A Comprehensive Examination of the Role of Epigenetic Factors in Multiple Sclerosis
Abstract
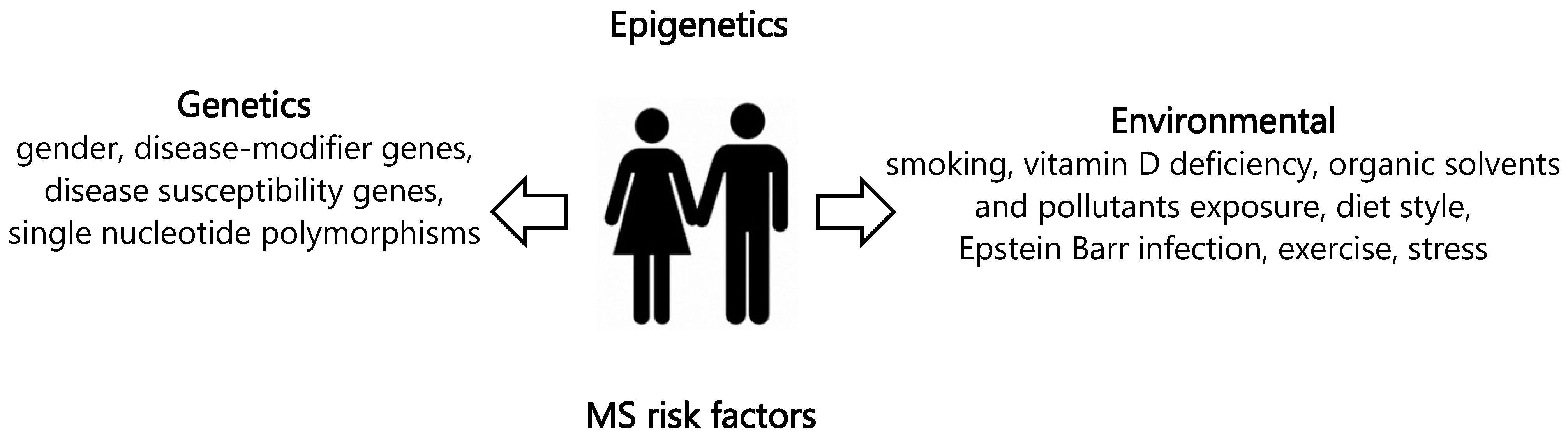
:1. Introduction
2. DNA Methylation: Contribution in Patients with MS
3. Post-Translational Histone Modifications: Contribution to MS
4. miRNAs: Contribution in Patients with MS
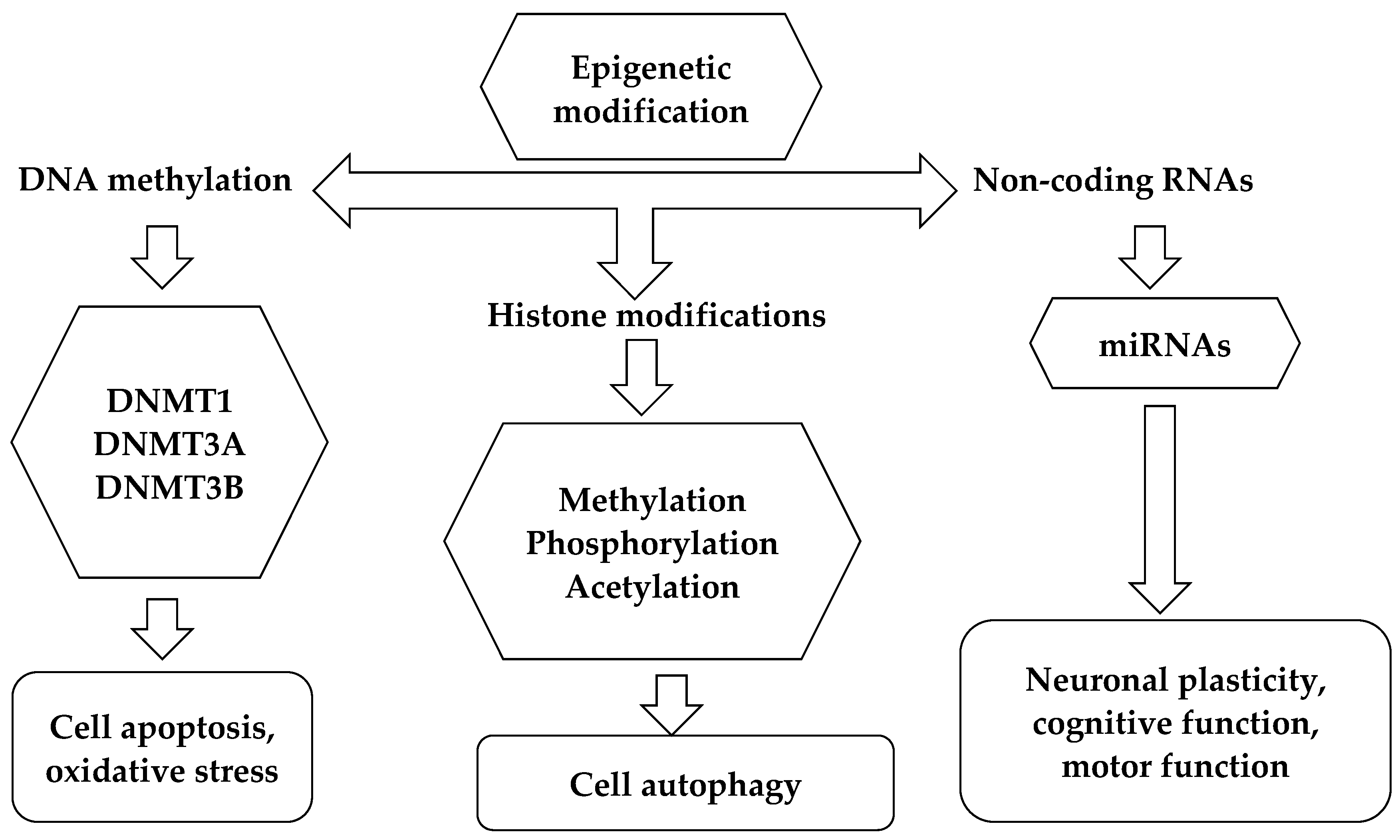
5. Remarks on the Interaction of Epigenetic Changes
6. Conclusions and Future Prospective
Author Contributions
Funding
Conflicts of Interest
References
- Dendrou, C.A.; Fugger, L.; Friese, M.A. Immunopathology of multiple sclerosis. Nat. Rev. Immuno. 2015, 15, 545–558. [Google Scholar] [CrossRef]
- Franklin, R.J.; Ffrench-Constant, C. Remyelination in the CNS: From biology to therapy. Nat. Rev. Neurosci. 2008, 9, 839–855. [Google Scholar] [CrossRef]
- Yamout, B.I.; Alroughani, R. Multiple Sclerosis. Semin. Neurol. 2018, 38, 212–225. [Google Scholar]
- Kuhlmann, T.; Moccia, M.; Coetzee, T.; Cohen, J.A.; Correale, J.; Graves, J.; Marrie, R.A.; Montalban, X.; Yong, V.W.; Thompson, A.J.; et al. International Advisory Committee on Clinical Trials in Multiple Sclerosis. Multiple sclerosis progression: Time for a new mechanism-driven framework. Lancet Neurol. 2023, 22, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Zettl, U.K.; Stuve, O.; Patejd, R. Immune-mediated CNS diseases: A review on nosological classification and clinical features. Autoimmun. Rev. 2012, 11, 167–173. [Google Scholar] [CrossRef]
- Lassmann, H.; van Horssen, J.; Mahad, D. Progressive multiple sclerosis: Pathology and pathogenesis. Nat. Rev. Neurol. 2012, 8, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.H.; Leary, S.M. Primary-progressive multiple sclerosis. Lancet Neurol. 2007, 6, 903–912. [Google Scholar] [CrossRef]
- Olsson, T.; Barcellos, L.F.; Alfredsson, L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat. Rev. Neurol. 2017, 13, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Briggs, F.B.; Acuna, B.; Shen, L.; Ramsay, P.; Quach, H.; Bernstein, A.; Bellesis, K.H.; Kockum, I.S.; Hedström, A.K.; Alfredsson, L.; et al. Smoking and risk of multiple sclerosis: Evidence of modification by NAT1 variants. Epidemiology 2015, 25, 605–614. [Google Scholar] [CrossRef]
- Scazzone, C.; Agnello, L.; Bivona, G.; Lo Sasso, B.; Ciaccio, M. Vitamin D and Genetic Susceptibility to Multiple Sclerosis. Biochem. Genet. 2021, 59, 1–30. [Google Scholar] [CrossRef]
- Agnello, L.; Scazzone, C.; Sasso, B.L.; Vidali, M.; Giglio, R.V.; Ciaccio, A.M.; Ragonese, P.; Salemi, G.; Ciaccio, M. Role of Multiple Vitamin D-Related Polymorphisms in Multiple Sclerosis Severity: Preliminary Findings. Genes 2022, 13, 1307. [Google Scholar] [CrossRef] [PubMed]
- Scazzone, C.; Agnello, L.; Lo Sasso, B.; Salemi, G.; Gambino, C.M.; Ragonese, P.; Candore, G.; Ciaccio, A.M.; Giglio, R.V.; Bivona, G.; et al. FOXP3 and GATA3 Polymorphisms, Vitamin D3 and Multiple Sclerosis. Brain Sci. 2021, 11, 415. [Google Scholar] [CrossRef] [PubMed]
- Scazzone, C.; Agnello, L.; Sasso, B.L.; Ragonese, P.; Bivona, G.; Realmuto, S.; Iacolino, G.; Gambino, C.M.; Bellia, C.; Salemi, G.; et al. Klotho and vitamin D in multiple sclerosis: An Italian study. Arch. Med. Sci. 2019, 16, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Agnello, L.; Scazzone, C.; Lo Sasso, B.; Bellia, C.; Bivona, G.; Realmuto, S.; Brighina, F.; Schillaci, R.; Ragonese, P.; Salemi, G.; et al. VDBP, CYP27B1, and 25-Hydroxyvitamin D Gene Polymorphism Analyses in a Group of Sicilian Multiple Sclerosis Patients. Biochem. Genet. 2017, 55, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Sawcer, S.; Hellenthal, G.; Pirinen, M.; Spencer, C.C.A.; Patsopoulos, N.A.; Moutsianas, L.; Dilthey, A.; Su, Z.; Freeman, C.; Hunt, S.E.; et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011, 476, 214–219. [Google Scholar]
