Transcriptome Profiles Reveal Key Regulatory Networks during Single and Multifactorial Stresses Coupled with Melatonin Treatment in Pitaya (Selenicereus undatus L.)
Abstract
:1. Introduction
2. Results
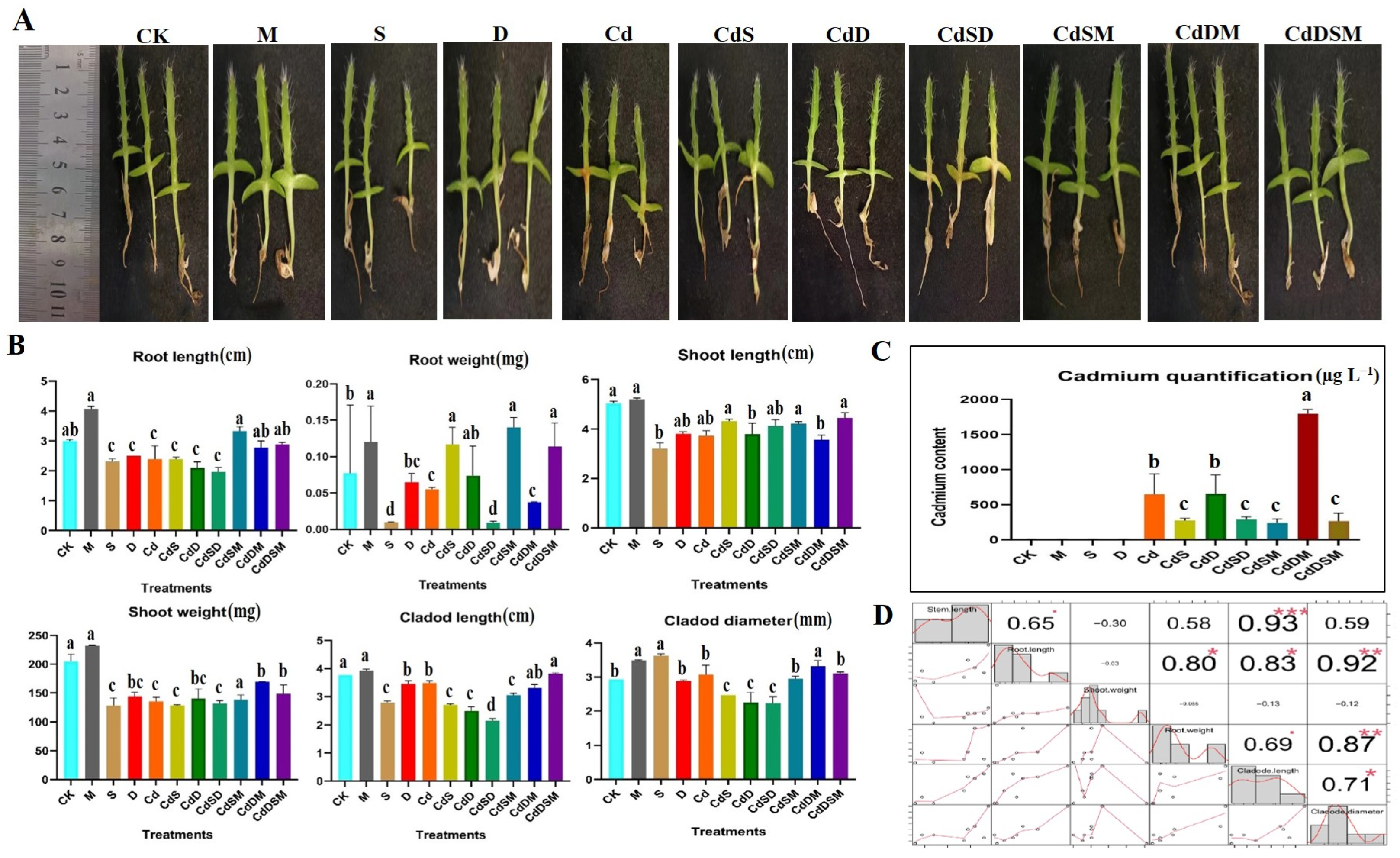
2.1. Phenotypic Characteristics and Plant Material Under Single and Combination Stressors
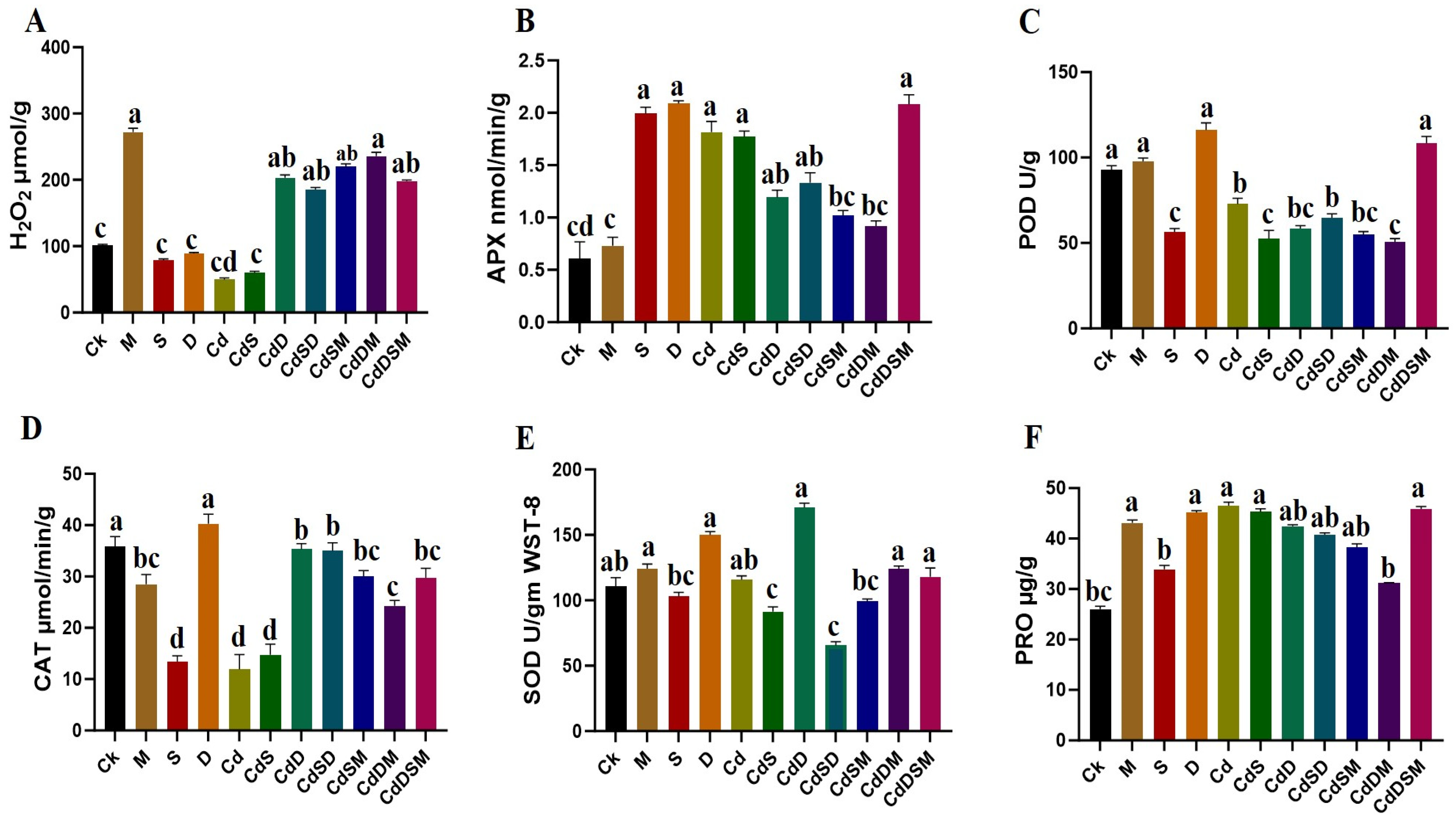
2.2. Biochemical Responses to Single and Combined Stresses
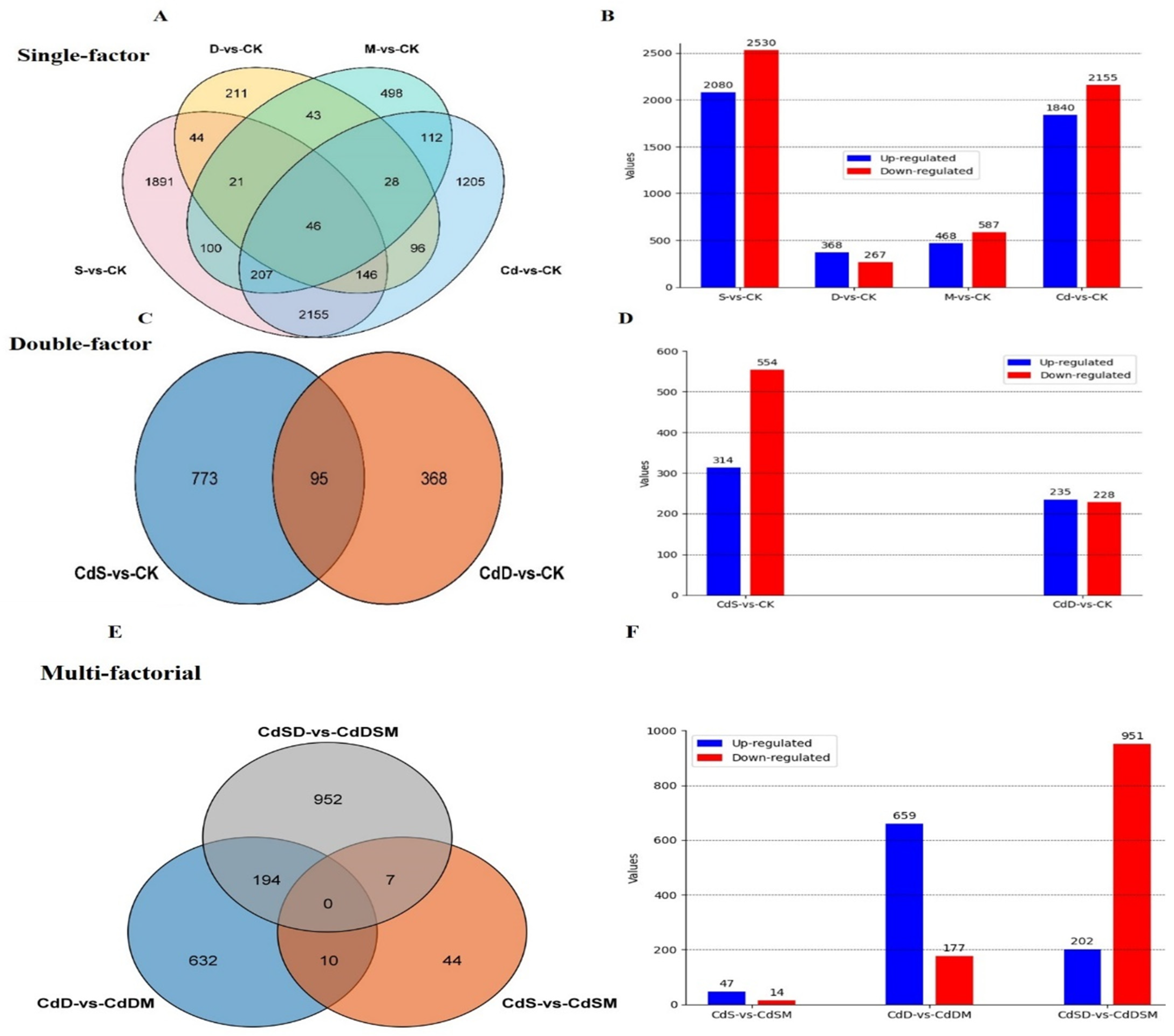
2.3. Transcriptome Outcome of Pitaya
2.4. Gene Expression Analysis of Pitaya Seedlings to Single, Double, and Multifactorial Stresses
2.5. Gene GO & KEGG Pathway Enrichment Analysis of DEGs
2.6. Identification of DEGs Associated with Plant Hormones and Signal Transduction
2.7. Expression of Genes Involved in Metabolism and Biosynthesis
2.8. Photosynthesis-Related Genes Under Single and Combined Stresses
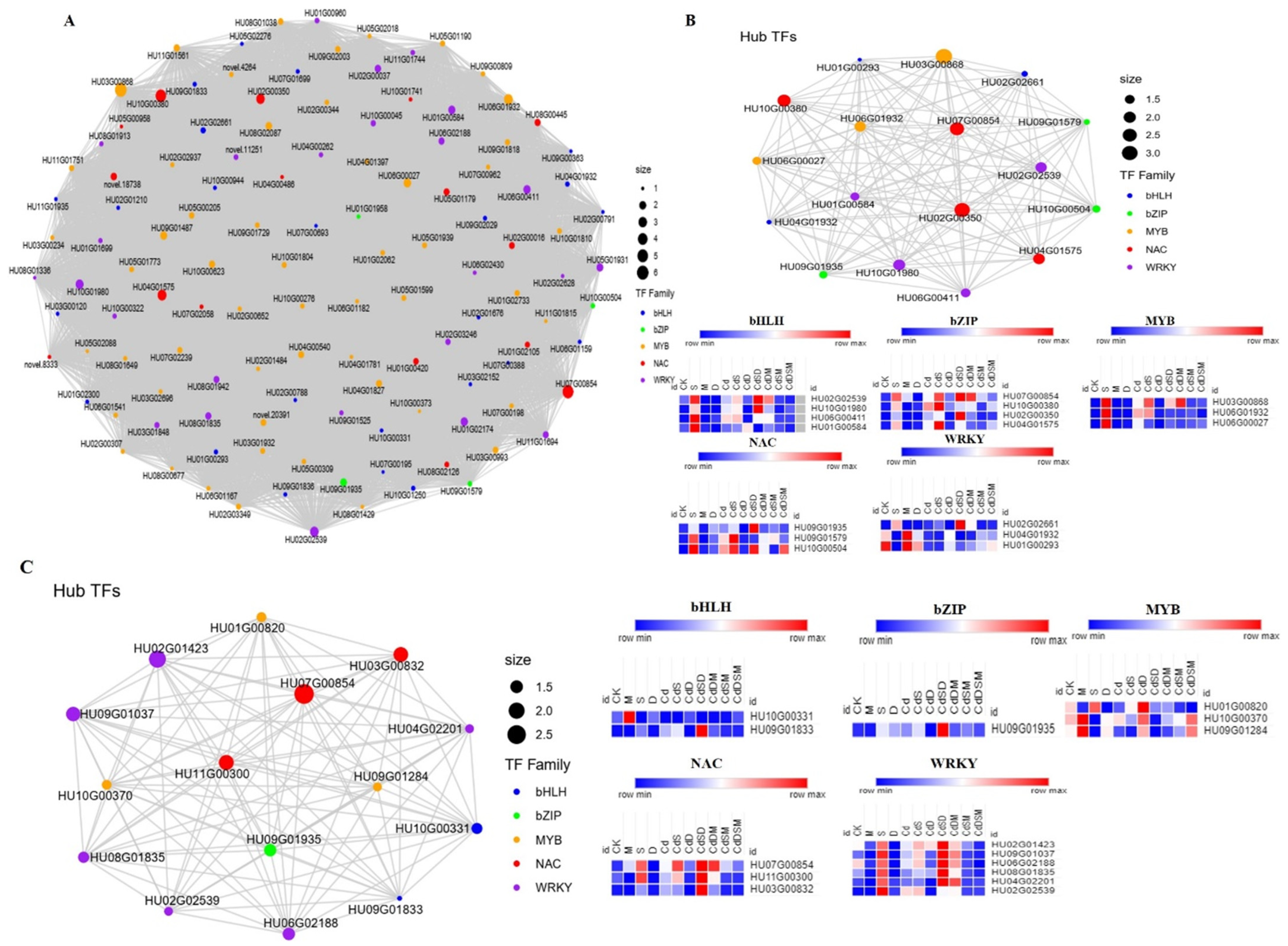
2.9. Transcription Factors Responding to Single, Double, and Multifactorial Stresses
2.10. Melatonin Induces ROS-Scavenging and Circadian Clock Genes
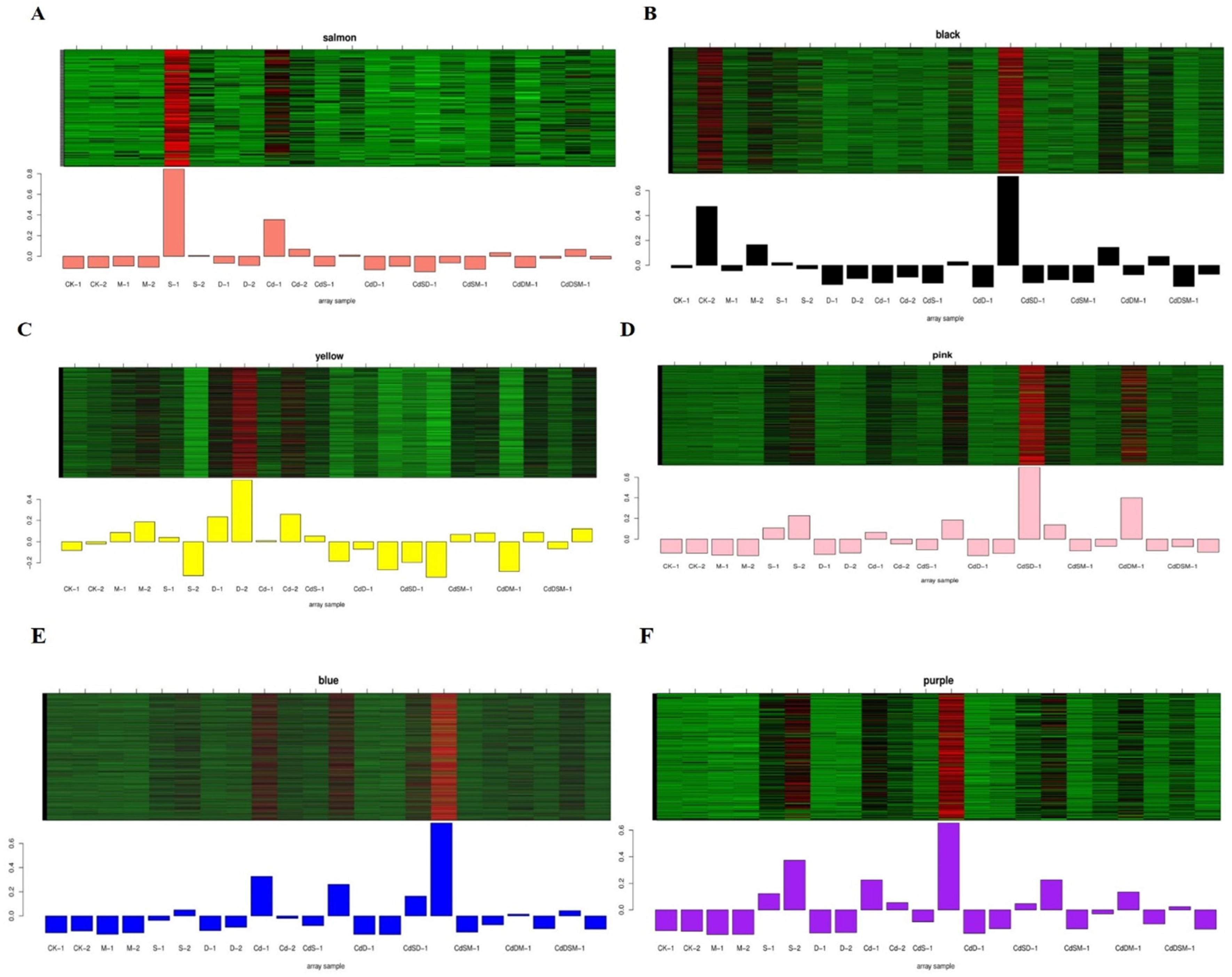
2.11. Co-Expression Network Construction and Identification of WGCNA Modules
2.12. Identification of Hub TFs and Network Construction
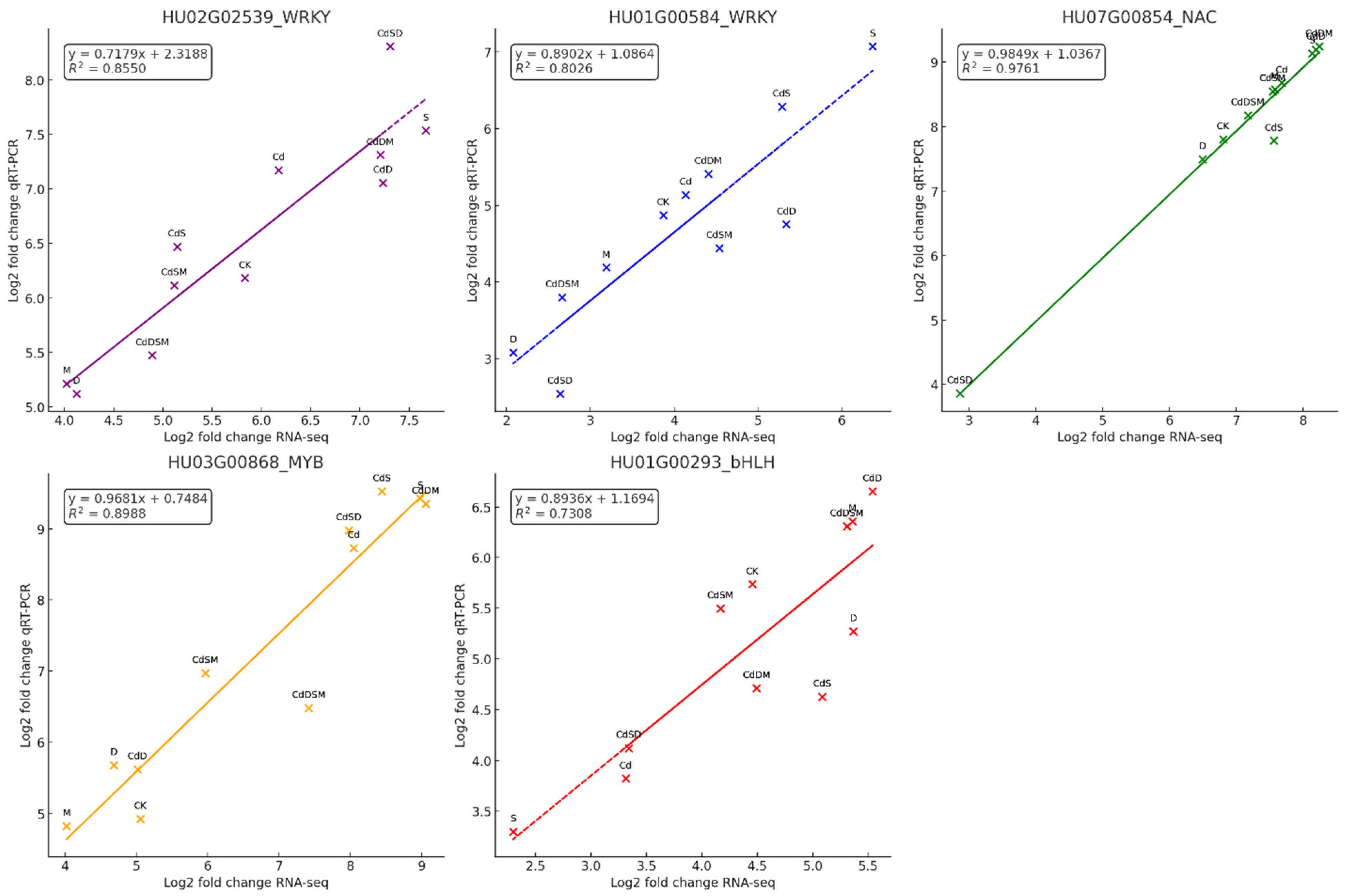
2.13. Validation of the DEGs by qRT-PCR
3. Discussion
3.1. The Effect of Stress on Plant Performance Is Enhanced by Melatonin
3.2. The Common and Unique Transcriptome Response and Cross-Talk of Single and Multiple Stresses
3.3. Enhanced Antioxidant Activities in Pitaya Under Combined-Stress Conditions
3.4. Transcription Factors Involved in the Response to Single and Combined Stress
3.5. Phytohormone Signals Under Single and Combined Stresses
3.6. Phytohormone Signals Under Single and Combined Stresses
3.7. Signaling Pathway Mediates Single- and Combined-Stress Responses
3.8. Metabolism Response to Single and Combined Stresses
4. Materials and Methods
4.1. Plant Materials and Stress Treatments and Melatonin Application
4.2. Phenotyping and Quantification of Cd Content
4.3. RNA Extraction, cDNA Library Construction, and Sequencing
4.4. De Novo Assembly, and Annotation
4.5. Identification of Differential Gene Expression (DEGs)
4.6. Enrichment Analysis of DEGs
4.7. Identification of Hub Genes and Sub-Networks Associated with Treatments
4.8. Enzymatic Assay
4.9. RNA Extraction and qRT-PCR
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| M | melatonin |
| D | drought |
| S | salinity |
| Cd | cadmium |
| Ck | control |
| CdD | cadmium + drought |
| CdS | cadmium + salinity |
| CdSD | cadmium + salinity + drought |
| CdDSM | cadmium + drought + salinity + melatonin |
| CdSM | cadmium + salinity + melatonin |
| CdDM | cadmium + drought + melatonin |
| CdDSM | cadmium + drought + salinity + melatonin |
| S. undatus | Selenicereus undatus |
| POD | peroxidase |
| CAT | catalase |
| APX | ascorbate peroxidase |
| SOD | superoxide dismutase |
| GR | glutathione reductase |
| H2O2 | hydrogen peroxide |
| WRKY | WRKY transcription factor |
| MYB | myeloblastosis transcription factor |
| bHLH | basic-helix–loop-helix transcription factor |
