Bacterial Production of CDKL5 Catalytic Domain: Insights in Aggregation, Internal Translation and Phosphorylation Patterns
Abstract
1. Introduction
2. Results
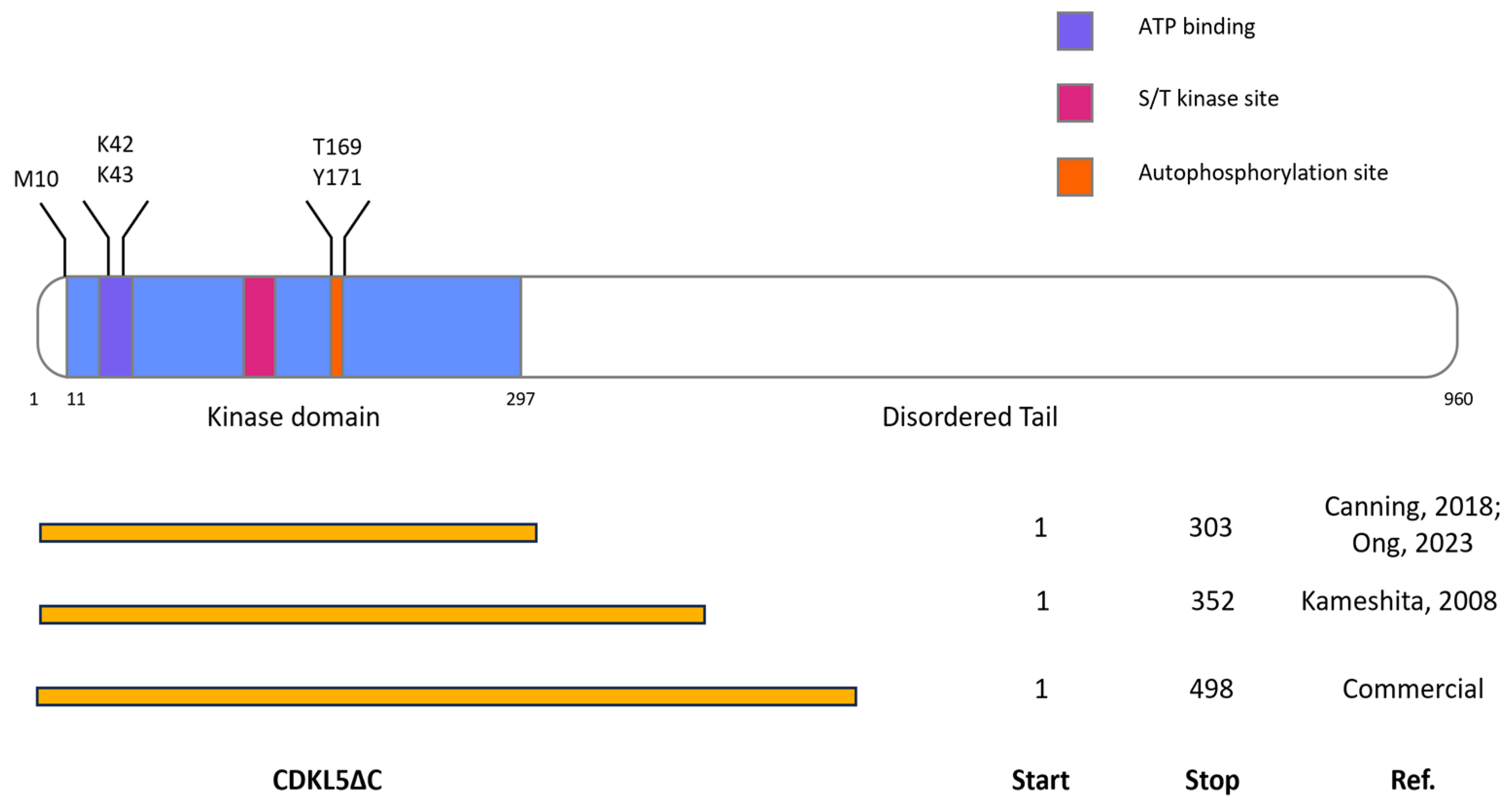
2.1. Evaluation of Multiple Variables for the Recombinant Production of CDKL5ΔC in E. coli
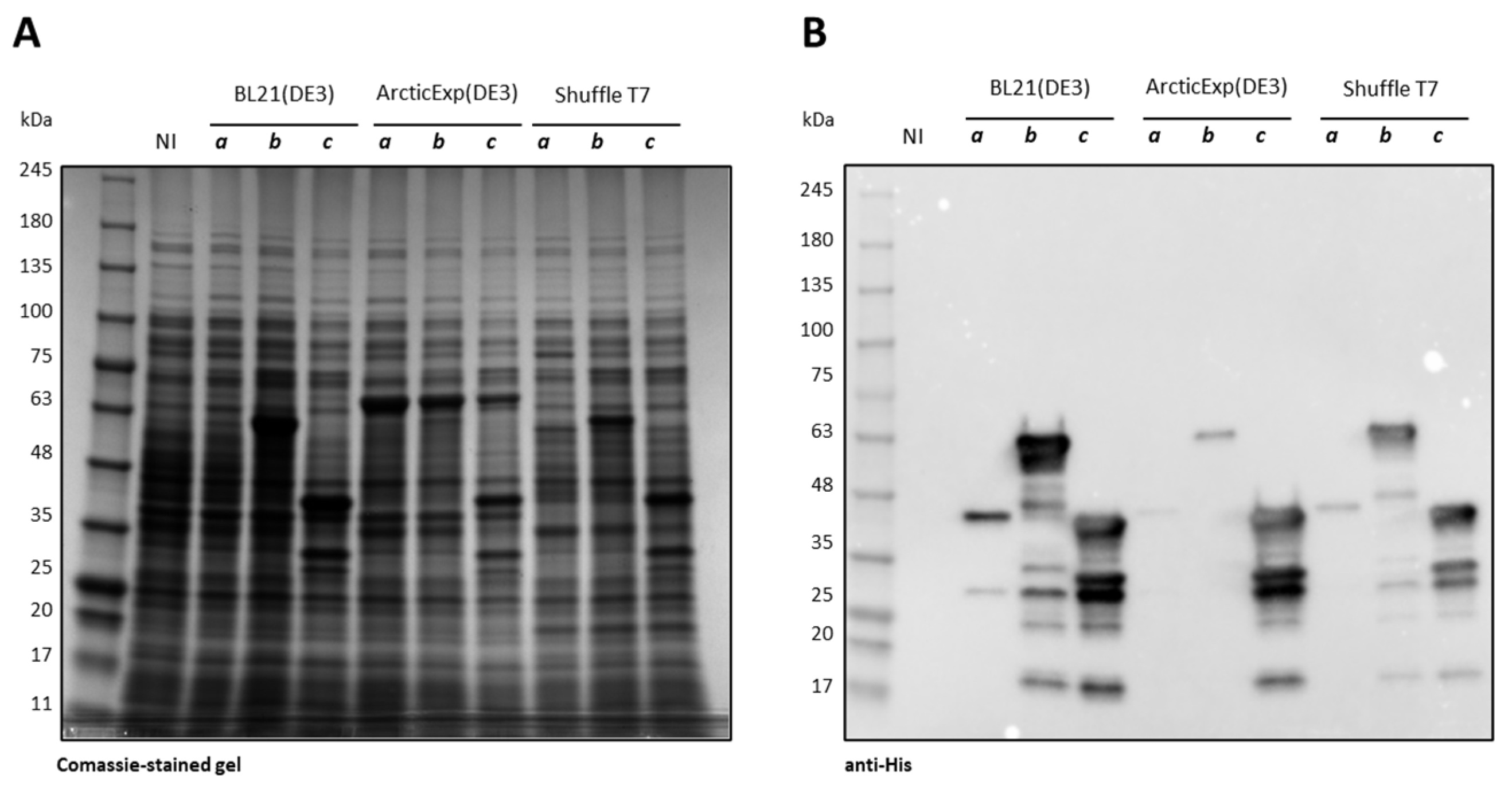
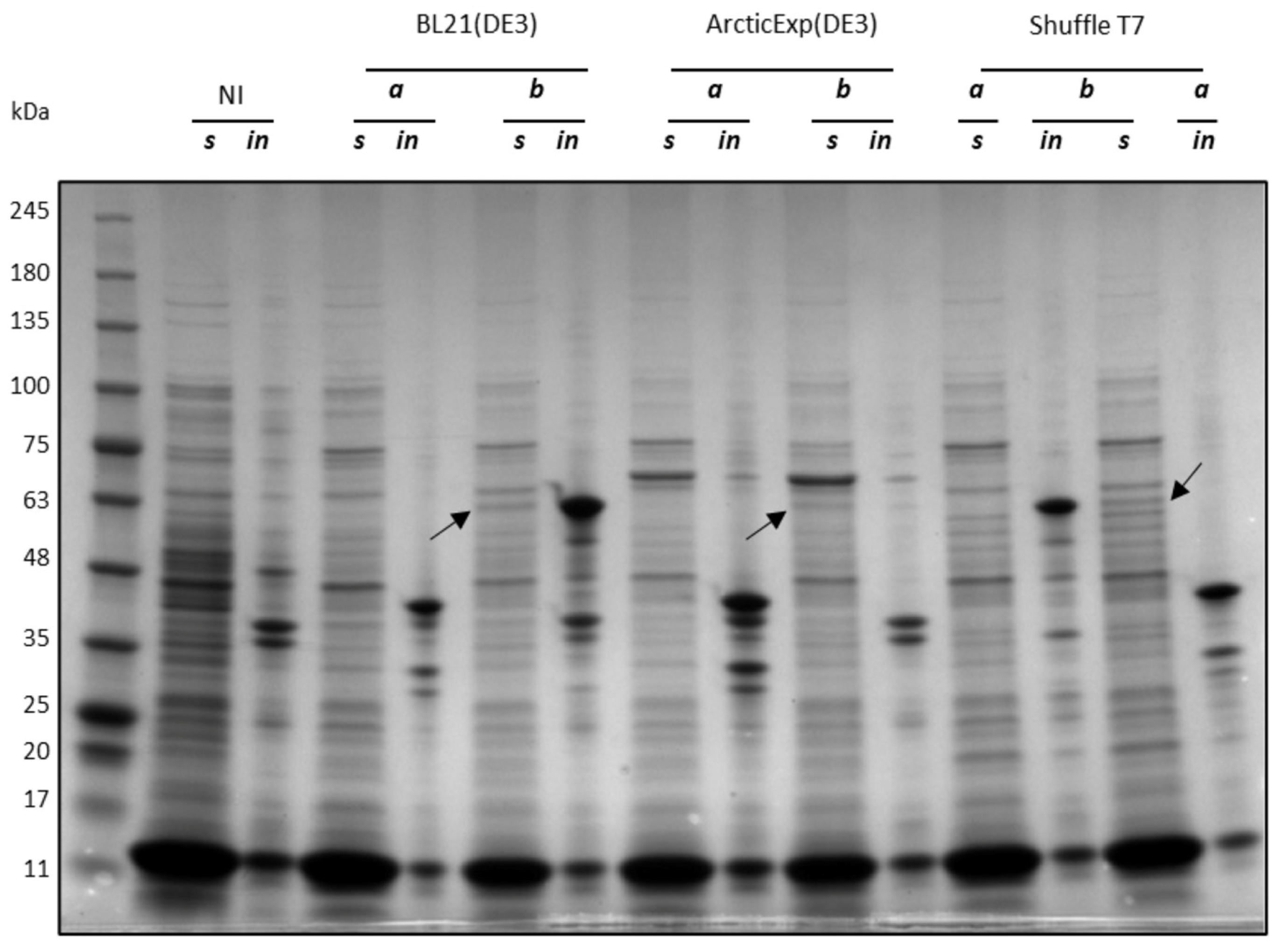
2.2. Influence of Codon Composition and Sumo Tag on E. coli Expression of CDKL5ΔC
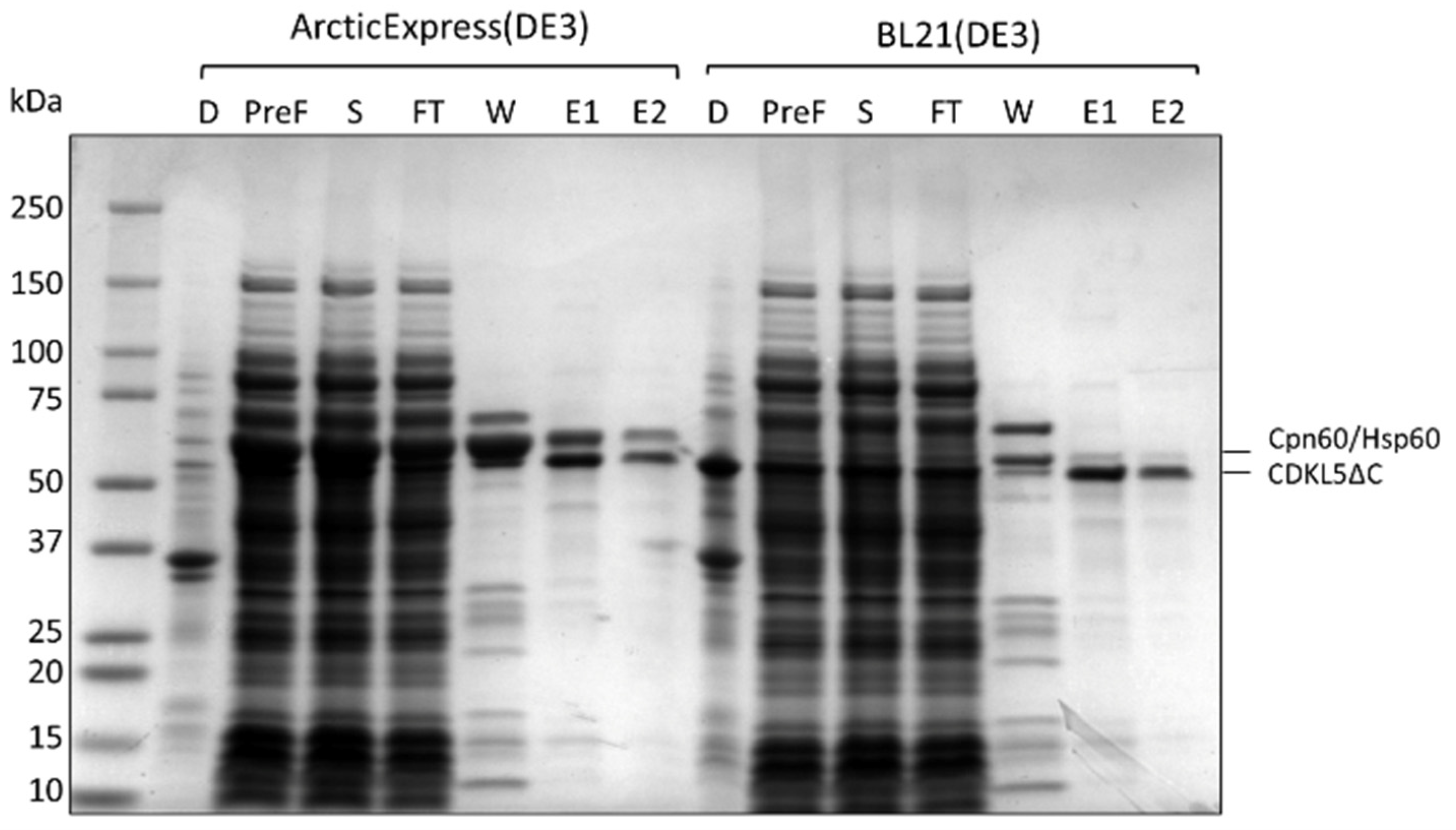
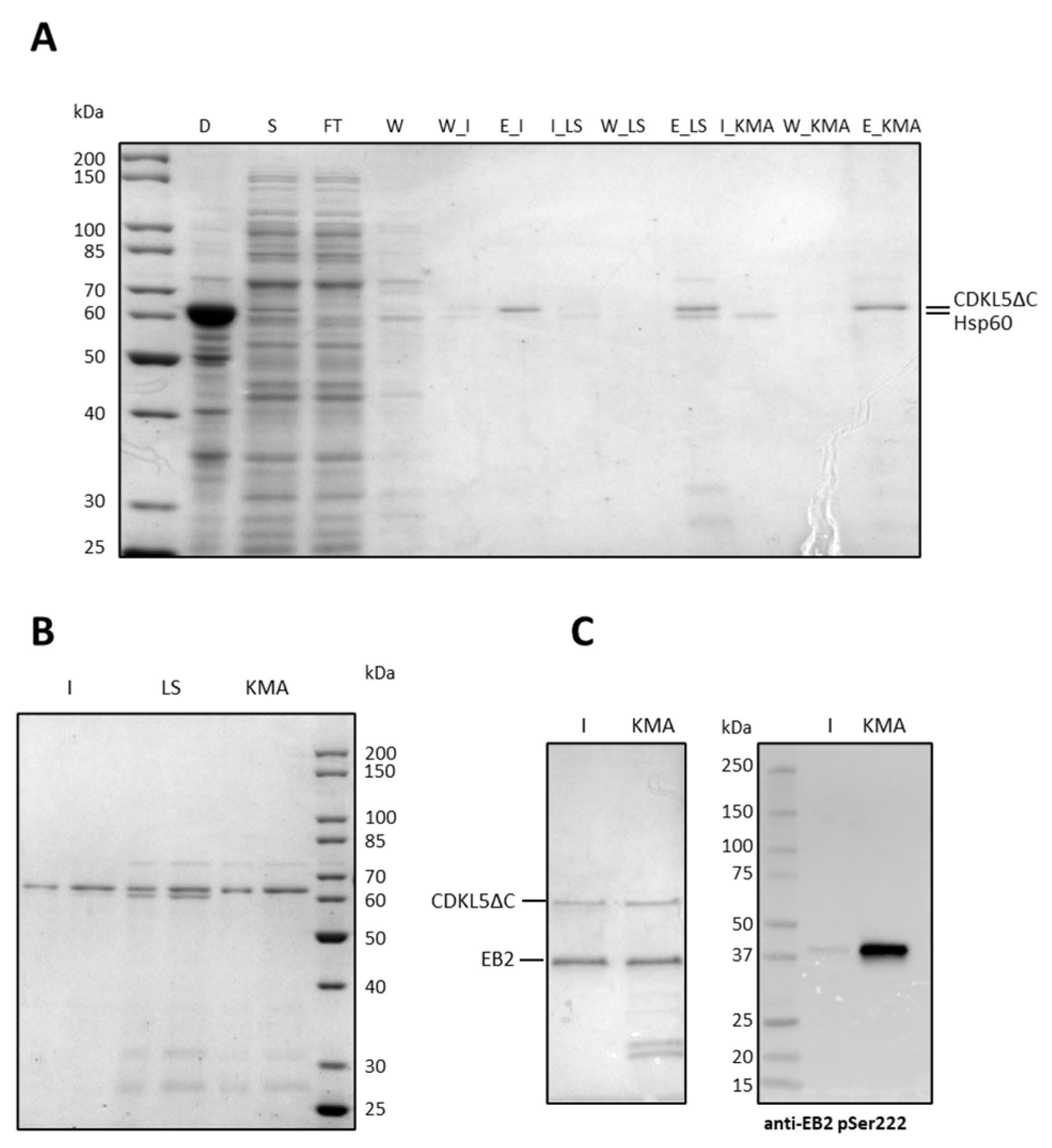
2.3. Purification of Ph_Sumo-Tat-CDKL5(1-352)-His M10V from BL21(DE3) and ArcticExpress(DE3) Strains in Native Conditions
2.4. Effect of the Co-Expression of E. coli Molecular Chaperons on CDKL5ΔC Solubility
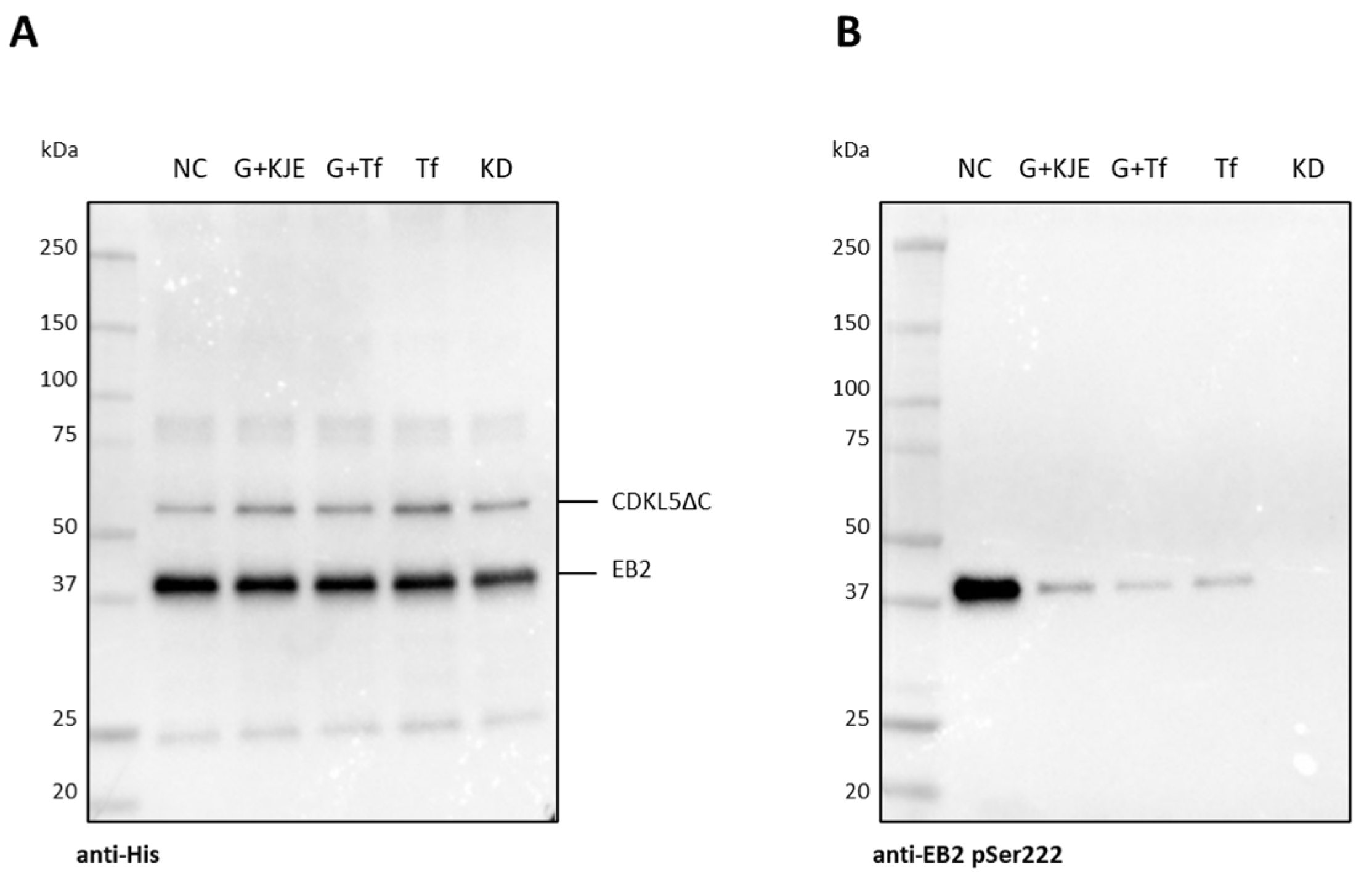
2.5. Effect of the Co-Expression of E. coli Molecular Chaperons on CDKL5ΔC Activity
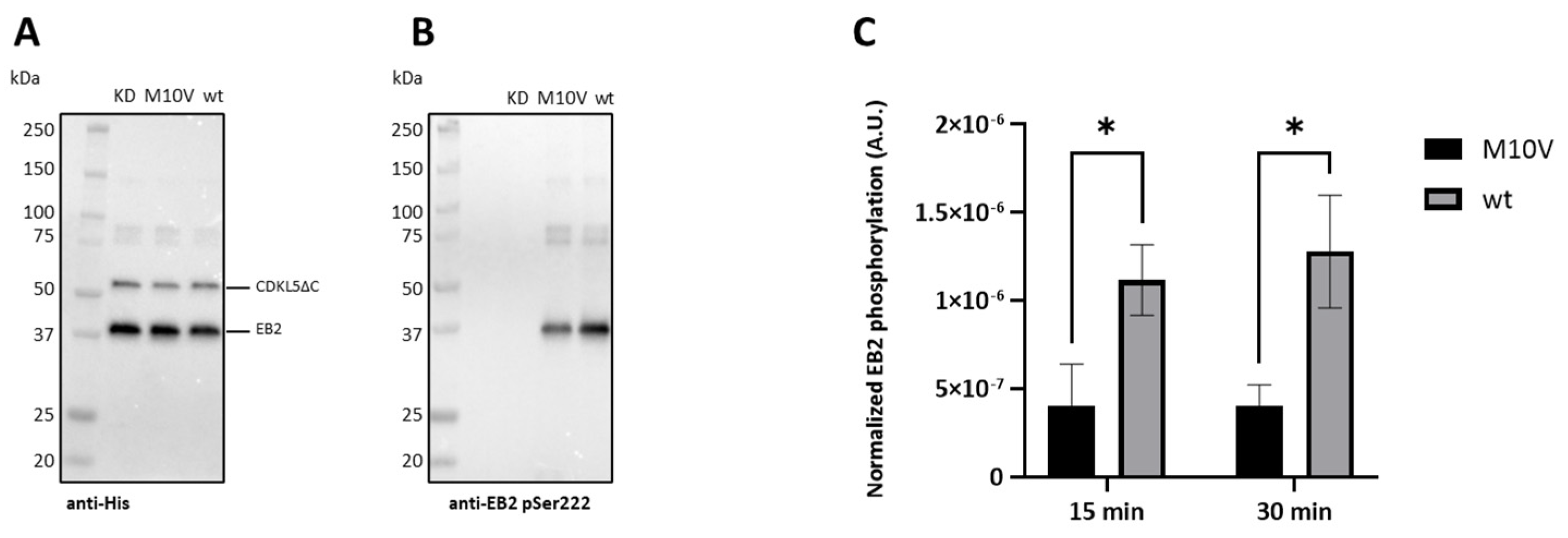
2.6. Analysis of CDKL5ΔC Autophosphorylation by Western Blotting
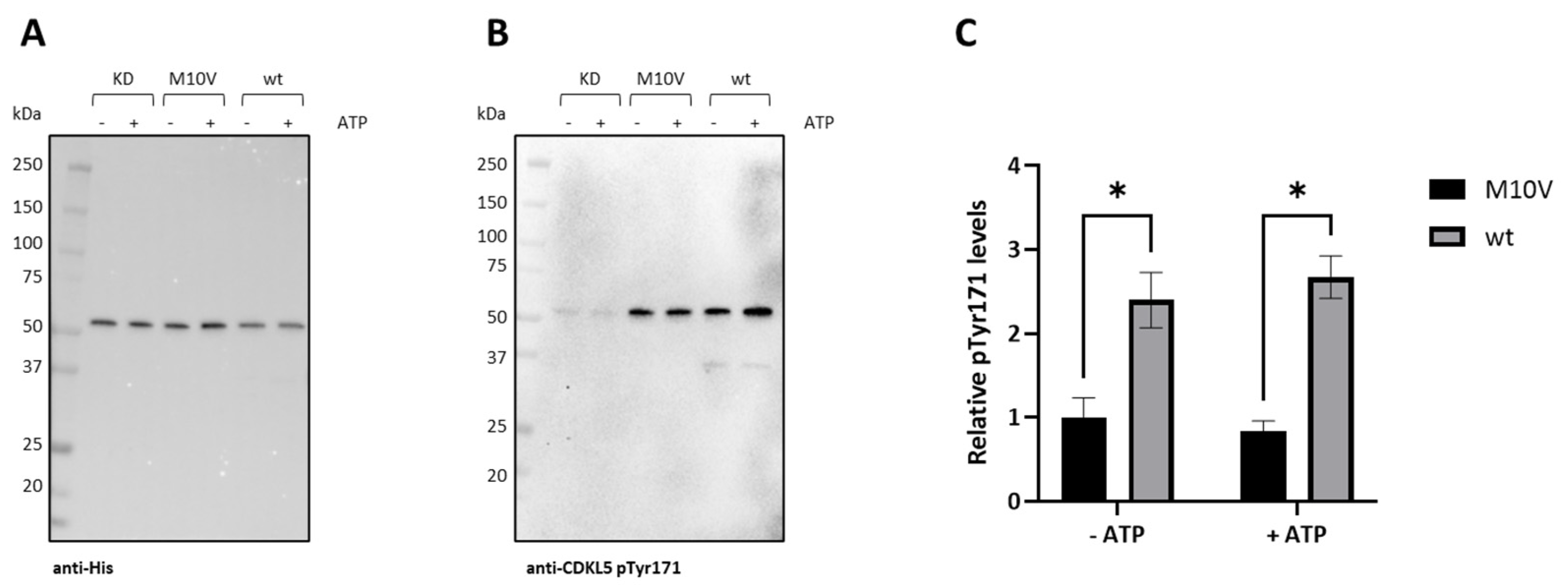
2.7. Analysis of CDKL5ΔC Autophosphorylation by Mass Spectrometry
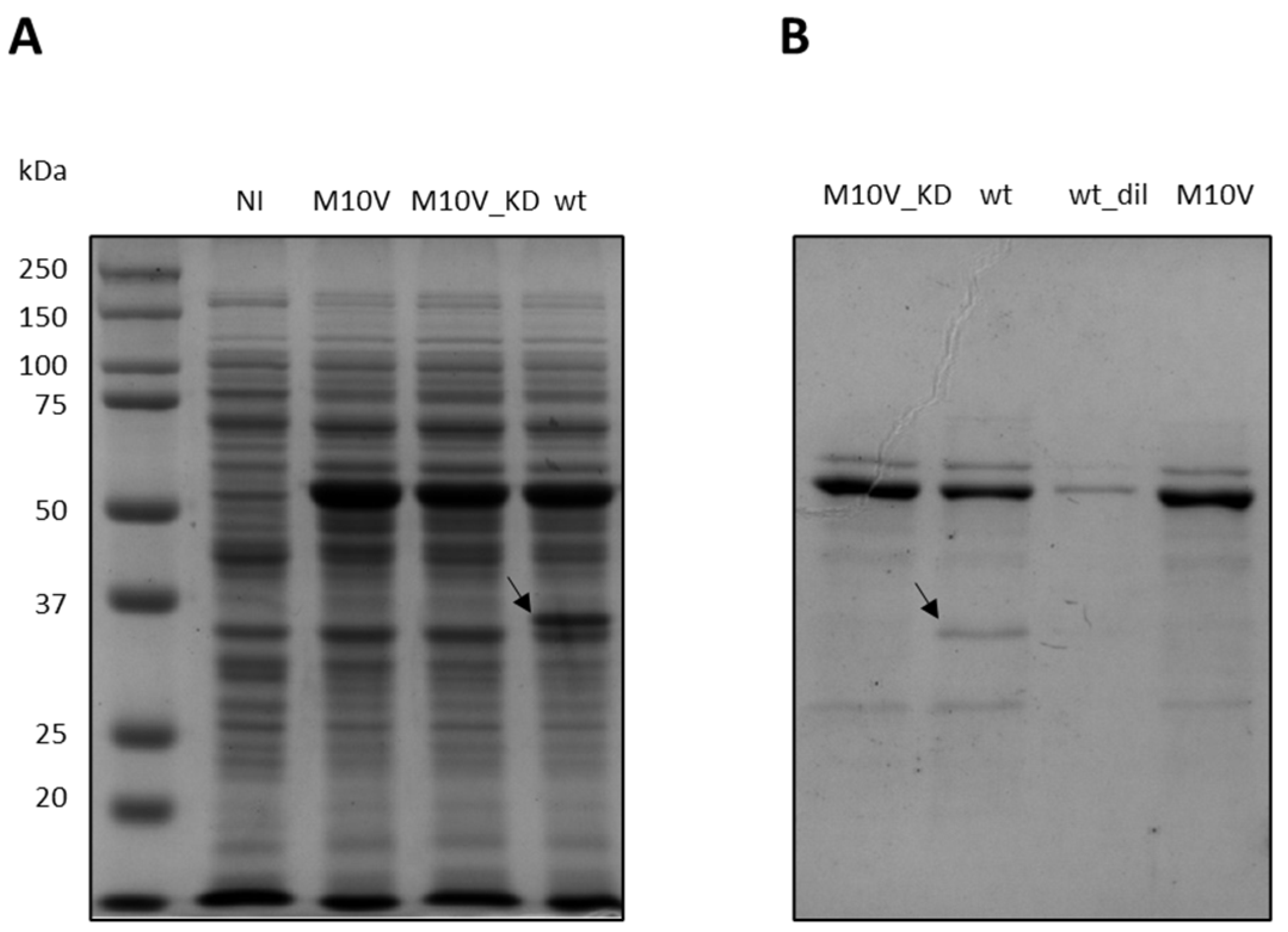
2.8. Effect of M10V Mutation on E. coli Expression of CDKL5ΔC and Catalytic Activity
2.9. Resolution of the Internal Translation Start with a Silent Mutation
2.10. Elimination of Hsp60 from CDKL5ΔC Preparations
3. Discussion
4. Materials and Methods
4.1. Cloning Procedures
4.2. Expression of CDKL5ΔC Variants in E. coli Strains
4.3. Analysis of the Solubility of CDKL5ΔC Variants
4.4. CDKL5ΔC Variants Purification via Native IMAC
4.5. Purification of Ec_Tat-CDKL5(1-352)-His from E. coli Aggregates
4.6. EB2 Recombinant Production and Purification
4.7. In Vitro Kinase Assays
4.8. Western Blot Analyses
4.9. Mass Spectrometry Analysis for Investigation of Recombinant Proteins Phosphorylation State
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Canning, P.; Park, K.; Gonçalves, J.; Li, C.; Howard, C.J.; Sharpe, T.D.; Holt, L.J.; Pelletier, L.; Bullock, A.N.; Leroux, M.R. CDKL family kinases have evolved distinct structural features and ciliary function. Cell Rep. 2018, 22, 885–894. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Franco, B.; Rosner, M.R. CDKL5/Stk9 kinase inactivation is associated with neuronal developmental disorders. Hum. Mol. Genet. 2005, 14, 3775–3786. [Google Scholar] [CrossRef] [PubMed]
