Metabolomic Profiling of Breast Cancer Patients Undergoing Neoadjuvant Chemotherapy for Predicting Disease-Free and Overall Survival
Abstract
:1. Introduction
2. Results
2.1. Differences in the Metabolic Profiles of Patients before and after NACT
2.2. The MRSS Helps to Determine the Contributions of Metabolites and Clinical Covariates in NACT Patients
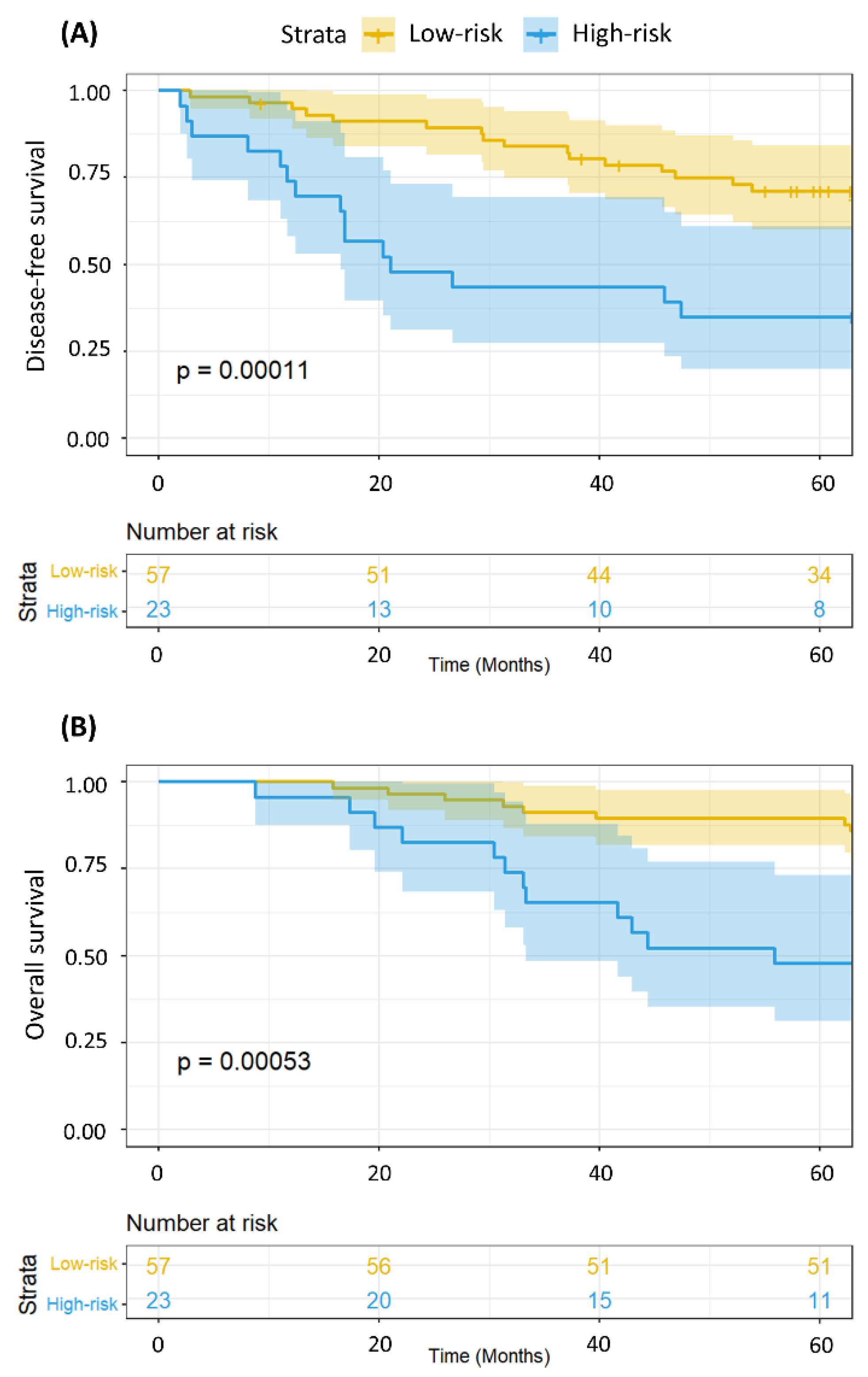
2.3. The MRSS Can Distinguish between Patients with Low- and High-Risk of Relapse and Even Overall Survival
2.4. Metabolomics Data Can Provide Additional Survival Information Even after Adjustment for Disease Stage and Patient Age
3. Discussion
4. Materials and Methods
4.1. Patient Selection, Accrual, and Sample Processing
4.2. Clinical Follow-Up
4.3. Metabolite Normalization
4.4. Metabolite Differential Fold-Changes
4.5. Metabolite Contribution Using Multivariate Cox Proportional Hazards Selected by the Variation Inflation Factor (VIF)
4.6. Metabolite-Related Survival Score (MRSS)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Haddad, T.C.; Goetz, M.P. Landscape of Neoadjuvant Therapy for Breast Cancer. Ann. Surg. Oncol. 2015, 22, 1408–1415. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Tumour-Infiltrating Lymphocytes and Prognosis in Different Subtypes of Breast Cancer: A Pooled Analysis of 3771 Patients Treated with Neoadjuvant Therapy. Lancet. Oncol. 2018, 19, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.; Kin, T. Clinical Perspectives in Addressing Unsolved Issues in (Neo)Adjuvant Therapy for Primary Breast Cancer. Cancers 2021, 13, 926. [Google Scholar] [CrossRef] [PubMed]
- Debik, J.; Euceda, L.R.; Lundgren, S.; Gythfeldt, H.V.D.L.; Garred, Ø.; Borgen, E.; Engebraaten, O.; Bathen, T.F.; Giskeødegård, G.F. Assessing Treatment Response and Prognosis by Serum and Tissue Metabolomics in Breast Cancer Patients. J. Proteome Res. 2019, 18, 3649–3660. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Xu, R.; Mao, S.; Zhang, Y.; Dai, Y.; Guo, Q.; Song, X.; Zhang, Q.; Li, L.; Chen, Q. Metabolic Biomarker Signature for Predicting the Effect of Neoadjuvant Chemotherapy of Breast Cancer. Ann. Transl. Med. 2019, 7, 670. [Google Scholar] [CrossRef]
- Salvador-Coloma, C.; Santaballa, A.; Sanmartín, E.; Calvo, D.; García, A.; Hervás, D.; Cordón, L.; Quintas, G.; Ripoll, F.; Panadero, J.; et al. Immunosuppressive Profiles in Liquid Biopsy at Diagnosis Predict Response to Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer. Eur. J. Cancer 2020, 139, 119–134. [Google Scholar] [CrossRef]