| bZIP | basic leucine zipper transcription factor |
| AP2/ERF | APETALA2/ethylene-responsive factor transcription factor |
| NAC | NAM, ATAF, and CUC |
| ABA | abscisic acid |
| JA | jasmonic acid |
| FPKM | fragments per kilobase of transcript per million mapped reads |
| GA | gibberellic acid |
| SA | Salicylic acid |
| DEGs | differentially expressed genes |
| GO | gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| WGCNA | weighted gene co-expression network analysis |
| TFs | transcription factors |
References
- Angulo-Bejarano, P.I.; Puente-Rivera, J.; Cruz-Ortega, R. Metal and metalloid toxicity in plants: An overview on molecular aspects. Plants 2021, 10, 635. [Google Scholar] [CrossRef]
- Bostock, R.M.; Pye, M.F.; Roubtsova, T.V. Predisposition in plant disease: Exploiting the nexus in abiotic and biotic stress perception and response. Annu. Rev. Phytopathol. 2014, 52, 517–549. [Google Scholar] [CrossRef]
- Chen, C.; Li, F.; Xie, F.; Chen, J.; Hua, Q.; Chen, J.; Wu, Z.; Zhang, Z.; Zhang, R.; Zhao, J.; et al. Pitaya Genome and Multiomics Database (PGMD): A Comprehensive and Integrative Resource of Selenicereus undatus. Genes 2022, 13, 745. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.P.; Leach, J.E. Abiotic and biotic stresses induce a core transcriptome response in rice. Sci. Rep. 2019, 9, 6273. [Google Scholar] [CrossRef] [PubMed]
- Zandalinas, S.I.; Fritschi, F.B.; Mittler, R. Global warming, climate change, and environmental pollution: Recipe for a multifactorial stress combination disaster. Trends Plant Sci. 2021, 26, 588–599. [Google Scholar] [CrossRef]
- Zaman, Q.U.; Garg, V.; Raza, A.; Nazir, M.F.; Hui, L.; Khan, D.; Varshney, R.K. Unique regulatory network of dragon fruit simultaneously mitigates the effect of vanadium pollutant and environmental factors. Physiol. Plant. 2024, 176, e14416. [Google Scholar] [CrossRef]
- Woldesemayat, A.A.; Modise, D.M.; Gemeildien, J.; Ndimba, B.K.; Christoffels, A. Cross-species multiple environmental stress responses: An integrated approach to identify candidate genes for multiple stress tolerance in sorghum (Sorghum bicolor (L.) Moench) and related model species. PLoS ONE 2018, 13, e0192678. [Google Scholar]
- Khokhar, A.A.; Hui, L.; Khan, D.; Hussain, M.A.; Lv, W.; Zaman, Q.U.; Wang, H.-F. Comprehensive characterization of SBP genes revealed their role under multifactorial stress combinations in dragon fruit (Selenicereus undatus L.). Plant Stress 2023, 10, 100294. [Google Scholar] [CrossRef]
- Xu, L.; Cao, M.; Wang, Q.; Xu, J.; Liu, C.; Ullah, N.; Li, J.; Hou, Z.; Liang, Z.; Zhou, W. Insights into the plateau adaptation of Salvia castanea by comparative genomic and WGCNA analyses. J. Adv. Res. 2022, 42, 221–235. [Google Scholar] [CrossRef]
- Khan, R.; Reddy, P.; Gupta, R. (Eds.) Advancements in Developing Abiotic Stress-Resilient Plants: Basic Mechanisms to Trait Improvements; CRC Press: Boca Raton, FL, USA, 2022. [Google Scholar] [CrossRef]
- Kumar, N.; Verma, S.; Kumar, A.; Dasila, H.; Suyal, D.C.; Kumari, G.; Kumar, A. Molecular insights into stress-responsive genes in the mitigation of environmental stresses. In Unravelling Plant-Microbe Synergy; Academic Press: Cambridge, MA, USA, 2023; pp. 87–117. [Google Scholar]
