the androgen receptor and sequence analysis of its DNA-binding domain in canine prostate cancer. Vet. J. 2009, 181, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Smith, A.; Ong, M.; de Bono, J.S. Novel drugs targeting the androgen receptor pathway in prostate cancer. Cancer Metastasis Rev. 2014, 33, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Brand, L.J.; Dehm, S.M. Androgen receptor gene rearrangements: New perspectives on prostate cancer progression. Curr. Drug Targets 2013, 14, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Hohl, A.; Marcelli, M. Androgen Receptor in Health and Disease. In Testosterone: From Basic to Clinical Aspects; Hohl, A., Ed.; Springer Nature Switzerland AG: Cham, Switzerland, 2017; pp. 21–76. [Google Scholar]
- Chandrasekar, T.; Yang, J.C.; Gao, A.C.; Evans, C.P. Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl. Androl. Urol. 2015, 4, 365–380. [Google Scholar]
- Messner, E.A.; Steele, T.M.; Tsamouri, M.M.; Hejazi, N.; Gao, A.C.; Mudryj, M.; Ghosh, P.M. The Androgen Receptor in Prostate Cancer: Effect of Structure, Ligands and Spliced Variants on Therapy. Biomedicines 2020, 8, 422. [Google Scholar] [CrossRef]
- Ramalingam, S.; Ramamurthy, V.P.; Njar, V.C.O. Dissecting major signaling pathways in prostate cancer development and progression: Mechanisms and novel therapeutic targets. J. Steroid Biochem. Mol. Biol. 2017, 166, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Berchuck, J.E.; Viscuse, P.V.; Beltran, H.; Aparicio, A. Clinical considerations for the management of androgen indifferent prostate cancer. Prostate Cancer Prostatic Dis. 2021, 24, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Katzenwadel, A.; Wolf, P. Androgen deprivation of prostate cancer: Leading to a therapeutic dead end. Cancer Lett. 2015, 367, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Altuwaijri, S.; Lai, K.P.; Wu, C.T.; Ricke, W.A.; Messing, E.M.; Yao, J.; Yeh, S.; Chang, C. Androgen receptor is a tumor suppressor and proliferator in prostate cancer. Proc. Natl. Acad. Sci. USA 2008, 105, 12182–12187. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, J.T. Resolving the Coffey Paradox: What does the androgen receptor do in normal vs. malignant prostate epithelial cells? Am. J. Clin. Exp. Urol. 2018, 6, 55–61. [Google Scholar] [PubMed]
- Sena, L.A.; Kumar, R.; Sanin, D.E.; Thompson, E.A.; Rosen, D.M.; Dalrymple, S.L.; Antony, L.; Yang, Y.; Gomes-Alexandre, C.; Hicks, J.L.; et al. Androgen receptor activity in prostate cancer dictates efficacy of bipolar androgen therapy through MYC. J. Clin. Investig. 2022, 132, e162396. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Gao, Y.; He, H.H.; Han, D.; Han, W.; Avery, A.; Macoska, J.A.; Liu, X.; Chen, S.; Ma, F.; et al. Androgen Receptor Tumor Suppressor Function Is Mediated by Recruitment of Retinoblastoma Protein. Cell Rep. 2016, 17, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Formaggio, N.; Rubin, M.A.; Theurillat, J.P. Loss and revival of androgen receptor signaling in advanced prostate cancer. Oncogene 2021, 40, 1205–1216. [Google Scholar] [CrossRef]
- Scaccianoce, E.; Festuccia, C.; Dondi, D.; Guerini, V.; Bologna, M.; Motta, M.; Poletti, A. Characterization of prostate cancer DU145 cells expressing the recombinant androgen receptor. Oncol. Res. 2003, 14, 101–112. [Google Scholar] [CrossRef]
- Bryan, J.N.; Keeler, M.R.; Henry, C.J.; Bryan, M.E.; Hahn, A.W.; Caldwell, C.W. A population study of neutering status as a risk factor for canine prostate cancer. Prostate 2007, 67, 1174–1181. [Google Scholar] [CrossRef]