- Bertani, I.; Rusconi, L.; Bolognese, F.; Forlani, G.; Conca, B.; De Monte, L.; Badaracco, G.; Landsberger, N.; Kilstrup-Nielsen, C. Functional consequences of mutations in CDKL5, an X-linked gene involved in infantile spasms and mental retardation. J. Biol. Chem. 2006, 281, 32048–32056. [Google Scholar] [CrossRef] [PubMed]
- Broekhuis, J.R.; Verhey, K.J.; Jansen, G. Regulation of cilium length and intraflagellar transport by the RCK-kinases ICK and MOK in renal epithelial cells. PLoS ONE 2014, 9, e108470. [Google Scholar] [CrossRef] [PubMed]
- Tam, L.W.; Ranum, P.T.; Lefebvre, P.A. CDKL5 regulates flagellar length and localizes to the base of the flagella in Chlamydomonas. Mol. Biol. Cell 2013, 24, 588–600. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Li, C.; Tsiropoulou, S.; Gonçalves, J.; Kondratev, C.; Pelletier, L.; Blacque, O.E.; Leroux, M.R. CDKL kinase regulates the length of the ciliary proximal segment. Curr. Biol. 2021, 31, 2359–2373.e7. [Google Scholar] [CrossRef] [PubMed]
- Maurya, A.K.; Sengupta, P. The DYF-5 RCK and CDKL-1 CDKL5 kinases contribute differentially to shape distinct sensory neuron cilia morphologies in C. elegans. microPublication Biol. 2022, 2022. [Google Scholar] [CrossRef]
- Silvestre, M.; Dempster, K.; Mihaylov, S.R.; Claxton, S.; Ultanir, S.K. Cell type-specific expression, regulation and compensation of CDKL5 activity in mouse brain. Mol. Psychiatry 2024, 1–13. [Google Scholar] [CrossRef]
- Berginski, M.E.; Moret, N.; Liu, C.; Goldfarb, D.; Sorger, P.K.; Gomez, S.M. The Dark Kinase Knowledgebase: An online compendium of knowledge and experimental results of understudied kinases. Nucleic Acids Res. 2021, 49, D529–D535. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Hector, R.D.; Kalscheuer, V.M.; Hennig, F.; Leonard, H.; Downs, J.; Clarke, A.; Benke, T.A.; Armstrong, J.; Pineda, M.; Bailey, M.E.S.; et al. CDKL5 variants. Neurol. Genet. 2017, 3, e200. [Google Scholar] [CrossRef]