- Nong, Q.D.; Zhang, M.Y.; Chen, J.T.; Zhang, M.; Cheng, H.P.; Jian, S.G.; Lu, H.F.; Xia, K.F. RNA-Seq De Novo Assembly of Red Pitaya (Hylocereus polyrhizus) Roots and Differential Transcriptome Analysis in Response to Salt Stress. Trop. Plant Biol. 2019, 12, 55–66. [Google Scholar] [CrossRef]
- Gomez-Cano, F.; Rodriguez, J.; Zhou, P.; Chu, Y.-H.; Magnusson, E.; Gomez-Cano, L.; Krishnan, A.; Springer, N.M.; de Leon, N.; Grotewold, E. Prioritizing Metabolic Gene Regulators through Multi-Omic Network Integration in Maize. bioRxiv 2024. [Google Scholar] [CrossRef]
- Romero-Puertas, M.C.; Terrón-Camero, L.C.; Peláez-Vico, M.Á.; Molina-Moya, E.; Sandalio, L.M. An update on redox signals in plant responses to biotic and abiotic stress crosstalk: Insights from cadmium and fungal pathogen interactions. J. Exp. Bot. 2021, 72, 5857–5875. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, I.; Ahmad, S.; Shen, W. Melatonin-mediated molecular responses in plants: Enhancing stress tolerance and mitigating environmental challenges in cereal crop production. Int. J. Mol. Sci. 2024, 25, 4551. [Google Scholar] [CrossRef] [PubMed]
- Simonini, S.; Bencivenga, S.; Trick, M.; Østergaard, L. Auxin-Induced Modulation of ETTIN Activity Orchestrates Gene Expression in Arabidopsis. Plant Cell 2017, 29, 1864–1882. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Parvin, K.; Bardhan, K.; Nahar, K.; Anee, T.I.; Masud, A.A.C.; Fotopoulos, V. Biostimulants for the regulation of reactive oxygen species metabolism in plants under abiotic stress. Cells 2021, 10, 2537. [Google Scholar] [CrossRef]
- Iwaniuk, P.; Kaczyński, P.; Pietkun, M.; Łozowicka, B. Evaluation of Titanium and Silicon Role in Mitigation of Fungicides Toxicity in Wheat Expressed at the Level of Biochemical and Antioxidant Profile. Chemosphere 2022, 308, 136284. [Google Scholar] [CrossRef]
- Iwaniuk, P.; Łuniewski, S.; Kaczyński, P.; Łozowicka, B. The Influence of Humic Acids and Nitrophenols on Metabolic Compounds and Pesticide Behavior in Wheat under Biotic Stress. Agronomy 2023, 13, 1378. [Google Scholar] [CrossRef]
- Ellouzi, H.; Ben Hamed, K.; Asensi-Fabado, M.A.; Müller, M.; Abdelly, C.; Munné-Bosch, S. Drought and cadmium may be as effective as salinity in conferring subsequent salt stress tolerance in Cakile maritima. Planta 2013, 237, 1311–1323. [Google Scholar] [CrossRef]
- Liu, M.; Shi, M.; Gao, H.; Zheng, Y.; Yi, L. Nitrogen Addition Alleviates Cadmium Toxicity in Eleocarpus glabripetalus Seedlings. Forests 2023, 14, 1264. [Google Scholar] [CrossRef]
- Khaled, B.; Rachid, R.; Mohamed, R.D. Cadmium phytotoxicity on seed germination, early growth, and the differential antioxidant response of guaiacol peroxidase in Phragmites australis seedling organs. Pol. J. Environ. Stud. 2023, 32, 1531–1544. [Google Scholar] [CrossRef]
- Chmur, M.; Bajguz, A. Melatonin involved in protective effects against cadmium stress in Wolffia arrhiza. Int. J. Mol. Sci. 2023, 24, 1178. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Xue, X.; Yan, Y.; Zhao, X.; Li, L.; Sheng, K.; Zhang, Z. Silicon combined with melatonin reduces Cd absorption and translocation in maize. Plants 2023, 12, 3537. [Google Scholar] [CrossRef] [PubMed]