- He, B.; Kemppainen, J.A.; Wilson, E.M. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem. 2000, 275, 22986–22994. [Google Scholar] [CrossRef] [PubMed]
- Dubbink, H.J.; Hersmus, R.; Verma, C.S.; van der Korput, H.A.; Berrevoets, C.A.; van Tol, J.; der Made, A.C.Z.-V.; Brinkmann, A.O.; Pike, A.C.; Trapman, J. Distinct recognition modes of FXXLF and LXXLL motifs by the androgen receptor. Mol. Endocrinol. 2004, 18, 2132–2150. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, C.M.; Lowe, L.P.; Lee, H.; Hayes, M.G.; Dyer, A.R.; Metzger, B.E.; Lowe, W.L.; Urbanek, M. Ethnic variation in allele distribution of the androgen receptor (AR) (CAG)n repeat. J. Androl. 2012, 33, 210–215. [Google Scholar] [CrossRef]
- Albrizio, M.; Siniscalchi, M.; Sasso, R.; Quaranta, A. Effects of the environment on dog semen parameters and testosterone concentration. Theriogenology 2013, 80, 800–804. [Google Scholar] [CrossRef] [PubMed]
- Gloyna, R.E.; Siiteri, P.K.; Wilson, J.D. Dihydrotestosterone in prostatic hypertrophy. II. The formation and content of dihydrotestosterone in the hypertrophic canine prostate and the effect of dihydrotestosterone on prostate growth in the dog. J. Clin. Investig. 1970, 49, 1746–1753. [Google Scholar] [CrossRef] [PubMed]
- Izumi, K.; Mizokami, A. Suppressive Role of Androgen/Androgen Receptor Signaling via Chemokines on Prostate Cancer Cells. J. Clin. Med. 2019, 8, 354. [Google Scholar] [CrossRef] [PubMed]
- Bonaccorsi, L.; Muratori, M.; Carloni, V.; Zecchi, S.; Formigli, L.; Forti, G.; Baldi, E. Androgen receptor and prostate cancer invasion. Int. J. Androl. 2003, 26, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Vasilatis, D.M.; Ghosh, P.M. Clinicopathologic Characterization of Prostatic Cancer in Dogs. Animals 2024, 14, 588. [Google Scholar] [CrossRef] [PubMed]
- Fonseca-Alves, C.E.; Rodrigues, M.M.; de Moura, V.M.; Rogatto, S.R.; Laufer-Amorim, R. Alterations of C-MYC, NKX3.1, and E-cadherin expression in canine prostate carcinogenesis. Microsc. Res. Tech. 2013, 76, 1250–1256. [Google Scholar] [CrossRef]
- Sever, R.; Glass, C.K. Signaling by nuclear receptors. Cold Spring Harb. Perspect. Biol. 2013, 5, a016709. [Google Scholar] [CrossRef]
- Lei, Q.; Jiao, J.; Xin, L.; Chang, C.J.; Wang, S.; Gao, J.; Gleave, M.E.; Witte, O.N.; Liu, X.; Wu, H. NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell 2006, 9, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Wang, Z.A. Transcriptional regulation of the Nkx3.1 gene in prostate luminal stem cell specification and cancer initiation via its 3′ genomic region. J. Biol. Chem. 2017, 292, 13521–13530. [Google Scholar] [CrossRef] [PubMed]
- Gurel, B.; Ali, T.Z.; Montgomery, E.A.; Begum, S.; Hicks, J.; Goggins, M.; Eberhart, C.G.; Clark, D.P.; Bieberich, C.J.; Epstein, J.I.; et al. NKX3.1 as a marker of prostatic origin in metastatic tumors. Am. J. Surg. Pathol. 2010, 34, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.J.; Smith-Jones, P.M.; Wongvipat, J.; Navarro, V.; Kim, S.; Bander, N.H.; Larson, S.M.; Sawyers, C.L. Noninvasive measurement of androgen receptor signaling with a positron-emitting radiopharmaceutical that targets prostate-specific membrane antigen. Proc. Natl. Acad. Sci. USA 2011, 108, 9578–9582. [Google Scholar] [CrossRef] [PubMed]
- Sommer, U.; Siciliano, T.; Ebersbach, C.; Beier, A.K.; Stope, M.B.; Jöhrens, K.; Baretton, G.B.; Borkowetz, A.; Thomas, C.; Erb, H.H.H. Impact of Androgen Receptor Activity on Prostate-Specific Membrane Antigen Expression in Prostate Cancer Cells. Int. J. Mol. Sci. 2022, 23, 1046. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Maibaum, D.; Pich, A.; Nolte, I.; Escobar, H.M. Verification of a canine PSMA (FolH1) antibody. Anticancer Res. 2015, 35, 145–148. [Google Scholar] [PubMed]
