Abstract
Cytokinins (CKs) are a group of phytohormones that are involved in plant growth, development, and disease resistance. The isopentenyl transferase (IPT) and cytokinin oxidase/dehydrogenase (CKX) families comprise key enzymes controlling CK biosynthesis and degradation. However, an integrated analysis of these two gene families in radish has not yet been explored. In this study, 13 RsIPT and 12 RsCKX genes were identified and characterized, most of which had four copies in Brassica napus and two copies in radish and other diploid Brassica species. Promoter analysis indicated that the genes contained at least one phytohormone or defense and stress responsiveness cis-acting element. RsIPTs and RsCKXs were expanded through segmental duplication. Moreover, strong purifying selection drove the evolution of the two gene families. The expression of the RsIPT and RsCKX genes distinctly showed diversity in different tissues and developmental stages of the root. Expression profiling showed that RsCKX1-1/1-2/1-3 was significantly upregulated in club-resistant materials during primary infection, suggesting their vital function in clubroot resistance. The interaction network of CKX proteins with similar 3D structures also reflected the important role of RsCKX genes in disease resistance. This study provides a foundation for further functional study on the IPT and CKX genes for clubroot resistance improvement in Raphanus.
1. Introduction
Radish (Raphanus sativus L. 2n = 18), belonging to the Brassicaceae, is an important vegetable and oil crop cultivated widely around the world. Clubroot disease, which is caused by biotrophic protist Plasmodiophora brassicae Woronin, leads to serious losses in radish, Chinese cabbage (Brassica rapa L. ssp. pekinensis Lour., 2n = 20), and other Cruciferous crops and has gradually evolved into a catastrophic disease worldwide [1,2,3]. Plasmodiophora brassicae has a complex life cycle, while the infection process comprises two distinct phases. The two phases occur in different parts of plant roots and have different infection routes. The primary phase starts with infection by primary zoospores, whose resting spores germinate in root hair and epidermal cells [4]. Then, the primary zoospores form primary plasmodia, further split into zoosporangia, and produce secondary zoospores in epidermal cells. A sign of the secondary phase is that the secondary zoospores infect cortical tissue. Subsequently, secondary zoospores develop into secondary plasmodia and then release resting spores into the soil. It is difficult to manage and control clubroot disease due to the persistent vitality and diverse transmission of P. brassicae [5,6].
Cytokinins (CKs) are essential phytohormones that are involved in various biological processes related to plant growth and development, such as cell division in roots and shoots, apical dominance, shoot or root branching, leaf development, bud growth, seed germination, photosynthesis, senescence, and nutrient metabolism [7]. Furthermore, CKs participate in the response to multiple stress factors and have a significant role in plant immunity against microorganisms [8,9,10,11,12]. Thus far, CKs have been considered connected to disease symptoms and abnormal morphogenesis, such as the formation of nodules or galls, fasciation, senescence, and the generation of green islands [13,14]. Some studies have shown that CKs mediate plant susceptibility to gall-causing pathogens and biotrophic fungi [15,16,17,18].
Typically, a host plant has its own stable source–sink relationship; pathogen invasion can affect the reconfiguration of plant metabolism, such as by altering the hormone balance, leading to a new source–sink relationship [17,19]. Previous studies have shown that the alteration of plant root hormone homeostasis plays a prominent part in clubroot formation [20,21,22,23]. In particular, the alteration of CK homeostasis driven by P. brassicae induces cell division and hypertrophy by reconfiguring existing meristematic activity, resulting in root nodules [24,25,26]. In Arabidopsis thaliana L. (2n = 10), research has shown that the CK concentration increases in root galls after infection with P. brassicae [27,28,29]. Further research has shown that the CK concentration increases in the early phase but decreases in the later period of root hypertrophy in Arabidopsis [22,30]. In turnips (Brassica rapa subsp. Rapa L., 2n = 20) and rapeseed (Brassica napus L., 2n = 38), the CK content is higher in infected roots than in non-infected roots [23,31]. CK-depleted Arabidopsis plants are more resistant to clubroot disease [21,23]. Therefore, previous studies have clearly indicated that CKs are a key factor in plant resistance to P. brassicae.
Although multiple CK biosynthesis routes have been proposed, CK biosynthesis is mainly catalyzed by the isopentenyl transferase (IPT) family [8,32,33,34]. The synthesis of isopentenyladenine(ip)- and trans-zeatin(tz)-type CKs is controlled by ATP/ADP IPTs, and the synthesis of cis-zeatin-type CKs is controlled by tRNA IPTs. The degradation processes of CKs and their derivatives are mainly catalyzed by CK oxidase/dehydrogenases (CKXs).
IPT and CKX gene families have been analyzed in crucifer crops such as Arabidopsis thaliana, Brassica rapa L. (2n = 20), Brassica oleracea L. (2n = 18), and Brassica napus L. (2n = 38) [32,35,36,37,38,39]. In Arabidopsis, nine IPT members (AtIPT1–AtIPT9) and seven CKX members (AtCKX1–AtCKX7) have been identified [32,40]. In B. rapa, 13 BrIPT genes and 12 BrCKX genes have been identified [36]. In B. oleracea, 36 CKX genes have been identified [38]. In B. napus, a total of twenty-six IPT genes were identified [39]. Further function analysis has been carried out on these two family members in Arabidopsis. The overexpression of one IPT gene, AtIPT4, which has unique isopentenyltransferase activity for DMAPP: ATP/ADP in its purified form, led to CK-independent shoot formation on Arabidopsis calli [41]. The overexpression of AtCKX genes in transgenic tobacco and its own plants displays various degrees of reduction of cytokinin or CK-deficiency traits [35,42]. Compared to genes related to CK biosynthesis and degradation, which have been extensively studied in Arabidopsis, research on these genes has been less frequently reported in other crucifer crops. In particular, genome-wide identification of IPT and CKX gene family members has still not been reported in radish. It is unclear what role IPT and CKX genes play in the radish-P. brassicae interaction.
2. Results
2.1. Genome-Wide Identification and Distribution of CKX and IPT Genes in Radish
CKX and IPT genes were identified based on BLAST and hmmsearch among Cruciferous crops. A total of 13 IPT and 12 CKX genes were identified in radish. Based on homologous genes in A. thaliana (AtCKX and AtIPT), these genes were named the RsIPT and RsCKX genes. Furthermore, IPT and CKX genes were identified in other Cruciferous crops. Specifically, 9, 13, 13, 15, and 30 IPT genes and 7, 12, 12, 12, and 25 CKX genes were identified in Arabidopsis thaliana (2n = 10), Brassica rapa (2n = 20), Brassica oleracea (2n = 18), Brassica nigra L. (2n = 16), and Brassica napus (2n = 38), respectively (Table S1). The number of CKX and IPT genes showed obvious diversity among species with different ploidy and increased when ploidies doubled.
The physicochemical properties of the IPT and CKX genes were predicted (Table 1 and Table 2). The proteins of 13 RsIPT genes ranged from 113 (RsIPT7-2) to 499 (RsIPT9-1) amino acids in length, while 12 RsCKX genes ranged from 387 (RsCKX1-2) to 1033 (RsCKX3-1) amino acids in length. The relative molecular weight (MW) of the RsIPT genes varied from 12,356.3 Da (RsIPT7-2) to 56,375.9 Da (RsIPT9-1), while the MW of the RsCKX genes varied from 43,085.8 Da (RsCKX1-2) to 116,376 Da (RsCKX3-1). The PI of the RsIPT genes ranged from 5.28 (RsIPT10) to 10.35 (RsIPT7-2), while the PI of RsCKX ranged from 4.38 (RsCKX7-2) to 9.35 (RsCKX1-2). The predicted subcellular locations of RsIPT genes showed that most were in the chloroplast, and a few were located in the cytoskeleton, nucleus, and mitochondrion. Among the RsCKX genes, four were in the mitochondrion, two in the chloroplast, extracell, and cytoplasm, and one was in the vacuole and plasma membrane. The RsCKX genes showed more diversity in subcellular localization than the RsIPT genes. The paralogous genes, namely RsIPT9-1/9-2, RsIPT7-1/7-2, RsCKX1-1/1-2/1-3, and RsCKX7-1/7-2, were in the same subcellular location, while other paralogs were distributed in different locations.

