The AtMINPP Gene, Encoding a Multiple Inositol Polyphosphate Phosphatase, Coordinates a Novel Crosstalk between Phytic Acid Metabolism and Ethylene Signal Transduction in Leaf Senescence
Abstract
:1. Introduction
2. Results
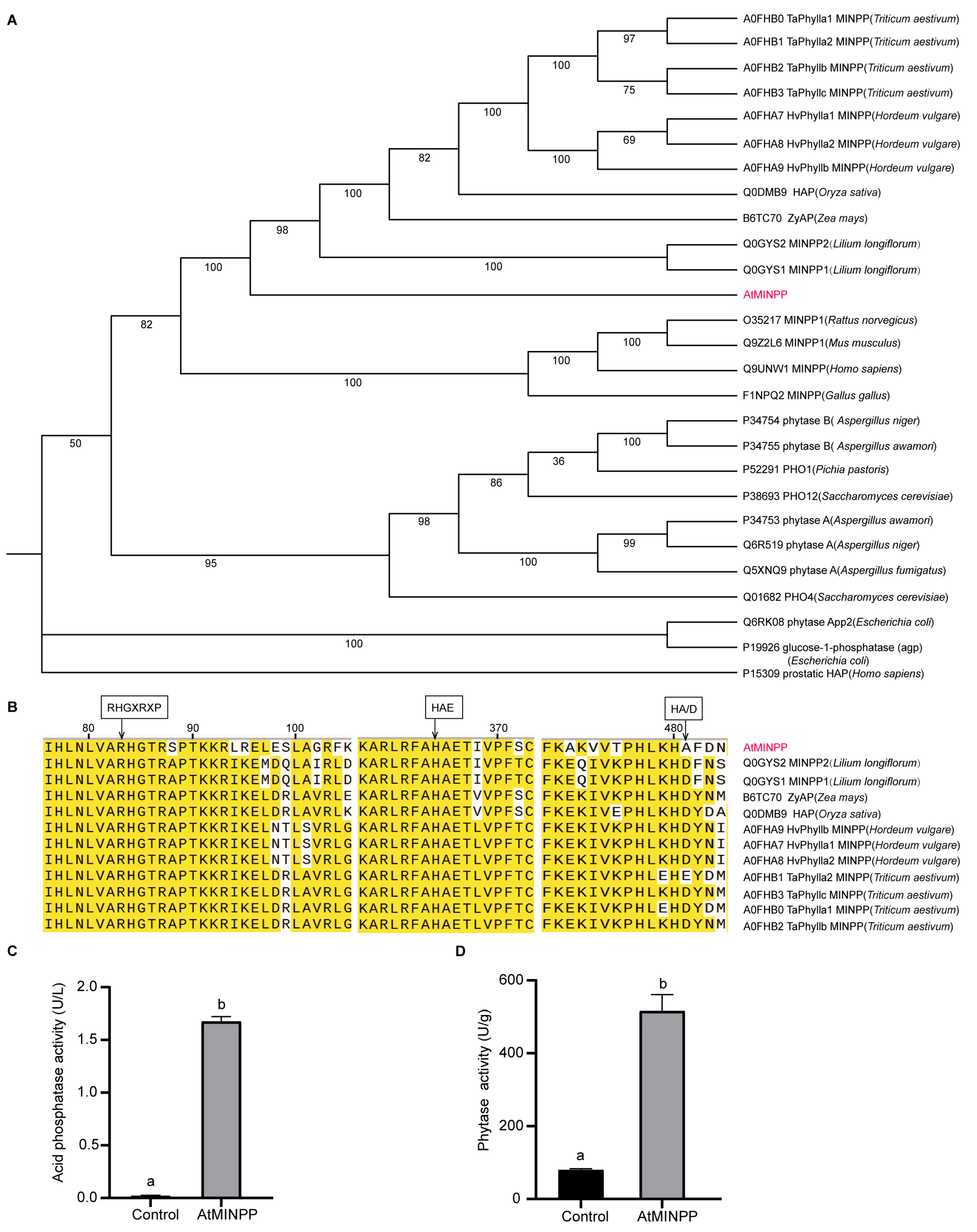
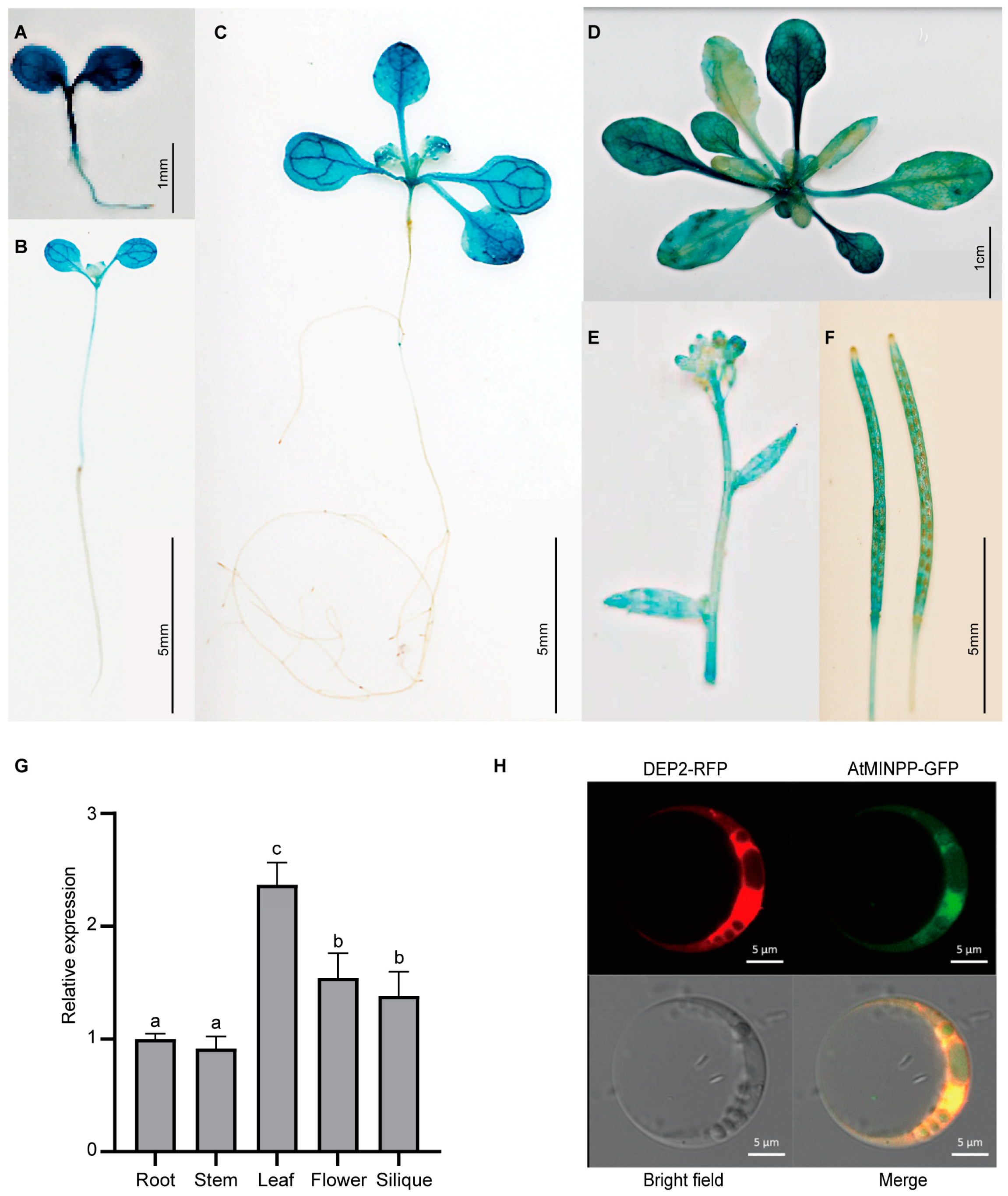
2.1. Isolation and Identification of the Multiple Inositol Polyphosphate Phosphatase Gene AtMINPP in Arabidopsis
2.2. AtMINPP Involves the Regulation of Leaf Senescence
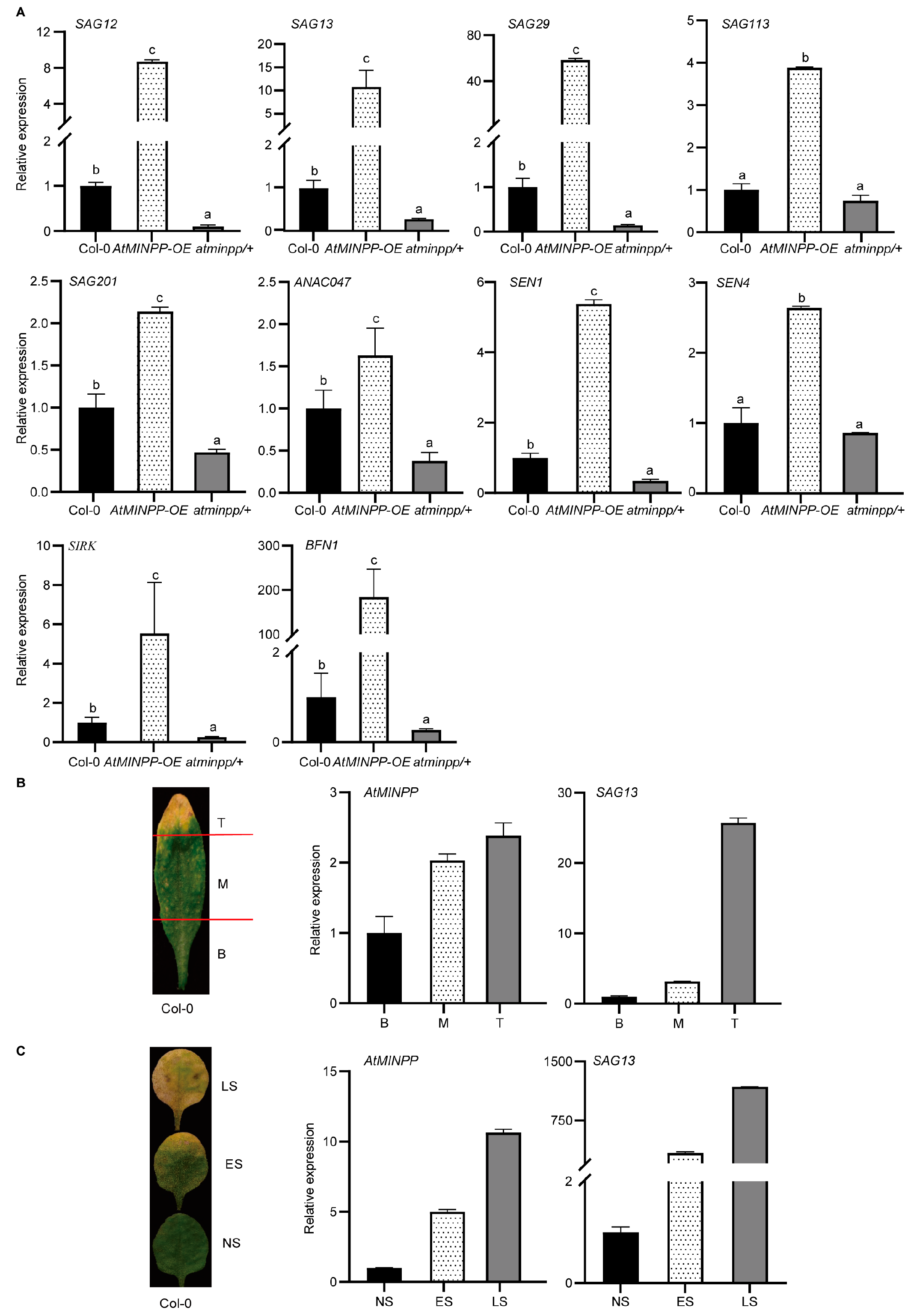
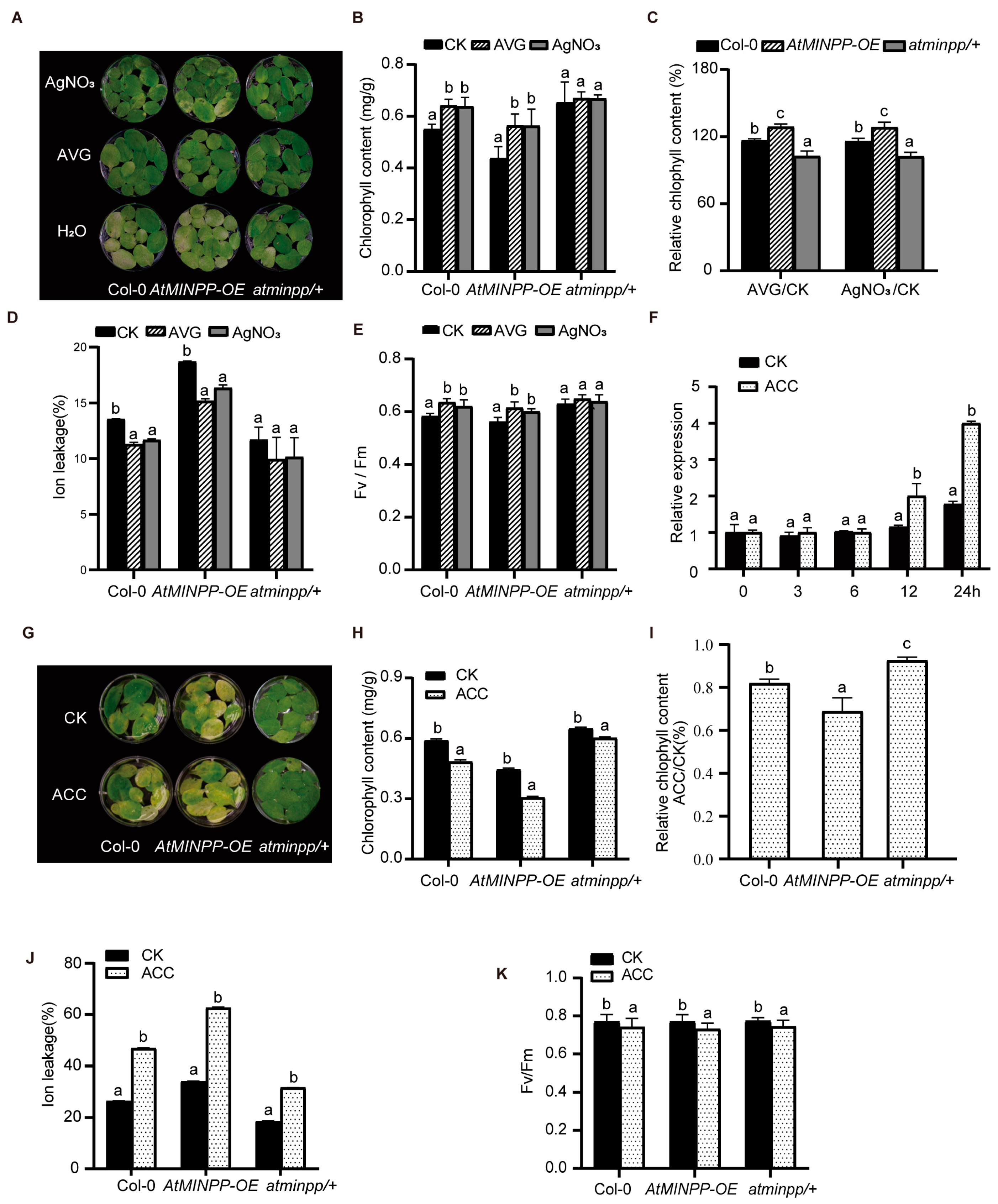
2.3. AtMINPP Mediates the Ethylene Signaling in Leaf Senescence
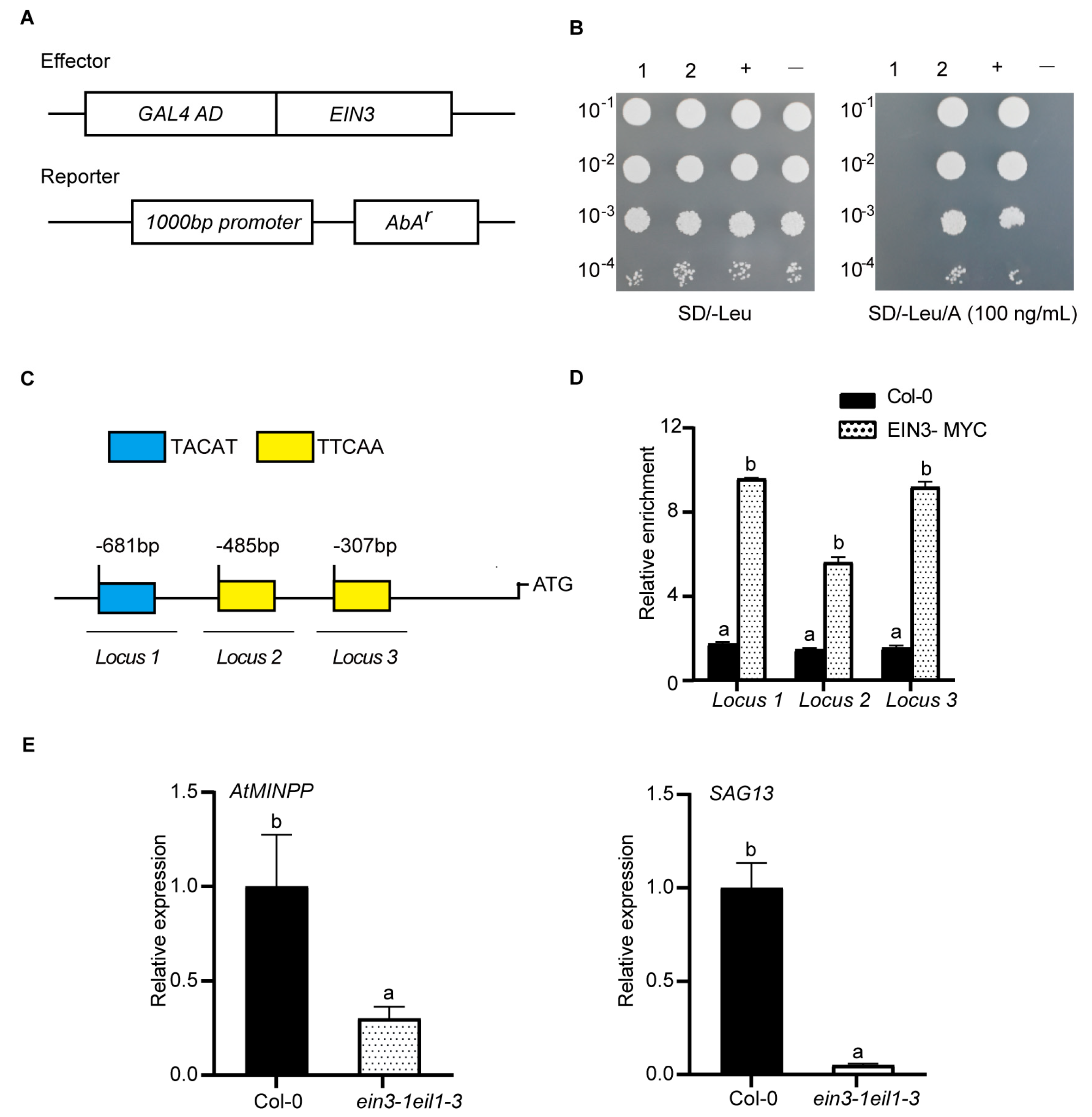
2.4. EIN3 Binds to the Promoter of AtMINPP Both In Vitro and In Vivo
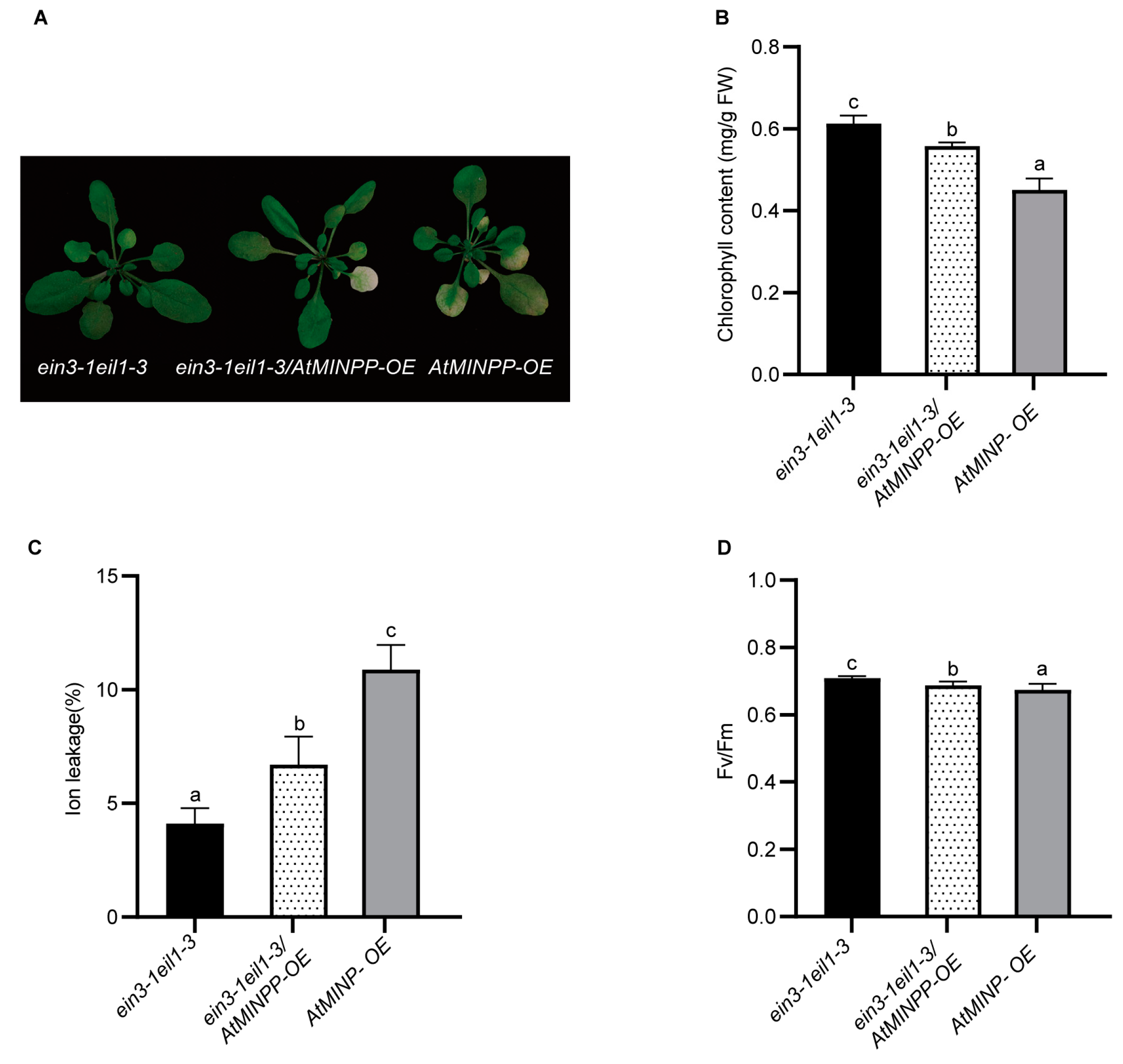
2.5. AtMINPP Mediates Ethylene Signaling to Accelerate Leaf Senescence
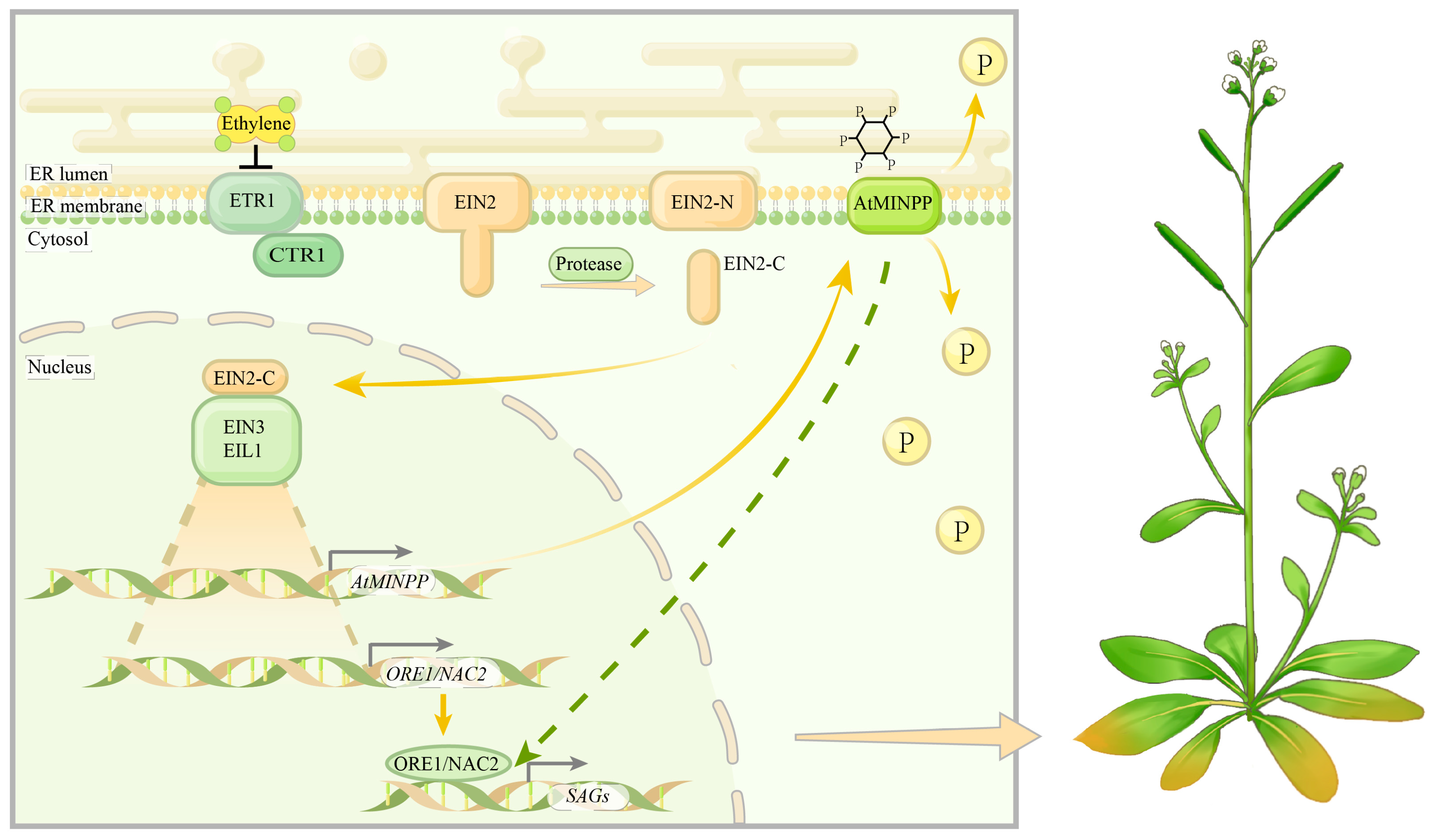
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Sequence Analysis and Alignment
4.3. Acid Phosphatase Activity, Phytase Activity, and Phytic Acid Content Assay
4.4. Measurement of Chlorophyll Content
4.5. Chlorophyll Fluorescence Measurement of Fv/Fm
4.6. Measurement of Ion Leakage
4.7. Real-Time Quantitative PCR Analysis
4.8. Yeast One-Hybrid Library Screening, Yeast One-Hybrid, and ChIP Assay
4.9. Treatment with ACC, AgNO3, and AVG
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gan, S.S.; Amasino, R.M. Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol. 1997, 113, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf senescence: Systems and dynamics aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef] [PubMed]
- Sarwat, M.; Naqvi, A.R.; Ahmad, P.; Ashraf, M.; Akram, N.A. Phytohormones and microRNAs as sensors and regulators of leaf senescence: Assigning macro roles to small molecules. Biotechnol. Adv. 2013, 31, 1153–1171. [Google Scholar] [CrossRef] [PubMed]
- Buchanan-Wollaston, V.; Earl, S.; Harrison, E.; Mathas, E.; Navabpour, S.; Page, T.; Pink, D. The molecular analysis of leaf senescence–A genomics approach. Plant Biotechnol. J. 2003, 1, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Eric, G.; Rainer, S.; Anja, S.; Marcelo, D.; Ulf-Ingo, F.; Reinhard, K. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 2006, 141, 776–792. [Google Scholar]