- He, X.; Gu, J.; Zou, D.; Yang, H.; Zhang, Y.; Ding, Y.; Teng, L. NMR-Based Metabolomics Analysis Predicts Response to Neoadjuvant Chemotherapy for Triple-Negative Breast Cancer. Front. Mol. Biosci. 2021, 8, 708052. [Google Scholar] [CrossRef]
- Díaz, C.; González-Olmedo, C.; Díaz-Beltrán, L.; Camacho, J.; Mena García, P.; Martín-Blázquez, A.; Fernández-Navarro, M.; Ortega-Granados, A.L.; Gálvez-Montosa, F.; Marchal, J.A.; et al. Predicting Dynamic Response to Neoadjuvant Chemotherapy in Breast Cancer: A Novel Metabolomics Approach. Mol. Oncol. 2022, 16, 2658–2671. [Google Scholar] [CrossRef]
- Zapater-Moros, A.; Díaz-Beltrán, L.; Gámez-Pozo, A.; Trilla-Fuertes, L.; Lumbreras-Herrera, M.I.; López-Camacho, E.; González-Olmedo, C.; Espinosa, E.; Zamora, P.; Sánchez-Rovira, P.; et al. Metabolomics Unravels Subtype-Specific Characteristics Related to Neoadjuvant Therapy Response in Breast Cancer Patients. Metabolomics 2023, 19, 60. [Google Scholar] [CrossRef]
- Vignoli, A.; Ghini, V.; Meoni, G.; Licari, C.; Takis, P.G.; Tenori, L.; Turano, P.; Luchinat, C. High-Throughput Metabolomics by 1D NMR. Angew. Chem. Int. Ed. Engl. 2019, 58, 968–994. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, M.R.; Silva, A.A.R.; Talarico, M.C.R.; Sanches, P.H.G.; Sforça, M.L.; Rocco, S.A.; Rezende, L.M.; Quintero, M.; Costa, T.B.B.C.; Viana, L.R.; et al. Metabolomics by NMR Combined with Machine Learning to Predict Neoadjuvant Chemotherapy Response for Breast Cancer. Cancers 2022, 14, 5055. [Google Scholar] [CrossRef] [PubMed]
- Debik, J.; Sangermani, M.; Wang, F.; Madssen, T.S.; Giskeødegård, G.F. Multivariate Analysis of NMR-Based Metabolomic Data. NMR Biomed. 2022, 35, e4638. [Google Scholar] [CrossRef] [PubMed]
- Clish, C.B. Metabolomics: An Emerging but Powerful Tool for Precision Medicine. Cold Spring Harb. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Suman, S.; Sharma, R.K.; Kumar, V.; Sinha, N.; Shukla, Y. Metabolic Fingerprinting in Breast Cancer Stages through (1)H NMR Spectroscopy-Based Metabolomic Analysis of Plasma. J. Pharm. Biomed. Anal. 2018, 160, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Gumà, J.; Adriá-Cebrián, J.; Ruiz-Aguado, B.; Albacar, C.; Girona, J.; Rodríguez-Calvo, R.; Martínez-Micaelo, N.; Lam, E.W.F.; Masana, L.; Guaita-Esteruelas, S. Altered Serum Metabolic Profile Assessed by Advanced 1H-NMR in Breast Cancer Patients. Cancers 2021, 13, 4281. [Google Scholar] [CrossRef] [PubMed]
