Effects of Pulsed Electromagnetic Field Treatment on Skeletal Muscle Tissue Recovery in a Rat Model of Collagenase-Induced Tendinopathy: Results from a Proteome Analysis
Abstract
1. Introduction
2. Results
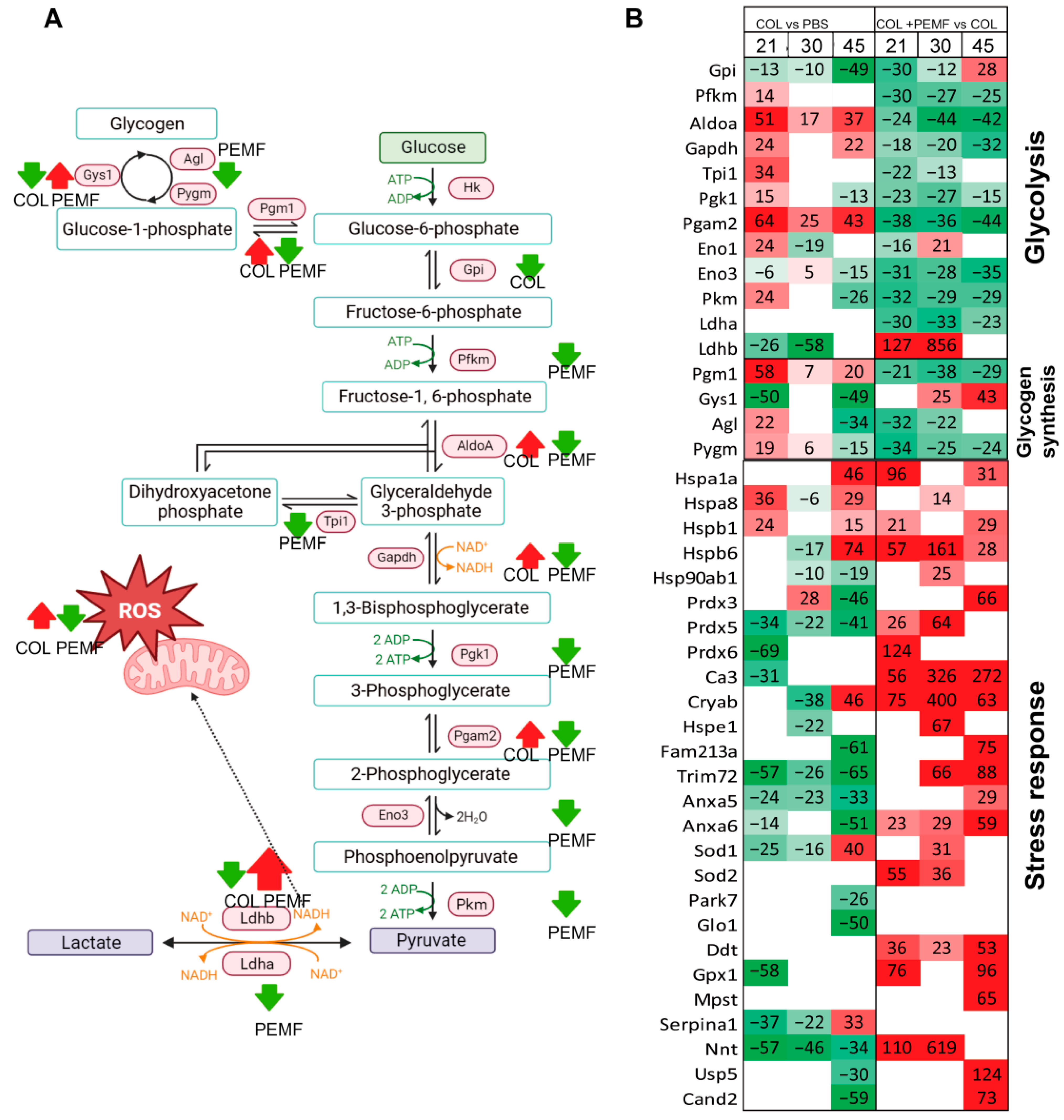
2.1. Glucose Metabolism and Stress Response
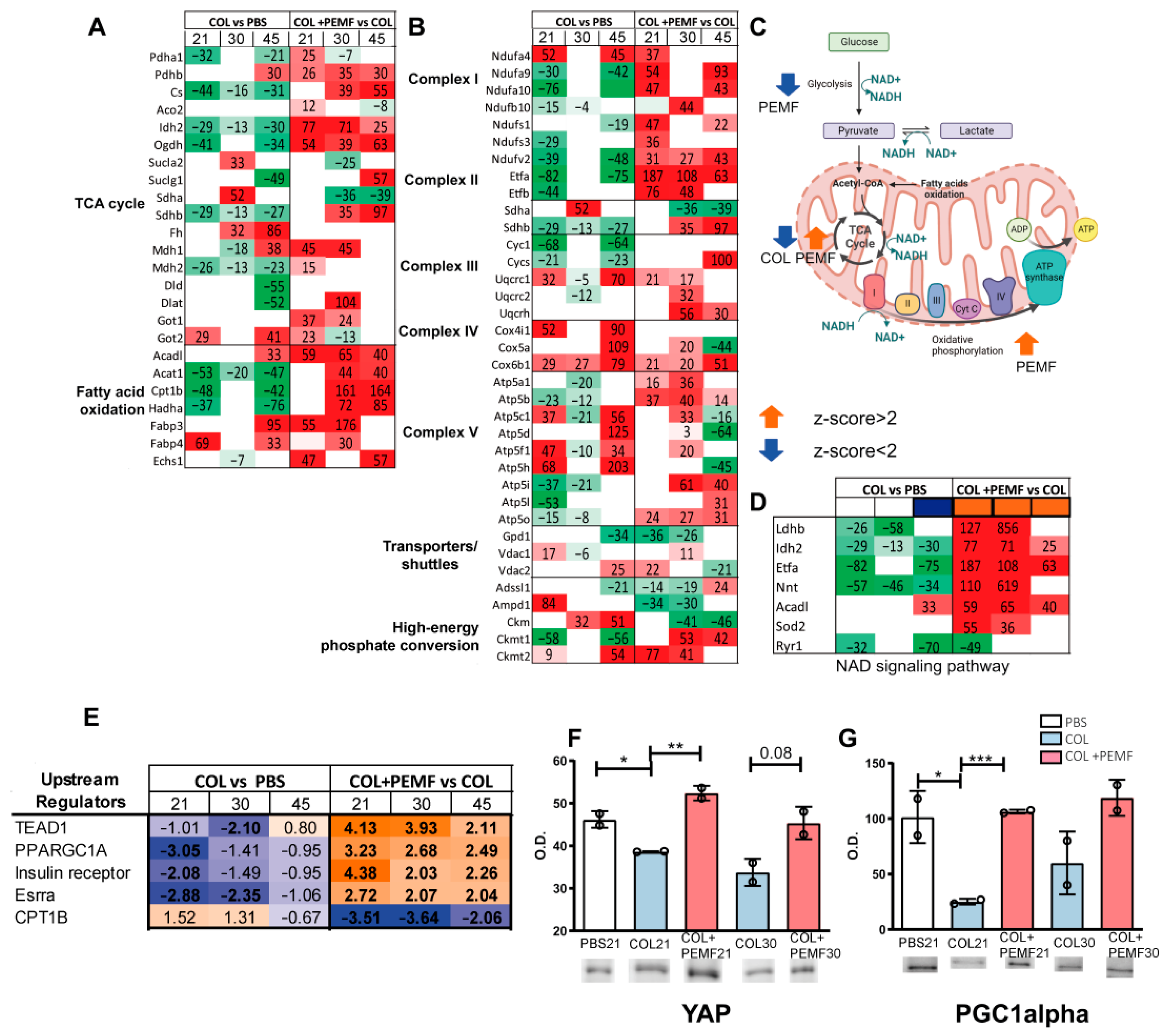
2.2. TCA Cycle, Fatty Acid Oxidation, and Oxidative Phosphorylation Pathways
2.3. Bioinformatic Analysis of Metabolic Pathways
2.4. Muscle Fiber Characterization
2.5. Contractile Proteins
2.6. Structural Proteins
2.7. Calcium Binding Proteins
2.8. Diseases and Biofunctions
3. Discussion
4. Materials and Methods
4.1. Animal Housing and Ethic Statements
4.2. Study Design
4.3. Protein Extraction
4.4. LC-MS/MS Analysis
4.5. Bioinformatic Analysis
4.6. Immunoblotting
4.7. MyHC Fiber Composition
4.8. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Edom-Vovard, F.; Duprez, D. Signals regulating tendon formation during chick embryonic development. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2004, 229, 449–457. [Google Scholar] [CrossRef]
- Charvet, B.; Ruggiero, F.; Le Guellec, D. The development of the myotendinous junction. A review. Muscles Ligaments Tendons J. 2012, 2, 53–63. [Google Scholar] [PubMed]
- Schweitzer, R.; Zelzer, E.; Volk, T. Connecting muscles to tendons: Tendons and musculoskeletal development in flies and vertebrates. Development 2010, 137, 2807–2817. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.C.; Pirkl, C.; Pfirrmann, C.W.; Zanetti, M.; Gerber, C. Asymmetric atrophy of the supraspinatus muscle following tendon tear. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2005, 23, 254–258. [Google Scholar] [CrossRef]
- Meyer, D.C.; Hoppeler, H.; von Rechenberg, B.; Gerber, C. A pathomechanical concept explains muscle loss and fatty muscular changes following surgical tendon release. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2004, 22, 1004–1007. [Google Scholar] [CrossRef] [PubMed]
- Shupak, N.M.; Prato, F.S.; Thomas, A.W. Human exposure to a specific pulsed magnetic field: Effects on thermal sensory and pain thresholds. Neurosci. Lett. 2004, 363, 157–162. [Google Scholar] [CrossRef]
- Cadossi, R.; Massari, L.; Racine-Avila, J.; Aaron, R.K. Pulsed Electromagnetic Field Stimulation of Bone Healing and Joint Preservation: Cellular Mechanisms of Skeletal Response. J. Am. Acad. Orthop. Surgeons. Glob. Res. Rev. 2020, 4, e1900155. [Google Scholar] [CrossRef]
- Bagnato, G.L.; Miceli, G.; Marino, N.; Sciortino, D.; Bagnato, G.F. Pulsed electromagnetic fields in knee osteoarthritis: A double blind, placebo-controlled, randomized clinical trial. Rheumatology 2016, 55, 755–762. [Google Scholar] [CrossRef]
