Assessing the Efficacy of Protease Inactivation for the Preservation of Bioactive Amphibian Skin Peptides
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Reagents
3.2. Skin Secretions
3.3. Compliance with Ethical Standards
3.4. Mass Spectrometry
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zasloff, M. Antimicrobial Peptides of Multicellular Organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Sahl, H.-G. Antimicrobial and Host-Defense Peptides as New Anti-Infective Therapeutic Strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Lebedev, A.T.; Vasileva, I.D.; Samgina, T.Y. FT-MS in the de Novo Top-down Sequencing of Natural Nontryptic Peptides. Mass Spectrom. Rev. 2022, 41, 284–313. [Google Scholar] [CrossRef] [PubMed]
- Vasileva, I.D.; Samgina, T.Y.; Meng, Z.; Zubarev, R.A.; Lebedev, A.T. EThcD Benefits for the Sequencing Inside Intramolecular Disulfide Cycles of Amphibian Intact Peptides. J. Am. Soc. Mass Spectrom. 2023, 34, 1979–1988. [Google Scholar] [CrossRef] [PubMed]
- Samgina, T.Y.; Vasileva, I.D.; Trebse, P.; Torkar, G.; Surin, A.K.; Meng, Z.; Zubarev, R.A.; Lebedev, A.T. Mass Spectrometry Differentiation between Rana Arvalis Populations Based on Their Skin Peptidome Composition. J. Am. Soc. Mass Spectrom. 2022, 33, 1480–1491. [Google Scholar] [CrossRef]
- Artemenko, K.A.; Zubarev, A.R.; Samgina, T.Y.; Lebedev, A.T.; Savitski, M.M.; Zubarev, R.A. Two Dimensional Mass Mapping as a General Method of Data Representation in Comprehensive Analysis of Complex Molecular Mixtures. Anal. Chem. 2009, 81, 3738–3745. [Google Scholar] [CrossRef] [PubMed]
- Samgina, T.Y.; Kovalev, S.V.; Tolpina, M.D.; Trebse, P.; Torkar, G.; Lebedev, A.T. EThcD Discrimination of Isomeric Leucine/Isoleucine Residues in Sequencing of the Intact Skin Frog Peptides with Intramolecular Disulfide Bond. J. Am. Soc. Mass Spectrom. 2018, 29, 842–852. [Google Scholar] [CrossRef]