- Goll, M.G.; Bestor, T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005, 74, 481–514. [Google Scholar] [CrossRef]
- Lorincz, M.C.; Dickerson, D.R.; Schmitt, M.; Groudine, M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 2004, 11, 1068–1075. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Kavak, E.; Gregory, M.; Imashimizu, M.; Shutinoski, B.; Kashlev, M.; Oberdoerffer, P.; Sandberg, R.; Oberdoerffer, S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 2011, 479, 74–79. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Castillo-Aguilera, O.; Depreux, P.; Halby, L.; Arimondo, P.B.; Goossens, L. DNA Methylation Targeting: The DNMT/HMT Crosstalk Challenge. Biomolecules 2017, 7, 3. [Google Scholar] [CrossRef]
- Barau, J.; Teissandier, A.; Zamudio, N.; Roy, S.; Nalesso, V.; Hérault, Y.; Guillou, F.; Bourc’his, D. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 2016, 354, 909–912. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Yun, M.; Wu, J.; Workman, J.L.; Li, B. Readers of histone modifications. Cell Res. 2011, 21, 564–578. [Google Scholar] [CrossRef] [PubMed]
- Shahbazian, M.D.; Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007, 76, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Backes, C.; Meese, C.E.; Lenhof, H.P.; Keller, A. A dictionary on microRNAs and their putative target pathways. Nucleic Acids Res. 2010, 38, 4476–4486. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, G.; Perego, S.; Sansoni, V.; Banfi, G. Circulating miRNA as fine regulators of the physiological responses to physical activity: Pre-analytical warnings for a novel class of biomarkers. Clin. Biochem. 2016, 49, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Iyengar, B.R.; Choudhary, A.; Sarangdhar, M.A.; Venkatesh, K.V.; Gadgil, C.J.; Pillai, B. Non-coding RNA interact to regulate neuronal development and function. Front. Cell. Neurosci. 2014, 8, 47. [Google Scholar] [CrossRef]
- Jin, X.F.; Wu, N.; Wang, L.; Li, J. Circulating microRNAs: A novel class of potential biomarkers for diagnosing and prognosing central nervous system diseases. Cell. Mol. Neurobiol. 2013, 33, 601–613. [Google Scholar] [CrossRef]
- Liu, J.; Sandoval, J.; Doh, S.T.; Cai, L.; López-Rodas, G.; Casaccia, P. Epigenetic modifiers are necessary but not sufficient for reprogramming non-myelinating cells into myelin gene-expressing cells. PLoS ONE 2010, 5, e13023. [Google Scholar] [CrossRef]
- Mao, X.Y.; Yin, X.X.; Guan, Q.W.; Xia, Q.X.; Yang, N.; Zhou, H.H.; Liu, Z.Q.; Jin, W.L. Dietary nutrition for neurological disease therapy: Current status and future directions. Pharmacol. Ther. 2021, 226, 107861. [Google Scholar] [CrossRef]
- Copray, S.; Huynh, J.L.; Sher, F.; Casaccia-Bonnefil, P.; Boddeke, E. Epigenetic mechanisms facilitating oligodendrocyte development, maturation, and aging. Glia 2009, 57, 1579–1587. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Zhang, Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef]
- Xu, Z.; Li, X. DNA methylation in neurodegenerative disorders. Curr. Geriatr. Rep. 2012, 1, 199–205. [Google Scholar] [CrossRef]
- Landgrave-Gomez, J.; Mercado-Gomez, O.; Guevara-Guzman, R. Epigenetic mechanisms in neurological and neurodegenerative diseases. Front. Cell. Neurosci. 2015, 9, 58. [Google Scholar]
- Hojati, Z. Molecular Genetic and Epigenetic Basis of Multiple Sclerosis. Adv. Exp. Med. Biol. 2017, 958, 65–90. [Google Scholar] [PubMed]
- Kürtüncü, M.; Tüzün, E. Multiple sclerosis: Could it be an epigenetic disease? Med. Hypotheses 2008, 71, 945–947. [Google Scholar] [CrossRef]