- Christ, B.; Hörtensteiner, S. Mechanism and significance of chlorophyll breakdown. J. Plant Growth Regul. 2014, 33, 4–20. [Google Scholar] [CrossRef]
- Woo, H.R.; Koo, H.J.; Kim, J.; Jeong, H.; Yang, J.O.; Lee, I.H.; Jun, J.H.; Choi, S.H.; Park, S.J.; Kang, B.; et al. Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in Arabidopsis. Plant Physiol. 2016, 171, 452–467. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gan, S. Leaf senescence: Signals, execution, and regulation. Curr. Top. Dev. Biol. 2005, 71, 83–112. [Google Scholar]
- Wang, C.Q.; Dai, S.Y.; Zhang, Z.L.; Lao, W.Q.; Wang, Q.Y.; Meng, X.Q.; Zhou, X. Ethylene and salicylic acid synergistically accelerate leaf senescence in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 828–833. [Google Scholar] [CrossRef]
- Guo, Y.; Ren, G.; Zhang, K.; Li, Z.; Miao, Y.; Guo, H.W. Leaf senescence: Progression, regulation, and application. Mol. Hortic. 2021, 1, 5–30. [Google Scholar] [CrossRef] [PubMed]
- Abeles, F.B.; Dunn, L.J.; Morgens, P.; Callahan, A.; Dinterman, R.E.; Schmidt, J. Induction of 33-kD and 60-kD peroxidases during ethylene-induced senescence of cucumber cotyledons. Plant Physiol. 1988, 87, 609–615. [Google Scholar] [CrossRef]
- Wang, N.N.; Yang, S.F.; Charng, Y. Differential expression of 1-aminocyclopropane-1-carboxylate synthase genes during orchid flower senescence induced by the protein phosphatase inhibitor okadaic acid. Plant Physiol. 2001, 126, 253–260. [Google Scholar] [CrossRef]
- Li, Z.H.; Guo, H.W. Ethylene treatment in studying leaf senescence in Arabidopsis. In Plant Senescence: Methods in Molecular Biology; Humana Press: New York, NY, USA, 2018; Volume 1744, pp. 105–112. [Google Scholar]
- Wang, K.L.; Li, H.; Ecker, J.R. Ethylene biosynthesis and signaling networks. Plant Cell 2002, 14, S131–S151. [Google Scholar] [CrossRef] [PubMed]
- Chao, Q.; Rothenberg, M.; Solano, R.; Roman, G.; Terzaghi, W.; Ecker, J.R. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 1997, 89, 1133–1144. [Google Scholar] [CrossRef]