- Ghasemi, M.; Turnbull, T.; Sebastian, S.; Kempson, I. The MTT Assay: Utility, Limitations, Pitfalls, and Interpretation in Bulk and Single-Cell Analysis. Int. J. Mol. Sci. 2021, 22, 12827. [Google Scholar] [CrossRef]
- Bader, D.A.; McGuire, S.E. Tumour metabolism and its unique properties in prostate adenocarcinoma. Nat. Rev. Urol. 2020, 17, 214–231. [Google Scholar] [CrossRef]
- Siltari, A.; Syvälä, H.; Lou, Y.R.; Gao, Y.; Murtola, T.J. Role of Lipids and Lipid Metabolism in Prostate Cancer Progression and the Tumor’s Immune Environment. Cancers 2022, 14, 4293. [Google Scholar] [CrossRef]
- Berridge, M.V.; Tan, A.S. Characterization of the cellular reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT): Subcellular localization, substrate dependence, and involvement of mitochondrial electron transport in MTT reduction. Arch. Biochem. Biophys. 1993, 303, 474–482. [Google Scholar] [CrossRef]
- Leung, J.K.; Sadar, M.D. Non-Genomic Actions of the Androgen Receptor in Prostate Cancer. Front. Endocrinol. 2017, 8, 2. [Google Scholar] [CrossRef]
- Bonaccorsi, L.; Carloni, V.; Muratori, M.; Salvadori, A.; Giannini, A.; Carini, M.; Serio, M.; Forti, G.; Baldi, E. Androgen receptor expression in prostate carcinoma cells suppresses alpha6beta4 integrin-mediated invasive phenotype. Endocrinology 2000, 141, 3172–3182. [Google Scholar] [CrossRef]
- Tang, X.; Sui, X.; Weng, L.; Liu, Y. SNAIL1: Linking Tumor Metastasis to Immune Evasion. Front. Immunol. 2021, 12, 724200. [Google Scholar] [CrossRef]
- Packeiser, E.M.; Taher, L.; Kong, W.; Ernst, M.; Beck, J.; Hewicker-Trautwein, M.; Brenig, B.; Schütz, E.; Escobar, H.M.; Nolte, I. RNA-seq of nine canine prostate cancer cell lines reveals diverse therapeutic target signatures. Cancer Cell Int. 2022, 22, 54. [Google Scholar] [CrossRef]
- Winkler, S.; Escobar, H.M.; Eberle, N.; Reimann-Berg, N.; Nolte, I.; Bullerdiek, J. Establishment of a cell line derived from a canine prostate carcinoma with a highly rearranged karyotype. J. Hered. 2005, 96, 782–785. [Google Scholar] [CrossRef] [PubMed]
- Packeiser, E.M.; Hewicker-Trautwein, M.; Thiemeyer, H.; Mohr, A.; Junginger, J.; Schille, J.T.; Escobar, H.M.; Nolte, I. Characterization of six canine prostate adenocarcinoma and three transitional cell carcinoma cell lines derived from primary tumor tissues as well as metastasis. PLoS ONE 2020, 15, e0230272. [Google Scholar] [CrossRef]
- Thudi, N.K.; Shu, S.T.; Martin, C.K.; Lanigan, L.G.; Nadella, M.V.; Van Bokhoven, A.; Werbeck, J.L.; Simmons, J.K.; Murahari, S.; Kisseberth, W.C.; et al. Development of a brain metastatic canine prostate cancer cell line. Prostate 2011, 71, 1251–1263. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, V.; Hitte, C.; Kidd, J.M.; Masterson, P.; Murphy, T.D.; Emery, S.; Davis, B.; Buckley, R.M.; Liu, Y.H.; Zhang, X.Q.; et al. Dog10K_Boxer_Tasha_1.0: A Long-Read Assembly of the Dog Reference Genome. Genes 2021, 12, 847. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. SDS-Polyacrylamide Gel Electrophoresis of Proteins. CSH Protoc. 2006, 2006, pdb.prot4540. [Google Scholar] [CrossRef]