- Samgina, T.Y.; Vasileva, I.D.; Zubarev, R.A.; Lebedev, A.T. EThcD as a Unique Tool for the Top-Down De Novo Sequencing of Intact Natural Ranid Amphibian Peptides. Anal. Chem. 2024, 96, 12057–12064. [Google Scholar] [CrossRef]
- Simmaco, M.; Mignogna, G.; Barra, D. Antimicrobial Peptides from Amphibian Skin: What Do They Tell Us? Biopolymers 1998, 47, 435–450. [Google Scholar] [CrossRef]
- Xu, X.; Lai, R. The Chemistry and Biological Activities of Peptides from Amphibian Skin Secretions. Chem. Rev. 2015, 115, 1760–1846. [Google Scholar] [CrossRef]
- Rollins-Smith, L.A. The Importance of Antimicrobial Peptides (AMPs) in Amphibian Skin Defense. Dev. Comp. Immunol. 2023, 142, 104657. [Google Scholar] [CrossRef]
- Tennessen, J.A. Molecular Evolution of Animal Antimicrobial Peptides: Widespread Moderate Positive Selection. J. Evol. Biol. 2005, 18, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Tennessen, J.A. Adaptive Diversity and Divergence at Frog Antimicrobial Peptide Loci; Oregon State University: Corvallis, OR, USA, 2009. [Google Scholar]
- Tennessen, J.A.; Woodhams, D.C.; Chaurand, P.; Reinert, L.K.; Billheimer, D.; Shyr, Y.; Caprioli, R.M.; Blouin, M.S.; Rollins-Smith, L.A. Variations in the Expressed Antimicrobial Peptide Repertoire of Northern Leopard Frog (Rana Pipiens) Populations Suggest Intraspecies Differences in Resistance to Pathogens. Dev. Comp. Immunol. 2009, 33, 1247–1257. [Google Scholar] [CrossRef]
- Çevikbaş, A. Antibacterial Activity in the Skin Secretion of the Frog Rana Ridibunda. Toxicon 1978, 16, 195–197. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Barra, D.; Simmaco, M. Amphibian Skin: A Promising Resource for Antimicrobial Peptides. Trends Biotechnol. 1995, 13, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Indriani, S.; Karnjanapratum, S.; Nirmal, N.P.; Nalinanon, S. Amphibian Skin and Skin Secretion: An Exotic Source of Bioactive Peptides and Its Application. Foods 2023, 12, 1282. [Google Scholar] [CrossRef]