- Karumannil, S.; Khan, T.A.; Kappachery, S.; Gururani, M.A. Impact of exogenous melatonin application on photosynthetic machinery under abiotic stress conditions. Plants 2023, 12, 2948. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wu, M.; Jin, W.; Lei, T.; Li, Y.; Wu, X.; Fu, S. Gene identification and transcriptome analysis of cadmium stress in tomato. Front. Sustain. Food Syst. 2023, 7, 1303753. [Google Scholar] [CrossRef]
- Moravčíková, D.; Žiarovská, J. The Effect of Cadmium on Plants in Terms of the Response of Gene Expression Level and Activity. Plants 2023, 12, 1848. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin: A growth-stimulating compound present in lupin tissues. Planta 2014, 220, 140–144. [Google Scholar]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Rasmussen, S.; Barah, P.; Suarez-Rodriguez, M.C.; Bressendorff, S.; Friis, P.; Costantino, P.; Bones, A.M.; Nielsen, H.B.; Mundy, J.; Petersen, M. Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol. 2013, 161, 1783–1794. [Google Scholar] [CrossRef]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the point of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 97–103. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Shi, K.; Xia, X.J.; Zhou, Y.H.; Yu, J.Q. Hydrogen Peroxide is Involved in the Cold Acclimation-Induced Chilling Tolerance of Tomato Plants. Plant Physiol. Biochem. 2020, 49, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. ROS are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Ascorbate and Glutathione: The Heart of the Redox Hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive Oxygen Species and Antioxidant Machinery in Abiotic Stress Tolerance in Crop Plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Bowler, C.; Van Montagu, M.; Inzé, D. Superoxide Dismutase and Stress Tolerance. Annu. Rev. Plant Biol. 1992, 43, 83–116. [Google Scholar] [CrossRef]
- Szabados, L.; Savouré, A. Proline: A Multifunctional Amino Acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Gong, Y.; Zhang, L.; Hu, Y.; Wang, H.-Y. Exploring the Key Genes of Fucoxanthin Biosynthesis in Phaeodactylum tricornutum Under Phosphorus Deficiency, Red Light and Yellow Light Using WGCNA. Hereditas 2023, 45, 237–249. [Google Scholar] [PubMed]
- Wang, S.; Wu, G.-Q.; Wei, M. Functional Mechanisms of WRKY Transcription Factors in Regulating Plant Response to Abiotic Stresses. Chin. J. Biotechnol. 2024, 40, 35–52. [Google Scholar]
- Filiz, E.; Kurt, F. Expression and co-expression analyses of WRKY, MYB, bHLH and bZIP transcription factor genes in potato (Solanum tuberosum) under abiotic stress conditions: RNA-seq data analysis. Potato Res. 2021, 64, 721–741. [Google Scholar] [CrossRef]
- Sahu, S.; Rao, A.R.; Bansal, K.C.; Muthusamy, S.K.; Chinnusamy, V. Genome-Wide Analysis and Identification of Abiotic Stress Responsive Transcription Factor Family Genes and miRNAs in Bread Wheat (Triticum aestivum L.): Genomic Study of Bread Wheat. In Proceedings of the 2016 International Conference on Bioinformatics and Systems Biology (BSB), Allahabad, India, 4–6 March 2016; pp. 1–4. [Google Scholar]