- Varani, K.; Vincenzi, F.; Ravani, A.; Pasquini, S.; Merighi, S.; Gessi, S.; Setti, S.; Cadossi, M.; Borea, P.A.; Cadossi, R. Adenosine Receptors as a Biological Pathway for the Anti-Inflammatory and Beneficial Effects of Low Frequency Low Energy Pulsed Electromagnetic Fields. Mediat. Inflamm. 2017, 2017, 2740963. [Google Scholar] [CrossRef]
- Sakhrani, N.; Stefani, R.M.; Setti, S.; Cadossi, R.; Ateshian, G.A.; Hung, C.T. Pulsed Electromagnetic Field Therapy and Direct Current Electric Field Modulation Promote the Migration of Fibroblast-like Synoviocytes to Accelerate Cartilage Repair In Vitro. Appl. Sci. 2022, 12, 12406. [Google Scholar] [CrossRef]
- Stefani, R.M.; Barbosa, S.; Tan, A.R.; Setti, S.; Stoker, A.M.; Ateshian, G.A.; Cadossi, R.; Vunjak-Novakovic, G.; Aaron, R.K.; Cook, J.L.; et al. Pulsed electromagnetic fields promote repair of focal articular cartilage defects with engineered osteochondral constructs. Biotechnol. Bioeng. 2020, 117, 1584–1596. [Google Scholar] [CrossRef] [PubMed]
- Veronesi, F.; Cadossi, M.; Giavaresi, G.; Martini, L.; Setti, S.; Buda, R.; Giannini, S.; Fini, M. Pulsed electromagnetic fields combined with a collagenous scaffold and bone marrow concentrate enhance osteochondral regeneration: An in vivo study. BMC Musculoskelet. Disord. 2015, 16, 233. [Google Scholar] [CrossRef] [PubMed]
- Fini, M.; Torricelli, P.; Giavaresi, G.; Aldini, N.N.; Cavani, F.; Setti, S.; Nicolini, A.; Carpi, A.; Giardino, R. Effect of pulsed electromagnetic field stimulation on knee cartilage, subchondral and epyphiseal trabecular bone of aged Dunkin Hartley guinea pigs. Biomed. Pharmacother. 2008, 62, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Guo, H.; Ye, W.; Yang, L.; He, C. Pulsed Electromagnetic Field Attenuates Osteoarthritis Progression in a Murine Destabilization-Induced Model through Inhibition of TNF-alpha and IL-6 Signaling. Cartilage 2021, 13, 1665S–1675S. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; He, H.; Zhang, C.; Wang, H.; Gao, C.; Yu, X.; He, C. Effects of pulsed electromagnetic fields on postmenopausal osteoporosis. Bioelectromagnetics 2017, 38, 406–424. [Google Scholar] [CrossRef] [PubMed]
- Griffin, X.L.; Costa, M.L.; Parsons, N.; Smith, N. Electromagnetic field stimulation for treating delayed union or non-union of long bone fractures in adults. Cochrane Database Syst. Rev. 2011, 4, CD008471. [Google Scholar] [CrossRef]
- Hannemann, P.F.; Mommers, E.H.; Schots, J.P.; Brink, P.R.; Poeze, M. The effects of low-intensity pulsed ultrasound and pulsed electromagnetic fields bone growth stimulation in acute fractures: A systematic review and meta-analysis of randomized controlled trials. Arch. Orthop. Trauma Surg. 2014, 134, 1093–1106. [Google Scholar] [CrossRef] [PubMed]