- Lécuyer, L.; Victor Bala, A.; Deschasaux, M.; Bouchemal, N.; Nawfal Triba, M.; Vasson, M.-P.; Rossary, A.; Demidem, A.; Galan, P.; Hercberg, S.; et al. NMR Metabolomic Signatures Reveal Predictive Plasma Metabolites Associated with Long-Term Risk of Developing Breast Cancer. Int. J. Epidemiol. 2018, 47, 484–494. [Google Scholar] [CrossRef]
- Corona, G.; Di Gregorio, E.; Vignoli, A.; Muraro, E.; Steffan, A.; Miolo, G. 1H-NMR Plasma Lipoproteins Profile Analysis Reveals Lipid Metabolism Alterations in HER2-Positive Breast Cancer Patients. Cancers 2021, 13, 5845. [Google Scholar] [CrossRef]
- Jobard, E.; Dossus, L.; Baglietto, L.; Fornili, M.; Lécuyer, L.; Mancini, F.R.; Gunter, M.J.; Trédan, O.; Boutron-Ruault, M.-C.; Elena-Herrmann, B.; et al. Investigation of Circulating Metabolites Associated with Breast Cancer Risk by Untargeted Metabolomics: A Case-Control Study Nested within the French E3N Cohort. Br. J. Cancer 2021, 124, 1734–1743. [Google Scholar] [CrossRef] [PubMed]
- Asiago, V.M.; Alvarado, L.Z.; Shanaiah, N.; Gowda, G.A.N.; Owusu-Sarfo, K.; Ballas, R.A.; Raftery, D. Early Detection of Recurrent Breast Cancer Using Metabolite Profiling. Cancer Res. 2010, 70, 8309–8318. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Baek, H.-M.; Kim, S.S.I.S.; Kim, M.J.; Youk, J.H.; Moon, H.J.; Kim, E.-K.; Han, K.H.; Kim, D.-H.; Kim, S.S.I.S.; et al. HR-MAS MR Spectroscopy of Breast Cancer Tissue Obtained with Core Needle Biopsy: Correlation with Prognostic Factors. PLoS ONE 2012, 7, e51712. [Google Scholar] [CrossRef] [PubMed]
- Zidi, O.; Souai, N.; Raies, H.; Ben Ayed, F.; Mezlini, A.; Mezrioui, S.; Tranchida, F.; Sabatier, J.-M.; Mosbah, A.; Cherif, A.; et al. Fecal Metabolic Profiling of Breast Cancer Patients during Neoadjuvant Chemotherapy Reveals Potential Biomarkers. Molecules 2021, 26, 2266. [Google Scholar] [CrossRef] [PubMed]
- Vignoli, A.; Risi, E.; McCartney, A.; Migliaccio, I.; Moretti, E.; Malorni, L.; Luchinat, C.; Biganzoli, L.; Tenori, L. Precision Oncology via NMR-Based Metabolomics: A Review on Breast Cancer. Int. J. Mol. Sci. 2021, 22, 4687. [Google Scholar] [CrossRef] [PubMed]
- Hart, C.D.; Vignoli, A.; Tenori, L.; Uy, G.L.; Van To, T.; Adebamowo, C.; Hossain, S.M.; Biganzoli, L.; Risi, E.; Love, R.R.; et al. Serum Metabolomic Profiles Identify ER-Positive Early Breast Cancer Patients at Increased Risk of Disease Recurrence in a Multicenter Population. Clin. Cancer Res. 2017, 23, 1422–1431. [Google Scholar] [CrossRef] [PubMed]