- Krishnaraj, R.; Ho, G.; Christodoulou, J. RettBASE: Rett syndrome database update. Hum. Mutat. 2017, 38, 922–931. [Google Scholar] [CrossRef]
- Rusconi, L.; Salvatoni, L.; Giudici, L.; Bertani, I.; Kilstrup-Nielsen, C.; Broccoli, V.; Landsberger, N. CDKL5 expression is modulated during neuronal development and its subcellular distribution is tightly regulated by the C-terminal tail. J. Biol. Chem. 2008, 283, 30101–30111. [Google Scholar] [CrossRef]
- Chen, Q.; Zhu, Y.; Yu, J.; Miao, S.; Zheng, J.; Xu, L.; Zhou, Y.; Li, D.; Zhang, C.; Tao, J.; et al. CDKL5, a Protein Associated with Rett Syndrome, Regulates Neuronal Morphogenesis via Rac1 Signaling. J. Neurosci. 2010, 30, 12777–12786. [Google Scholar] [CrossRef]
- Hector, R.D.; Dando, O.; Landsberger, N.; Kilstrup-Nielsen, C.; Kind, P.C.; Bailey, M.E.S.; Cobb, S.R.C. Characterisation of CDKL5 Transcript Isoforms in Human and Mouse. PLoS ONE 2016, 11, e0157758. [Google Scholar] [CrossRef]
- Ricciardi, S.; Kilstrup-Nielsen, C.; Bienvenu, T.; Jacquette, A.; Landsberger, N.; Broccoli, V. CDKL5 influences RNA splicing activity by its association to the nuclear speckle molecular machinery. Hum. Mol. Genet. 2009, 18, 4590–4602. [Google Scholar] [CrossRef]
- Khanam, T.; Muñoz, I.; Weiland, F.; Carroll, T.; Morgan, M.; Borsos, B.N.; Pantazi, V.; Slean, M.; Novak, M.; Toth, R.; et al. CDKL5 kinase controls transcription-coupled responses to DNA damage. EMBO J. 2021, 40, e108271. [Google Scholar] [CrossRef]
- Ricciardi, S.; Ungaro, F.; Hambrock, M.; Rademacher, N.; Stefanelli, G.; Brambilla, D.; Sessa, A.; Magagnotti, C.; Bachi, A.; Giarda, E.; et al. CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat. Cell Biol. 2012, 14, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Kontaxi, C.; Ivanova, D.; Davenport, E.C.; Kind, P.C.; Cousin, M.A. Epilepsy-Related CDKL5 Deficiency Slows Synaptic Vesicle Endocytosis in Central Nerve Terminals. J. Neurosci. 2023, 43, 2002–2020. [Google Scholar] [CrossRef]
