Transcription Factors Sox8 and Sox10 Contribute with Different Importance to the Maintenance of Mature Oligodendrocytes
Abstract
1. Introduction
2. Results
2.1. Ablation of Sox8 and Sox10 in Adult Oligodendrocytes is Efficient
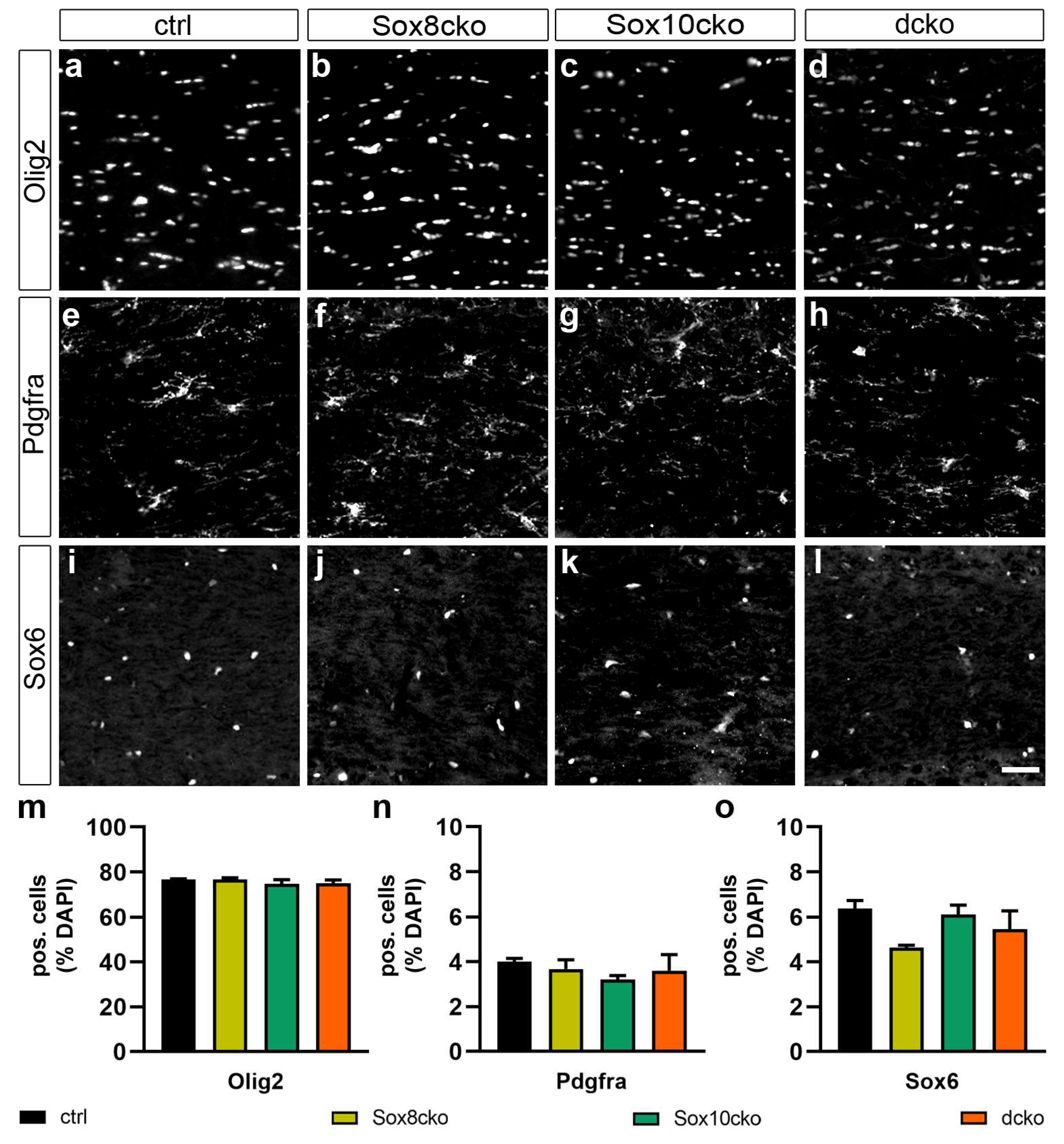
2.2. The Number and State of Oligodendroglial Cells Remain Unchanged upon Loss of Sox8 and Sox10
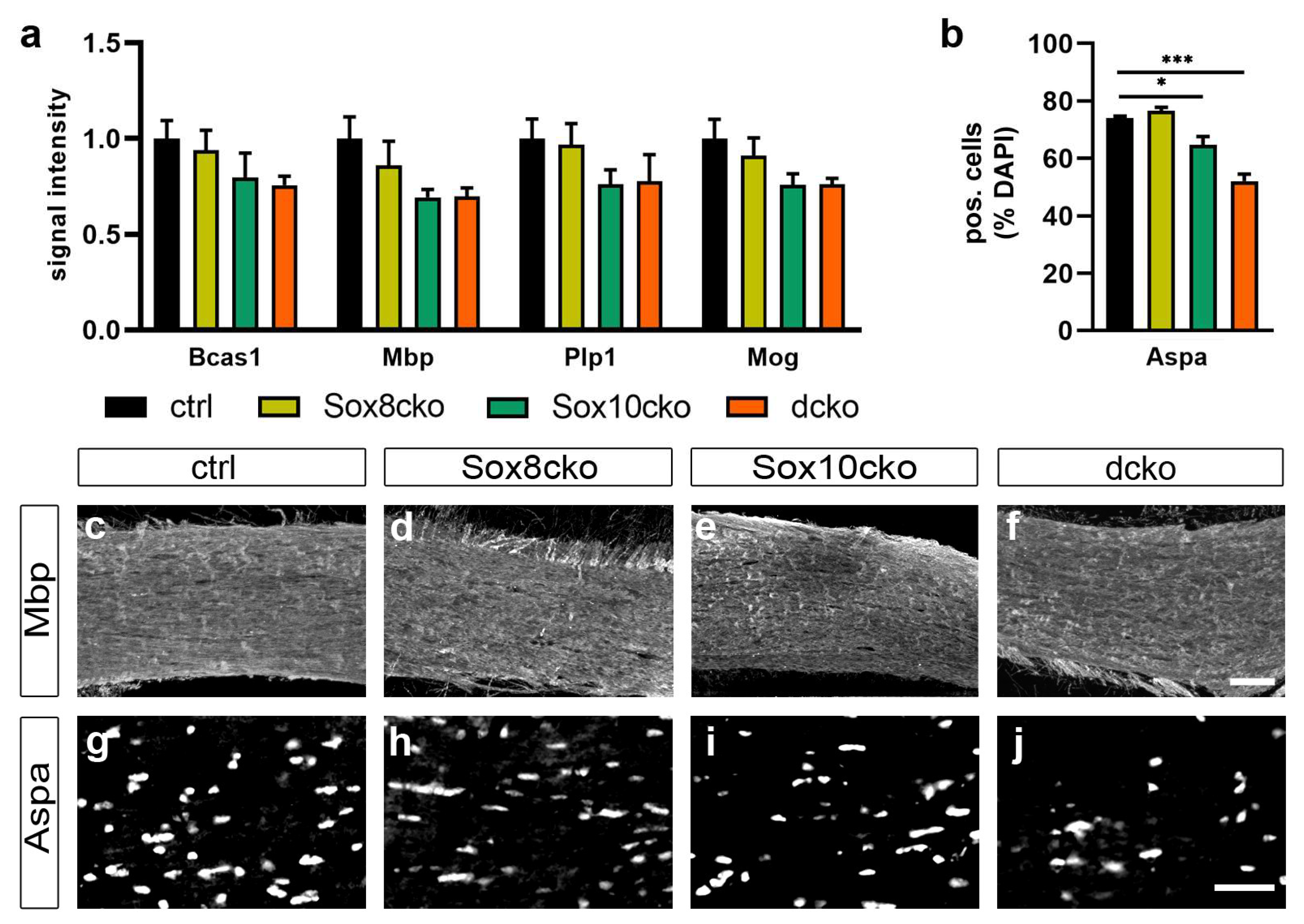
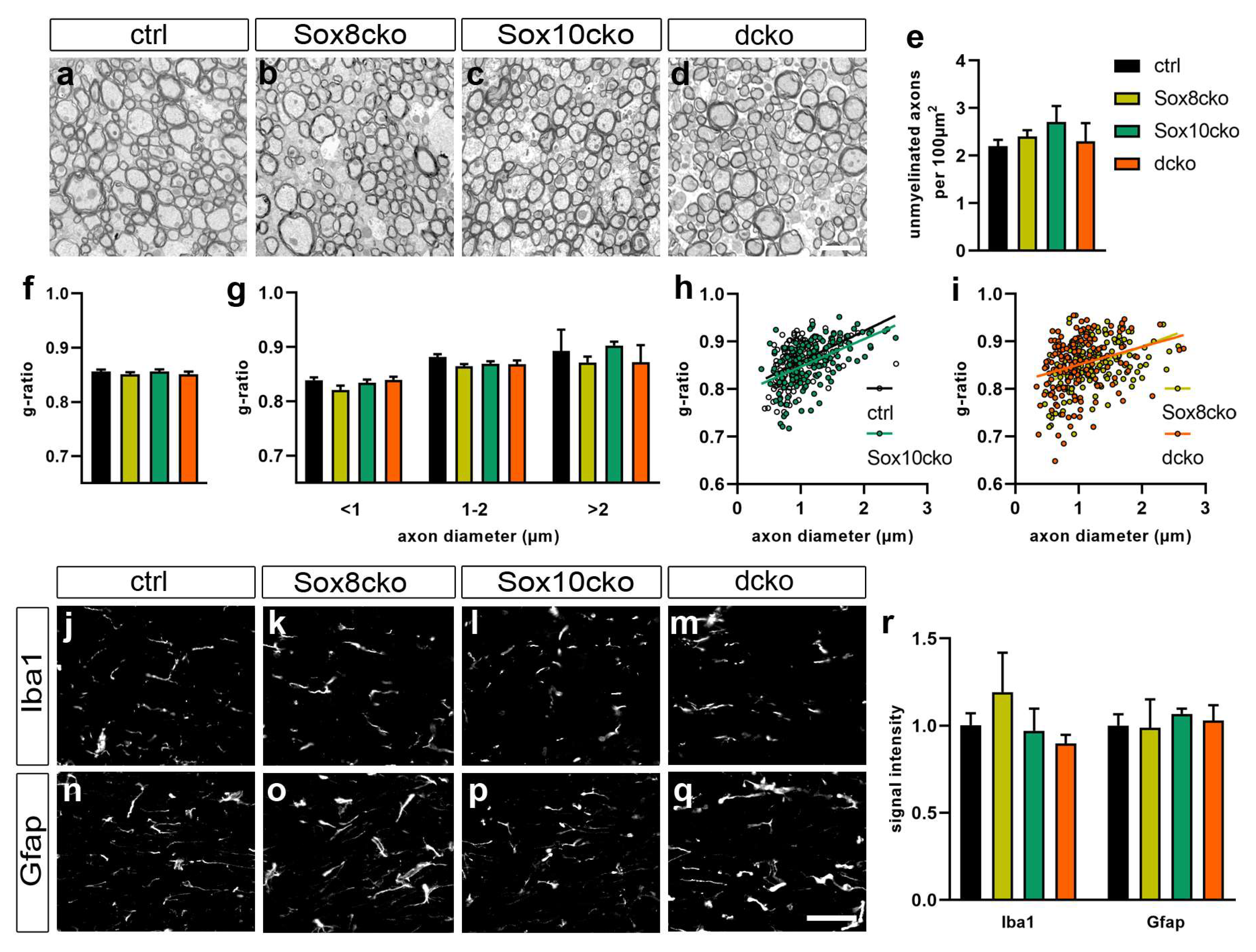
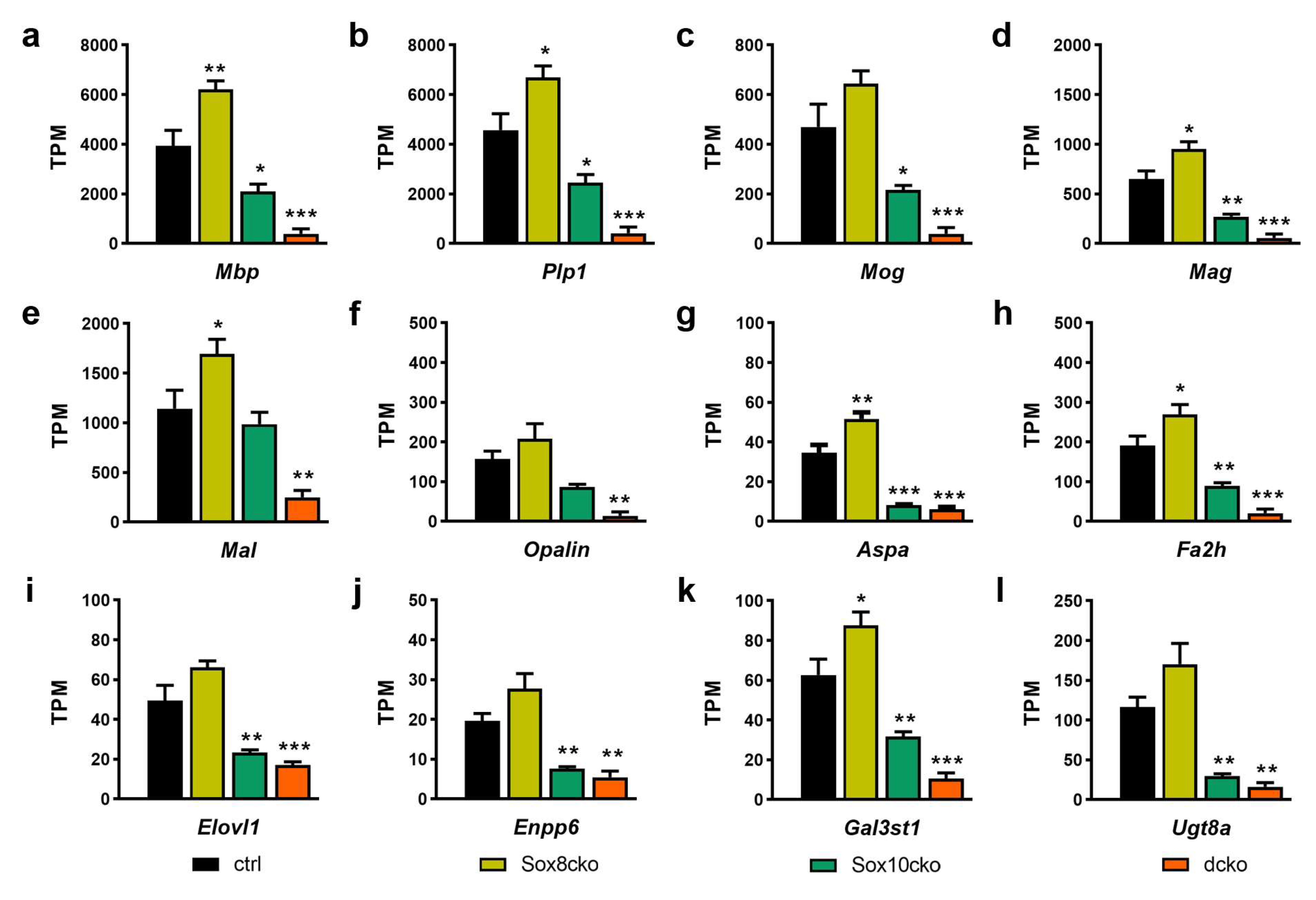
2.3. Changes in Myelin Are Subtle after the Deletion of Sox8 and Sox10 in Adult Oligodendrocytes
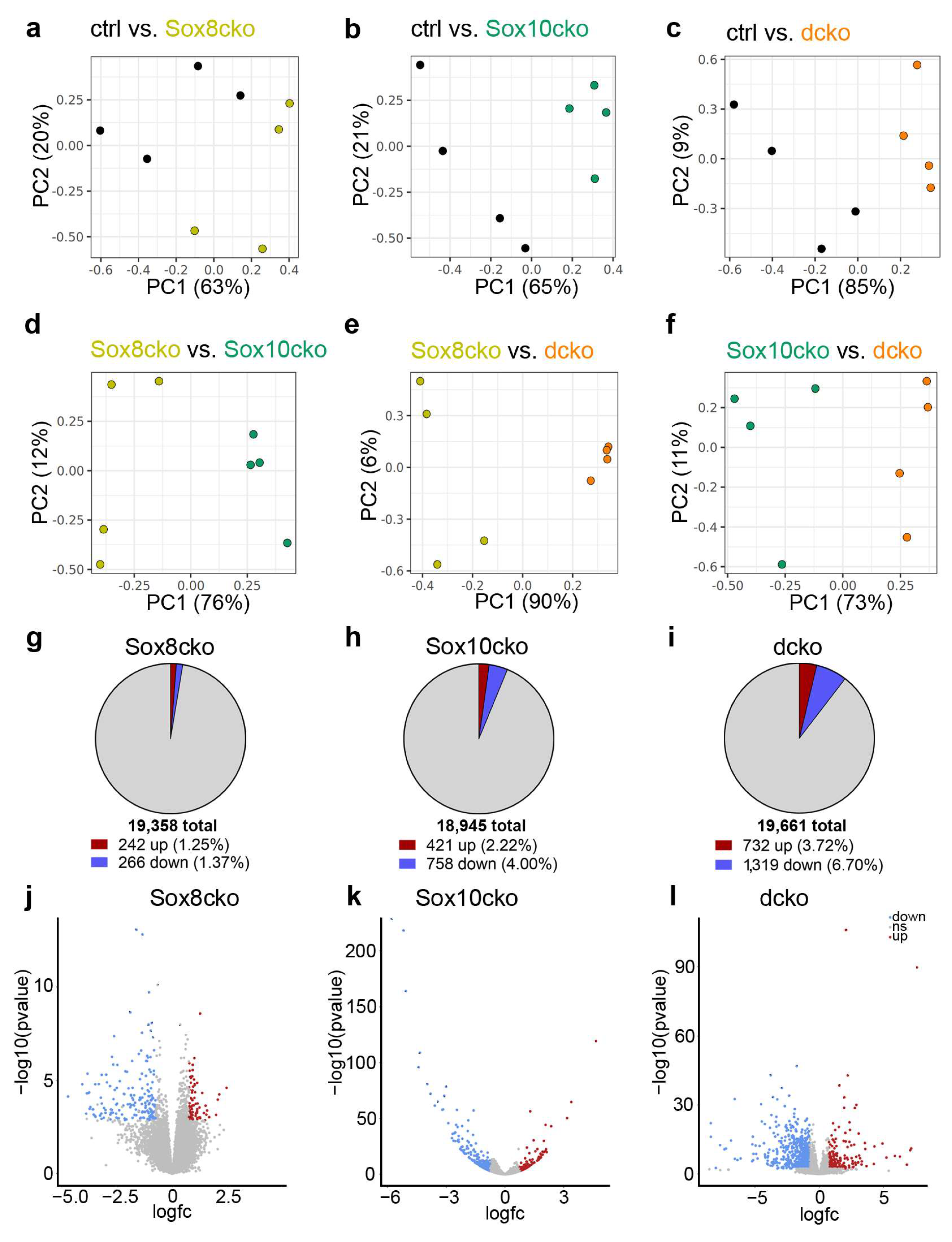
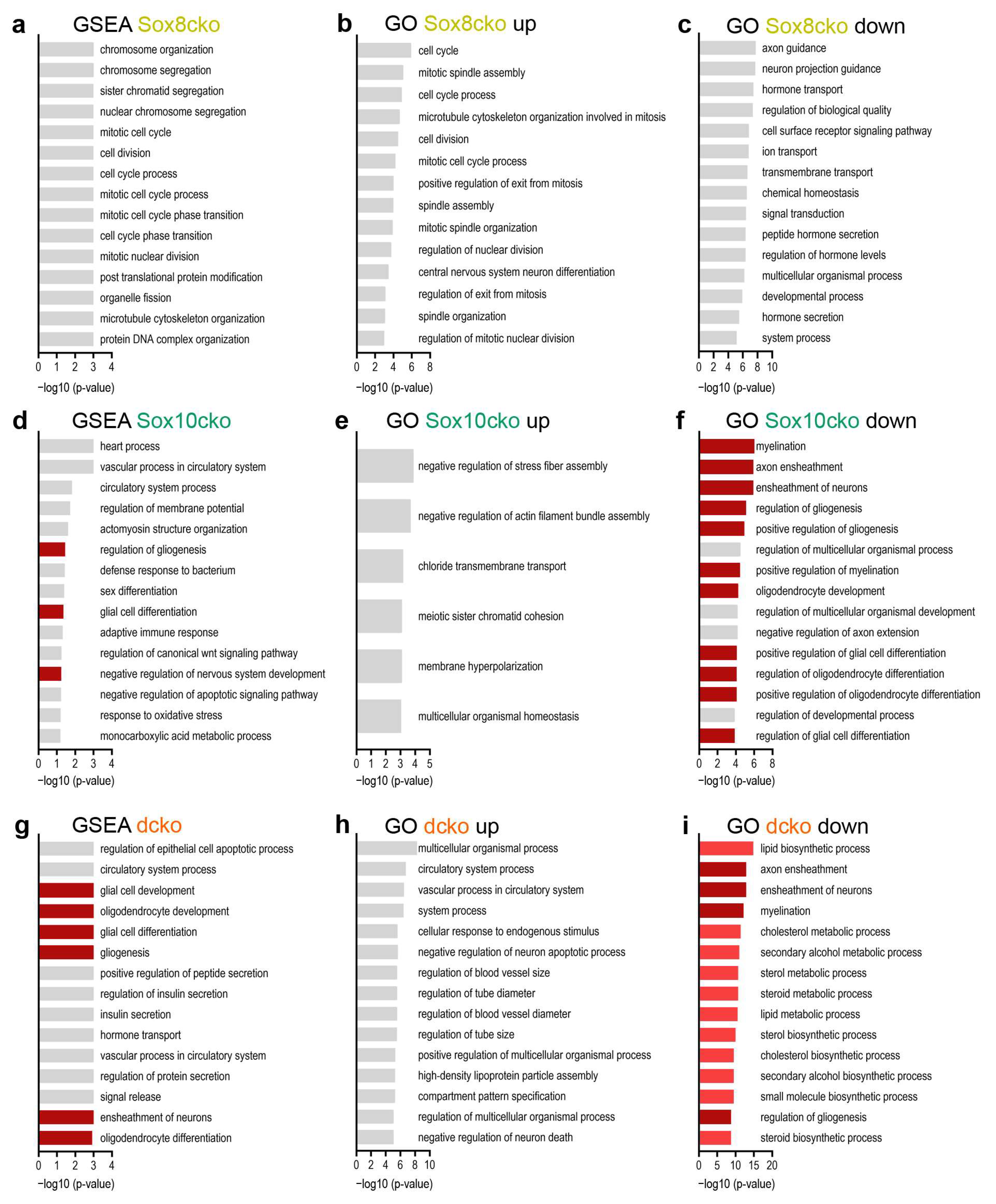
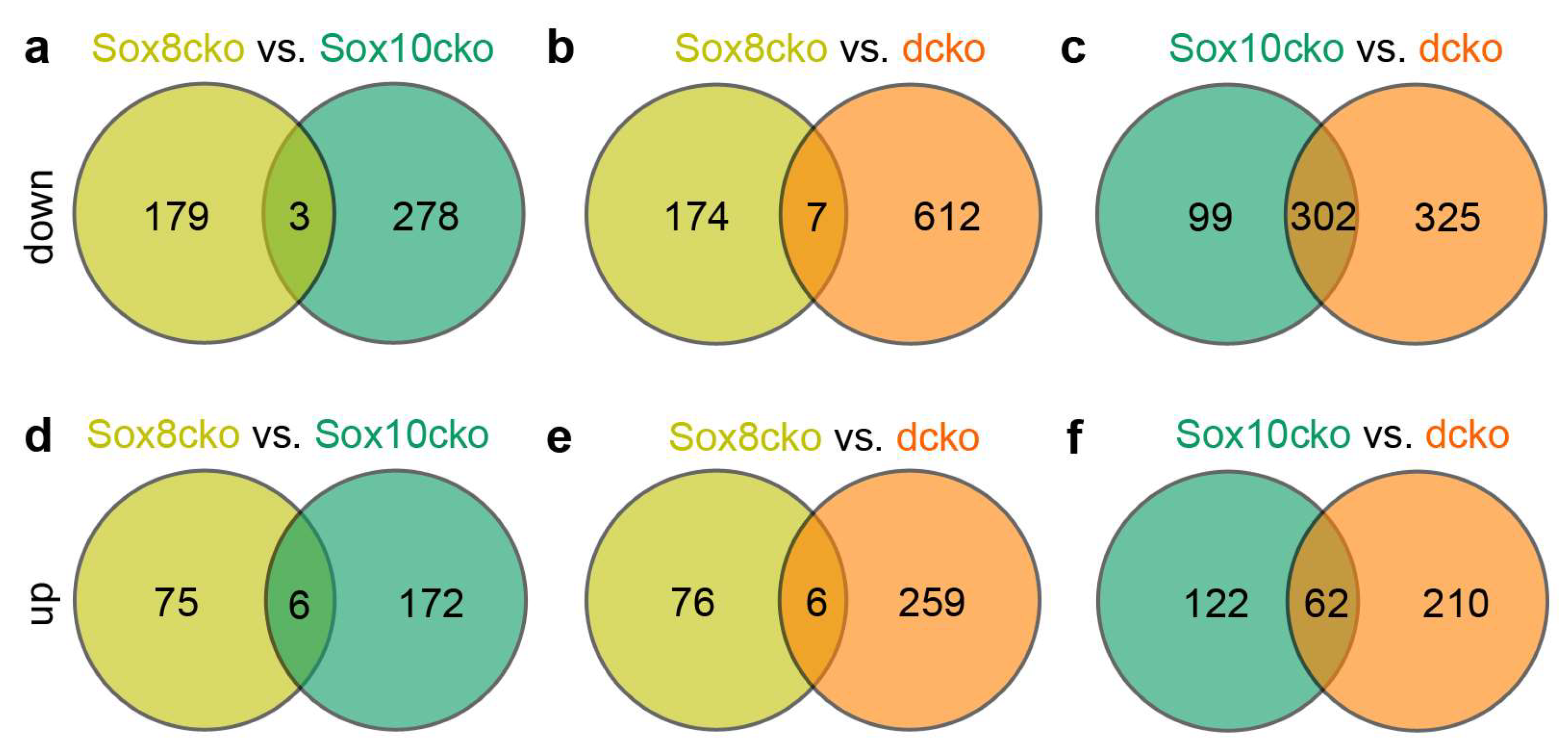
2.4. Gene Expression Changes Substantially in the Adult Oligodendrocytes of Sox10cko and Dcko Mice
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Tissue Stainings
4.3. Electron Microscopy
4.4. RNA Sequencing
4.5. Quantitative RT-PCR (qRT-PCR)
4.6. Quantifications and Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rowitch, D.H. Glial specification in the vertebrate neural tube. Nat. Rev. Neurosci. 2004, 5, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Cristobal, C.D.; Lee, H.K. Development of myelinating glia: An overview. Glia 2022, 70, 2237–2259. [Google Scholar] [CrossRef] [PubMed]