- Yao, W.; Zhou, B.; Zhang, X.; Zhao, K.; Cheng, Z.; Jiang, T. Transcriptome analysis of transcription factor genes under multiple abiotic stresses in Populus simonii × P. nigra. Gene 2019, 707, 189–197. [Google Scholar] [CrossRef]
- Sun, S.; Liu, X.; Zhang, T.; Yu, B. Functional Characterisation of the Transcription Factor GsWRKY23 Gene from Glycine soja in Overexpressed Soybean Composite Plants and Arabidopsis under Salt Stress. Plants 2023, 12, 3030. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Wang, E.; Xu, X.; Tong, C.; Zhao, X.; Song, X.; Wang, J.; Wang, H.-L.; Xia, X.-L.; Zhao, Y. Beat the Heat: Signaling Pathway-Mediated Strategies for Plant Thermotolerance. Forests 2023, 14, 2026. [Google Scholar] [CrossRef]
- Zhang, S.; Li, C.; Wang, R.; Chen, Y.; Shu, S.; Huang, R.; Yang, C. The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol. 2017, 173, 2294–2307. [Google Scholar] [CrossRef] [PubMed]
- Arraño-Salinas, P.; Domínguez-Figueroa, J.; Herrera-Vásquez, A.; Zavala, D.; Medina, J.; Vicente-Carbajosa, J.; Blanco-Herrera, F. WRKY7,-11 and-17 transcription factors are modulators of the bZIP28 branch of the unfolded protein response during PAMP-triggered immunity in Arabidopsis thaliana. Plant Sci. 2018, 277, 242–250. [Google Scholar] [CrossRef]
- Zhao, K.; Zhang, D.; Lv, K.; Zhang, X.; Cheng, Z.; Li, R.; Zhou, B.; Jiang, T. Functional characterization of poplar WRKY75 in salt and osmotic tolerance. Plant Sci. 2019, 289, 110259. [Google Scholar] [CrossRef]
- Khan, I.; Asaf, S.; Jan, R.; Bilal, S.; Lubna; Khan, A.L.; Kim, K.-M.; Al-Harrasi, A. Genome-wide annotation and expression analysis of WRKY and bHLH transcriptional factor families reveal their involvement under cadmium stress in tomato (Solanum lycopersicum L.). Front. Plant Sci. 2023, 14, 1100895. [Google Scholar] [CrossRef] [PubMed]
- Goel, P.; Bhuria, M.; Sinha, R.; Sharma, T.R.; Singh, A.K. Promising transcription factors for salt and drought tolerance in plants. In Molecular Approaches in Plant Biology and Environmental Challenges; Springer: Cham, Switzerland, 2019; pp. 7–50. [Google Scholar]
- Baloglu, M.C.; Inal, B.; Kavas, M.; Unver, T. Diverse expression pattern of wheat transcription factors against abiotic stresses in wheat species. Gene 2014, 550, 117–122. [Google Scholar] [CrossRef]
- Zhang, B.X.; Li, P.S.; Wang, Y.Y.; Wang, J.J.; Liu, X.L.; Wang, X.Y.; Hu, X.M. Characterization and synthesis of indole-3-acetic acid in plant growth promoting Enterobacter sp. RSC Adv. 2021, 11, 31601–31607. [Google Scholar] [CrossRef]
- Rehman, A.; Azhar, M.T.; Hinze, L.; Qayyum, A.; Li, H.; Peng, Z.; Du, X. Insight into abscisic acid perception and signaling to increase plant tolerance to abiotic stress. J. Plant Interact. 2021, 16, 222–237. [Google Scholar] [CrossRef]
- Wang, F.; Cui, X.; Sun, Y.; Dong, C.-H. Ethylene signaling and regulation in plant growth and stress responses. Plant Cell Rep. 2013, 32, 1099–1109. [Google Scholar] [CrossRef]