- Zhu, Y.C.; Li, D.; Wang, L.; Lu, B.; Zheng, J.; Zhao, S.L.; Zeng, R.; Xiong, Z.Q. Palmitoylation-dependent CDKL5-PSD-95 interaction regulates synaptic targeting of CDKL5 and dendritic spine development. Proc. Natl. Acad. Sci. USA 2013, 110, 9118–9123. [Google Scholar] [CrossRef] [PubMed]
- Baltussen, L.L.; Negraes, P.D.; Silvestre, M.; Claxton, S.; Moeskops, M.; Christodoulou, E.; Flynn, H.R.; Snijders, A.P.; Muotri, A.R.; Ultanir, S.K. Chemical genetic identification of CDKL 5 substrates reveals its role in neuronal microtubule dynamics. EMBO J. 2018, 37, e99763. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, I.M.; Morgan, M.E.; Peltier, J.; Weiland, F.; Gregorczyk, M.; Brown, F.C.; Macartney, T.; Toth, R.; Trost, M.; Rouse, J. Phosphoproteomic screening identifies physiological substrates of the CDKL 5 kinase. EMBO J. 2018, 37, e99559. [Google Scholar] [CrossRef] [PubMed]
- Katayama, S.; Inazu, T. Straightforward and rapid method for detection of cyclin-dependent kinase-like 5 activity. Anal. Biochem. 2019, 566, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Colarusso, A.; Lauro, C.; Calvanese, M.; Parrilli, E.; Tutino, M.L. Active human full-length CDKL5 produced in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Microb. Cell Fact. 2022, 21, 211. [Google Scholar] [CrossRef] [PubMed]
- Olson, H.E.; Demarest, S.T.; Pestana-Knight, E.M.; Swanson, L.C.; Iqbal, S.; Lal, D.; Leonard, H.; Cross, J.H.; Devinsky, O.; Benke, T.A. Cyclin-Dependent Kinase-Like 5 Deficiency Disorder: Clinical Review. Pediatr. Neurol. 2019, 97, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Kameshita, I.; Sekiguchi, M.; Hamasaki, D.; Sugiyama, Y.; Hatano, N.; Suetake, I.; Tajima, S.; Sueyoshi, N. Cyclin-dependent kinase-like 5 binds and phosphorylates DNA methyltransferase 1. Biochem. Biophys. Res. Commun. 2008, 377, 1162–1167. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, M.; Katayama, S.; Hatano, N.; Shigeri, Y.; Sueyoshi, N.; Kameshita, I. Identification of amphiphysin 1 as an endogenous substrate for CDKL5, a protein kinase associated with X-linked neurodevelopmental disorder. Arch. Biochem. Biophys. 2013, 535, 257–267. [Google Scholar] [CrossRef]