- Chen, X.; Liu, S.; Fang, J.; Zheng, S.; Wang, Z.; Jiao, Y.; Xia, P.; Wu, H.; Ma, Z.; Hao, L. Peptides Isolated from Amphibian Skin Secretions with Emphasis on Antimicrobial Peptides. Toxins 2022, 14, 722. [Google Scholar] [CrossRef]
- Bowie, J.H.; Separovic, F.; Tyler, M.J. Host-Defense Peptides of Australian Anurans. Part 2. Structure, Activity, Mechanism of Action, and Evolutionary Significance. Peptides 2012, 37, 174–188. [Google Scholar] [CrossRef]
- Demori, I.; Rashed ZEl Corradino, V.; Catalano, A.; Rovegno, L.; Queirolo, L.; Salvidio, S.; Biggi, E.; Zanotti-Russo, M.; Canesi, L.; Catenazzi, A.; et al. Peptides for Skin Protection and Healing in Amphibians. Molecules 2019, 24, 347. [Google Scholar] [CrossRef]
- Wang, G. Bioinformatic Analysis of 1000 Amphibian Antimicrobial Peptides Uncovers Multiple Length-Dependent Correlations for Peptide Design and Prediction. Antibiotics 2020, 9, 491. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Ohsuye, K.; Wada, Y.; Fuchimura, K.; Tanaka, S.; Matsuo, H. Cloning and Sequence of CDNA Encoding a Peptide C-Terminal α-Amidating Enzyme from Xenopuslaevis. Biochem. Biophys. Res. Commun. 1987, 148, 546–552. [Google Scholar] [CrossRef]
- Mollay, C.; Vilas, U.; Hutticher, A.; Kreil, G. Isolation of a Dipeptidyl Aminopeptidase, a Putative Processing Enzyme, from Skin Secretion of Xenopus Laevis. Eur. J. Biochem. 1986, 160, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, M.G.; Poulter, L.; Gibson, B.W.; Williams, D.H. Biosynthesis and Degradation of Peptides Derived from Xenopus Laevis Prohormones. Biochem. J. 1987, 243, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Darby, N.J.; Lackey, D.B.; Smyth, D.G. Purification of a Cysteine Endopeptidase Which Is Secreted with Bioactive Peptides from the Epidermal Glands of Xenopus Laevis. Eur. J. Biochem. 1991, 195, 65–70. [Google Scholar] [CrossRef]