- Stadelmann, C.; Timmler, S.; Barrantes-Freer, A.; Simons, M. Myelin in the Central Nervous System: Structure, Function, and Pathology. Physiol. Rev. 2019, 99, 1381–1431. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, J.; da Costa, N.M.; Cheadle, L. Emerging roles of oligodendrocyte precursor cells in neural circuit development and remodeling. Trends Neurosci. 2023, 46, 628–639. [Google Scholar] [CrossRef] [PubMed]
- Sock, E.; Wegner, M. Using the lineage determinants Olig2 and Sox10 to explore transcriptional regulation of oligodendrocyte development. Dev. Neurobiol. 2021, 81, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Elbaz, B.; Popko, B. Molecular Control of Oligodendrocyte Development. Trends Neurosci. 2019, 42, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Emery, B.; Lu, Q.R. Transcriptional and Epigenetic Regulation of Oligodendrocyte Development and Myelination in the Central Nervous System. Cold Spring Harb. Perspect. Biol. 2015, 7, a020461. [Google Scholar] [CrossRef] [PubMed]
- Wittstatt, J.; Reiprich, S.; Kuspert, M. Crazy Little Thing Called Sox-New Insights in Oligodendroglial Sox Protein Function. Int. J. Mol. Sci. 2019, 20, 2713. [Google Scholar] [CrossRef]
- Stolt, C.C.; Lommes, P.; Friedrich, R.P.; Wegner, M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development 2004, 131, 2349–2358. [Google Scholar] [CrossRef]
- Turnescu, T.; Arter, J.; Reiprich, S.; Tamm, E.R.; Waisman, A.; Wegner, M. Sox8 and Sox10 jointly maintain myelin gene expression in oligodendrocytes. Glia 2018, 66, 279–294. [Google Scholar] [CrossRef]
- Leone, D.P.; Genoud, S.; Atanasoski, S.; Grausenburger, R.; Berger, P.; Metzger, D.; Macklin, W.B.; Chambon, P.; Suter, U. Tamoxifen-inducible glia-specific Cre mice for somatic mutagenesis in oligodendrocytes and Schwann cells. Mol. Cell Neurosci. 2003, 22, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Bremer, M.; Fröb, F.; Kichko, T.; Reeh, P.; Tamm, E.R.; Suter, U.; Wegner, M. Sox10 is required for Schwann cell homeostasis and myelin maintenance in the adult peripheral nerve. Glia 2011, 59, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Britsch, S.; Li, L.; Kirchhoff, S.; Theuring, F.; Brinkmann, V.; Birchmeier, C.; Riethmacher, D. The