- Katayama, S.; Sueyoshi, N.; Kameshita, I. Critical determinants of substrate recognition by cyclin-dependent kinase-like 5 (CDKL5). Biochemistry 2015, 54, 2975–2987. [Google Scholar] [CrossRef]
- Fu, Z.; Larson, K.A.; Chitta, R.K.; Parker, S.A.; Turk, B.E.; Lawrence, M.W.; Kaldis, P.; Galaktionov, K.; Cohn, S.M.; Shabanowitz, J.; et al. Identification of Yin-Yang Regulators and a Phosphorylation Consensus for Male Germ Cell-Associated Kinase (MAK)-Related Kinase. Mol. Cell. Biol. 2006, 26, 8639–8654. [Google Scholar] [CrossRef]
- Albanese, S.K.; Parton, D.L.; Işlk, M.; Rodríguez-Laureano, L.; Hanson, S.M.; Behr, J.M.; Gradia, S.; Jeans, C.; Levinson, N.M.; Seeliger, M.A.; et al. An Open Library of Human Kinase Domain Constructs for Automated Bacterial Expression. Biochemistry 2018, 57, 4675–4689. [Google Scholar] [CrossRef] [PubMed]
- Piserchio, A.; Cowburn, D.; Ghose, R. Expression and purification of Src-family kinases for solution NMR studies. Methods Mol. Biol. 2012, 831, 111–131. [Google Scholar] [PubMed]
- Ong, H.W.; Liang, Y.; Richardson, W.; Lowry, E.R.; Wells, C.I.; Chen, X.; Silvestre, M.; Dempster, K.; Silvaroli, J.A.; Smith, J.L.; et al. Discovery of a Potent and Selective CDKL5/GSK3 Chemical Probe That Is Neuroprotective. ACS Chem. Neurosci. 2023, 14, 1672–1685. [Google Scholar] [CrossRef] [PubMed]
- Trazzi, S.; De Franceschi, M.; Fuchs, C.; Bastianini, S.; Viggiano, R.; Lupori, L.; Mazziotti, R.; Medici, G.; Martire, V.L.; Ren, E.; et al. CDKL5 protein substitution therapy rescues neurological phenotypes of a mouse model of CDKL5 disorder. Hum. Mol. Genet. 2018, 27, 1572–1592. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, Y.; Morii, A.; Fujino, Y.; Nagase, M.; Kitano, A.; Ueno, S.; Takeuchi, K.; Yamashita, R.; Inazu, T. Comprehensive In Silico Functional Prediction Analysis of CDKL5 by Single Amino Acid Substitution in the Catalytic Domain. Int. J. Mol. Sci. 2022, 23, 12281. [Google Scholar] [CrossRef]