- Diniz, P.; Soejima, K.; Ito, G. Nitric oxide mediates the effects of pulsed electromagnetic field stimulation on the osteoblast proliferation and differentiation. Nitric Oxide Biol. Chem. 2002, 7, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Pall, M.L. Electromagnetic fields act via activation of voltage-gated calcium channels to produce beneficial or adverse effects. J. Cell. Mol. Med. 2013, 17, 958–965. [Google Scholar] [CrossRef]
- Petecchia, L.; Sbrana, F.; Utzeri, R.; Vercellino, M.; Usai, C.; Visai, L.; Vassalli, M.; Gavazzo, P. Electro-magnetic field promotes osteogenic differentiation of BM-hMSCs through a selective action on Ca2+-related mechanisms. Sci. Rep. 2015, 5, 13856. [Google Scholar] [CrossRef]
- de Girolamo, L.; Stanco, D.; Galliera, E.; Vigano, M.; Colombini, A.; Setti, S.; Vianello, E.; Corsi Romanelli, M.M.; Sansone, V. Low frequency pulsed electromagnetic field affects proliferation, tissue-specific gene expression, and cytokines release of human tendon cells. Cell Biochem. Biophys. 2013, 66, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Vincenzi, F.; Targa, M.; Corciulo, C.; Gessi, S.; Merighi, S.; Setti, S.; Cadossi, R.; Goldring, M.B.; Borea, P.A.; Varani, K. Pulsed electromagnetic fields increased the anti-inflammatory effect of A(2)A and A(3) adenosine receptors in human T/C-28a2 chondrocytes and hFOB 1.19 osteoblasts. PLoS ONE 2013, 8, e65561. [Google Scholar] [CrossRef] [PubMed]
- Colombini, A.; Perucca Orfei, C.; Vincenzi, F.; De Luca, P.; Ragni, E.; Vigano, M.; Setti, S.; Varani, K.; de Girolamo, L. A2A adenosine receptors are involved in the reparative response of tendon cells to pulsed electromagnetic fields. PLoS ONE 2020, 15, e0239807. [Google Scholar] [CrossRef] [PubMed]
- Perucca Orfei, C.; Lovati, A.B.; Lugano, G.; Vigano, M.; Bottagisio, M.; D’Arrigo, D.; Sansone, V.; Setti, S.; de Girolamo, L. Pulsed electromagnetic fields improve the healing process of Achilles tendinopathy: A pilot study in a rat model. Bone Jt. Res. 2020, 9, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, M.C.; Krishna, L.; Pannir Selvan, R.M.; Tai, Y.K.; Kit Wong, C.J.; Yin, J.N.; Toh, S.J.; Torta, F.; Triebl, A.; Frohlich, J.; et al. Magnetic field therapy enhances muscle mitochondrial bioenergetics and attenuates systemic ceramide levels following ACL reconstruction: Southeast Asian randomized-controlled pilot trial. J. Orthop. Transl. 2022, 35, 99–112. [Google Scholar] [CrossRef]
- Jeon, H.S.; Kang, S.Y.; Park, J.H.; Lee, H.S. Effects of pulsed electromagnetic field therapy on delayed-onset muscle soreness in biceps brachii. Phys. Ther. Sport Off. J. Assoc. Chart. Physiother. Sports Med. 2015, 16, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Trofe, A.; Piras, A.; Muehsam, D.; Meoni, A.; Campa, F.; Toselli, S.; Raffi, M. Effect of Pulsed Electromagnetic Fields (PEMFs) on Muscular Activation during Cycling: A Single-Blind Controlled Pilot Study. Healthcare 2023, 11, 922. [Google Scholar] [CrossRef] [PubMed]