- On, N.H.; Kiptoo, P.; Siahaan, T.J.; Miller, D.W. Modulation of blood-brain barrier permeability in mice using synthetic E-cadherin peptide. Mol. Pharm. 2014, 11, 974–981. [Google Scholar] [CrossRef]
- Liggettt, T.; Melnikov, A.; Tilwalli, S.; Yi, Q.; Chen, H.; Replogle, C.; Feng, X.; Reder, A.; Stefoski, D.; Balabanov, R.; et al. Methylation patterns of cell-free plasma DNA in relapsing-remitting multiple sclerosis. J. Neurol. Sci. 2010, 290, 16–21. [Google Scholar] [CrossRef]
- Tiane, A.; Schepers, M.; Reijnders, R.A.; van Veggel, L.; Chenine, S.; Rombaut, B.; Dempster, E.; Verfaillie, C.; Wasner, K.; Grünewald, A.; et al. From methylation to myelination: Epigenomic and transcriptomic profiling of chronic inactive demyelinated multiple sclerosis lesions. Acta Neuropathol. 2023, 146, 283–299. [Google Scholar] [CrossRef]
- Tiane, A.; Somers, V.; Hellings, N.; van den Hove, D.L.A.; Vanmierlo, T. The Impact of Sample Storage on Blood Methylation: Towards Assessing Myelin Gene Methylation as a Biomarker for Progressive Multiple Sclerosis. Int. J. Mol. Sci. 2024, 25, 3468. [Google Scholar] [CrossRef]
- Sokratous, M.; Dardiotis, E.; Tsouris, Z.; Bellou, E.; Michalopoulou, A.; Siokas, V.; Arseniou, S.; Stamati, T.; Tsivgoulis, G.; Bogdanos, D.; et al. Deciphering the role of DNA methylation in multiple sclerosis: Emerging issues. Autoimmun. Highlights 2016, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Levenson, V.V.; Melnikov, A.A. DNA methylation as clinically useful biomarkers-light at the end of the tunnel. Pharmaceuticals 2012, 5, 94. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, C.; Kalman, B.; Middleton, F.A.; Vyshkina, T.; Massa, P.T. Increased promoter methylation of the immune regulatory gene SHP-1 in leukocytes of multiple sclerosis subjects. J. Neuroimmunol. 2012, 246, 51–57. [Google Scholar] [CrossRef]
- Janson, P.C.; Linton, L.B.; Bergman, E.A.; Marits, P.; Eberhardson, M.; Piehl, F.; Malmström, V.; Winqvist, O. Profiling of CD4+ T cells with epigenetic immune lineage analysis. J. Immunol. 2011, 186, 92–102. [Google Scholar] [CrossRef]
- Koch, M.W.; Metz, L.M.; Kovalchuk, O. Epigenetic changes in patients with multiple sclerosis. Nat. Rev. Neurol. 2013, 9, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Mastronardi, F.G.; Noor, A.; Wood, D.D.; Paton, T.; Moscarello, M.A. Peptidyl arginine deiminase 2 CpG island in multiple sclerosis white matter is hypomethylated. J. Neurosci. Res. 2007, 85, 2006–21016. [Google Scholar] [CrossRef]
- Ransohoff, R.M. How neuroinflammation contributes to neurodegeneration. Science 2016, 353, 777–783. [Google Scholar] [CrossRef]
- Von Bartheld, C.S.; Bahney, J.; Herculano-Houzel, S. The search for true numbers of neurons and glial cells in the human brain: A review of 150 years of cell counting. J. Comp. Neurol. 2016, 524, 3865–3895. [Google Scholar] [CrossRef]
- Huynh, J.L.; Garg, P.; Thin, T.H.; Yoo, S.; Dutta, R.; Trapp, B.D.; Haroutunian, V.; Zhu, J.; Donovan, M.J.; Sharp, A.J.; et al. Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat. Neurosci. 2014, 17, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Chomyk, A.M.; Volsko, C.; Tripathi, A.; Deckard, S.A.; Trapp, B.D.; Fox, R.J.; Dutta, R. DNA methylation in demyelinated multiple sclerosis hippocampus. Sci. Rep. 2017, 7, 8696. [Google Scholar] [CrossRef] [PubMed]
- Kular, L.; Needhamsen, M.; Adzemovic, M.Z.; Kramarova, T.; Gomez-Cabrero, D.; Ewing, E.; Piket, E.; Tegnér, J.; Beck, S.; Piehl, F.; et al. Neuronal methylome reveals CREB-associated neuro-axonal impairment in multiple sclerosis. Clin. Epigenet. 2019, 11, 86. [Google Scholar] [CrossRef] [PubMed]