- Barnes, T.; Bell, K.; DiSebastiano, K.M.; Vance, V.; Hanning, R.; Russell, C.; Dubin, J.A.; Bahl, M.; Califaretti, N.; Campbell, C.; et al. Plasma Amino Acid Profiles of Breast Cancer Patients Early in the Trajectory of the Disease Differ from Healthy Comparison Groups. Appl. Physiol. Nutr. Metab. 2014, 39, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Brosnan, M.E.; Brosnan, J.T. Histidine Metabolism and Function. J. Nutr. 2020, 150, 2570S–2575S. [Google Scholar] [CrossRef]
- Moro, J.; Tomé, D.; Schmidely, P.; Demersay, T.-C.; Azzout-Marniche, D. Histidine: A Systematic Review on Metabolism and Physiological Effects in Human and Different Animal Species. Nutrients 2020, 12, 1414. [Google Scholar] [CrossRef]
- Kanarek, N.; Keys, H.R.; Cantor, J.R.; Lewis, C.A.; Chan, S.H.; Kunchok, T.; Abu-Remaileh, M.; Freinkman, E.; Schweitzer, L.D.; Sabatini, D.M. Histidine Catabolism Is a Major Determinant of Methotrexate Sensitivity. Nature 2018, 559, 632–636. [Google Scholar] [CrossRef]
- Giskeødegård, G.F.; Lundgren, S.; Sitter, B.; Fjøsne, H.E.; Postma, G.; Buydens, L.M.C.; Gribbestad, I.S.; Bathen, T.F. Lactate and Glycine-Potential MR Biomarkers of Prognosis in Estrogen Receptor-Positive Breast Cancers. NMR Biomed. 2012, 25, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Euceda, L.R.; Haukaas, T.H.; Giskeødegård, G.F.; Vettukattil, R.; Engel, J.; Silwal-Pandit, L.; Lundgren, S.; Borgen, E.; Garred, Ø.; Postma, G.; et al. Evaluation of Metabolomic Changes during Neoadjuvant Chemotherapy Combined with Bevacizumab in Breast Cancer Using MR Spectroscopy. Metabolomics 2017, 13, 37. [Google Scholar] [CrossRef]
- Cao, M.D.; Giskeødegård, G.F.; Bathen, T.F.; Sitter, B.; Bofin, A.; Lønning, P.E.; Lundgren, S.; Gribbestad, I.S. Prognostic Value of Metabolic Response in Breast Cancer Patients Receiving Neoadjuvant Chemotherapy. BMC Cancer 2012, 12, 39. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Vousden, K.H. Serine and One-Carbon Metabolism in Cancer. Nat. Rev. Cancer 2016, 16, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Geeraerts, S.L.; Heylen, E.; De Keersmaecker, K.; Kampen, K.R. The Ins and Outs of Serine and Glycine Metabolism in Cancer. Nat. Metab. 2021, 3, 131–141. [Google Scholar] [CrossRef]