- Venugobal, S.; Tai, Y.K.; Goh, J.; Teh, S.; Wong, C.; Goh, I.; Maier, A.B.; Kennedy, B.K.; Franco-Obregon, A. Brief, weekly magnetic muscle therapy improves mobility and lean body mass in older adults: A Southeast Asia community case study. Aging 2023, 15, 1768–1790. [Google Scholar] [CrossRef]
- Maiullari, S.; Cicirelli, A.; Picerno, A.; Giannuzzi, F.; Gesualdo, L.; Notarnicola, A.; Sallustio, F.; Moretti, B. Pulsed Electromagnetic Fields Induce Skeletal Muscle Cell Repair by Sustaining the Expression of Proteins Involved in the Response to Cellular Damage and Oxidative Stress. Int. J. Mol. Sci. 2023, 24, 16631. [Google Scholar] [CrossRef]
- Tai, Y.K.; Ng, C.; Purnamawati, K.; Yap, J.L.Y.; Yin, J.N.; Wong, C.; Patel, B.K.; Soong, P.L.; Pelczar, P.; Frohlich, J.; et al. Magnetic fields modulate metabolism and gut microbiome in correlation with Pgc-1alpha expression: Follow-up to an in vitro magnetic mitohormetic study. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 11143–11167. [Google Scholar] [CrossRef]
- Capitanio, D.; Fania, C.; Torretta, E.; Vigano, A.; Moriggi, M.; Bravata, V.; Caretti, A.; Levett, D.Z.H.; Grocott, M.P.W.; Samaja, M.; et al. TCA cycle rewiring fosters metabolic adaptation to oxygen restriction in skeletal muscle from rodents and humans. Sci. Rep. 2017, 7, 9723. [Google Scholar] [CrossRef] [PubMed]
- Urbanska, K.; Orzechowski, A. Unappreciated Role of LDHA and LDHB to Control Apoptosis and Autophagy in Tumor Cells. Int. J. Mol. Sci. 2019, 20, 2085. [Google Scholar] [CrossRef]
- Liang, X.; Liu, L.; Fu, T.; Zhou, Q.; Zhou, D.; Xiao, L.; Liu, J.; Kong, Y.; Xie, H.; Yi, F.; et al. Exercise Inducible Lactate Dehydrogenase B Regulates Mitochondrial Function in Skeletal Muscle. J. Biol. Chem. 2016, 291, 25306–25318. [Google Scholar] [CrossRef] [PubMed]
- Mammoto, A.; Muyleart, M.; Kadlec, A.; Gutterman, D.; Mammoto, T. YAP1-TEAD1 signaling controls angiogenesis and mitochondrial biogenesis through PGC1alpha. Microvasc. Res. 2018, 119, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Brooks, G.A. Lactate as a fulcrum of metabolism. Redox Biol. 2020, 35, 101454. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wang, Y.; Li, P.F. Mutual regulation of lactate dehydrogenase and redox robustness. Front. Physiol. 2022, 13, 1038421. [Google Scholar] [CrossRef]
- Kashihara, T.; Mukai, R.; Oka, S.I.; Zhai, P.; Nakada, Y.; Yang, Z.; Mizushima, W.; Nakahara, T.; Warren, J.S.; Abdellatif, M.; et al. YAP mediates compensatory cardiac hypertrophy through aerobic glycolysis in response to pressure overload. J. Clin. Investig. 2022, 132, e150595. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Sun, L.; Fan, X.; Yin, B.; Kang, Y.; An, S.; Tang, L. Pulsed electromagnetic fields alleviate streptozotocin-induced diabetic muscle atrophy. Mol. Med. Rep. 2018, 18, 1127–1133. [Google Scholar] [CrossRef]