- Kular, L.; Ewing, E.; Needhamsen, M.; Pahlevan Kakhki, M.; Covacu, R.; Gomez-Cabrero, D.; Brundin, L.; Jagodic, M. DNA methylation changes in glial cells of the normal-appearing white matter in Multiple Sclerosis patients. Epigenetics 2022, 17, 1311–1330. [Google Scholar] [CrossRef]
- Celarain, N.; Tomas-Roig, J. Aberrant DNA methylation profile exacerbates inflammation and neurodegeneration in multiple sclerosis patients. J. Neuroinflamm. 2020, 17, 21. [Google Scholar] [CrossRef]
- Kiselev, I.S.; Kulakova, O.G.; Baturina, O.A.; Kabilov, M.R.; Boyko, A.N.; Favorova, O.O. DNA Methylation Profile of CD14+ Monocytes Changes in Primary Progressive Multiple Sclerosis. Mol. Biol. 2023, 57, 819–826. [Google Scholar] [CrossRef]
- Maltby, V.E.; Lea, R.A.; Ribbons, K.A.; Sanders, K.A.; Kennedy, D.; Min, M.; Scott, R.J.; Lechner-Scott, J. DNA methylation changes in CD4+ T cells isolated from multiple sclerosis patients on dimethyl fumarate. Mult. Scler. J. Exp. Transl. Clin. 2018, 4, 2055217318787826. [Google Scholar] [CrossRef]
- Ewing, E.; Kular, L.; Fernandes, S.J.; Karathanasis, N.; Lagani, V.; Ruhrmann, S.; Tsamardinos, I.; Tegner, J.; Piehl, F.; Gomez-Cabrero, D.; et al. Combining evidence from four immune cell types identifies DNA methylation patterns that implicate functionally distinct pathways during multiple sclerosis progression. EBioMedicine 2019, 43, 411–423. [Google Scholar] [CrossRef]
- Kiselev, I.; Danilova, L.; Baulina, N.; Baturina, O.; Kabilov, M.; Boyko, A.; Kulakova, O.; Favorova, O. Genome-wide DNA methylation profiling identifies epigenetic changes in CD4+ and CD14+ cells of multiple sclerosis patients. Mult. Scler. Relat. Disord. 2022, 60, 103714. [Google Scholar] [CrossRef]
- Graves, M.C.; Benton, M.; Lea, R.A.; Boyle, M.; Tajouri, L.; Macartney-Coxson, D.; Scott, R.J.; Lechner-Scott, J. Methylation differences at the HLA-DRB1 locus in CD4+ T-Cells are associated with multiple sclerosis. Mult. Scler. 2014, 20, 1033–1041. [Google Scholar] [CrossRef]
- Kular, L.; Liu, Y.; Ruhrmann, S.; Zheleznyakova, G.; Marabita, F.; Gomez-Cabrero, D.; James, T.; Ewing, E.; Lindén, M.; Górnikiewicz, B.; et al. DNA methylation as a mediator of HLA-DRB1*15:01 and a protective variant in multiple sclerosis. Nat. Commun. 2018, 9, 2397. [Google Scholar] [CrossRef] [PubMed]
- Xavier, A.; Maltby, V.E.; Ewing, E.; Campagna, M.P.; Burnard, S.M.; Tegner, J.N.; Slee, M.; Butzkueven, H.; Kockum, I.; Kular, L.; et al. DNA Methylation Signatures of Multiple Sclerosis Occur Independently of Known Genetic Risk and Are Primarily Attributed to B Cells and Monocytes. Int. J. Mol. Sci. 2023, 24, 12576. [Google Scholar] [CrossRef]
- Ramazi, S.; Allahverdi, A.; Zahiri, J. Evaluation of post-translational modifications in histone proteins: A review on histone modification defects in developmental and neurological disorders. J. Biosci. 2020, 45, 135. [Google Scholar] [CrossRef]
- Huynh, J.L.; Casaccia, P. Epigenetic mechanisms in multiple sclerosis: Implications for pathogenesis and treatment. Lancet Neurol. 2013, 12, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Niller, H.H.; Wolf, H.; Minarovits, J. Epigenetic dysregulation of the host cell genome in Epstein-Barr virus-associated neoplasia. Semin. Cancer Biol. 2009, 19, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Wan, E.S.; Qiu, W.; Baccarelli, A.; Carey, V.J.; Bacherman, H.; Rennard, S.I.; Agusti, A.; Anderson, W.; Lomas, D.A.; Demeo, D.L. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum. Mol. Genet. 2012, 21, 3073–3082. [Google Scholar] [CrossRef]