- Resnick, N.M.; Maloy, W.L.; Guy, H.R.; Zasloff, M. A Novel Endopeptidase from Xenopus That Recognizes α-Helical Secondary Structure. Cell 1991, 66, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Magainins, a Class of Antimicrobial Peptides from Xenopus Skin: Isolation, Characterization of Two Active Forms, and Partial CDNA Sequence of a Precursor. Proc. Natl. Acad. Sci. USA 1987, 84, 5449–5453. [Google Scholar] [CrossRef]
- Samgina, T.Y.; Tolpina, M.D.; Hakalehto, E.; Artemenko, K.A.; Bergquist, J.; Lebedev, A.T. Proteolytic Degradation and Deactivation of Amphibian Skin Peptides Obtained by Electrical Stimulation of Their Dorsal Glands. Anal. Bioanal. Chem. 2016, 408, 3761–3768. [Google Scholar] [CrossRef] [PubMed]
- Bermúdez-Puga, S.; Dias, M.; Reis, I.L.; de Oliveira, T.F.; de Almeida, S.R.Y.; Mendes, M.A.; Moore, S.J.; Almeida, J.R.; Proaňo-Bolaňos, C.; de Souza Oliveira, R.P. Microscopic and metabolomics analysis of the anti-Listeria activity of natural and engineered cruzioseptins. Biochimie 2024, 225, 168–175. [Google Scholar] [CrossRef]
- Hans-Dieter Jakubke, H.-D.; Jeschkeit, H. Aminosäuren, Peptide, Proteine; Akademie-Verlag: Berlin, Germany, 1981; 301p. [Google Scholar]
- Kinter, M.; Sherman, N.E. Protein Sequencing and Identification Using Tandem Mass Spectrometry. In Wiley-Interscience Series on Mass Spectrometry; Desiderio, D.M., Nibbering, N.M.M., Eds.; John Wiley & Sons Inc.: New York, NY, USA, 2000; 320p. [Google Scholar]
- Steinborner, S.T.; Currie, G.J.; Bowie, J.H.; Wallace, J.C.; Tyler, M.J. New Antibiotic Caerin 1 Peptides from the Skin Secretion of the Australian Tree Frog Litoria Chloris. Comparison of the Activities of the Caerin 1 Peptides from the Genus Litoria. J. Pept. Res. 2009, 51, 121–126. [Google Scholar] [CrossRef]
- Vasileva, I.D.; Samgina, T.Y.; Lebedev, A.T. Mass Spectrometric De Novo Sequencing of Natural Peptides. In Peptidomics: Methods and Strategies, Methods in Molecular Biology; Schrader, M., Fricker, L.D., Eds.; Humana: New York, NY, USA, 2024; Volume 2758. [Google Scholar] [CrossRef]
- Simmaco, M.; Mignogna, G.; Barra, D.; Bossa, F. Antimicrobial peptides from skin secretion of R esculenta. J. Biol. Chem. 1944, 269, 11956–11961. [Google Scholar] [CrossRef]