- Li, A.M.; Ye, J. Reprogramming of Serine, Glycine and One-Carbon Metabolism in Cancer. Biochim. Biophys. Acta—Mol. Basis Dis. 2020, 1866, 165841. [Google Scholar] [CrossRef] [PubMed]
- Reina-Campos, M.; Linares, J.F.; Duran, A.; Cordes, T.; L’Hermitte, A.; Badur, M.G.; Bhangoo, M.S.; Thorson, P.K.; Richards, A.; Rooslid, T.; et al. Increased Serine and One-Carbon Pathway Metabolism by PKCλ/ι Deficiency Promotes Neuroendocrine Prostate Cancer. Cancer Cell 2019, 35, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Possemato, R.; Marks, K.M.; Shaul, Y.D.; Pacold, M.E.; Kim, D.; Birsoy, K.; Sethumadhavan, S.; Woo, H.-K.; Jang, H.G.; Jha, A.K.; et al. Functional Genomics Reveal That the Serine Synthesis Pathway Is Essential in Breast Cancer. Nature 2011, 476, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Nees, J.; Schafferer, S.; Yuan, B.; Tang, Q.; Scheffler, M.; Hartkopf, A.; Golatta, M.; Schneeweiß, A.; Burwinkel, B.; Wallwiener, M. How Previous Treatment Changes the Metabolomic Profile in Patients with Metastatic Breast Cancer. Arch. Gynecol. Obstet. 2022, 306, 2115–2122. [Google Scholar] [CrossRef]
- Cox, T.R. The Matrix in Cancer. Nat. Rev. Cancer 2021, 21, 217–238. [Google Scholar] [CrossRef]
- Matés, J.M.; Segura, J.A.; Alonso, F.J.; Márquez, J. Oxidative Stress in Apoptosis and Cancer: An Update. Arch. Toxicol. 2012, 86, 1649–1665. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xia, S.; He, J.; Lu, G.; Xie, Z.; Han, H. Roles of Taurine in Cognitive Function of Physiology, Pathologies and Toxication. Life Sci. 2019, 231, 116584. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lu, H.; Wang, Y.; Liu, C.; Zhu, W.; Zheng, S.; Wan, F. Taurine Induces the Apoptosis of Breast Cancer Cells by Regulating Apoptosis-Related Proteins of Mitochondria. Int. J. Mol. Med. 2015, 35, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Li, Q.; Hou, R.; Liang, H.; Zhang, Y.; Yang, Y. An Integrated Metabonomics Study to Reveal the Inhibitory Effect and Metabolism Regulation of Taurine on Breast Cancer. J. Pharm. Biomed. Anal. 2022, 214, 114711. [Google Scholar] [CrossRef] [PubMed]
- Slupsky, C.M.; Steed, H.; Wells, T.H.; Dabbs, K.; Schepansky, A.; Capstick, V.; Faught, W.; Sawyer, M.B. Urine Metabolite Analysis Offers Potential Early Diagnosis of Ovarian and Breast Cancers. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 5835–5841. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.L.; Olival, A.; Perestrelo, R.; Silva, P.; Tomás, H.; Câmara, J.S. Untargeted Urinary 1H NMR-Based Metabolomic Pattern as a Potential Platform in Breast Cancer Detection. Metabolites 2019, 9, 269. [Google Scholar] [CrossRef] [PubMed]