- Pereira, F.; Barbáchano, A.; Singh, P.K.; Campbell, M.J.; Muñoz, A.; Larriba, M.J. Vitamin D has wide regulatory effects on histone demethylase genes. Cell Cycle 2012, 11, 1081–1089. [Google Scholar] [CrossRef]
- Gray, S.G.; Dangond, F. Rationale for the use of histone deacetylase inhibitors as a dual therapeutic modality in multiple sclerosis. Epigenetics 2006, 1, 67–75. [Google Scholar] [CrossRef]
- Pedre, X.; Mastronardi, F.; Bruck, W.; López-Rodas, G.; Kuhlmann, T.; Casaccia, P. Changed histone acetylation patterns in normal-appearing white matter and early multiple sclerosis lesions. J. Neurosci. 2011, 31, 3435–3445. [Google Scholar] [CrossRef]
- Singhal, N.K.; Li, S.; Arning, E.; Alkhayer, K.; Clements, R.; Sarcyk, Z.; Dassanayake, R.S.; Brasch, N.E.; Freeman, E.J.; Bottiglieri, T.; et al. Changes in Methionine Metabolism and Histone H3 Trimethylation Are Linked to Mitochondrial Defects in Multiple Sclerosis. J. Neurosci. 2015, 35, 15170–15186. [Google Scholar] [CrossRef]
- Singhal, N.K.; Freeman, E.; Arning, E.; Wasek, B.; Clements, R.; Sheppard, C.; Blake, P.; Bottiglieri, T.; McDonough, J. Dysregulation of methionine metabolism in multiple sclerosis. Neurochem. Int. 2018, 112, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Singhal, N.K.; Huang, H.; Li, S.; Clements, R.; Gadd, J.; Daniels, A.; Kooijman, E.E.; Bannerman, P.; Burns, T.; Guo, F.; et al. The neuronal metabolite NAA regulates histone H3 methylation in oligodendrocytes and myelin lipid composition. Exp. Brain Res. 2017, 235, 279–292. [Google Scholar] [CrossRef]
- Li, S.; Clements, R.; Sulak, M.; Gregory, R.; Freeman, E.; McDonough, J. Decreased NAA in gray matter is correlated with decreased availability of acetate in white matter in postmortem multiple sclerosis cortex. Neurochem. Res. 2013, 38, 2385–2396. [Google Scholar] [CrossRef]
- Sternbach, S.; West, N.; Singhal, N.K.; Clements, R.; Basu, S.; Tripathi, A.; Dutta, R.; Freeman, E.J.; McDonough, J. The BHMT-betaine methylation pathway epigenetically modulates oligodendrocyte maturation. PLoS ONE 2021, 16, e0250486. [Google Scholar] [CrossRef]
- Mastronardi, F.G.; Wood, D.D.; Mei, J.; Raijmakers, R.; Tseveleki, V.; Dosch, H.M.; Probert, L.; Casaccia-Bonnefil, P.; Moscarello, M.A. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: A role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. J. Neurosci. 2006, 26, 11387–11396. [Google Scholar] [CrossRef]
- Zhang, J.; Jing, L.; Li, M.; He, L.; Guo, Z. Regulation of histone arginine methylation/demethylation by methylase and demethylase (Review). Mol. Med. Rep. 2019, 19, 3963–3971. [Google Scholar] [CrossRef]
- Faigle, W.; Cruciani, C.; Wolski, W.; Roschitzki, B.; Puthenparampil, M.; Tomas-Ojer, P.; Sellés-Moreno, C.; Zeis, T.; Jelcic, I.; Schaeren-Wiemers, N.; et al. Brain Citrullination Patterns and T Cell Reactivity of Cerebrospinal Fluid-Derived CD4+ T Cells in Multiple Sclerosis. Front. Immunol. 2019, 10, 540. [Google Scholar] [CrossRef] [PubMed]
- Dubey, S.K.; Dubey, R.; Kleinman, M.E. Unraveling Histone Loss in Aging and Senescence. Cells 2024, 13, 320. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Liu, R.; Wu, J.; Guo, H.; Yao, W.; Li, S.; Lu, Y.; Jia, Y.; Liang, X.; Tang, J.; Zhang, H. Post-translational modifications of histones: Mechanisms, biological functions, and therapeutic targets. MedComm 2023, 4, e292. [Google Scholar] [CrossRef] [PubMed]
- Sidoli, S.; Bhanu, N.V.; Karch, K.R.; Wang, X.; Garcia, B.A. Complete Workflow for Analysis of Histone Post-translational Modifications Using Bottom-up Mass Spectrometry: From Histone Extraction to Data Analysis. J. Vis. Exp. 2016, 111, 54112. [Google Scholar]