- Yow, A.G.; Laosuntisuk, K.; Young, R.A.; Doherty, C.J.; Gillitt, N.; Perkins-Veazie, P.; Jenny Xiang, Q.Y.; Iorizzo, M. Comparative Transcriptome Analysis Reveals Candidate Genes for Cold Stress Response and Early Flowering in Pineapple. Sci. Rep. 2023, 13, 18890. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Yin, Y.; Fei, S.Z. Brassinosteroid signaling network: Implications on yield and stress tolerance. Plant Cell Rep. 2013, 32, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Bulgakov, V.P.; Avramenko, T.V. Linking brassinosteroid and ABA signaling in the context of stress acclimation. Int. J. Mol. Sci. 2020, 21, 5108. [Google Scholar] [CrossRef]
- Lozano-Elena, F.; Fàbregas, N.; Coleto-Alcudia, V.; Caño-Delgado, A.I. Analysis of Metabolic Dynamics During Drought Stress in Arabidopsis Plants. Sci. Data 2022, 9, 90. [Google Scholar] [CrossRef]
- Jha, U.C.; Nayyar, H.; Jha, R.; Khurshid, M.; Zhou, M.; Mantri, N.; Siddique, K.H. Long non-coding RNAs: Emerging players regulating plant abiotic stress response and adaptation. BMC Plant Biol. 2020, 20, 466. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Feng, H.; Bu, Y.; Lu, Y.; Ji, N.; Zhao, S. Comparative transcriptome analysis reveals molecular mechanism and regulatory network of Rosa chinensis ‘Old Blush’ leaves in response to drought stress. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Qu, Y.; Nong, Q.; Jian, S.; Lu, H.; Zhang, M.; Xia, K. An AP2/ERF Gene, HuERF1, from Pitaya (Hylocereus undatus) Positively Regulates Salt Tolerance. Int. J. Mol. Sci. 2020, 21, 4586. [Google Scholar] [CrossRef]
- Oltehua-López, O.; Arteaga-Vázquez, M.A.; Sosa, V. Stem transcriptome screen for selection in wild and cultivated pitahaya (Selenicereus undatus): An epiphytic cactus with edible fruit. PeerJ 2023, 11, e14581. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.J.; Yan, F.X.; Qiao, G.; Zhang, B.X.; Wen, X.P. Identification of differentially-expressed genes potentially implicated in drought response in pitaya (Hylocereus undatus) by suppression subtractive hybridization and cDNA microarray analysis. Gene 2014, 533, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Oono, Y.; Yazawa, T.; Kawahara, Y.; Kanamori, H.; Kobayashi, F.; Sasaki, H.; Matsumoto, T. Genome-wide transcriptome analysis reveals that cadmium stress signaling controls the expression of genes in drought stress signal pathways in rice. PLoS ONE 2014, 9, e96946. [Google Scholar] [CrossRef] [PubMed]
- Xuan, H.; Huang, Y.; Zhou, L.; Deng, S.; Wang, C.; Xu, J.; Wang, H.; Zhao, J.; Guo, N.; Xing, H. Key soybean seedlings drought-responsive genes and pathways revealed by comparative transcriptome analyses of two cultivars. Int. J. Mol. Sci. 2022, 23, 2893. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Peng, L.; Liu, Y.; Yao, R.; Liu, Z.; Li, X.; Wang, J. Arabidopsis calcium-dependent protein kinase AtCPK1 plays a positive role in salt/drought-stress response. Biochem. Biophys. Res. Commun. 2018, 498, 92–98. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef] [PubMed]