- Sakurai, T.; Satake, A.; Sumi, S.; Inoue, K.; Miyakoshi, J. An extremely low frequency magnetic field attenuates insulin secretion from the insulinoma cell line, RIN-m. Bioelectromagnetics 2004, 25, 160–166. [Google Scholar] [CrossRef]
- Henique, C.; Mansouri, A.; Fumey, G.; Lenoir, V.; Girard, J.; Bouillaud, F.; Prip-Buus, C.; Cohen, I. Increased mitochondrial fatty acid oxidation is sufficient to protect skeletal muscle cells from palmitate-induced apoptosis. J. Biol. Chem. 2010, 285, 36818–36827. [Google Scholar] [CrossRef]
- Valle, I.; Alvarez-Barrientos, A.; Arza, E.; Lamas, S.; Monsalve, M. PGC-1alpha regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc. Res. 2005, 66, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C.; et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Ward, W.F. PGC-1alpha: A key regulator of energy metabolism. Adv. Physiol. Educ. 2006, 30, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.H.; Kim, J.Y. Role of PGC-1alpha in the Mitochondrial NAD(+) Pool in Metabolic Diseases. Int. J. Mol. Sci. 2021, 22, 4558. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.S.; Tracy, C.M.; Miller, M.R.; Makaju, A.; Szulik, M.W.; Oka, S.I.; Yuzyuk, T.N.; Cox, J.E.; Kumar, A.; Lozier, B.K.; et al. Histone methyltransferase Smyd1 regulates mitochondrial energetics in the heart. Proc. Natl. Acad. Sci. USA 2018, 115, E7871–E7880. [Google Scholar] [CrossRef] [PubMed]
- Shao, D.; Zhai, P.; Del Re, D.P.; Sciarretta, S.; Yabuta, N.; Nojima, H.; Lim, D.S.; Pan, D.; Sadoshima, J. A functional interaction between Hippo-YAP signalling and FoxO1 mediates the oxidative stress response. Nat. Commun. 2014, 5, 3315. [Google Scholar] [CrossRef]
- Liu, J.; Wen, T.; Dong, K.; He, X.; Zhou, H.; Shen, J.; Fu, Z.; Hu, G.; Ma, W.; Li, J.; et al. TEAD1 protects against necroptosis in postmitotic cardiomyocytes through regulation of nuclear DNA-encoded mitochondrial genes. Cell Death Differ. 2021, 28, 2045–2059. [Google Scholar] [CrossRef]
- LaFramboise, W.A.; Jayaraman, R.C.; Bombach, K.L.; Ankrapp, D.P.; Krill-Burger, J.M.; Sciulli, C.M.; Petrosko, P.; Wiseman, R.W. Acute molecular response of mouse hindlimb muscles to chronic stimulation. Am. J. Physiol. Cell Physiol. 2009, 297, C556–C570. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.; Edstrom, L.; Lindegren, B.; Gorza, L.; Schiaffino, S. MHC composition and enzyme-histochemical and physiological properties of a novel fast-twitch motor unit type. Am. J. Physiol. 1991, 261, C93–C101. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef]