- Salis, H.M.; Mirsky, E.A.; Voigt, C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009, 27, 946–950. [Google Scholar] [CrossRef] [PubMed]
- Bolanos-Garcia, V.M.; Davies, O.R. Structural analysis and classification of native proteins from E. coli commonly co-purified by immobilised metal affinity chromatography. Biochim. Biophys. Acta-Gen. Subj. 2006, 1760, 1304–1313. [Google Scholar] [CrossRef] [PubMed]
- Roobol, A.; Grantham, J.; Whitaker, H.C.; Carden, M.J. Disassembly of the cytosolic chaperonin in mammalian cell extracts at intracellular levels of K+ and ATP. J. Biol. Chem. 1999, 274, 19220–19227. [Google Scholar] [CrossRef] [PubMed]
- Leonard, H.; Whitehouse, A.; Jacoby, P.; Benke, T.; Demarest, S.; Saldaris, J.; Wong, K.; Reddihough, D.; Williams, K.; Downs, J. Quality of life beyond diagnosis in intellectual disability—Latent profiling. Res. Dev. Disabil. 2022, 129, 104322. [Google Scholar] [CrossRef] [PubMed]
- de Marco, A.; Deuerling, E.; Mogk, A.; Tomoyasu, T.; Bukau, B. Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 2007, 7, 32. [Google Scholar] [CrossRef]
- Joseph, R.E.; Andreotti, A.H. Bacterial expression and purification of Interleukin-2 Tyrosine kinase: Single step separation of the chaperonin impurity. Protein Expr. Purif. 2008, 60, 194–197. [Google Scholar] [CrossRef]
- Haacke, A.; Fendrich, G.; Ramage, P.; Geiser, M. Chaperone over-expression in Escherichia coli: Apparent increased yields of soluble recombinant protein kinases are due mainly to soluble aggregates. Protein Expr. Purif. 2009, 64, 185–193. [Google Scholar] [CrossRef]
- Kim, J.Y.; Bai, Y.; Jayne, L.A.; Hector, R.D.; Persaud, A.K.; Ong, S.S.; Rojesh, S.; Raj, R.; Feng, M.J.H.H.; Chung, S.; et al. A kinome-wide screen identifies a CDKL5-SOX9 regulatory axis in epithelial cell death and kidney injury. Nat. Commun. 2020, 11, 1924. [Google Scholar] [CrossRef]
- Gracia, L.; Lora, G.; Blair, L.J.; Jinwal, U.K. Therapeutic Potential of the Hsp90/Cdc37 Interaction in Neurodegenerative Diseases. Front. Neurosci. 2019, 13, 1–8. [Google Scholar] [CrossRef]