- Zaprasis, A.; Bleisteiner, M.; Kerres, A.; Hoffmann, T.; Bremer, E. Uptake of amino acids and their metabolic conversion into the compatible solute proline confers osmoprotection to Bacillus subtilis. Appl. Environ. Microbiol. 2015, 81, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Tietel, Z.; Wikoff, W.R.; Kind, T.; Ma, Y.; Fiehn, O. Hyperosmotic stress in Chlamydomonas induces metabolomic changes in biosynthesis of complex lipids. Eur. J. Phycol. 2020, 55, 11–29. [Google Scholar] [CrossRef]
- Weiss, I.; Mizrahi, Y.; Raveh, E. Effect of elevated CO2 on vegetative and reproductive growth characteristics of the CAM plants Hylocereus undatus and Selenicereus megalanthus. Sci. Hortic. 2010, 123, 531–536. [Google Scholar] [CrossRef]
- Wu, T.; Fu, Y.; Shi, Y.; Li, Y.; Kou, Y.; Mao, X.; Liu, J. Functional Characterization of Long-Chain Acyl-CoA Synthetase Gene Family from the Oleaginous Alga Chromochloris zofingiensis. J. Agric. Food Chem. 2020, 68, 4473–4484. [Google Scholar] [CrossRef]
- Lin, Y.; Liu, S.; Fang, X.; Ren, Y.; You, Z.; Xia, J.; Hakeem, A.; Yang, Y.; Wang, L.; Fang, J.; et al. The Physiology of Drought Stress in Two Grapevine Cultivars: Photosynthesis, Antioxidant System, and Osmotic Regulation Responses. Physiol. Plant. 2023, 175, e14005. [Google Scholar] [CrossRef]
- Shomali, A.; Das, S.; Arif, N.; Sarraf, M.; Zahra, N.; Yadav, V.; Aliniaeifard, S.; Chauhan, D.K.; Hasanuzzaman, M. Diverse Physiological Roles of Flavonoids in Plant Environmental Stress Responses and Tolerance. Plants 2022, 11, 3158. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Liu, A.; Li, Z.; Guo, T.; Chen, S.; Ahammed, G.J. Anthocyanin synthesis is critical for melatonin-induced chromium stress tolerance in tomato. J. Hazard. Mater. 2023, 453, 131456. [Google Scholar] [CrossRef]
- Liang, T.; Shi, C.; Peng, Y.; Tan, H.; Xin, P.; Yang, Y.; Liu, H. Brassinosteroid-activated BRI1-EMS-SUPPRESSOR 1 inhibits flavonoid biosynthesis and coordinates growth and UV-B stress responses in plants. Plant Cell 2020, 32, 3224–3239. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The Water Culture Method for Growing Plants Without Soil. Calif. Agric. Exp. Stn. Circ. 1938, 347, 32. [Google Scholar]
- Song, W.; Li, L.; Huang, H.; Jiang, K.; Zhang, F.; Wang, L.; Zhao, M.; Ma, L. Tissue-based transcriptomics of Chionodraco hamatus: Sequencing, de novo assembly, annotation and marker discovery. J. Fish Biol. 2019, 94, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, M.-C.; Konaté, M.M.; Chen, L.; Das, B.; Karlovich, C.; Williams, P.M.; Evrard, Y.A.; Doroshow, J.H.; McShane, L.M. TPM, FPKM, or Normalized Counts? A Comparative Study of Quantification Measures for the Analysis of RNA-seq Data from the NCI Patient-Derived Models Repository. J. Transl. Med. 2021, 19, 269. [Google Scholar] [CrossRef]
- Konishi, T. Parametric Analysis of RNA-Seq Expression Data. Genes Cells 2016, 21, 639–647. [Google Scholar] [CrossRef]
- Ye, F.; Li, Y.; Li, L.; Wang, X.; Chen, Z. Identification of Specific Modules and Significant Genes Associated with Colon Cancer by Weighted Gene Co-Expression Network Analysis. Mol. Med. Rep. 2019, 20, 693–700. [Google Scholar]