- De Benedittis, S.; Gaspari, M.; Magariello, A.; Spadafora, P.; Citrigno, L.; Romeo, N.; Qualtieri, A. LC-MALDI-TOF ISD MS analysis is an effective, simple and rapid method of investigation for histones characterization: Application to EBV lymphoblastoid cell lines. J. Mass Spectrom. 2021, 56, e4712. [Google Scholar] [CrossRef] [PubMed]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [PubMed]
- Raisch, J.; Darfeuille-Michaud, A.; Nguyen, H.T. Role of microRNAs in the immune system, inflammation and cancer. World, J. Gastroenterol. 2013, 19, 2985–2996. [Google Scholar] [CrossRef]
- Mohammed, E.M. Environmental Influencers, MicroRNA, and Multiple Sclerosis. J. Cent. Nerv. Syst. Dis. 2020, 12, 1179573519894955. [Google Scholar] [CrossRef] [PubMed]
- Piket, E.; Zheleznyakova, G.Y.; Kular, L.; Jagodic, M. Small non-coding RNAs as important players, biomarkers and therapeutic targets in multiple sclerosis: A comprehensive overview. J. Autoimmun. 2019, 101, 17–25. [Google Scholar] [CrossRef] [PubMed]
- de Faria, O., Jr.; Moore, C.S.; Kennedy, T.E.; Antel, J.P.; Bar-Or, A.; Dhaunchak, A.S. MicroRNA dysregulation in multiple sclerosis. Front. Genet. 2013, 3, 311. [Google Scholar]
- Cox, M.B.; Cairns, M.J.; Gandhi, K.S.; Carroll, A.P.; Moscovis, S.; Stewart, G.J.; Broadley, S.; Scott, R.J.; Booth, D.R.; Lechner-Scott, J. ANZgene Multiple Sclerosis Genetics Consortium. MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS ONE 2010, 5, e12132. [Google Scholar] [CrossRef]
- Lindberg, R.L.; Hoffmann, F.; Mehling, M.; Kuhle, J.; Kappos, L. Altered expression of miR-17-5p in CD4+ lymphocytes of relapsing-remitting multiple sclerosis patients. Eur. J. Immunol. 2010, 40, 888–898. [Google Scholar] [CrossRef]
- Juntilla, M.M.; Koretzky, G.A. Critical roles of the PI3K/Akt signaling pathway in T cell development. Immunol. Lett. 2008, 116, 104–110. [Google Scholar] [CrossRef]
- Fruman, D.A.; Bismuth, G. Fine tuning the immune response with PI3K. Immunol. Rev. 2009, 228, 253–272. [Google Scholar] [CrossRef] [PubMed]
- Guerau-de-Arellano, M.; Smith, K.M.; Godlewski, J.; Liu, Y.; Winger, R.; Lawler, S.E.; Whitacre, C.C.; Racke, M.K.; Lovett-Racke, A.E. Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain 2011, 134 Pt 12, 3578–3589. [Google Scholar] [CrossRef]
- Mahad, D.H.; Trapp, B.D.; Lassmann, H. Pathological mechanisms in progressive multiple sclerosis. Lancet Neurol. 2015, 14, 183–193. [Google Scholar] [CrossRef]
- Yousuf, A.; Qurashi, A. Non-coding RNAs in the Pathogenesis of Multiple Sclerosis. Front. Genet. 2021, 12, 717922. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Y.; Lin, Y.C.; Li, J.; Huang, K.Y.; Shrestha, S.; Hong, H.C.; Tang, Y.; Chen, Y.G.; Jin, C.N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef] [PubMed]
- Welch, W.J. Heat shock proteins functioning as molecular chaperones: Their roles in normal and stressed cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1993, 339, 327–333. [Google Scholar]
- Nimmagadda, V.K.; Makar, T.K.; Chandrasekaran, K.; Sagi, A.R.; Ray, J.; Russell, J.W.; Bever, C.T., Jr. SIRT1 and NAD+ precursors: Therapeutic targets in multiple sclerosis a review. J. Neuroimmunol. 2017, 304, 29–34. [Google Scholar] [CrossRef]
- Piotrzkowska, D.; Miller, E.; Kucharska, E.; Niwald, M.; Majsterek, I. Association of miRNA and mRNA Levels of the Clinical Onset of Multiple Sclerosis Patients. Biology 2021, 10, 554. [Google Scholar] [CrossRef]
- Erkal, B.; Vural Korkut, S. Identification of miRNAs and their potential effects on multiple sclerosis related pathways using ın silico analysis. Mult. Scler. Relat. Disord. 2022, 59, 103642. [Google Scholar] [CrossRef]