- Rivera-Calderón, L.G.; Fonseca-Alves, C.E.; Kobayashi, P.E.; Carvalho, M.; Drigo, S.A.; de Oliveira Vasconcelos, R.; Laufer-Amorim, R. Alterations in PTEN, MDM2, TP53 and AR protein and gene expression are associated with canine prostate carcinogenesis. Res. Vet. Sci. 2016, 106, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Zheng, H.; Liu, X.; Xie, G. The Analysis of E-Cadherin, N-Cadherin, Vimentin, HER-2, CEA, CA15-3 and SF Expression in the Diagnosis of Canine Mammary Tumors. Animals 2022, 12, 3050. [Google Scholar] [CrossRef] [PubMed]
- Sammarco, A.; Gomiero, C.; Beffagna, G.; Cavicchioli, L.; Ferro, S.; Michieletto, S.; Orvieto, E.; Patruno, M.; Zappulli, V. Epithelial-to-Mesenchymal Transition and Phenotypic Marker Evaluation in Human. Canine, and Feline Mammary Gland Tumors. Animals 2023, 13, 878. [Google Scholar] [CrossRef]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.C.; Park, A.Y.; Guan, J.L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef] [PubMed]
- van de Merbel, A.F.; van der Horst, G.; Buijs, J.T.; van der Pluijm, G. Protocols for Migration and Invasion Studies in Prostate Cancer. In Prostate Cancer: Methods and Protocols; Culig, Z., Ed.; Springer Science+Business Media: New York, NY, USA, 2018; pp. 67–79. [Google Scholar]
| Aggressive Behavior | 1508 | Leo | 1258 |
|---|---|---|---|
| Colony Formation | ↓ | ↓ | No Change |
| MTT Metabolism | ↓ | ↑ | No Change |
| Migration | ↓ | No Change | ↑ |
| Invasion | ↓ | No Change | ↑ |
| Vimentin | No Change | ↑ | ↑ |
| N-cadherin | No Change | ↑ | ↓ |
| SNAIL1 | No Change | No Change | ↑ |
| Gene | siRNA Sequence (5′—3′) |
|---|---|
| Vimentin | Sense: GAAACUACAUGAUGAGGAAUU |
| Antisense: UUCCUCAUCAUGUAGUUUCUU | |
| N-cadherin | Sense: GAGAAGAAGACCAGGGAUUAUU |
| Antisense: UAAUCCUGGUCUUCUUCUCUU | |
| SNAIL1 | Sense: GGACGAGGACAGUGGGAAAUU |
| Antisense: UUUCCCACUGUCCUCGUCCUU |
| Gene | Sequence (5′—3′) |
|---|---|
| AR a | F: CGCCCCTGACCTGGTTT |
| R: GGCTGTACATCCGGGACTTG | |
| NKX3.1 a | F: TGAGGTGGTTGGAGGTTTGC |
| R: TTTCATTGGCCCATCACTGA | |
| FOLH1 b | F: GTGTTTGGTGGCATTGACC |
| R: TTCTGCATCCCAGCTTGC | |
| Vimentin c | F: TACGCCAGCAATATGAAAGCG |
| R: AGGGCATCATTGTTCCGGTTA | |
| N-cadherin c | F: AGCACCCTCCTCAGTCAACG |
| R: TGTCAACATGGTCCCAGCA | |
| SNAIL1 d | F: ACTGCAGCCGTGCCTTTG |
| R: AAGGTTCGGGAACAGGTCTTG | |
| HPRT1 a | F: AGCTTGCTGGTGAAAAGGAC |
| R: TTATAGTCAAGGGCATATCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasilatis, D.M.; Batra, N.; Lucchesi, C.A.; Abria, C.J.; Packeiser, E.-M.; Murua Escobar, H.; Ghosh, P.M. Alterations in Tumor Aggression Following Androgen Receptor Signaling Restoration in Canine Prostate Cancer Cell Lines. Int. J. Mol. Sci. 2024, 25, 8628. https://doi.org/10.3390/ijms25168628
Vasilatis DM, Batra N, Lucchesi CA, Abria CJ, Packeiser E-M, Murua Escobar H, Ghosh PM. Alterations in Tumor Aggression Following Androgen Receptor Signaling Restoration in Canine Prostate Cancer Cell Lines. International Journal of Molecular Sciences. 2024; 25(16):8628. https://doi.org/10.3390/ijms25168628
Chicago/Turabian StyleVasilatis, Demitria M., Neelu Batra, Christopher A. Lucchesi, Christine J. Abria, Eva-Maria Packeiser, Hugo Murua Escobar, and Paramita M. Ghosh. 2024. "Alterations in Tumor Aggression Following Androgen Receptor Signaling Restoration in Canine Prostate Cancer Cell Lines" International Journal of Molecular Sciences 25, no. 16: 8628. https://doi.org/10.3390/ijms25168628
APA StyleVasilatis, D. M., Batra, N., Lucchesi, C. A., Abria, C. J., Packeiser, E.-M., Murua Escobar, H., & Ghosh, P. M. (2024). Alterations in Tumor Aggression Following Androgen Receptor Signaling Restoration in Canine Prostate Cancer Cell Lines. International Journal of Molecular Sciences, 25(16), 8628. https://doi.org/10.3390/ijms25168628