ErbB2 and ErbB3 receptors and their ligand, neuregulin-1, are essential for development of the sympathetic nervous system. Genes Dev. 1998, 12, 1825–1836. [Google Scholar] [CrossRef]
- Finzsch, M.; Schreiner, S.; Kichko, T.; Reeh, P.; Tamm, E.R.; Bösl, M.R.; Meijer, D.; Wegner, M. Sox10 is required for Schwann cell identity and progression beyond the immature Schwann cell stage. J. Cell Biol. 2010, 189, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Fard, M.K.; van der Meer, F.; Sanchez, P.; Cantuti-Castelvetri, L.; Mandad, S.; Jakel, S.; Fornasiero, E.F.; Schmitt, S.; Ehrlich, M.; Starost, L.; et al. BCAS1 expression defines a population of early myelinating oligodendrocytes in multiple sclerosis lesions. Sci. Transl. Med. 2017, 9, 419. [Google Scholar] [CrossRef]
- Finzsch, M.; Stolt, C.C.; Lommes, P.; Wegner, M. Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor {alpha} expression. Development 2008, 135, 637–646. [Google Scholar] [CrossRef]
- Hassel, L.A.; Fröb, F.; Küspert, M.; Hillgärtner, S.; Arnold, P.; Huang, W.; Kirchhoff, F.; Wegner, M. Differential activity of transcription factor Sox9 in early and adult oligodendroglial progenitor cells. Glia 2023, 71, 1890–1905. [Google Scholar] [CrossRef]