Table 1.
Sequence characteristics and physicochemical properties of RsIPT genes in radish.

Table 2.
Sequence characteristics and physicochemical properties of RsCKX genes in radish.
The RsIPT and RsCKX genes were mapped to radish chromosomes (Figure 1). RsIPT3-2 was unmapped on a chromosome due to its location on scaffold 638. The overall distribution of the RsIPT and RsCKX genes on nine chromosomes was uneven. Chromosome 2 (R02) had the most RsIPT and RsCKX genes, followed by R01, R05, and R07. Specifically, five genes were located on R02, four genes were located on R01, R05, and R07, and three genes were located on R04. Both R03 and R09 had two genes. However, no RsIPT or RsCKX genes were identified on R06 and R08. No RsIPT genes were identified on R09.
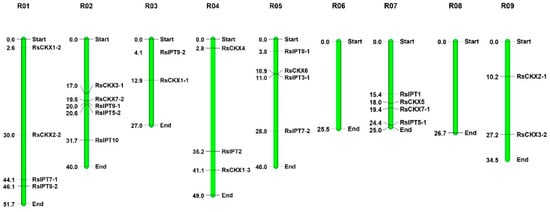
Figure 1.
Chromosomal mapping of the RsIPT and RsCKX genes in radish. The green bars represent chromosomes. Values corresponding to the RsIPT and RsCKX genes represent the physical distance (Mb).
2.2. Phylogenetic Analysis of RsIPT and RsCKX Gene Families
To study the evolutionary relationships and identify the subfamilies in the RsIPT and RsCKX gene families, phylogenetic analysis of the IPT and CKX genes was performed in A. thaliana, B. rapa, B. oleracea, B. nigra, B. napus, and R. sativus using MEGA with protein sequence information. Based on the phylogenetic tree, the RsIPT genes were classified into four subgroups (Figure 2): Group I (RsIPT1, 8-1, 8-2), Group II (RsIPT2), Group III (RsIPT3-1, 3-2, 5-1, 5-2, 7-1, 7-2), and Group IV (RsIPT9-1, 9-2). This classification of RsIPT genes was consistent with their homologues in other genetically close species. Most IPT genes had one corresponding gene in Arabidopsis, two in R. sativus, B. rapa, B. oleracea, and B. nigra, and four in B. napus, while RsIPT2 had two in B. napus and only one copy in Arabidopsis and other diploid species. There were no corresponding genes for AtIPT4 and AtIPT6 in radish or other Brassica species, suggesting that these genes might have been lost in the divergence process from Arabidopsis. RsIPT10 was not clustered into any group, suggesting that this gene may be unique to radish. The RsCKX genes were classified into four subgroups (Figure 3): Group I (RsCKX1-1, 1-2, 1-3, 6), Group II (RsCKX2-1, 2-2, 3-1, 3-2, 4), Group III (RsCKX5), and Group IV (RsCKX7-1, 7-2). Similar to IPT genes, most CKX genes also had one corresponding gene in Arabidopsis, two in radish and other diploid species, and four in B. napus. Moreover, the homologues of AtCKX1 contained three paralogous genes in diploid species and six in B. napus. Although there were various degrees of gene loss and gene duplication in the process of speciation in Arabidopsis, the number of IPT and CKX genes in Raphanus and Brassica species was approximately twice as high as in Arabidopsis.
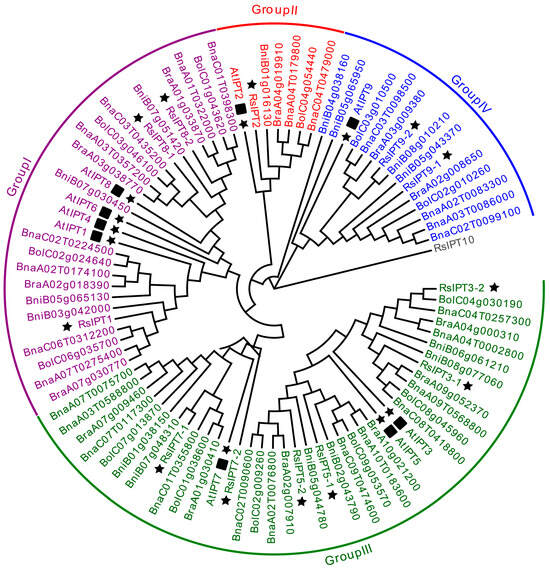
Figure 2.
Phylogenetic tree of IPT genes from Arabidopsis thaliana, Brassica rapa, B. oleracea, B. nigra, B. napus, and Raphanus sativus. IPT genes are divided into four groups according to phylogenetic analysis. Different groups are indicated with corresponding colors. The gene RsIPT10 which not belong to any group was presented by gray color. The IPT genes of R. sativus are highlighted by black stars, and the IPT genes of A. thaliana are highlighted by black stars and squares. The A. thaliana, B. rapa, B. oleracea, B. nigra, B. napus, and R. sativus genes are marked with “At”, “Bra”, “Bol”, “Bni”, “Bna”, and “Rs”, respectively.
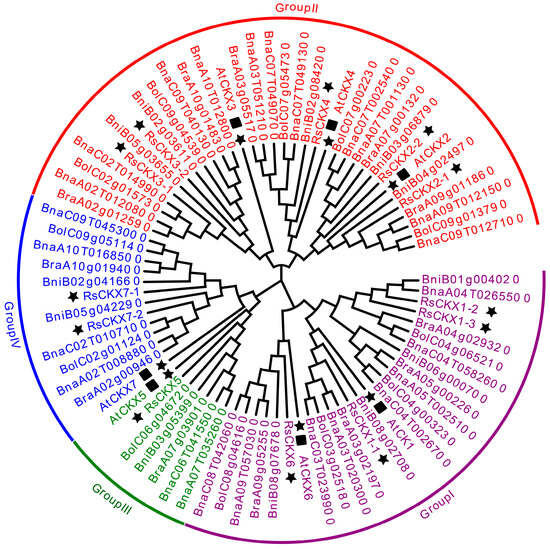
Figure 3.
Phylogenetic tree of CKX genes from Arabidopsis thaliana, Brassica rapa, B. oleracea, B. nigra, B. napus, and Raphanus sativus. CKX genes are divided into four groups according to phylogenetic analysis. Different groups are indicated with corresponding colors. The IPT genes of R. sativus are highlighted by black stars, and the IPT genes of A. thaliana are highlighted by black stars and squares. The A. thaliana, B. rapa, B. oleracea, B. nigra, B. napus, and R. sativus genes are marked with “At”, “Bra”, “Bol”, “Bni”, “Bna”, and “Rs”, respectively.
2.3. Gene Structure and Conserved Motif Analysis of the RsIPT and RsCKX Genes
To further understand the IPT and CKX genes, the exon-intron structure of these genes was analyzed. Most RsIPT genes (66.67%) had one or no introns (RsIPT1, 3, 5, 7, 8), while another one-third had more than nine introns (RsIPT2 and RsIPT9) (Figure 4b). Compared to the IPT family, the CKX family presented more diversity (Figure 5b,c). Not only did most RsCKX genes have three or four introns, but the length of the intron also varied among groups. However, the genes that were classified into the same group had similar structures, especially the number and length of exons that were relatively conserved.
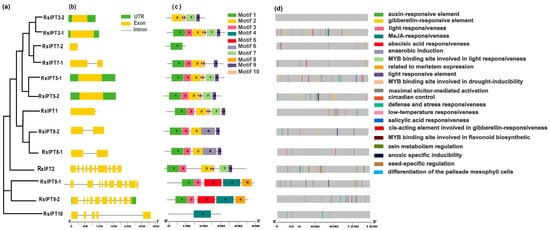
Figure 4.