- Taipale, M.; Krykbaeva, I.; Koeva, M.; Kayatekin, C.; Westover, K.D.; Karras, G.I.; Lindquist, S. Quantitative analysis of Hsp90-client interactions reveals principles of substrate recognition. Cell 2012, 150, 987–1001. [Google Scholar] [CrossRef]
- Verba, K.A.; Agard, D.A. How Hsp90 and Cdc37 Lubricate Kinase Molecular Switches. Trends Biochem. Sci. 2017, 42, 799–811. [Google Scholar] [CrossRef]
- Murtaza, N.; Cheng, A.A.; Brown, C.O.; Meka, D.P.; Hong, S.; Uy, J.A.; El-Hajjar, J.; Pipko, N.; Unda, B.K.; Schwanke, B.; et al. Neuron-specific protein network mapping of autism risk genes identifies shared biological mechanisms and disease-relevant pathologies. Cell Rep. 2022, 41, 111678. [Google Scholar] [CrossRef] [PubMed]
- Butturini, E.; Gotte, G.; Dell’Orco, D.; Chiavegato, G.; Marino, V.; Canetti, D.; Cozzolino, F.; Monti, M.; Pucci, P.; Mariotto, S. Intermolecular disulfide bond influences unphosphorylated STAT3 dimerization and function. Biochem. J. 2016, 473, 3205–3219. [Google Scholar] [CrossRef]
- Zanca, C.; Cozzolino, F.; Quintavalle, C.; Di Costanzo, S.; Ricci-Vitiani, L.; Santoriello, M.; Monti, M.; Pucci, P.; Condorelli, G. PED interacts with Rac1 and regulates cell migration/invasion processes in human non-small cell lung cancer cells. J. Cell. Physiol. 2010, 225, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Di Sanzo, M.; Cozzolino, F.; Battaglia, A.M.; Aversa, I.; Monaco, V.; Sacco, A.; Biamonte, F.; Palmieri, C.; Procopio, F.; Santamaria, G.; et al. Ferritin Heavy Chain Binds Peroxiredoxin 6 and Inhibits Cell Proliferation and Migration. Int. J. Mol. Sci. 2022, 23, 12987. [Google Scholar] [CrossRef]
- Salvia, R.; Cozzolino, F.; Scieuzo, C.; Grimaldi, A.; Franco, A.; Vinson, S.B.; Monti, M.; Falabella, P. Identification and Functional Characterization of Toxoneuron nigriceps Ovarian Proteins Involved in the Early Suppression of Host Immune Response. Insects 2022, 13, 144. [Google Scholar] [CrossRef] [PubMed]