- Men, Y.; Li, L.; Zhang, F.; Kong, X.; Zhang, W.; Hao, C.; Wang, G. Evaluation of Heavy Metals and Metabolites in the Urine of Patients with Breast Cancer. Oncol. Lett. 2020, 19, 1331–1337. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.; Monette, G. Generalized Collinearity Diagnostics. J. Am. Stat. Assoc. 1992, 87, 178–183. [Google Scholar] [CrossRef]
- Lausen, B.; Schumacher, M. Maximally Selected Rank Statistics. Biometrics 1992, 48, 73–85. [Google Scholar] [CrossRef]
- Team R Core R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria; Available online: https://www.r-project.org/ (accessed on 3 February 2024).




| Disease-Free Survival † | ||||
|---|---|---|---|---|
| Metabolite | VIF | HR | (CI 95%) | p-Value |
| Alanine | 2.88 | 2.88 | (0.44–18.83) | 0.27 |
| Arginine | 2.92 | 1.28 | (0.31–5.23) | 0.73 |
| Ascorbate | 2.09 | 0.85 | (0.46–1.59) | 0.62 |
| Asparagine | 2.21 | 0.97 | (0.41–2.33) | 0.95 |
| Aspartate | 2.24 | 1.51 | (0.69–3.30) | 0.30 |
| Betaine | >3 | - | - | - |
| Carnitine | >3 | - | - | - |
| Choline | 2.77 | 0.47 | (0.14–1.56) | 0.22 |
| Citrate | 2.32 | 1.66 | (0.40–6.90) | 0.49 |
| Creatine | 1.89 | 0.59 | (0.26–1.32) | 0.20 |
| Creatinine | 2.07 | 3.81 | (0.65–22.12) | 0.14 |
| Formate | >3 | - | - | - |
| Glucose | >3 | - | - | - |
| Glutamate | 2.77 | 1.65 | (0.41–6.59) | 0.48 |
| Glutamine | 2.20 | 2.37 | (0.30–18.75) | 0.41 |
| Glycerol | 2.08 | 0.58 | (0.25–1.32) | 0.19 |
| Glycine | >3 | - | - | - |
| Histidine | 2.75 | 0.32 | (0.11–0.95) | 0.04 |
| Isoleucine | >3 | - | - | - |
| Lactate | 2.48 | 0.32 | (0.09–1.11) | 0.07 |
| Leucine | >3 | - | - | - |
| Lysine | >3 | - | - | - |
| Methionine | >3 | - | - | - |
| myo-Inositol | 2.34 | 1.27 | (0.36–4.40) | 0.71 |
| N,N-Dimethylglycine | 2.05 | 0.53 | (0.18–1.51) | 0.23 |
| Pantothenate | 2.83 | 0.84 | (0.26–2.74) | 0.77 |
| Phenylalanine | 2.24 | 0.97 | (0.49–1.92) | 0.94 |
| Proline | 2.26 | 0.58 | (0.17–2.01) | 0.39 |
| Serine | 2.81 | 5.06 | (1.45–17.70) | 0.01 |
| sn-Glycero-3-phosphocholine | 2.76 | 2.89 | (0.47–17.81) | 0.25 |
| Taurine | 1.63 | 2.51 | (1.07–5.91) | 0.03 |
| Threonine | >3 | - | - | - |
| Tyrosine | >3 | - | - | - |
| Urea | >3 | - | - | - |