- Chapman, T.W.; Hill, R.A. Myelin plasticity in adulthood and aging. Neurosci. Lett. 2020, 715, 134645. [Google Scholar] [CrossRef]
- Sampaio-Baptista, C.; Johansen-Berg, H. White Matter Plasticity in the Adult Brain. Neuron 2017, 96, 1239–1251. [Google Scholar] [CrossRef]
- Franklin, R.J.M.; Simons, M. CNS remyelination and inflammation: From basic mechanisms to therapeutic opportunities. Neuron 2022, 110, 3549–3565. [Google Scholar] [CrossRef]
- Schepers, G.E.; Taesdale, R.D.; Koopman, P. Twenty pairs of Sox: Extent, homology, and nomenclature of the mouse and human Sox transcription factor families. Dev. Cell 2002, 3, 167–170. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Marie, C.; Zhao, C.; Kim, B.; Wang, J.; Deng, Y.; Clavairoly, A.; Frah, M.; Wang, H.; He, X.; et al. Chd7 cooperates with Sox10 and regulates the onset of CNS myelination and remyelination. Nat. Neurosci. 2016, 19, 678–689. [Google Scholar] [CrossRef]
- Parras, C.; Marie, C.; Zhao, C.; Lu, Q.R. Chromatin remodelers in oligodendroglia. Glia 2020, 68, 1604–1618. [Google Scholar] [CrossRef] [PubMed]