- Li, Z.H.; Peng, J.; Wen, X.; Guo, H.W. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 2013, 25, 3311–3328. [Google Scholar] [CrossRef] [PubMed]
- Belgaroui, N.; Berthomieu, P.; Rouached, H.; Hanin, M. The secretion of the bacterial phytase PHY-US417 by Arabidopsis roots reveals its potential for increasing phosphate acquisition and biomass production during co-growth. Plant Biotechnol. J. 2014, 14, 1914–1924. [Google Scholar] [CrossRef]
- Cominelli, E.; Pilu, R.; Sparvoli, F. Phytic Acid and Transporters: What can we learn from low phytic acid Mutants? Plants 2020, 9, 69. [Google Scholar] [CrossRef]
- Takagi, D.; Miyagi, A.; Tazoe, Y.; Suganami, M.; Kawai-Yamada, M.; Ueda, A.; Suzuki, Y.; Noguchi, K.; Hirotsu, N.; Makino, A. Phosphorus toxicity disrupts rubisco activation and reactive oxygen species defence systems by phytic acid accumulation in leaves. Plant Cell Environ. 2020, 43, 2033–2053. [Google Scholar] [CrossRef]
- Raboy, V. Myo-inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 2003, 64, 1033–1043. [Google Scholar] [CrossRef]
- Alkarawi, H.H.; Zotz, G. Phytic acid in green leaves of herbaceous plants-temporal variation in situ and response to different nitrogen/phosphorus fertilizing regimes. Aob Plants 2014, 6, 48. [Google Scholar] [CrossRef] [PubMed]
- Raboy, V. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 2009, 177, 281–296. [Google Scholar] [CrossRef]
- Brinch-Pedersen, H.; Madsen, C.K.; Holme, I.B.; Dionisio, G. Increased understanding of the cereal phytase complement for better mineral bio-availability and resource management. J. Cereal Sci. 2014, 59, 373–381. [Google Scholar] [CrossRef]
- Sparvoli, F.; Cominelli, E. Seed biofortification and phytic acid reduction: A conflict of interest for the plant? Plants 2015, 4, 728–755. [Google Scholar] [CrossRef]
- Yamaji, N.; Takemoto, Y.; Miyaji, T.; Mitani-Ueno, N.; Yoshida, K.T.; Ma, J.F. Reducing phosphorus accumulation in rice grains with an impaired transporter in the node. Nature 2017, 541, 92–95. [Google Scholar] [CrossRef]
- Lott, J.N.A.; Ockenden, I.; Raboy, V.; Batten, G.D. Phytic acid and phosphorus in crop seeds and fruits: A global estimate. Seed Sci. Res. 2000, 10, 11–33. [Google Scholar] [CrossRef]