- Waschbisch, A.; Atiya, M.; Linker, R.A.; Potapov, S.; Schwab, S.; Derfuss, T. Glatiramer acetate treatment normalizes deregulated microRNA expression in relapsing remitting multiple sclerosis. PLoS ONE 2011, 6, e24604. [Google Scholar] [CrossRef]
- Han, L.; Witmer, P.D.; Casey, E.; Valle, D.; Sukumar, S. DNA methylation regulates MicroRNA expression. Cancer Biol. Ther. 2007, 6, 1284–1288. [Google Scholar] [CrossRef] [PubMed]
- Scott, G.K.; Mattie, M.D.; Berger, C.E.; Benz, S.C.; Benz, C.C. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006, 66, 1277–1281. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, Y.; Liang, Y.; Zhao, M.; Long, H.; Ding, S.; Yin, H.; Lu, Q. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2011, 63, 1376–1386. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Chen, Y.; Zhou, X. The roles of microRNAs in epigenetic regulation. Curr. Opin. Chem. Biol. 2019, 51, 11–17. [Google Scholar] [CrossRef] [PubMed]
- van der Star, B.J.; Vogel, D.Y.; Kipp, M.; Puentes, F.; Baker, D.; Amor, S. In vitro and in vivo models of multiple sclerosis. CNS Neurol. Disord. Drug Targets 2012, 11, 570–588. [Google Scholar] [CrossRef] [PubMed]
- Lalive, P.H.; Neuhaus, O.; Benkhoucha, M.; Burger, D.; Hohlfeld, R.; Zamvil, S.S.; Weber, M.S. Glatiramer acetate in the treatment of multiple sclerosis: Emerging concepts regarding its mechanism of action. CNS Drugs 2011, 25, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Blair, H.A. Ofatumumab: A Review in Relapsing Forms of Multiple Sclerosis. Drugs 2022, 82, 55–62. [Google Scholar] [CrossRef]
| Tissue | Cohort | Method | Findings | Reference |
|---|---|---|---|---|
| cell-free plasma DNA | RRMS in remission vs. HC; RRMS in exacerbation vs. HC; and RRMS in exacerbation vs. remission | microarray-based assay | RR-MS patients have an ICAM1 hypermethylation pattern in response to clinical remission, the observed differences mostly correlated with the disease process. | Ligget et al., 2010 [40] |
| Brain tissue | Matched NAWM and chronically demyelinated MS lesions | pyrosequencing | DNA methylation status in the promoter region of MBP, may be used to bidirectionally influence myelination and cellular differentiation in vitro. | Tiane et al., 2023 [41] |
| Blood | RRMS and SPMS vs. NNC | pyrosequencing | MBP, MAG, and CNTN2 showed a significant difference in methylation pattern between the control and MS sample. | Tiane et al., 2024 [42] |
| Buffy coat | 69 MS (7 PPMS, 50 RRMS, 12 SPMS; 49 F, 20 M), 19 HC | cloning BS-sequencing | Increased DNA methylation level of SHP-1 promoter 2 in MS compared with HC. No relationships between DNA methylation and MS clinical parameters (EDSS, disease duration, phase). | Kumagai et al., 2012 [45] |
| CD4+ T cells | 17 RRMS (10 MS tysabri, 3 glatiramer acetate, 2 IFN-1b treated, 2 nontreated), 7 HC | cloning BS-sequencing | No difference in DNA methylation level of FOX3P and IL-17A between MS tysabri-treated patients and HC. Demethylation of FOX3P and IL-17A loci in tysabri untreated patients compared with HC. | Janson et al., 2011 [46] |
| Brain NAWM | DC: 28 MS; VC: 10 MS, 20 NNC | Illumina 450 K array | 220 hypomethylated DMRs (1235 CpGs) and 319 hypermethylated DMRs (1292 CpGs) at oligodendrocyte specific genes (BCL2L2, HAGHL, NDRG1, CTSZ, andLGMN). Correlation with expression of a portion of corresponding genes. | Huynh et al., 2014 [51] |
| Hippocampus | myelinated (n = 8) or demyelinated MS (n = 7) | Illumina 450 K array | Hyper-methylation in 10 genes and hypo-methylation in 6 genes. | Chomyk et al., 2017 [52] |