| Valine | >3 | - | - | - |
| HER-2 | 2.17 | 0.80 | (0.28–2.31) | 0.68 |
| Hormonal Receptor | 1.59 | 1.45 | (0.53–3.96) | 0.47 |
| Characteristic | n (%) | Low Risk n = 57 (71%) | High Risk n = 23 (29%) | p-Value | |
|---|---|---|---|---|---|
| Age (years) | ≥50 | 46 (57.0) | 34 (59.6) | 12 (52.2) | 0.72 |
| <50 | 34 (43.0) | 23 (40.4) | 11 (47.8) | ||
| Disease Stage | I | 4 (5.0) | 3 (5.3) | 1 (4.3) | 0.46 |
| II | 48 (60.0) | 37 (64.9) | 11 (47.8) | ||
| III | 22 (27.5) | 13 (22.8) | 9 (39.1) | ||
| IV | 6 (7.5) | 4 (7.0) | 2 (8.7) | ||
| Race | Caucasian | 69 (86.3) | 48 (84.2) | 21 (91.3) | 0.63 |
| Noncaucasian | 11 (13.8) | 9 (15.8) | 2 (8.7) | ||
| Age of menarche (years) | >12 | 42 (52.5) | 30 (52.6) | 12 (52.2) | 1.00 |
| ≤12 | 38 (47.5) | 27 (47.4) | 11 (47.8) | ||
| Menopause | No | 36 (45.0) | 24 (42.1) | 12 (52.2) | 0.57 |
| Yes | 44 (55.0) | 33 (57.9) | 11 (47.8) | ||
| Hormone replacement therapy | No | 68 (85.0) | 48 (84.2) | 20 (87.0) | 1.00 |
| Yes | 12 (15.0) | 9 (15.8) | 3 (13.0) | ||
| Previous pregnancy | Yes | 73 (91.3) | 55 (96.5) | 18 (78.3) | 0.03 |
| No | 7 (8.7) | 2 (3.5) | 5 (21.7) | ||
| Lactation * | Yes | 63 (78.8) | 46 (80.7) | 17 (73.9) | 0.71 |
| No | 17 (21.2) | 11 (19.3) | 6 (26.1) | ||
| Smoking | Yes | 17 (21.2) | 13 (22.8) | 4 (17.4) | 0.81 |
| No | 63 (78.8) | 44 (77.2) | 19 (82.6) | ||
| BMI categories | Normal weight | 24 (30.0) | 14 (24.6) | 10 (43.5) | 0.21 |
| Overweight | 21 (26.3) | 17 (29.8) | 4 (17.4) | ||
| Obese | 35 (43.7) | 26 (45.6) | 9 (39.1) | ||
| Diabetes | No | 71 (88.7) | 50 (87.7) | 21 (91.3) | 0.94 |
| Yes | 9 (11.3) | 7 (12.3) | 2 (8.7) | ||
| Family history of breast or ovarian cancer | No | 59 (73.8) | 41 (71.9) | 18 (78.3) | 0.76 |
| Yes | 21 (26.2) | 16 (28.1) | 5 (21.7) | ||
| Relapse/progression | No | 47 (58.7) | 40 (70.2) | 7 (30.4) | 0.002 |
| Yes | 33 (42.3) | 17 (29.3) | 16 (69.6) | ||
| Death | No | 55 (68.7) | 45 (78.9) | 10 (43.5) | 0.004 |
| Yes | 25 (32.3) | 12 (21.1) | 13 (56.3) | ||
| Final status by the end of follow-up | Alive without disease | 47 (58.8) | 40 (70.2) | 7 (30.4) | 0.003 |
| Alive with disease | 8 (10.0) | 5 (8.8) | 3 (13.0) | ||
| Deceased | 25 (31.2) | 12 (21.1) | 13 (56.5) | ||
| Characteristic | n (%) | Low Risk n = 57 (71%) | High Risk n = 23 (29%) | p-Value | |
|---|---|---|---|---|---|
| Histological grade | 1/2 | 39 (48.75) | 29 (50.9) | 10 (43.5) | 0.72 |
| 3 | 41 (51.25) | 28 (49.1) | 13 (56.5) | ||