- Federico, A.; Sepe, R.; Cozzolino, F.; Piccolo, C.; Iannone, C.; Iacobucci, I.; Pucci, P.; Monti, M.; Fusco, A. The complex CBX7-PRMT1 has a critical role in regulating E-cadherin gene expression and cell migration. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, F.; Iacobucci, I.; Monaco, V.; Angrisano, T.; Monti, M. Lysines Acetylome and Methylome Profiling of H3 and H4 Histones in Trichostatin A-Treated Stem Cells. Int. J. Mol. Sci. 2021, 22, 2063. [Google Scholar] [CrossRef] [PubMed]



| Boundaries | Codon Composition | Mutations | Solubility Tags | Chaperones | Expression Temperature | Lysate Fraction |
|---|---|---|---|---|---|---|
| 1–303 1–352 1–498 | Escherichia coli (Ec) Pseudoalteromonas haloplanktis TAC125 (Ph) Manual optimization | M10V KK42,43RR | Sumo GST | Cpn60/10 DsbC GroELS DnaK DnaJ GrpE Trigger factor | 15–37 °C | Soluble Insoluble |
| Chaperones | Volumetric Yield (mg Protein/L of Culture) |
|---|---|
| None | 6.21 |
| GroELS, DnaK, DnaJ, GrpE | 13.75 |
| GroELS, Tf | 7.31 |
| Tf | 20.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colarusso, A.; Lauro, C.; Canè, L.; Cozzolino, F.; Tutino, M.L. Bacterial Production of CDKL5 Catalytic Domain: Insights in Aggregation, Internal Translation and Phosphorylation Patterns. Int. J. Mol. Sci. 2024, 25, 8891. https://doi.org/10.3390/ijms25168891
Colarusso A, Lauro C, Canè L, Cozzolino F, Tutino ML. Bacterial Production of CDKL5 Catalytic Domain: Insights in Aggregation, Internal Translation and Phosphorylation Patterns. International Journal of Molecular Sciences. 2024; 25(16):8891. https://doi.org/10.3390/ijms25168891
Chicago/Turabian StyleColarusso, Andrea, Concetta Lauro, Luisa Canè, Flora Cozzolino, and Maria Luisa Tutino. 2024. "Bacterial Production of CDKL5 Catalytic Domain: Insights in Aggregation, Internal Translation and Phosphorylation Patterns" International Journal of Molecular Sciences 25, no. 16: 8891. https://doi.org/10.3390/ijms25168891
APA StyleColarusso, A., Lauro, C., Canè, L., Cozzolino, F., & Tutino, M. L. (2024). Bacterial Production of CDKL5 Catalytic Domain: Insights in Aggregation, Internal Translation and Phosphorylation Patterns. International Journal of Molecular Sciences, 25(16), 8891. https://doi.org/10.3390/ijms25168891