Phylogenetic relationship (a), gene structure (b), conserved motifs (c), and cis-acting elements in promoters (d) of IPT genes in radish. The yellow rectangles and black lines represent exons and introns of genes, respectively, and their lengths are presented proportionally. The 10 conserved motifs and 21 cis-acting elements are indicated in different colors.
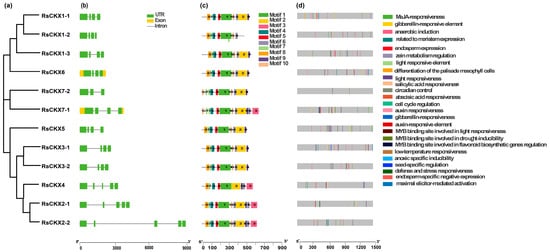
Figure 5.
Phylogenetic relationship (a), gene structure (b), conserved motifs (c), and cis-acting elements in promoters (d) of CKX genes in radish. The green rectangles and black lines represent exons and introns of genes, respectively, and their lengths are presented proportionally. The 10 conserved motifs and 25 cis-acting elements are indicated in different colors.
Then, to further reveal protein functional diversification, 10 conserved motifs in the RsIPT and RsCKX genes were predicted using MEME software. The results showed similar motif compositions in the same groups (Figure 4d and Figure 5d). For instance, group I (RsIPT) contained motifs 1, 2, 3, 6, and 9; group III (RsIPT) contained motifs 1, 3, 2, 10, 7, and 9; and group II (RsCKX) contained motifs 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10. However, some genes lacked motifs in the identical group. In group III (RsIPT), RsIPT3-2 lacked motifs 1 and 3, RsIPT7-1 lacked motif 7, and RsIPT7-2 only had motif 1. In group I (RsCKX), RsCKX1-2 lacked motifs 2 and 9. In group II (RsCKX), RsCKX3-1/3-2 lacked motif 3. Taken together, the motif composition in all RsCKX genes was very similar, except in RsCKX 7-1/7-2, which lacked motif 8 (common motifs 1, 4, 5, 6, 7, 9, and 10), while the common motifs were only motifs 1 and 3 in most RsIPT genes. The RsCKX genes exhibited a similar motif composition, suggesting that these proteins might have similar functions. In general, the variation trend of the exon–intron structure and motifs of the RsIPT and RsCKX genes aligned with their phylogenetic classification.
2.4. Cis-Acting Elements in the Promoters of RsIPT and RsCKX Genes
To obtain the potential functions and transcriptional regulation of RsIPT and RsCKX genes, the cis-regulatory elements in the promoters were identified. In total, 21 cis-regulatory elements were obtained in the RsIPT genes (Figure 4d). Cis-regulatory elements related to light responsiveness, such as GT1-motif, AAAC-motif, Sp1, G-box, and MRE, were most widely distributed. Among phytohormone-related elements, auxin-responsive elements (TGAs) were identified in five RsIPT genes; salicylic acid (SA) responsiveness (TCA) was identified in four RsIPT genes. Abscisic acid responsiveness (ABRE) and gibberellin-responsive elements, such as the P-box and GARE motif, were identified in seven RsIPT genes. Methyl jasmonate (MeJA) responsiveness (TGACG-motif and CGTCA-motif) was identified in eight RsIPT genes. At least one of the phytohormone-related elements was present in each RsIPT gene. This result shows that the RsIPT genes are extensively associated with phytohormone metabolism and signal transduction, playing an important role in plant growth and development. Stress-related elements, such as anaerobic induction (ARE), low-temperature responsiveness (LTR), MYB binding site involved in drought inducibility (MBS), MYB binding site involved in flavonoid biosynthetic (MBSI), and defense and stress responsiveness (TC-rich repeats), were also widely distributed among RsIPT genes. This suggests that RsIPT genes are responsive to multiple stimuli and play a part in defense to abiotic and biotic stress. In addition, identified elements related to differentiation of the palisade mesophyll cells (HD-Zip 1), seed-specific regulation (RY-element), meristem expression (CAT-box), and zein metabolism regulation (O2-site) further illustrated the functional diversification.
Compared to the RsIPT genes, 25 cis-regulatory elements were identified in the RsCKX genes (Figure 5d). Auxin responsiveness (AuxRR-core), cell cycle regulation (MSA-like), endosperm expression (GCN4_motif), and endosperm-specific negative expression (AACA_motif) were only predicted in the RsCKX genes. All genes contained light-responsive elements, but the types of elements differed from the RsIPT genes. Specifically, the 3-AF1 binding site and ACE elements were unique to the RsCKX genes, and the AAAC motif was only identified in the RsIPT genes. The hormone-related cis-elements were widely distributed, indicating that the RsCKX genes also play a vital role in plant growth and development processes in which hormones participate. Similarly, the RsCKX genes play an important part in the stress response due to their stress-related elements.
2.5. Gene Duplication and Divergence of the RsIPT and RsCKX Genes
To comprehend the expansion of the RsIPT and RsCKX genes and evolutionary mechanisms, gene duplication events were analyzed. Generally, tandem and segmental duplications are the major patterns of gene duplication. In this study, MCScanX and BLAST were used to search for tandem and segmental duplications. The results are shown in Figure 6 and Figure 7. Although the result revealed no tandem duplicated genes, segmental duplication existed in the two gene families. In the RsIPT genes, three segmental duplicate pairs were obtained: RsIPT5-1/RsIPT5-2, RsIPT8-1/RsIPT8-2, and RsIPT9-1/RsIPT9-2. Moreover, there were eight gene pairs with segmental duplication in RsCKX genes: RsCKX1-1/RsCKX1-3, RsCKX2-1/RsCKX2-2, RsCKX2-1/RsCKX4, RsCKX3-1/RsCKX3-2, RsCKX3-1/RsCKX4, RsCKX3-2/RsCKX4, RsCKX4/RsCKX8, and RsCKX7-1/RsCKX7-2. Six genes also presented trinal pairs: RsCKX3-1, RsCKX3-2, and RsCKX4; and RsCKX2-1, RsCKX2-2, and RsCKX4. Based on these results, segmental duplication was the major type of gene duplication causing expansion and played a key role in RsIPT and RsCKX gene evolution.
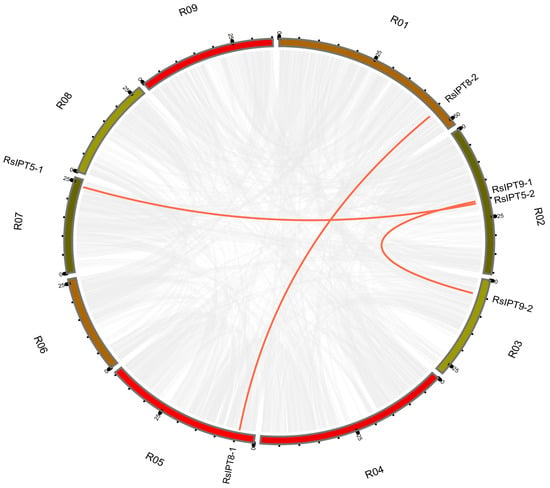
Figure 6.
Collinearity analysis of RsIPT genes among radish chromosomes. Different chromosomes are indicated with corresponding number and colors in different regions of circle. The genes connected by the red line are segmental duplication pairs.
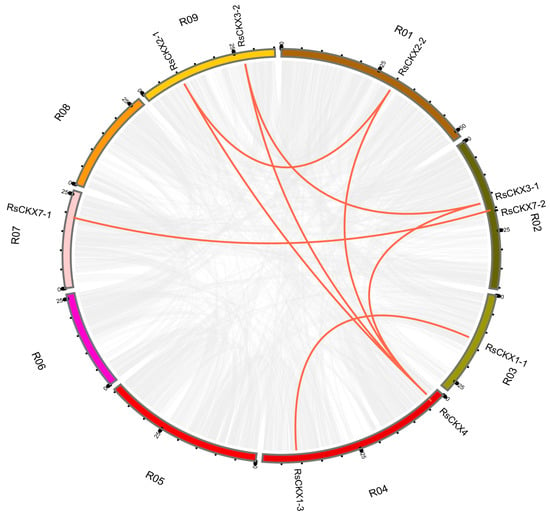
Figure 7.