| Brain tissue | active lesion, chronic active lesion, chronic inactive lesion and NAWM vs. HC | Illumina 450 K array | Decreased activity of the transcription factor CREB, linked to neuro-axonal dysfunction in MS patients vs. controls. | Kular et al., 2019 [53] |
| Brain tissue | active lesion, chronic active lesion, chronic inactive lesion and NAWM vs. HC | Illumina 450 K array | A total of 1226 significantly differentiably methylated sites between MS and NNC. | Kular et al., 2022 [54] |
| CD14+ monocytes | PPMS vs. HC | Illumina 450 K array | A total of 169 DMPs, 90.5% of which were hypermethylated in PPMS patients. | Kiselev et al., 2020 [55] |
| CD4+ and CD8+ T cells, CD14+ monocytes and CD19+ B cells | RRMS, SPMS and HC | EWAS | A total of 1511, 666, and 30 significant DMPs in CD19+, CD14+, and CD8+ cells, respectively, between RRMS, SPMS, and HC. | Ewing et al., 2019 [58] |
| CD4+ T lymphocytes and CD14+ monocytes | treatment-naïve RR-MS patients and HC | EWAS | In CD4+ cells of MS patients, most of the DMPs were hypomethylated, while in CD14+ cells they were hypermethylated. | Kiselev et al., 2022 [59] |
| CD4+ T cells | 30 RRMS (interferons, glatiramer acetate, natalizumab or fingolimod treated), 28 HC | Illumina 450 K array | Correlation of HLA-DRB1 DNA methylation status and HLA-DRB1*15:01 haplotype. Differently methylated CpG sites (19) inside of MHC region and 55 outside. | Graves et al., 2014 [60] |
| Blood, | MS vs. HC | EWAS | Negative correlation between gene expression, and methylation at DMR-2 | Xavier et al., 2023 [62]. |
| Sources | Methodology | Cohort | Upregulated miRNAs | Downregulated miRNAs | Reference |
|---|---|---|---|---|---|
| N.A. | comprehensive literature review (61 papers) | N.A. | miR-142-3p, miR-146a/b, miR-145, miR-155, miR-22, miR223/-3p, miR-326, miR-584 | miR-103, miR-15, miR-548, let-7, miR-140 | Piket et al., 2019 [86] |
| Whole blood | microarray analysis | 59 treatment naïve MS patients (18 PPMS, 17 SPMS, 24 RRMS) vs. 37 HC | miR-17 and miR-20a | COX et al., 2010 [88] | |
| CD4+ | qRT-PCR | 8 RRMS patients vs. 10 HC (discovery cohort) 23 RRMS patients vs. 20 HC (validation cohort) | miR-485-3p, miR-376a, miR-497, miR-193a, miR-126, miR-17-5p | miR-34a | Lindberg et al., 2010 [89] |
| naïve CD4+ T cells, memory CD4+ T cells | qRT-PCR | 22 treatment naïve MS patients (5 PPMS, 5 SPMS, 12 RRMS) vs. 16 HC | miR-128, miR-27b (naïve CD4+ T cells), miR-340 (memory CD4+ T cells) | Guerau-de-Arellano et al., 2011 [92] | |
| PBMC | qRT-PCR | 28 MS patients (15 SPMS, 13 RRMS) vs. 33 HC | miR-132, miR-21-5p, miR-181b-5p, miR-34a, miR-132-3p | miR-134 | Piotrzkowska et al., 2021 [98] |
| PBMC | Datasets analysis (from GEO) | 66 MS patients vs. 185 HC | mir-142-3p, mir-98, mir-629 | hsa-mir-212 | Erkal et al., 2022 [99] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manna, I.; De Benedittis, S.; Porro, D. A Comprehensive Examination of the Role of Epigenetic Factors in Multiple Sclerosis. Int. J. Mol. Sci. 2024, 25, 8921. https://doi.org/10.3390/ijms25168921
Manna I, De Benedittis S, Porro D. A Comprehensive Examination of the Role of Epigenetic Factors in Multiple Sclerosis. International Journal of Molecular Sciences. 2024; 25(16):8921. https://doi.org/10.3390/ijms25168921
Chicago/Turabian StyleManna, Ida, Selene De Benedittis, and Danilo Porro. 2024. "A Comprehensive Examination of the Role of Epigenetic Factors in Multiple Sclerosis" International Journal of Molecular Sciences 25, no. 16: 8921. https://doi.org/10.3390/ijms25168921
APA StyleManna, I., De Benedittis, S., & Porro, D. (2024). A Comprehensive Examination of the Role of Epigenetic Factors in Multiple Sclerosis. International Journal of Molecular Sciences, 25(16), 8921. https://doi.org/10.3390/ijms25168921