- Agarkova, I.; Ehler, E.; Lange, S.; Schoenauer, R.; Perriard, J.C. M-band: A safeguard for sarcomere stability? J. Muscle Res. Cell Motil. 2003, 24, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Schoenauer, R.; Lange, S.; Hirschy, A.; Ehler, E.; Perriard, J.C.; Agarkova, I. Myomesin 3, a novel structural component of the M-band in striated muscle. J. Mol. Biol. 2008, 376, 338–351. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.H.; Del Re, D.P.; Sussman, M.A. The Rac and Rho hall of fame: A decade of hypertrophic signaling hits. Circ. Res. 2006, 98, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Nulali, J.; Zhan, M.; Zhang, K.; Tu, P.; Liu, Y.; Song, H. Osteoglycin: An ECM Factor Regulating Fibrosis and Tumorigenesis. Biomolecules 2022, 12, 1674. [Google Scholar] [CrossRef] [PubMed]
- Carnovali, M. High-Intensity Low Frequency Pulsed Electromagnetic Fields Treatment Stimulates Fin Regeneration in Adult Zebrafish—A Preliminary Report. Appl. Sci. 2022, 12, 7768. [Google Scholar] [CrossRef]
- Headrick, J.P.; Willems, L.; Ashton, K.J.; Holmgren, K.; Peart, J.; Matherne, G.P. Ischaemic tolerance in aged mouse myocardium: The role of adenosine and effects of A1 adenosine receptor overexpression. J. Physiol. 2003, 549, 823–833. [Google Scholar] [CrossRef]
- Gnad, T.; Navarro, G.; Lahesmaa, M.; Reverte-Salisa, L.; Copperi, F.; Cordomi, A.; Naumann, J.; Hochhauser, A.; Haufs-Brusberg, S.; Wenzel, D.; et al. Adenosine/A2B Receptor Signaling Ameliorates the Effects of Aging and Counteracts Obesity. Cell Metab. 2020, 32, 56–70.e7. [Google Scholar] [CrossRef] [PubMed]
- Espinal, J.; Challiss, R.A.; Newsholme, E.A. Effect of adenosine deaminase and an adenosine analogue on insulin sensitivity in soleus muscle of the rat. FEBS Lett. 1983, 158, 103–106. [Google Scholar] [CrossRef]
- Johnston-Cox, H.; Koupenova, M.; Yang, D.; Corkey, B.; Gokce, N.; Farb, M.G.; LeBrasseur, N.; Ravid, K. The A2b adenosine receptor modulates glucose homeostasis and obesity. PLoS ONE 2012, 7, e40584. [Google Scholar] [CrossRef]
- Perucca Orfei, C.; Lovati, A.B.; Vigano, M.; Stanco, D.; Bottagisio, M.; Di Giancamillo, A.; Setti, S.; de Girolamo, L. Dose-Related and Time-Dependent Development of Collagenase-Induced Tendinopathy in Rats. PLoS ONE 2016, 11, e0161590. [Google Scholar] [CrossRef]


| Group Name | Muscle Group Name | Number of Samples | Type I Collagenase Injection | PEMF Treatment | Start of PEMF Treatment (Days after Collagenase Injection) | Length of Treatment (Days) | Sacrifice and Muscle Extraction |
|---|---|---|---|---|---|---|---|
| PBS21 | 3 | NO | NO | -- | -- | Day 21 | |
| PBS | PBS30 | 3 | NO | NO | -- | -- | Day 30 |
| PBS45 | 3 | NO | NO | -- | -- | Day 45 | |
| COL21 | 3 | YES | NO | -- | -- | Day 21 | |
| COL | COL30 | 3 | YES | NO | -- | -- | Day 30 |
| COL45 | 3 | YES | NO | -- | -- | Day 45 | |
| PBS + PEMF21 | 3 | NO | YES | Day 7 | 15 | Day 21 | |