Collinearity analysis of RsCKX genes among radish chromosomes. Different chromosomes are indicated with corresponding number and colors in different regions of circle. The genes connected by the red line are segmental duplication pairs.
To understand the divergence of IPT and CKX genes in Raphanus, the Ka and Ks values of orthologous and paralogous genes of both gene families were obtained (Table 3 and Table S1). The Ks values of orthologous genes between A. thaliana and R. sativus ranged from 0.32 to 0.62, suggesting that the divergence time of the RsIPT and RsCKX genes from A. thaliana was 10–21 MYA (Table 3). Comparative analysis indicated that the divergence times of two orthologous copies of the IPT and CKX genes in A. thaliana were nearly consistent. The Ks values of orthologous genes between B. rapa and R. sativus ranged from 0.08 to 0.51 (Table S2a, but the Ks value among orthologous copies of the same RsIPT or RsCKX genes varied distinctly. Therefore, the divergence times were classified into two periods: 2–10 and 10–17 MYA (Table S2b). Compared with the corresponding orthologous genes, the Ks values of paralogous genes in radish were lower, suggesting that the duplication of IPT and CKX genes in radish was due to divergence from A. thaliana. In addition, the Ka/Ks of orthologous and paralogous genes were significantly smaller than 1, suggesting that the genes of these two families experienced purifying selection during evolution.

Table 3.
Ka and Ks analyses of IPT and CKX genes in Arabidopsis thaliana and Raphanus sativus.
2.6. Expression Profiles of RsIPT and RsCKX Genes in Tissues and Growth Stages of Radish Root
To determine the expression of RsIPT and RsCKX genes among different tissues and root development stages, the early transcriptional data of tissues, including roots, stems (the bolting stems), leaves, flowers, siliques, and callus and seedling stage (ESS), splitting stage (SS), early expanding stage (EES), rapid expanding stage (RES), and mature stage (MS) of roots, were used to detect the expression profiles of RsIPT and RsCKX genes. The RsIPT and RsCKX genes presented diversified expression profiles (Figure 8). Many genes in the same group presented similar expression patterns. Groups II (RsIPT2) and IV (RsIPT9-1, RsIPT9-2) were highly expressed in all tissues and root development stages, while group I (RsIPT1, RsIPT8-1, and RsIPT8-2) was almost not expressed in any tissues or root stages of root except RsIPT1, which had only low expression levels in the bolting stems and flowers. However, the group III genes showed differentiated modes of expression. RsIPT3-1 and RsIPT3-2 were highly expressed in roots but possessed low expression levels in calli. RsIPT3-2 was also significantly expressed in the bolting stems. RsIPT5-1 had a high expression level in roots, seedpods, and stems, while RsIPT5-2 was only expressed in roots. RsIPT7-1 and RsIPT7-2 also exhibited organ-specific expression in leaves and roots, respectively. In addition, RsIPT10 was mainly expressed in roots, flowers, seedpods, and calli. Compared with XB-36-2 (white freshy root), RsIPT3-1, RsIPT3-2, and RsIPT5-1 were only expressed in EES-Xinlimei (red freshy root). These genes may play an important part in the early expansion of freshy roots in Xinlimei. As shown in Figure 8b, the number of expressed RsCKX genes was lower than that of RsIPT genes. The expression of genes that belonged to the same groups presented diversity. RsCKX1-1 was expressed only in the roots, specifically in the root splitting stage. RsCKX6 was expressed in leaves and roots and was mainly expressed in the early growth phases before expanding in the roots. However, RsCKX1-2 and RsCKX1-3 of the same group were not expressed in any tissues or root growth stages, and neither were RsCKX2-1, RsCKX2-2, or RsCKX4 of group II. Another two genes (RsCKX3-1 and RsCKX3-2) of group II were expressed in three tissue types (flowers, calli, seedpods, or leaves). Group III only contained one gene, RsCKX5, that was expressed in all tissues except stems. Similarly, RsCKX7-2 was also expressed in all tissues except leaves. Only RsCKX7-1 (group IV) was ubiquitously expressed in all tissues and root growth phases.
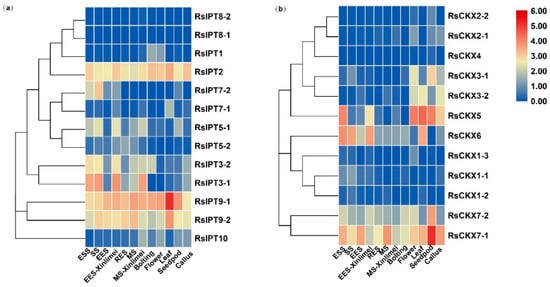
Figure 8.
Expression profiles of (a) RsIPT and (b) RsCKX genes in different tissues and root growth stages in XYB36-2 and Xinlimei radish. ESS: seedling stage; SS: splitting stage; EES: early expanding stage; RES: rapid expanding stage; MS: mature stage. Color scale indicates the relative expression of genes by log2 transformation compared with the controls.
2.7. Expression Divergences of RsIPT and RsCKX Genes after Infection with P. brassicae
To investigate the response of RsIPT and RsCKX genes in P. brassicae-radish interactions in two infection periods, transcriptional data from eight treatments with primary infection (7 days after inoculation (dai)) and secondary infection (34 dai) of two different resistance materials were used to detect the expression patterns of RsIPT and RsCKX genes (Figure 9). RsIPT1 and RsIPT8-2 of group I were not expressed in roots; that is, there was no reaction after stimulation with P. brassicae. However, RsIPT8-1, another gene of group I, was upregulated in P1d7 (susceptible materials) compared with the corresponding control. RsIPT2, the only group II gene, had a fairly low expression level in both P1 and P2 at 7 and 34 dai. Similarly, most genes in Group III were also indistinctly expressed during primary infection (7 dai). However, during secondary infection, most of these genes were significantly downregulated in P1d34 but upregulated in P2d34. In group IV, although two genes (RsIPT9-1 and RsIPT9-2) had a relatively low expression level in most treatments of the two periods compared with their corresponding control, RsIPT9-1 was upregulated in P2d34, while RsIPT9-2 was downregulated in P2d7. RsIPT9-2 was the only gene that showed unapparent expression in P1d7 but significant downregulation in P2d7.
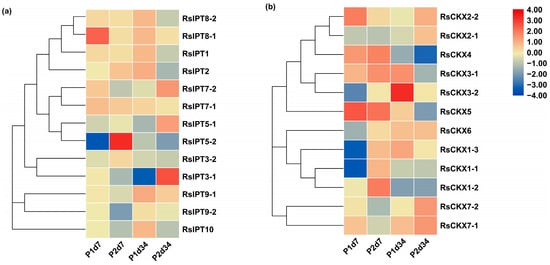
Figure 9.
Expression profiles of (a) RsIPT and (b) RsCKX genes during different infection periods after inoculation with Plasmodiophora brassicae in susceptible (P1, YS-472) and resistant varieties (P2, YR-456). P1d7: susceptible variety 7 days after inoculation/corresponding control with no infection; P2d7: resistant variety 7 days after inoculation/corresponding control with no infection; P1d34: susceptible variety 34 days after inoculation/corresponding control with no infection; P2d34: resistant variety 34 days after inoculation/corresponding control with no infection. Color scale indicates the relative expression of genes by log2 transformation compared with the controls.