- Bohn, L.; Meyer, A.S.; Rasmussen, S.S. Phytate: Impact on environment and human nutrition. A challenge for molecular breeding. J. Zhejiang Univ. Sci. B 2008, 9, 165–191. [Google Scholar] [CrossRef] [PubMed]
- Duong, Q.H.; Clark, K.D.; Lapsley, K.G.; Pegg, R.B. Quantification of inositol phosphates in almond meal and almond brown skins by HPLC/ESI/MS. Food Chem. 2017, 229, 84–92. [Google Scholar] [CrossRef]
- Colombo, F.; Paolo, D.; Cominelli, E.; Sparvoli, F.; Pilu, R. MRP transporters and low phytic acid mutants in major crops: Main pleiotropic effects and future perspectives. Front. Plant Sci. 2020, 11, 1301. [Google Scholar] [CrossRef]
- Frittelli, A.; Botticella, E.; Palombieri, S.; Masci, S.; Celletti, S.; Fontanella, M.C.; Astolfi, S.; Vita, P.D.; Volpato, M.; Sestili, F. The suppression of TdMRP3 genes reduces the phytic acid and increases the nutrient accumulation in durum wheat grain. Front. Plant Sci. 2023, 14, 1079559. [Google Scholar] [CrossRef] [PubMed]
- Brinch-Pedersen, H.; Sorensen, L.D.; Holm, P.B. Engineering crop plants: Getting a handle on phosphate. Trends Plant Sci. 2002, 7, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Madsen, C.K.; Brinch-Pedersen, H. Molecular advances on phytases in barley and wheat. Int. J. Mol. Sci. 2019, 20, 2459–2469. [Google Scholar] [CrossRef]
- Konietzny, U.; Greiner, R. Molecular and catalytic properties of phytate-degrading enzymes (phytases). Int. J. Food Sci. Technol. 2002, 37, 791–812. [Google Scholar] [CrossRef]
- Oh, B.C.; Choi, W.C.; Park, S.; Kim, Y.O.; Oh, T.K. Biochemical properties and substrate specificities of alkaline and histidine acid phytases. Appl. Microbiol. Biotechnol. 2004, 63, 362–372. [Google Scholar] [CrossRef]
- Dionisio, G.; Holm, P.B.; Brinch-Pedersen, H. Wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.) multiple inositol polyphosphate phosphatases (MINPPs) are phytases expressed during grain filling and germination. Plant Biotechnol. J. 2007, 5, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.B.; Tiller, G.E.; Dasouki, M.J.; Romano, P.R.; Wang, J.; O’Keefe, R.J.; Puzas, J.E.; Rosier, R.N.; Reynolds, P.R. Multiple inositol polyphosphate phosphatase: Evolution as a distinct group within the histidine phosphatase family and chromosomal localization of the human and mouse genes to chromosomes 10q23 and 19. Genomics 1999, 56, 324–336. [Google Scholar] [CrossRef]