- Molin, A.N.; Contentin, R.; Angelozzi, M.; Karvande, A.; Kc, R.; Haseeb, A.; Voskamp, C.; de Charleroy, C.; Lefebvre, V. Skeletal growth is enhanced by a shared role for SOX8 and SOX9 in promoting reserve chondrocyte commitment to columnar proliferation. Proc. Natl. Acad. Sci. USA 2024, 121, e2316969121. [Google Scholar] [CrossRef] [PubMed]
- Barrionuevo, F.; Scherer, G. SOX E genes: SOX9 and SOX8 in mammalian testis development. Int. J. Biochem. Cell Biol. 2010, 42, 433–436. [Google Scholar] [CrossRef]
- Koopman, P. Sex determination: A tale of two Sox genes. Trends Genet. 2005, 21, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Ming, Z.; Vining, B.; Bagheri-Fam, S.; Harley, V. SOX9 in organogenesis: Shared and unique transcriptional functions. Cell. Mol. Life Sci. 2022, 79, 522. [Google Scholar] [CrossRef] [PubMed]
- Bergsland, M.; Werme, M.; Malewicz, M.; Perlmann, T.; Muhr, J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006, 20, 3475–3486. [Google Scholar] [CrossRef] [PubMed]
- Bylund, M.; Andersson, E.; Novitch, B.G.; Muhr, J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 2003, 6, 1162–1168. [Google Scholar] [CrossRef]