As shown in Figure 9b, the expression of RsCKX genes in group I presented a contrasting tendency at 7 dai (primary infection), while a consistent tendency was observed at 34 dai (secondary infection). Specifically, the genes were upregulated in P2d7 but downregulated in P1d7 after infection with P. brassicae. The genes of group I showed differential expression between susceptible and resistant materials during primary infection. In particular, RsCKX1-2 was downregulated in susceptible P1d7 but was significantly upregulated in resistant P2d7. Two other genes (RsCKX1-1 and RsCKX1-3) showed significant downregulation in susceptible P1d7 but near-significant upregulation in resistant P2d7. These genes may play a part in clubroot resistance in resistant materials by regulating CK degradation. The genes of group II had a similar expression tendency during primary infection and an inverse tendency during secondary infection. Although two genes (RsCKX3-1 and RsCKX4) were significantly upregulated in P2d7, they were also similarly upregulated in P1d7 with no obvious distinction. However, RsCKX2-2 showed differential expression between P1d34 and P2d34 and revealed a near-significant upregulation in P2d34 and a slight downregulation in P1d34. Similarly, RsCKX7-1 and RsCKX7-2 were also upregulated in P2d34. These genes may take part in the clubroot resistance of P2 and play a more important role in secondary infection. Overall, the RsCKX genes were more expressed than the RsIPT genes and may play a more important role in resistance to P. brassicae.
To validate the expression of RsIPT and RsCKX genes after infection with P. brassicae, genes that might be related to clubroot resistance were selected to investigate their expression level based on qRT-PCR. As shown in Figure 10, the expression levels of these genes were consistent with previous transcriptional data. Four genes (RsCKX1-1, RsCKX1-2, RsCKX1-3, and RsCKX6), which all belonged to group I, showed a higher expression level in the resistant variety and a lower expression level in the susceptible variety during the primary infection period.
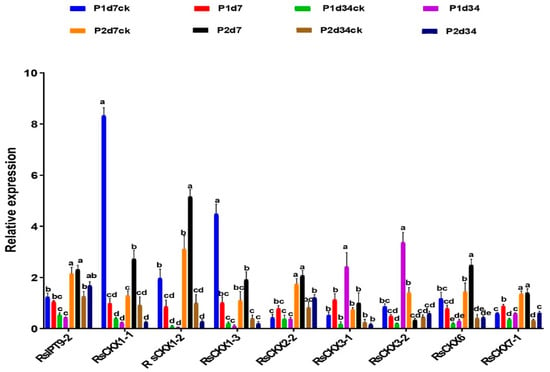
Figure 10.
Expression levels of selected RsIPT and RsCKX genes in eight treatments of susceptible and resistant materials under Plasmodiophora brassicae stress by qRT-PCR analysis. P1d7: susceptible variety 7 days after inoculation; P1d7CK: susceptible variety 7 days with no infection; P2d7: resistant variety 7 days after inoculation; P2d7CK: resistant variety after 7 days with no infection; P1d34: susceptible variety 34 days after inoculation; P1d34CK: susceptible variety after 34 days with no infection; P2d34: resistant variety 34 days after inoculation; P2d34CK: resistant variety after 34 days with no infection. Different lowercase letters indicate significant differences (p < 0.05).
2.8. Predicted Secondary Structure and 3D Structure of RsCKX Proteins
The RsCKX genes might be more involved in clubroot resistance than the RsIPT genes. Thus, the high-level protein structures of these genes were further predicted due to their similar biological functions. According to the SOMPA secondary structure prediction method, nine states of secondary structures were forecasted (Table 4). The secondary structure of all RsCKX proteins mainly included an alpha helix (Hh), an extended strand (Ee), a beta turn (Tt), and a random coil (Cc). The random coil had the highest proportion among the four types, followed by the α-helix and the extended strand. In comparison, the β-turn was the least. From the perspective of the subfamily, there were similar types of secondary structures, and no significant differences in the proportion of different states were observed among the four subfamilies.

Table 4.
Predicted secondary structure contents of RsCKX proteins.
The 3D structure of these proteins was further developed. In terms of the result of PSI-BLAST, the sequence identity between RsCKX proteins and their target templates, which were searched from the PDB database, was greater than 30%. Therefore, the 3D structure of RsCKX proteins was predicted based on the homology modeling method. As shown in Figure 11 by the 3D structure of RsCKX proteins, the secondary structure contents of the modeled 3D structure were generally consistent with those above, consisting mainly of random coil, alpha helix, and extended strand. Further analysis revealed that the RMSD value of alignment between different 3D structures of RsCKX proteins was less than 1 Å, suggesting that all RsCKX proteins had similar 3D structures (Table 5). Moreover, the RMSD values between the proteins belonging to the same group were closer to 0 than the proteins belonging to different groups. Thus, the 3D structure of the RsCKX proteins that were classified into the same group was more similar.
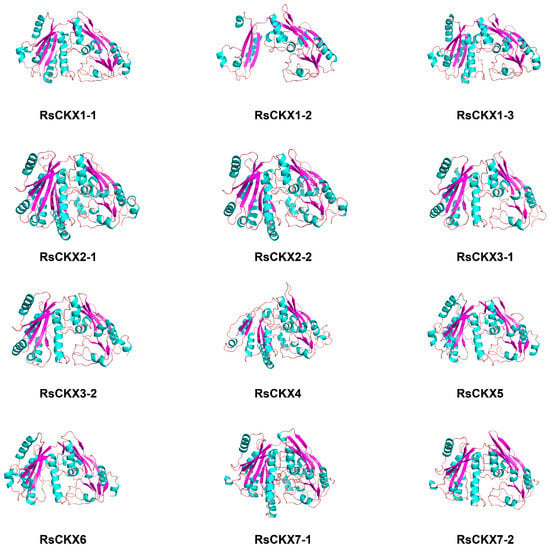
Figure 11.
3D structure of 12 RsCKX proteins. The secondary structure in green represents an alpha helix, purple represents an extended strand, and pink represents a random coil.

Table 5.
RMSD values of pairwise alignment among the 3D structures of RsCKX proteins.
2.9. Analysis of the Interaction Networks of RsCKX Proteins
To better determine the regulatory network of CKX proteins and their biological function, their protein-protein interaction relationships were predicted using an orthology-based method [43,44]. Seven RsCKX genes have orthologous relationships with Arabidopsis. Furthermore, 20 interactors were found for these CKX proteins in Arabidopsis. Unlike Arabidopsis, only 17 interactors were identified for corresponding CKX proteins based on orthologous relationships (Figure 12). There were no interactions between the IPT4, IPT6, and HK3 proteins and CKX proteins in radish.
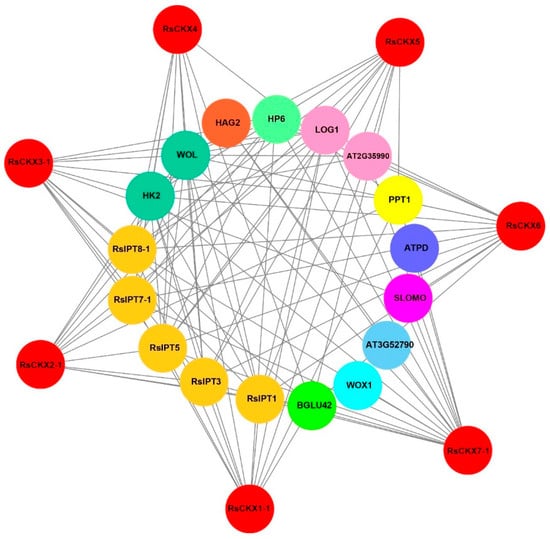
Figure 12.
Protein-protein interaction networks of RsCKX proteins with other proteins predicted using STRING. The red circles represent RsCKXs, and the inner circles represent proteins interacting with RsCKXs. The different colors indicate that the proteins belong to different families. Different colors including brown, lime, aqua, turquoise, purple, blue, yellow, pink, spring green, orange, and light green represent cytokinin synthase, beta-glucosidase, wuschel related homeobox 1, peptidoglycan-binding LysM domain-containing protein, F-box/LRR-repeat protein 15, ATP synthase, Polyprenyltransferase 1, cytokinin riboside 5′-monophosphate phosphoribohydrolase, histidine-containing phosphotransfer protein, histone acetyltransferase, and cytokinin receptors. The gray lines indicate interactions.