- Mehta, B.D.; Jog, S.P.; Johnson, S.C.; Murthy, P.P. Lily pollen alkaline phytase is a histidine phosphatase similar to mammalian multiple inositol polyphosphate phosphatase (MINPP). Phytochemistry 2006, 67, 1874–1886. [Google Scholar] [CrossRef]
- Pua, E.C.; Sim, G.E.; Chi, G.L.; Kong, L.F. Synergistic effect of ethylene inhibitors and putrescine on shoot regeneration from hypocotyl explants of Chinese radish (Raphanus sativus L. var. Longipinnatus Bailey) in vitro. Plant Cell Rep. 1996, 15, 685–690. [Google Scholar] [CrossRef]
- Madsen, C.K.; Brinch-Pedersen, H. Globoids and phytase: The mineral storage and release system in seeds. Int. J. Mol. Sci. 2020, 21, 7519. [Google Scholar] [CrossRef]
- Winkler, U.; Zotz, G. Highly efficient uptake of phosphorus in epiphytic bromeliads. Ann. Bot. 2009, 103, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Riya, P.; Kumar, S.S.; Giridhar, P. Phytoconstituents, GC-MS characterization of omega fatty acids, and antioxidant potential of less-known plant Rivina humilis L. ACS Omega 2023, 8, 28519–28530. [Google Scholar] [CrossRef] [PubMed]
- Laboure, A.M.; Gagnon, J.; Lescure, A.M. Purification and characterization of a phytase (myo-inositol-hexakisphosphate phosphohydrolase) accumulated in maize (Zea mays) seedlings during germination. Biochem. J. 1993, 295, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Hubel, F.; Beck, E. Maize Root Phytase (purification, characterization, and localization of enzyme activity and its putative substrate). Plant Physiol. 1996, 112, 1429–1436. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.Y.; Gruszewski, H.A.; Chevone, B.I.; Nessler, C.L. An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiol. 2008, 146, 323–324. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.F.; Qian, W.Q.; Lu, X.Z.; Li, D.P.; Liu, X.; Liu, K.F.; Wang, D.W. Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominantly transcribed in flower. Plant Mol. Biol. 2005, 59, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Kuang, R.B.; Chan, K.H.; Yeung, E.; Lim, B.L. Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiol. 2009, 151, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Hobbie, L.; McGovern, M.; Hurwitz, L.R.; Pierro, A.; Liu, N.Y.; Bandyopadhyay, A.; Estelle, M. The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 2000, 127, 23–32. [Google Scholar] [CrossRef]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.S.; Penfold, C.A.; Jenkins, D.; et al. High resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef]
- Kim, J.H.; Woo, H.R.; Kim, J.; Lim, P.O.; Lee, I.C.; Choi, S.H.; Hwang, D.; Nam, H.G. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057. [Google Scholar] [CrossRef]
- Kim, H.J.; Hong, S.H.; Kim, Y.W.; Lee, I.H.; Jun, J.H.; Phee, B.K.; Rupak, T.; Jeong, H.; Lee, Y.; Hong, B.S.; et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 2014, 65, 4023–4036. [Google Scholar] [CrossRef] [PubMed]
- Park, H.L.; Seo, D.H.; Lee, H.Y.; Bakshi, A.; Park, C.; Chien, Y.C.; Kieber, J.J.; Binder, B.M.; Yoon, G.M. Ethylene-triggered subcellular trafficking of CTR1 enhances the response to ethylene gas. Nat. Commun. 2023, 14, 365. [Google Scholar] [CrossRef]
- Robinson, W.D.; Carson, I.; Ying, S.; Ellis, K.; Plaxton, W.C. Eliminating the purple acid phosphatase AtPAP26 in Arabidopsis thaliana delays leaf senescence and impairs phosphorus remobilization. New Phytol. 2012, 196, 1024–1029. [Google Scholar] [CrossRef]
- Gao, W.W.; Lu, L.H.; Qiu, W.M.; Wang, C.; Shou, H.X. OsPAP26 encodes a major purple acid phosphatase and regulates phosphate remobilization in rice. Plant Cell Physiol. 2017, 58, 885–892. [Google Scholar] [CrossRef]
- Chen, L.G.; Xiang, S.Y.; Chen, Y.L.; Li, D.B.; Yu, D.Q. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant. 2017, 10, 1174–1189. [Google Scholar] [CrossRef]