- Basir, Y.J.; Conlon, J.M. Peptidomic analysis of the skin secretions of the pickerel frog R palustris identifies six novel families of structurally-related peptides. Peptides 2003, 24, 379–383. [Google Scholar] [CrossRef]
- Mangony, M.L.; Papo, N.; Mignigna, J.D.; Andreu, D.; Shai, Y.; Barra, D.; Simmaco, M. Ranacyclins, a new family of short cyclic AMPs: Biological function, mode of action, and parameters involved in target specificity. Biochemistry 2003, 42, 14023–14035. [Google Scholar] [CrossRef]
- Conlon, J.M.; Kolodziejek, J.; Mechkarska, M.; Coquet, L.; Leprince, J.; Jouenne, T.; Vaudry, H.; Nielsen, P.F.; Nowotny, N.; King, J.D. Host-defence peptides from Lithobates forreri, Hylarana luctuosa, and Hylarana signata (Ranidae): Phylogenetic relationships inferred from primary structures of ranatuerin-2 and brevinin-2 peptides. Comp. Biochem. Physiol. Part D 2014, 9, 49–57. [Google Scholar] [CrossRef]
- Conlon, J.M.; Mechkarska, M.; Coquet, L.; Leprince, J.; Jouenne, T.; Vaudry, H.; Measey, G.J. Evidence from peptidomic analysis of skin secretions that allopatric populations of Xenopus gilli (Anura: Pipidae) constitute distinct lineages. Peptides 2015, 63, 118–125. [Google Scholar] [CrossRef]
- Conlon, J.M.; Mechkarska, M.; Kolodziejek, J.; Leprince, J.; Coquet, L.; Jouenne, T.; Vaudry, H.; Nowotny, N.; King, J.D. Host-defense and trefoil factor family peptides in skin secretions of the Mawa clawed frog Xenopus boumbaensis (Pipidae). Peptides 2015, 72, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Clark, V.C. Collecting Arthropod and Amphibian Secretions for Chemical Analyses. In Behavioral and Chemical Ecology; Zhang, W., Liu, H., Eds.; Nova Science Publishers: Hauppauge, NY, USA, 2009; pp. 1–46. [Google Scholar]
- Samgina, T.Y.; Artemenko, K.A.; Gorshkov, V.A.; Ogourtsov, S.V.; Zubarev, R.A.; Lebedev, A.T. De Novo Sequencing of Peptides Secreted by the Skin Glands of the Caucasian Green Frog Rana Ridibunda. Rapid Commun. Mass Spectrom. 2008, 22, 3517–3525. [Google Scholar] [CrossRef] [PubMed]