Specifically, interactors with CKX proteins were divided into 11 categories in radish (Figure 12). There were five interacting proteins belonging to the IPT family among the CKX interactors. Other families had only one or two members. The functions of proteins interacting with CKX proteins were further analyzed. These proteins had abundant functions involved in CK biosynthesis (RsIPTs, LOG1, AT2G35990), initiating vascular primordium (WOX1), cell wall macromolecule catabolic process (AT3G52790), coenzyme Q (CoQ) biosynthesis (PPT1), inhibitor of CK signaling (HP6), auxin homeostasis (SLOMO), and transmission of the stress signal (HK2, WOL). Specifically, CKXs interacted with the BGLU42 protein, which was involved in the secretion of root-derived phenolics upon iron ion (Fe) depletion and promoted disease resistance.
3. Discussion
An increasing number of studies have found that CKs play a very important role in plant growth, development, and resistance [12,38,45]. IPTs and CKXs are key enzymes catalyzing CK synthesis and degradation, respectively. In this study, 13 IPT and 12 CKX genes were identified in R. sativus (2n = 18). Moreover, the number of genes was similar in diploid Brassica species and double in allotetraploid B. napus (2n = 38). These identified IPT and CKX genes were essentially consistent with previous studies in Arabidopsis (2n = 10) and B. rapa (2n = 20) [32,36,40,41].
Previous studies have revealed that angiosperms, including Brassicaceae species, have undergone multiple rounds of whole-genome duplication (WGD) [46,47,48,49,50,51,52]. Raphanus and Brassica species have both undergone whole-genome triplication (WGT) [50,52,53]. Moreover, the triplication processes were followed by diploidization, substantial genome rearrangements, and differential gene losses [54]. In our study, most IPT and CKX genes in radish were retained, maintaining around twice the number in Arabidopsis, suggesting that IPT and CKX genes also experienced a diploidization process following WGT events. Typically, the increase in genes was mainly by means of gene duplication, which occurred as tandem and segmental duplication. For the RsIPT and RsCKX genes identified in this study, gene expansion occurred through segmental duplication, with no tandem-duplicated genes. Similar results have been reported in B. rapa [36]. The varying degrees of gene losses are usually followed by polyploidization, except in the IPT and CKX families. For example, although most genes retained two copies, some genes, such as RsIPT2 and RsCKX5, only retained one copy. Two IPT genes (AtIPT4 and AtIPT6) were entirely lost and not identified in Raphanus and other Brassica species.
The synonymous substitution rate (Ks) between homologous genes was thought to be a measure to explore the timing of duplication events and the divergence of genes [55,56]. Based on the Ks values of orthologous genes, the comparative analysis of Arabidopsis with R. sativus or B. rapa indicated that the split from their common ancestor, Arabidopsis, occurred 13–29 MYA, and they shared a recent WGT event estimated at 10–21 MYA [52,54]. We investigated the divergence for orthologous genes of the IPT and CKX families between Arabidopsis and Raphanus and found that the divergence of these genes from their orthologs in Arabidopsis occurred 10–21 MYA, suggesting that the divergence of IPT and CKX genes occurred after the speciation of Raphanus from Arabidopsis and during the WGT event. In addition, the divergence of paralogous genes in radish occurred after the divergence of orthologous genes. The divergence time of orthologous genes between B. rapa and R. sativus was obviously later than the divergence time from Arabidopsis and nearly consistent with the split of Raphanus from Brassica species, which occurred during the WGT event, according to previous studies [52,57]. Therefore, the divergence of IPT and CKX genes between Raphanus and Brassica may be consistent with the divergence of the two species.
The exon–intron gene structure was an essential element that played an important role in evolution and was related to gene function and expression level [58,59,60]. In terms of the number of introns or exons, the RsIPT genes were classified into two gene categories. One group had one or no introns (RsIPT1, 3, 5, 7, 8), and the other category had more than nine introns (RsIPT2 and RsIPT9). Similar exon–intron structures of IPT genes were identified in Chinese cabbage, apples, and nine Rosaceae species [36,61,62]. The variation in gene structure resulted in a diversity of functions. According to their orthologous genes in Arabidopsis and B. rapa, two categories of exon–intron structures corresponded to different types of IPT genes: ATP/ADP IPT (RsIPT1, 3, 5, 7, 8) and tRNA IPT genes (RsIPT2 and RsIPT9) [32,33]. We found that the tRNA IPT genes, which were rich in introns, presented a higher level of expression in tissues than the ATP/ADP IPT genes. The highly expressed genes had more introns and were also present in rice and Arabidopsis [60,63]. In contrast, the gene structures of the RsCKX genes were relatively similar in the number of introns, whereas they also had fewer introns. The genes with few introns could rapidly respond to the challenges of stress in early studies [38,64,65]. Some stress-related gene families had a lower number of introns, such as the LRR family, Hsp20 family, and chitinase family [65,66,67]. In this study, most RsIPT and RsCKX genes contained few introns, suggesting that these genes might be kept ahead in transcription and activation of stress responses. In addition, the corresponding types of cis-elements in the promoters helped regulate transcription and expression to cope with stress. Many cis-elements related to phytohormones, defense, and stress, such as LTR (CCGAAA), ABRE (ACGTG), TGA (AACGAC), TCA (TCAGAAGAGG), CGTCA-motif, and TGACG-motif, were searched in the promoters of the RsIPT and RsCKX genes. These cis-elements have also been found to take part in resistance to biotic and abiotic stress [68,69,70]. The TGACG-motif binding factor (TGA) is a conserved transcription factor involved in the regulation of MeJA and SA signaling pathways as well as in the disease resistance response [71,72]. In Arabidopsis, CK can augment resistance to Pst (Pseudomonas syringae pv. tomato DC3000) by modulating the binding of ARR2/TGA3 with the PR-1 promoter [73]. A similar study has shown that CmTGA3, CmTGA8, and CmTGA9 also enhanced the resistance to Pst in melon [74]. Other studies have also revealed that the TGA gene is related to M7SB41-mediated host plant powdery mildew resistance [75]. The transcription factor PvNAC52 has been reported to specifically bind to ABRE and enhance transgenic Arabidopsis resistance to abiotic stress, such as salt and alkali stress [76]. The arrangement that responded to stress in the organization of these genes might also be conducive to P. brassicae.
Both the RsIPT and RsCKX genes had a wide variety of expression patterns among tissues. For example, RsIPT3-1, 3-2, 5-2, and RsCKX1-1, 6 were primarily expressed in nutritive organs, while RsIPT1 and RsCKX2-1, 2-2, 3-1, 3-2, 5 were primarily expressed in reproductive organs, which were similar to their orthologous genes in B. rapa [36]. Similarly, the genes RsIPT2 and RsIPT9-1/9-2, as well as AtIPT2, AtIPT9, BrIPT2, and BrIPT9-1/9-2, were ubiquitously expressed [33,36]. However, the BrCKX4 gene was highly expressed in the vegetative organs of Chinese cabbage, while the expression of RsCKX4 was barely detected in either the vegetative or reproductive organs. Expression differentiation was also presented in duplicate gene pairs. In Chinese cabbage, BrIPT5-1/5-2 and BrIPT7-1/7-2 were expressed in the same tissues, while the corresponding orthologous gene pairs were expressed in different tissues in radish. The expression of gene pair CKX1-1/1-2/1-3 also differed in Chinese cabbage and radish. The expression discrepancy of segmentally duplicated gene pairs of the IPT and CKX genes was also identified in other species [61,77]. Compared to B. rapa, the expression patterns of the corresponding genes in R. sativus were more abundant, suggesting that they might have more diversified functions.