| Ki67 (mean/SD) | Below 40% | 35 (43.7) | 27 (47.4) | 8 (34.8) | 0.44 |
| 40% or higher | 45 (57.5) | 30 (52.6) | 15 (65.2) | ||
| HER-2 | Negative | 46 (57.5) | 30 (52.6) | 16 (69.6) | 0.25 |
| Positive * | 34 (42.5) | 27 (47.4) | 7 (30.4) | ||
| Tumor size | T1/T2 | 53 (66.25) | 40 (70.2) | 13 (56.5) | 0.36 |
| T3/T4 | 27 (33.75) | 17 (29.8) | 10 (43.5) | ||
| Regional lymph node | N0 | 33 (41.25) | 25 (43.9) | 8 (34.8) | 0.62 |
| N1 or higher | 47 (58.75) | 32 (56.1) | 15 (65.2) | ||
| Metastasis | M0 | 74 (92.5) | 53 (93.0) | 21 (91.3) | 1.00 |
| M1 | 6 (7.5) | 4 (7.0) | 2 (8.7) | ||
| Hormonal Receptor | Negative | 21 (26.25) | 13 (22.8) | 8 (34.8) | 0.41 |
| Positive | 59 (73.75) | 44 (77.2) | 15 (65.2) | ||
| Disease-Free Survival | Overall Survival | ||||||
|---|---|---|---|---|---|---|---|
| Factor | n (%) | HR | (95%CI) | Adjusted p-Value * | HR | (95%CI) | Adjusted p-Value * |
| Age (years) | |||||||
| ≥50 | 46 (57%) | Ref. | Ref. | ||||
| <50 | 34 (43%) | 0.51 | (0.21–1.25) | 0.15 | 0.67 | (0.31–1.41) | 0.29 |
| Disease stage | |||||||
| I–II | 52 (65%) | Ref. | Ref. | ||||
| III–IV | 28 (35%) | 5.88 | (2.32–14.88) | <0.001 | 4.49 | (2.08–9.69) | <0.001 |
| MRSS | |||||||
| Low | 57 (71%) | Ref. | Ref. | ||||
| High | 23 (29%) | 3.42 | (1.51–7.74) | 0.003 | 3.34 | (1.64–6.80) | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Talarico, M.C.R.; Derchain, S.; da Silva, L.F.; Sforça, M.L.; Rocco, S.A.; Cardoso, M.R.; Sarian, L.O. Metabolomic Profiling of Breast Cancer Patients Undergoing Neoadjuvant Chemotherapy for Predicting Disease-Free and Overall Survival. Int. J. Mol. Sci. 2024, 25, 8639. https://doi.org/10.3390/ijms25168639
Talarico MCR, Derchain S, da Silva LF, Sforça ML, Rocco SA, Cardoso MR, Sarian LO. Metabolomic Profiling of Breast Cancer Patients Undergoing Neoadjuvant Chemotherapy for Predicting Disease-Free and Overall Survival. International Journal of Molecular Sciences. 2024; 25(16):8639. https://doi.org/10.3390/ijms25168639
Chicago/Turabian StyleTalarico, Maria Cecília Ramiro, Sophie Derchain, Lucas Ferreira da Silva, Maurício L. Sforça, Silvana A. Rocco, Marcella R. Cardoso, and Luís Otávio Sarian. 2024. "Metabolomic Profiling of Breast Cancer Patients Undergoing Neoadjuvant Chemotherapy for Predicting Disease-Free and Overall Survival" International Journal of Molecular Sciences 25, no. 16: 8639. https://doi.org/10.3390/ijms25168639