- Yu, Y.C.; Qi, Y.N.; Xu, J.P.; Dai, X.H.; Chen, J.C.; Dong, D.H.; Xiang, F.N. Arabidopsis WRKY71 regulates ethylene-mediated leaf senescence by directly activating EIN2, ORE1 and ACS2 genes. Plant J. 2021, 107, 1819–1836. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.J. Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in plant defense: Do we understand what they are whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef]
- Huang, P.X.; Dong, Z.; Guo, P.R.; Zhang, X.; Qiu, Y.P.; Li, B.S.; Wang, Y.C.; Guo, H.W. Salicylic Acid Suppresses Apical Hook Formation via NPR1-mediated repression of EIN3 and EIL1 in Arabidopsis. Plant Cell 2020, 32, 612–629. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.G.; Zhu, C.M.; Liu, Y.D.; Karthikeyan, A.S.; Bressan, R.A.; Raghothama, K.G.; Liu, D. Ethylene signalling is involved in regulation of phosphate starvation-induced gene expression and production of acid phosphases and anthocyanin in Arabidopsis. New Phytol. 2011, 189, 1084–1095. [Google Scholar] [CrossRef]
- Song, L.; Yu, H.P.; Dong, J.S.; Che, X.M.; Jiao, Y.L.; Liu, D. The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genet. 2016, 12, e1006194. [Google Scholar] [CrossRef]
- Otegui, M.; Capp, R.; Staehelin, L. Developing seeds of Arabidopsis store different minerals in two types of vacuoles and in the endoplasmic reticulum. Plant Cell 2002, 14, 1311–1327. [Google Scholar] [CrossRef]
- Himelblau, E.; Amasino, R.M. Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J. Plant Physiol. 2001, 158, 1317–1323. [Google Scholar] [CrossRef]
- Ulker, B.; Mukhtar, M.S.; Somssich, I.E. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 2007, 226, 125–137. [Google Scholar] [CrossRef]
- Avila-Ospina, L.; Moison, M.; Yoshimoto, K.; Masclaux, D.C. Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 2014, 65, 3799–3811. [Google Scholar] [CrossRef]
- Yan, B.F.; Hu, C.F.; Zhao, M.; Qiu, R.L.; Tang, Y.T. Characterizing the remobilization flux of cadmium from pre-anthesis vegetative pools in rice during grain filling using an improved stable isotope labeling method. Environ. Pollut. 2023, 331, 121891. [Google Scholar] [CrossRef]