Transcriptome analysis has shown the key role of CK in response to P. brassicae in Brassicaceae [21,38]. The levels of CK are mainly regulated by the IPT and CKX families. According to RNA-Seq in this study, although the RsIPT9 gene was downregulated in resistant materials, it was not significantly upregulated in susceptible materials. Further verification using qRT-PCR also showed that this gene had no significant expression differences between the two types of materials. This was also true for other RsIPT genes. Compared with the RsIPT genes, the RsCKX genes of group I presented differential expression between resistant and susceptible materials during primary infection. The RsCKX genes, but not the RsIPT genes, played a dominant role in regulating the CK level in response to pathogen infection. In particular, the RsCKX1-1/1-2/1-3 genes were upregulated in resistant materials but downregulated in susceptible materials during primary infection. Moreover, overexpression lines of the orthologous gene AtCKX1 have been shown to significantly alleviate clubroot symptoms and disease resistance in Arabidopsis thaliana [21]. Therefore, the RsCKX1-1/1-2/1-3 genes may play an important role in clubroot resistance in radish. Further promoter analysis showed that they all included two MeJA-responsiveness cis-acting elements (CGTCA/TGACG-motif), suggesting that they may participate in the interaction of CK signaling with the JA pathway. In addition, the MeJA-responsiveness elements were also identified in genes of the CKX family in B. oleracea [38]. Previous studies have indicated that CK takes part in SA-mediated resistance to the fungus Alternaria brassica in Arabidopsis [72]. Therefore, we assumed that the CK–JA interaction may have an important impact on clubroot resistance.
Previous studies have shown the 3D structure of TaPYL proteins in wheat is similar and rather conserved [44]. In our study, the 3D structures of RsCKX proteins were also similar (RMSD < 1 Å). The 3D structures of RsCKX proteins in the same group were closer (RMSD < 0.2 Å), especially the 3D structure of proteins of RsCKX1-1/1-2/1-3 (RMSD < 0.1 Å). Further, the 3D structures of RsCKX proteins were basically consistent with the results of phylogenetic, gene structure, and conserved motifs analysis. This indicated that the 3D structures of RsCKX proteins were also relatively conservative. The proteins that interacted with RsCKX proteins possessed multiple biological functions involving CK biosynthesis (RsIPTs, LOG1, AT2G35990), inhibitor of CK signaling (HP6), auxin homeostasis, transmission of the stress signal (HK2, WOL), and disease resistance (BGLU42). Previous studies have shown that the alteration of CKs and auxin homeostasis mediates clubroot formation [20,21,22,23,24,25,26]. The RsCKX proteins may play an important role by interacting with the proteins involved in the CK pathway and auxin homeostasis. In addition, the overexpression of β-glucosidase BGLU42, which was a MYB72-dependent key regulator, led to constitutive systemic resistance in Arabidopsis roots [78]. The RsCKX proteins interacted with BGLU42, which may be beneficial for participating in clubroot resistance.
4. Materials and Methods
4.1. Identifying IPT Genes and CKX Genes
The sequences of R. sativus, B. rapa (version 3.0), B. oleracea (JZS version 2.0), B. napus (ZS11H), and B. nigra (version 2.0) were downloaded from the Brassicaceae Database (BRAD, http://brassicadb.cn, accessed on 25 October 2023) [79]. IPT and CKX protein sequences of Arabidopsis, which were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/, accessed on 25 October 2023), were used as a query to search IPT and CKX gene families in the studied species using BLASTP (1 × 10−10). In addition, the hmmsearch program (HMMER 3.0) was utilized to screen protein sequences according to the hidden Markov model (HMM) of the CK-bind domain (Pfam: PF09265) and isopentenyl transferase domain (PF01745) with an E-value of 1 × 10−5 [80]. The results were integrated, and conserved domains were verified using CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 26 October 2023) and Pfam (http://pfam.xfam.org/, accessed on 26 October 2023) [81,82]. For the RsIPT and RsCKX genes, subcellular localization predictions were performed using WoLF PSORT (https://wolfpsort.hgc.jp/, accessed on 26 October 2023) [83].
4.2. Mapping IPT and CKX Genes to Chromosomes
Chromosome maps were constructed using the location of all CKX and IPT genes based on the chromosome length with MapChart software (version 2.32, https://www.wur.nl/en/show/Mapchart.htm accessed on 19 November 2023) [84]. The location of all genes on the chromosomes was visualized according to their start position.
4.3. Phylogenetic Tree Construction of RsIPT and RsCKX Gene Families
Multiple sequence alignments of all amino acid sequences were performed using MUSCLE program of MEGA (version 7.0, https://www.megasoftware.net accessed on 19 November 2023) [85]. Phylogenetic analysis was conducted using the neighbor-joining (NJ) method of the MEGA program [86]. The NJ tree was constructed using the Jones–Taylor–Thornton (JTT) model with 500 bootstrap replicates. The online tool Evolview (https://evolgenius.info//evolview-v2/#login, accessed on 29 October 2023) was used for further color-coding of branches and protein names to clearly classify proteins [87].
4.4. Analysis of Gene Structure and Conserved Motif
The exon–intron structures of these RsIPT and RsCKX genes were obtained using the genome annotation files downloaded from BRAD and analyzed using the Gene Structure Display Server (GSDS, https://gsds.gao-lab.org/ accessed on 19 November 2023) [88]. The motifs of the RsIPT and RsCKX genes were obtained from the MEME program (http://meme-suite.org/index.html, accessed on 5 November 2023) [89].
4.5. Analysis of the Promoters of RsIPT and RsCKX Genes
The 1500 base pair (bp) DNA sequences of initiation codons upstream of the RsIPT and RsCKX genes were extracted and submitted to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html, accessed on 6 November 2023) to identify cis-acting elements [90]. Then, the results were visualized together with the phylogenetic tree using TBtools (version 2.096) [91].
4.6. Analysis of Segmental Duplication, Tandem Duplication, and Collinearity between Raphanus and Arabidopsis or B. rapa
The tandem and segmental duplication events in radish were analyzed using the Multiple Collinearity Scan toolkit (MCScanX, https://github.com/wyp1125/MCScanX/, accessed on 10 November 2023) [92]. Gene duplication was confirmed using the BLAST program by following the criteria referred to by Zhao et al. [65]. The collinearity relationship between Raphanus and Arabidopsis, or B. rapa, was analyzed using MCscan (https://github.com/tanghaibao/jcvi/wiki/MCscan, accessed on 13 November 2023). The KaKs_Calculator was used to calculate the non-synonymous (Ka) and synonymous substitution rates (Ks) of duplicated RsIPT and RsCKX genes [93]. The divergence times of orthologous and paralogous genes were estimated using the formula T = Ks/2r, where r indicates the divergence rate of nuclear genes from plants. The r value is 1.5 × 10−8 synonymous substitutions per site per year for dicotyledonous plants [94].
4.7. Plant Materials, Cultivation, and Treatment with P. brassicae
Generally speaking, the fresh root of radish is composed of hypocotyl and root. The root was susceptible to P. brassicae. Two high-generation radish inbred lines, ‘YR-456’ (P2) and ‘YS-472’ (P1), were provided by the National Mid-term Genebank of Vegetable Germplasm Resources in Beijing (NMGVGR). ‘YR-456’ was highly resistant, while ‘YS-472’ was susceptible to P. brassicae race 4. Thirty germinated seeds were sown in plastic pots with a 10-cm diameter and filled with substrate, which consisted of a sterile peat–vermiculite mixture (1:1, v:v). All pots were placed in the identification chamber at 20–25 °C with a 14-h photoperiod. The treatment and control contained three biological replicates and 10 seedlings for each replicate (40 additional seedlings for each treatment were planted to observe the incidence of clubroot). Based on the previous research and the growth of seedlings, 20-day-old seedlings were inoculated with race 4 (2 × 108 spores/mL, 5 mL), and inoculation was performed following the two-stage combined inoculation method [3]. The control was inoculated with an equal volume of sterile water. Subsequently, three seedlings were observed every three days from the first day after inoculation. Finally, the roots of ‘YR-456’ (P2) and ‘YS-472’ (P1) materials at 7 (primary infection) and 34 dai (secondary infection) (eight treatments including the corresponding control) were collected for RNA extraction, transcriptional analysis, and qPCR analysis.