| PBS + PEMF | PBS + PEMF30 | 3 | NO | YES | Day 15 | 15 | Day 30 |
| PBS + PEMF45 | 3 | NO | YES | Day 15 | 30 | Day 45 | |
| COL + PEMF21 | 3 | YES | YES | Day 7 | 15 | Day 21 | |
| COL + PEMF | COL + PEMF30 | 3 | YES | YES | Day 15 | 15 | Day 30 |
| COL + PEMF45 | 3 | YES | YES | Day 15 | 30 | Day 45 |
| COL vs. PBS | COL + PEMF vs. COL | |||||||
|---|---|---|---|---|---|---|---|---|
| 21 d | 30 d | 45 d | 21 d | 30 d | 45 d | |||
| Extracellular matrix | P02454 | Col1a1 | 133.38 | −28.93 | 611.14 | 16.77 | −65.17 | −85.31 |
| F1LS40 | Col1a2 | 169.30 | 15.46 | 809.56 | 47.85 | −81.78 | −91.15 | |
| D3ZUL3 | Col6a1 | 28.86 | −21.04 | −21.69 | −24.96 | |||
| F1LNH3 | Col6a2 | −27.56 | 88.99 | −29.89 | ||||
| D3ZZT9 | Col14a1 | −31.25 | ||||||
| Q9EQP5 | Prelp | −49.97 | ||||||
| G3V6E7 | Fmod | −68.85 | −48.48 | |||||
| P51886 | Lum | −29.79 | ||||||
| P11762 | Lgals1 | −14.03 | ||||||
| D3ZVB7 | Ogn | −10.65 | −46.80 | −62.60 | 75.52 | 44.45 | ||
| Q5XIH1 | Aspn | 29.74 | −35.05 | |||||
| Q01129 | Dcn | −38.72 | 77.06 | −27.85 | ||||
| G3V9M6 | Fbn1 | 139.39 | −34.12 | |||||
| Cytoskeletal | P60711 | Actb | −21.87 | 151.82 | −29.33 | |||
| G3V8C3 | Vim | −66.07 | −40.97 | −26.47 | ||||
| F7F9U6 | Plec | −25.41 | 35.01 | −33.90 | 43.35 | 40.22 | ||
| Q6P725 | Des | 30.10 | 51.11 | 34.94 | 26.35 | |||
| Q6P9V9 | Tuba1b | −8.67 | −13.82 | |||||
| Q5XIF6 | Tuba4a | −20.32 | −18.15 | −25.49 | 15.73 | 27.77 | 56.35 | |
| G3V7C6 | Tubb4b | −21.14 | −19.46 | 27.07 | ||||
| A0A0G2JSM3 | Pdlim3 | 72.58 | 71.12 | −28.54 | ||||
| G3V6P7 | Myh9 | −39.31 | 9.20 | |||||
| G3V9G5 | Synm | −22.32 | 32.49 | |||||
| P11530 | Dmd | −50.43 | −35.62 | |||||
| COL vs. PBS | COL + PEMF vs. COL | |||||||
|---|---|---|---|---|---|---|---|---|
| 21 d | 30 d | 45 d | 21 d | 30 d | 45 d | |||
| Calcium binding | P02625 | Pvalb | −57.84 | 260.72 | 23.61 | 24.72 | −82.80 | −90.03 |
| F1LWG8 | Srl | −17.33 | −17.16 | 12.29 | ||||
| P19633 | Casq1 | −10.17 | −38.84 | −48.52 | ||||
| F1M944 | Casq2 | −53.03 | 60.38 | 124.76 | ||||
| F1LMY4 | Ryr1 | −31.99 | −69.65 | −49.24 | ||||
| Q6P9Y4 | Slc25a4 | 20.20 | 15.28 | 29.32 | ||||
| G3V6H5 | Slc25a11 | −43.44 | −51.29 | 39.97 | 39.16 | |||
| F1LX07 | Slc25a12 | −39.19 | −16.48 | −50.96 | ||||
| Q64578 | Atp2a1 | 88.39 | 17.38 | 73.89 | −28.80 | −55.07 | −56.95 | |
| P11507 | Atp2a2 | 14.99 | 77.83 | −36.49 | ||||
| A0A0G2K9N9 | Atp2a3 | −58.84 | −43.85 | 45.25 | ||||
| Diseases and Biofunctions | COL vs. PBS | COL + PEMF vs. COL | ||||
|---|---|---|---|---|---|---|
| 21 d | 30 d | 45 d | 21 d | 30 d | 45 d | |
| Cell death of muscle cells | 1.70 | 1.03 | 0.24 | −2.50 | −2.43 | −3.08 |
| Necrosis of muscle | 1.02 | 1.30 | 0.02 | −2.65 | −2.23 | −2.71 |
| Apoptosis of muscle cells | 1.53 | 1.19 | −0.18 | −2.58 | −2.16 | −2.75 |