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Li, L.; Li, B.; Zhu, S.R.; Wang, L.; Song, L.M.; Chen, J.; Ming, Z.H.; Liu, X.M.; Li, X.S.; Yu, F. TMK4 receptor kinase negatively modulates ABA signaling by phosphorylating ABI2 and enhancing its activity. J. Integr. Plant Biol. 2021, 63, 1161–1178. [Google Scholar] [CrossRef] [PubMed]
- Sartory, D.P.; Grobbelaar, J.U. Extraction of chlorophyll a from freshwater phytoplankton for spectrophotometric analysis. Hydrobiologia 1984, 114, 177–187. [Google Scholar] [CrossRef]
- Yang, H.J.; Zhou, Y.; Zhang, Y.N.; Wang, J.; Shi, H.Z. Identification of transcription factors of nitrate reductase gene promoters and NRE2 cis-element through yeast one-hybrid screening in Nicotiana tabacum. BMC Plant Biol. 2019, 19, 145. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.G.; Gao, L.; Dinh, T.T.; Shi, T.; Li, D.M.; Wang, R.Z.; Guo, L.; Xiao, L.T.; Chen, X.M. DNA topoisomerase Ι affects polycomb group protein-mediated epigenetic regulation and plant development by altering nucleosome distribution in Arabidopsis. Plant Cell 2014, 26, 2803–2817. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, X.; Li, H.; Xu, W.; Yang, Q.; Li, D.; Fan, T.; Li, B.; Ding, J.; Ku, W.; Deng, D.; et al. The AtMINPP Gene, Encoding a Multiple Inositol Polyphosphate Phosphatase, Coordinates a Novel Crosstalk between Phytic Acid Metabolism and Ethylene Signal Transduction in Leaf Senescence. Int. J. Mol. Sci. 2024, 25, 8969. https://doi.org/10.3390/ijms25168969
Peng X, Li H, Xu W, Yang Q, Li D, Fan T, Li B, Ding J, Ku W, Deng D, et al. The AtMINPP Gene, Encoding a Multiple Inositol Polyphosphate Phosphatase, Coordinates a Novel Crosstalk between Phytic Acid Metabolism and Ethylene Signal Transduction in Leaf Senescence. International Journal of Molecular Sciences. 2024; 25(16):8969. https://doi.org/10.3390/ijms25168969
Chicago/Turabian StylePeng, Xiaoyun, Haiou Li, Wenzhong Xu, Qian Yang, Dongming Li, Tingting Fan, Bin Li, Junhui Ding, Wenzhen Ku, Danyi Deng, and et al. 2024. "The AtMINPP Gene, Encoding a Multiple Inositol Polyphosphate Phosphatase, Coordinates a Novel Crosstalk between Phytic Acid Metabolism and Ethylene Signal Transduction in Leaf Senescence" International Journal of Molecular Sciences 25, no. 16: 8969. https://doi.org/10.3390/ijms25168969