- Samgina, T.Y.; Gorshkov, V.A.; Artemenko, K.A.; Vorontsov, E.A.; Klykov, O.V.; Ogourtsov, S.V.; Zubarev, R.A.; Lebedev, A.T. LC–MS/MS with 2D Mass Mapping of Skin Secretions’ Peptides as a Reliable Tool for Interspecies Identification inside Rana Esculenta Complex. Peptides 2012, 34, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Samgina, T.Y.; Gorshkov, V.A.; Vorontsov, E.A.; Artemenko, K.A.; Ogurtsov, S.V.; Zubarev, R.A.; Lebedev, A.T. Mass Spectrometric Study of Peptidome of the Skin Secretion of Frog Rana Lessonae. Mass-Spektrometria 2010, 7, 261–270. [Google Scholar]
- Samgina, T.Y.; Artemenko, K.A.; Gorshkov, V.A.; Lebedev, A.T.; Nielsen, M.L.; Savitski, M.M.; Zubarev, R.A. Electrospray Ionization Tandem Mass Spectrometry Sequencing of Novel Skin Peptides from Ranid Frogs Containing Disulfide Bridges. Eur. J. Mass Spectrom. 2007, 13, 155–163. [Google Scholar] [CrossRef]
- Chen, X.; Wang, H.; Shen, Y.; Wang, L.; Zhou, M.; Chen, T.; Shaw, C. Kunitzins: Prototypes of a New Class of Protease Inhibitor from the Skin Secretions of European and Asian Frogs. Biochem. Biophys. Res. Commun. 2016, 477, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Shi, D.; Ying, Y.; Xi, X.; Chen, X.; Wang, L.; Zhou, M.; Wu, Q.; Ma, C.; Chen, T. A Novel Kunitzin-Like Trypsin Inhibitor Isolated from Defensive Skin Secretion of Odorrana Versabilis. Biomolecules 2019, 9, 254. [Google Scholar] [CrossRef] [PubMed]
- Samgina, T.Y.; Tolpina, M.D.; Trebse, P.; Torkar, G.; Artemenko, K.A.; Bergquist, J.; Lebedev, A.T. LTQ Orbitrap Velos in Routine de Novo Sequencing of Non-Tryptic skin Peptides from the Frog Rana Latastei with Traditional and Reliable Manual Spectra Interpretation. Rapid Commun. Mass Spectrom. 2016, 30, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Iwakoshi-Ukena, E.; Soga, M.; Okada, G.; Fujii, T.; Sumida, M.; Ukena, K. Characterization of Novel Antimicrobial Peptides from the Skin of the Endangered Frog Odorrana Ishikawae by Shotgun CDNA Cloning. Biochem. Biophys. Res. Commun. 2011, 412, 673–677. [Google Scholar] [CrossRef]