- Zoppi, J.; Guillaume, J.-F.; Neunlist, M.; Chaffron, S. MiBiOmics: An interactive web application for multi-omics data exploration and integration. BMC Bioinform. 2021, 22, 6. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dong, M.; Tang, K.; Zhang, H.; Zeng, N.; Yang, H.; Liu, D. The effects of calcium fertilization on morphological and physio-biochemical characteristics in peanut seedlings under waterlogging stress. J. Plant Nutr. 2023, 46, 2865–2881. [Google Scholar] [CrossRef]
- Ukeda, H.; Kawana, D.; Maeda, S.; Sawamura, M. Spectrophotometric assay for superoxide dismutase based on the reduction of highly water-soluble tetrazolium salts by xanthine-xanthine oxidase. Biosci. Biotechnol. Biochem. 1999, 63, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Kochba, J.; Lavee, S.; Spiegel-Roy, P. Differences in peroxidase activity and isoenzymes in embryogenic ane non-embryogenic ‘Shamouti’ orange ovular callus lines. Plant Cell Physiol. 1977, 18, 463–467. [Google Scholar] [CrossRef]
- Aebi, H. Catalase in vitro. Methods Enzymol. 1984, 105, 121–126. [Google Scholar]
- Ullah, S.; Kolo, Z.; Egbichi, I.; Keyster, M.; Ludidi, N. Nitric oxide influences glycine betaine content and ascorbate peroxidase activity in maize. S. Afr. J. Bot. 2016, 105, 218–225. [Google Scholar] [CrossRef]
- Satterfield, C.N.; Bonnell, A.H. Interferences in titanium sulfate method for hydrogen peroxide. Anal. Chem. 1955, 27, 1174–1175. [Google Scholar] [CrossRef]
- Mansour, M.M.F.; Ali, E.F. Evaluation of proline functions in saline conditions. Phytochemistry 2017, 140, 52–68. [Google Scholar] [CrossRef]
- Campion, E.M.; Loughran, S.T.; Walls, D. Protein quantitation and analysis of purity. Methods Mol. Biol. 2011, 681, 229–258. [Google Scholar]
- Bokang, K.; Jeremy, M.; Van, R. RNA Sequencing of Pooled Samples Effectively Identifies Differentially Expressed Genes. Biology 2023, 12, 812. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.; Livak, K. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Zaman, Q.U.; Hussain, M.A.; Khan, L.U.; Hui, L.; Khan, D.; Khokhar, A.A.; Wang, H.F. Genome-Wide Identification and Expression Profiling of APX Gene Family under Multifactorial Stress Combinations and Melatonin-Mediated Tolerance in Pitaya. Sci. Hortic. 2023, 321, 112312. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khokhar, A.A.; Hui, L.; Khan, D.; You, Z.; Zaman, Q.U.; Usman, B.; Wang, H.-F. Transcriptome Profiles Reveal Key Regulatory Networks during Single and Multifactorial Stresses Coupled with Melatonin Treatment in Pitaya (Selenicereus undatus L.). Int. J. Mol. Sci. 2024, 25, 8901. https://doi.org/10.3390/ijms25168901
Khokhar AA, Hui L, Khan D, You Z, Zaman QU, Usman B, Wang H-F. Transcriptome Profiles Reveal Key Regulatory Networks during Single and Multifactorial Stresses Coupled with Melatonin Treatment in Pitaya (Selenicereus undatus L.). International Journal of Molecular Sciences. 2024; 25(16):8901. https://doi.org/10.3390/ijms25168901
Chicago/Turabian StyleKhokhar, Aamir Ali, Liu Hui, Darya Khan, Zhang You, Qamar U Zaman, Babar Usman, and Hua-Feng Wang. 2024. "Transcriptome Profiles Reveal Key Regulatory Networks during Single and Multifactorial Stresses Coupled with Melatonin Treatment in Pitaya (Selenicereus undatus L.)" International Journal of Molecular Sciences 25, no. 16: 8901. https://doi.org/10.3390/ijms25168901