| Necrosis | 1.49 | 0.95 | 0.67 | −2.75 | −0.81 | −2.03 |
| Apoptosis | 2.20 | 1.10 | −0.05 | −3.22 | −1.35 | −1.56 |
| Survival of stem cell lines | −1.13 | N/A | N/A | N/A | 2.83 | 2.24 |
| Quantity of reactive oxygen species | 2.62 | 1.53 | 1.10 | −2.95 | −2.82 | N/A |
| Synthesis of reactive oxygen species | 1.29 | 1.20 | 0.95 | −2.62 | −2.48 | −2.03 |
| Metabolism of reactive oxygen species | 0.61 | 0.93 | 0.46 | −2.08 | −1.87 | −1.65 |
| Concentration of lipid | 3.26 | 0.57 | 0.67 | −2.35 | −1.86 | −1.63 |
| Concentration of fatty acid | 2.51 | 0.89 | 0.96 | N/A | −3.05 | −2.47 |
| Oxidation of lipid | −1.43 | N/A | N/A | 2.15 | 0.92 | 1.52 |
| Fatty acid metabolism | 1.09 | −0.16 | 2.12 | −0.04 | 0.40 | −0.94 |
| Transmembrane potential of mitochondria | −2.60 | N/A | −2.43 | 1.39 | 1.53 | 1.25 |
| Respiration of mitochondria | −1.41 | N/A | −1.02 | N/A | 2.00 | 1.95 |
| Synthesis of ATP | −0.85 | −1.63 | −2.54 | 0.77 | 0.85 | −0.29 |
| Fibrosis | −0.66 | −0.35 | −1.63 | −3.23 | 0.18 | −1.49 |
| Production of lactic acid | 2.19 | N/A | N/A | −2.19 | N/A | N/A |
| Quantity of Ca2+ | N/A | N/A | N/A | 2.19 | N/A | 2.36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torretta, E.; Moriggi, M.; Capitanio, D.; Orfei, C.P.; Raffo, V.; Setti, S.; Cadossi, R.; de Girolamo, L.; Gelfi, C. Effects of Pulsed Electromagnetic Field Treatment on Skeletal Muscle Tissue Recovery in a Rat Model of Collagenase-Induced Tendinopathy: Results from a Proteome Analysis. Int. J. Mol. Sci. 2024, 25, 8852. https://doi.org/10.3390/ijms25168852
Torretta E, Moriggi M, Capitanio D, Orfei CP, Raffo V, Setti S, Cadossi R, de Girolamo L, Gelfi C. Effects of Pulsed Electromagnetic Field Treatment on Skeletal Muscle Tissue Recovery in a Rat Model of Collagenase-Induced Tendinopathy: Results from a Proteome Analysis. International Journal of Molecular Sciences. 2024; 25(16):8852. https://doi.org/10.3390/ijms25168852
Chicago/Turabian StyleTorretta, Enrica, Manuela Moriggi, Daniele Capitanio, Carlotta Perucca Orfei, Vincenzo Raffo, Stefania Setti, Ruggero Cadossi, Laura de Girolamo, and Cecilia Gelfi. 2024. "Effects of Pulsed Electromagnetic Field Treatment on Skeletal Muscle Tissue Recovery in a Rat Model of Collagenase-Induced Tendinopathy: Results from a Proteome Analysis" International Journal of Molecular Sciences 25, no. 16: 8852. https://doi.org/10.3390/ijms25168852
APA StyleTorretta, E., Moriggi, M., Capitanio, D., Orfei, C. P., Raffo, V., Setti, S., Cadossi, R., de Girolamo, L., & Gelfi, C. (2024). Effects of Pulsed Electromagnetic Field Treatment on Skeletal Muscle Tissue Recovery in a Rat Model of Collagenase-Induced Tendinopathy: Results from a Proteome Analysis. International Journal of Molecular Sciences, 25(16), 8852. https://doi.org/10.3390/ijms25168852