- Lebedev, A.T.; Damoc, E.; Makarov, A.A.; Samgina, T.Y. Discrimination of Leucine and Isoleucine in Peptides Sequencing with Orbitrap Fusion Mass Spectrometer. Anal. Chem. 2014, 86, 7017–7022. [Google Scholar] [CrossRef]
- Tyler, M.J.; Stone, D.J.M.; Bowie, J.H. A Novel Method for the Release and Collection of Dermal, Glandular Secretions from the Skin of Frogs. J. Pharmacol. Toxicol. Methods 1992, 28, 199–200. [Google Scholar] [CrossRef]
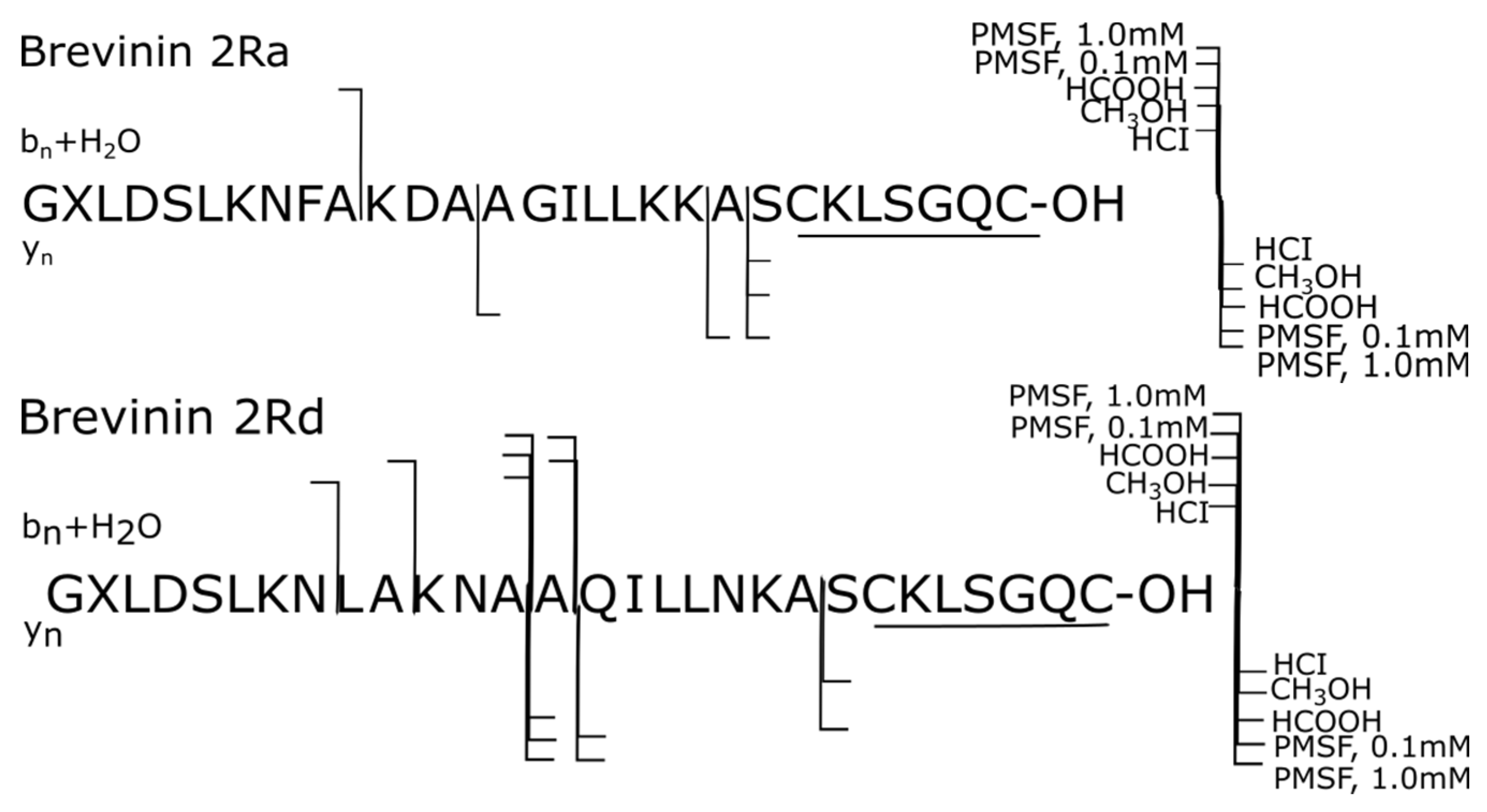

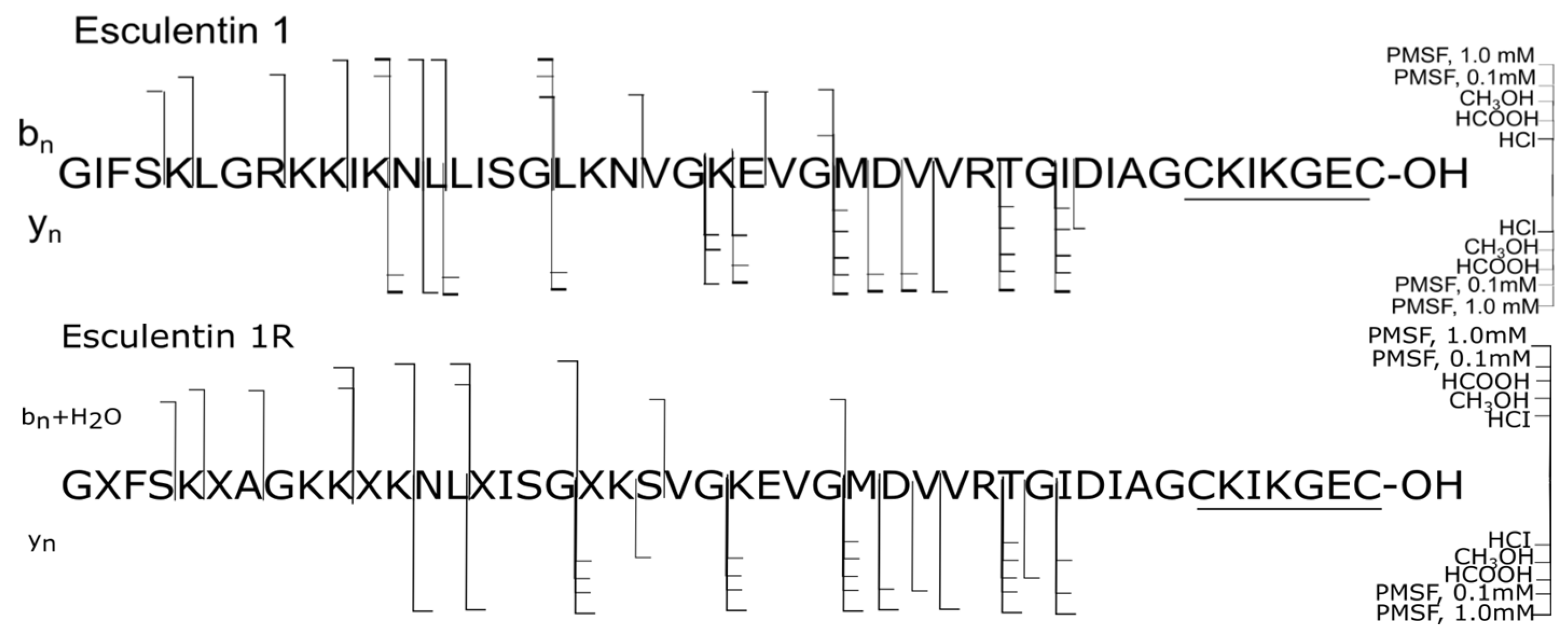
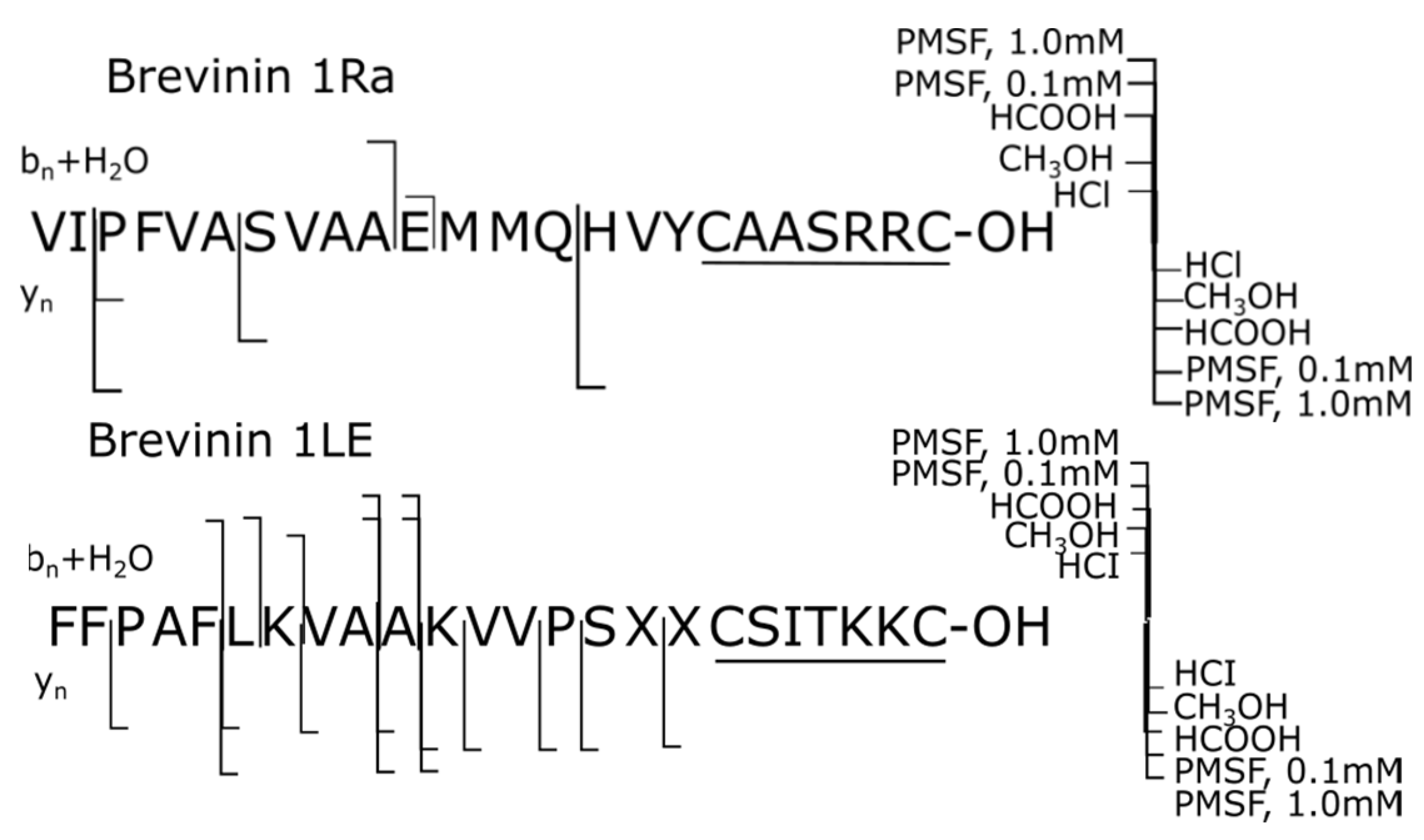
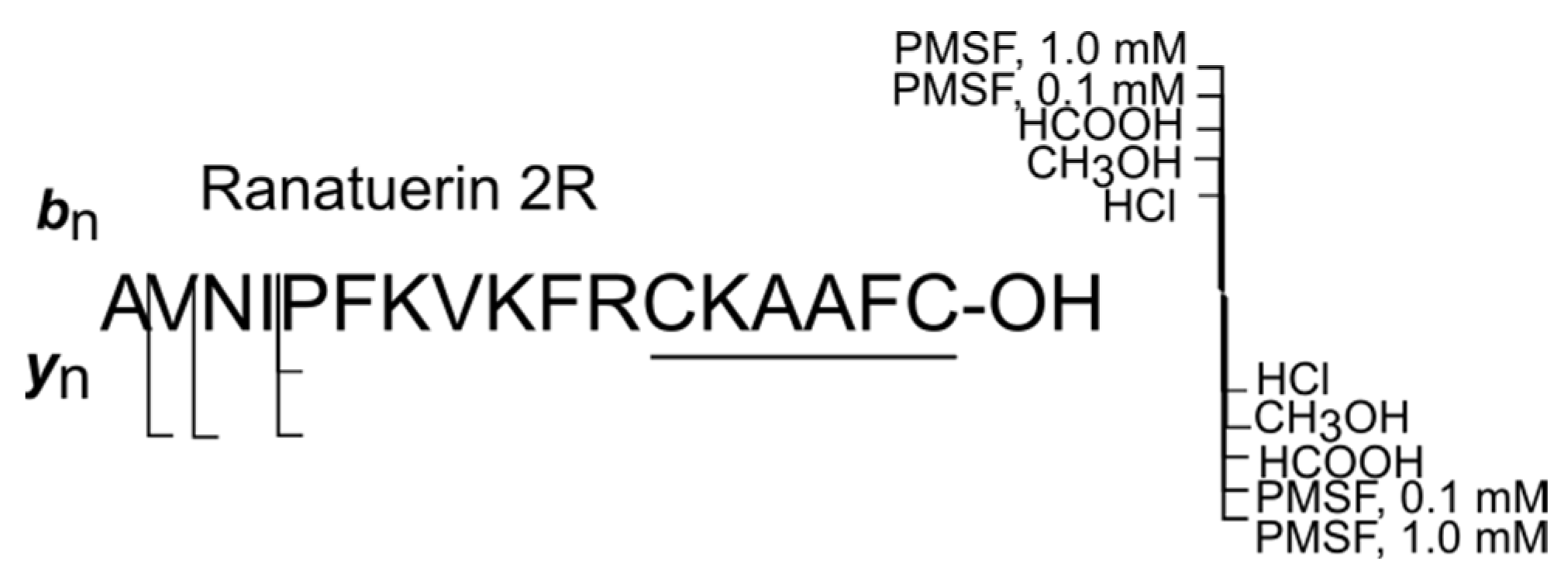
| № | Peptide | Mm, Da | Sequence |
|---|---|---|---|
| 1 | Esculentin 1 | 4882.7 | GIFSKLGRKKIKNLLISGLKNVGKEVGMDVVRTGIDIAGCKIKGEC-OH |
| 2 | Esculentin 1R | 4770.7 | GXFSKXAGKKXKNLXISGXKSVGKEVGMDVVRTGIDIAGCKIKGEC-OH |
| 3 | Brevinin 2L | 3242.8 | GXLSTLKNVAKTAGKGALQSLLNAASCKLSGQC-OH |
| 4 | Brevinin 2La | 3386.8 | GXWNTXKATGKSA ASNVAVTLLDKAKCK(LV)GEC-OH |
| 5 | Brevinin 2Ra | 2989.6 | GXLDSLKNFAKDAAGILLKKASCKLSGQC-OH |
| 6 | Brevinin 2Rd | 3011.6 | GXLDSLKNLAKNAAQILLNKASCKLSGQC-OH |
| 7 | Brevinin 1Ra | 2636.2 | VIPFVASVAAEMMQHVYCAASRRC-OH |
| 8 | Brevinin 1LE | 2607.5 | FFPAFLKVAAKVVPSXXCSITKKC-OH |
| 9 | Ranatuerin 2R | 1939.0 | AVNIPFKVKFRCKAAFC-OH |
| Peptides * | Escul 1 | Escul 1R | Brev 2L | Brev 2La | Brev 2Ra | Brev 2Rd | Brev 1Ra | Brev 1LE | Ranat 2R | |
|---|---|---|---|---|---|---|---|---|---|---|
| Forms | ||||||||||
| HCl (sample 1) | ||||||||||
| Intact, % | 38.61 | 64.86 | 98.79 | 30.98 | 98.98 | 99.61 | 93.51 | 100 | 85.21 | |
| Oxidized, % | 9.63 | 7.45 | - | - | - | - | 5.74 | - | - | |
| Proteoforms, % | 51.77 | 27.69 | 1.21 | 69.02 | 1.02 | 0.39 | 0.75 | - | 14.79 | |
| CH3OH (sample 2) | ||||||||||
| Intact, % | 79.99 | 77.85 | 95.34 | 100 | 100 | 100 | 98.27 | 100 | 100 | |