- Bhattaram, P.; Penzo-Méndez, A.; Sock, E.; Colmenares, C.; Kaneko, K.J.; DePamphilis, M.L.; Wegner, M.; Lefebvre, V. Organogenesis relies on Sox4, Sox11, and Sox12 for survival of neural and mesenchymal progenitor cells. Nat. Commun. 2010, 1, 9. [Google Scholar] [CrossRef]
- Stolt, C.C.; Schlierf, A.; Lommes, P.; Hillgärtner, S.; Werner, T.; Kosian, T.; Sock, E.; Kessaris, N.; Richardson, W.D.; Lefebvre, V.; et al. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev. Cell 2006, 11, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Wüst, H.M.; Wegener, A.; Frob, F.; Hartwig, A.C.; Wegwitz, F.; Kari, V.; Schimmel, M.; Tamm, E.R.; Johnsen, S.A.; Wegner, M.; et al. Egr2-guided histone H2B monoubiquitination is required for peripheral nervous system myelination. Nucleic Acids Res. 2020, 48, 8959–8976. [Google Scholar] [CrossRef] [PubMed]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Lill, C.M.; Schjeide, B.M.; Graetz, C.; Ban, M.; Alcina, A.; Ortiz, M.A.; Perez, J.; Damotte, V.; Booth, D.; Lopez de Lapuente, A.; et al. MANBA, CXCR5, SOX8, RPS6KB1 and ZBTB46 are genetic risk loci for multiple sclerosis. Brain 2013, 136, 1778–1782. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jörg, L.M.; Schlötzer-Schrehardt, U.; Lefebvre, V.; Sock, E.; Wegner, M. Transcription Factors Sox8 and Sox10 Contribute with Different Importance to the Maintenance of Mature Oligodendrocytes. Int. J. Mol. Sci. 2024, 25, 8754. https://doi.org/10.3390/ijms25168754
Jörg LM, Schlötzer-Schrehardt U, Lefebvre V, Sock E, Wegner M. Transcription Factors Sox8 and Sox10 Contribute with Different Importance to the Maintenance of Mature Oligodendrocytes. International Journal of Molecular Sciences. 2024; 25(16):8754. https://doi.org/10.3390/ijms25168754
Chicago/Turabian StyleJörg, Lisa Mirja, Ursula Schlötzer-Schrehardt, Véronique Lefebvre, Elisabeth Sock, and Michael Wegner. 2024. "Transcription Factors Sox8 and Sox10 Contribute with Different Importance to the Maintenance of Mature Oligodendrocytes" International Journal of Molecular Sciences 25, no. 16: 8754. https://doi.org/10.3390/ijms25168754
APA StyleJörg, L. M., Schlötzer-Schrehardt, U., Lefebvre, V., Sock, E., & Wegner, M. (2024). Transcription Factors Sox8 and Sox10 Contribute with Different Importance to the Maintenance of Mature Oligodendrocytes. International Journal of Molecular Sciences, 25(16), 8754. https://doi.org/10.3390/ijms25168754