| Oxidized, % | - | - | - | - | - | - | 1.13 | - | - | |
| Proteoforms, % | 20.01 | 22.15 | 4.66 | - | - | - | 0.60 | - | - | |
| HCOOH (sample 3) | ||||||||||
| Intact, % | 72.22 | 84.41 | 95.26 | 95.02 | 98.70 | 97.77 | 99.51 | 98.37 | 65.01 | |
| Oxidized, % | 10.79 | - | - | - | - | - | 0.19 | - | - | |
| Proteoforms, % | 16.99 | 15.59 | 4.74 | 4.98 | 1.30 | 2.23 | 0.30 | 1.63 | 34.99 | |
| PMSF (0.1 mM) (sample 4) | ||||||||||
| Intact, % | 3.32 | 2.42 | 0.36 | - | 11.80 | 1.55 | 77.80 | 7.27 | - | |
| Oxidized, % | 2.28 | 0.25 | - | - | - | - | 22.20 | - | - | |
| Proteoforms, % | 94.40 | 97.33 | 99.64 | - | 88.21 | 98.45 | - | 92.73 | - | |
| PMSF (1.0 mM) (sample 5) | ||||||||||
| Intact, % | 8.10 | 0.74 | 19.49 | - | 85.99 | 20.84 | 99.47 | 10.26 | 100 | |
| Oxidized, % | - | - | - | - | - | - | - | - | - | |
| Proteoforms, % | 91.90 | 99.26 | 80.51 | - | 14.01 | 79.16 | 0.53 | 89.74 | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samgina, T.Y.; Mazur, D.M.; Lebedev, A.T. Assessing the Efficacy of Protease Inactivation for the Preservation of Bioactive Amphibian Skin Peptides. Int. J. Mol. Sci. 2024, 25, 8759. https://doi.org/10.3390/ijms25168759
Samgina TY, Mazur DM, Lebedev AT. Assessing the Efficacy of Protease Inactivation for the Preservation of Bioactive Amphibian Skin Peptides. International Journal of Molecular Sciences. 2024; 25(16):8759. https://doi.org/10.3390/ijms25168759
Chicago/Turabian StyleSamgina, Tatiana Yu., Dmitrii M. Mazur, and Albert T. Lebedev. 2024. "Assessing the Efficacy of Protease Inactivation for the Preservation of Bioactive Amphibian Skin Peptides" International Journal of Molecular Sciences 25, no. 16: 8759. https://doi.org/10.3390/ijms25168759
APA StyleSamgina, T. Y., Mazur, D. M., & Lebedev, A. T. (2024). Assessing the Efficacy of Protease Inactivation for the Preservation of Bioactive Amphibian Skin Peptides. International Journal of Molecular Sciences, 25(16), 8759. https://doi.org/10.3390/ijms25168759