4.8. RNA Extraction, cDNA Synthesis, qRT-PCR, and Statistical Analysis
The total RNA of 24 radish samples was isolated using a plant RNA Kit (QIAGEN, Dusseldorf, North Rhine-Westphalia, Germany) following the manufacturer’s instructions. Then, the cDNAs were synthesized by reverse-transcription with the Servicebio®RT First Strand cDNA Synthesis Kit (Servicebio, Wuhan, China). The specific primers for the RsIPT and RsCKX genes selected were designed using Primer 5 (Table S3). The GAPDH gene was employed as the reference gene. qPCR analysis was performed on the StepOnePlusTM Real-Time PCR System (Bio-RAD, Hercules, California, USA) and using 2×SYBR Green qPCR Master Mix (High ROX) (Servicebio, Wuhan, China) with a 15 µL reaction containing 7.5 µL of 2×SYBR Green qPCR Master Mix (High ROX), 0.75 µL of each primer (2.5 µM), 2.0 µL of cDNA, and 4 µL ddH2O. The cycling conditions were as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 60 s. Analysis of the melting curve was carried out from 60 to 95 °C, with 0.3 °C every 15 s. The processing of expression levels was determined using the 2−ΔΔCT method. Statistical analysis was performed in SAS (version 9.3) and used factorial analysis of variance (ANOVA) with Duncan’s multiple range test (PROC GLM statistical procedure) (p < 0.05).
4.9. Expression Profile Analysis of the RsIPT and RsCKX Genes
Early RNA-seq data from different tissues and root growth stages of root in radish (XB-36-2 and Xinlimei) were obtained from the NCBI database [95,96]. Therefore, the data for Xinlimei only contained the early, expanding, and mature root stages. The expression data from different infection periods of roots infected by P. brassicae were supplied by the State Key Laboratory of Vegetable Biobreeding (Table S4). The fragments per kilobase of transcript per million mapped reads (FPKM) values were used to reflect transcript abundance, and log2(FPKM) represented the gene expression values. Heat maps with the phylogenetic tree were merged using TBtools [91].
4.10. Prediction and Analysis of Secondary Structure and 3D Structure of Proteins
The high-level structures of proteins contributed to understanding their functions. Based on protein sequences, the online analysis tool SOPMA (https://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html, accessed on 18 November 2023) was applied to predict the secondary structure [97]. Then, homology modeling methods were used to predict the 3D structure of proteins. Specifically, the most similar homologies of RsCKX proteins were acquired using the positive-specific interactive basic local alignment search tools (PSI-BLAST) (http://www.ncbi.nlm.nih.gov/BLAST/, accessed on 19 November 2023) in the PDB database (https://www.rcsb.org/, accessed on 19 November 2023) [98,99]. The Swiss-model (https://swissmodel.expasy.org/ accessed on 19 November 2023) was utilized to search templates and build models for CKX proteins [100]. The predicted 3D structures were verified using the SAVES server (https://saves.mbi.ucla.edu/, accessed on 20 November 2023) [101,102,103,104]. In addition, the 3D structures of RsCKX proteins were aligned with each other using Superpose (http://superpose.wishartlab.com/, accessed on 22 November 2023) and PyMol (https://www.pymol.org/, accessed on 25 November 2023) [105]. The alignment results were measured by the root-mean-square deviation (RMSD) values. The closer the value was to 0, the more similar the structure was.
4.11. Interaction Network Analysis of RsCKX Proteins
To further explore the biological function and interaction relationship of the RsCKX proteins involved, Ortho Venn2 (https://orthovenn2.bioinfotoolkits.net/home, accessed on 28 November 2023) was employed to perform orthologous analysis with an E-value of 1 × 10−2 and an inflation value of 1.50 to identify the ortholog pairs between RsCKX and AtCKX [106]. The interaction network of RsCKX proteins was identified based on the orthologous genes between Raphanus and Arabidopsis using the STRING (https://string-db.org/, accessed on 29 November 2023) database with medium confidence (0.40) and no more than 20 interactors (the maximum number of interactors) [107].
5. Conclusions
This study comprehensively reported the gene structure, conserved domains, cis-elements in promoters, evolution patterns, 3D structure, and interaction network of the IPT and CKX families in the radish genome. Moreover, the expression profiles of the RsIPT and RsCKX genes in different tissues and root development stages, as well as their response to infection with P. brassicae, were also characterized. A total of 13 IPT and 12 CKX genes were identified and classified into four groups. The expansion mode of the RsIPT and RsCKX genes was segmental duplication. The divergence with their orthologs occurred after the speciation of Raphanus from Arabidopsis and during WGT. Purifying selection impelled the evolution of the genes of the two families. Further analysis showed that promoters of the RsIPT and RsCKX genes contained various cis-acting elements related to phytohormones and stress responsiveness. The expression profiles indicated that RsCKX genes played a more important role in the infection of P. brassicae than RsIPT genes, especially RsCKX1-1/1-2/1-3, which were significantly upregulated in resistant materials but downregulated in susceptible materials. The 3D structure of the RsCKX genes was predicted based on homology modeling. The protein interaction network further indicated that the RsCKX proteins played a key role in the response to stress and disease resistance.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25168974/s1.
Author Contributions
Conceptualization, X.L. and X.Z.; methodology, X.L., H.Y. and Y.Y.; experiment, H.Y.; software, H.Y., X.W., W.L. and H.S.; data curation, H.Y. and X.L.; writing—original draft preparation, H.Y.; writing—review and editing, H.Y., X.L. and Y.Z.; supervision, X.L.; funding acquisition, X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the National Key Research and Development Program of China (2016YFD0100204-02) and the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS, CAAS-XTCX2016001, CAAS-XTCX2016016-4-4), the National Natural Science Foundation of China (31772303), and the Excellent Youth of Henan Academy of Agricultural Science (2024 YQ12).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article or Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hirai, M. Genetic analysis of clubroot resistance in brassica crops. Bred. Sci. 2006, 56, 223–229. [Google Scholar] [CrossRef]
- Wang, J.; Huang, Y.L.; Li, X.L.; Li, H.Z. Research progress in clubroot of crucifers. Plant Prot. 2011, 37, 153–158. [Google Scholar]
- Yang, H.; Yuan, Y.; Wei, X.; Zhang, X.; Wang, H.; Song, J.; Li, X. A new identification method reveals the resistance of an extensive-source radish collection to Plasmodiophora brassicae race 4. Agronomy 2021, 11, 792. [Google Scholar] [CrossRef]
- Ingram, D.S.; Tommerup, I.C.; Ingram, D.S.; Brian, P.W. The life history of Plasmodiophora bracssicae Woron. Proc. B Soc. Lond. B 1972, 180, 103–112. [Google Scholar]
- Wallenhammar, A.C. Prevalence of Plasmodiophora brassicae in a spring oilseed rape growing area in central sweden and factors influencing soil infestation levels. Plant Pathol. 2010, 45, 710–719. [Google Scholar] [CrossRef]
- Chai, A.L.; Xie, X.W.; Shi, Y.X.; Li, B.J. Research status of clubroot (Plasmodiophora brassicae) on cruciferous crops in China. Can. J. Plant Pathol. Rev. Can. Phytopathol. 2014, 36, 142–153. [Google Scholar] [CrossRef]
- Mok, M.C. Cytokinins and plant development—An overview. In Cytokinins—Chemistry, Activity, and Function; Mok, D.W.S., Mokeds, M.C., Eds.; CRC Press: Boca Raton, FL, USA, 1994; Volume 1, pp. 155–166. Available online: https://www.researchgate.net/publication/312888917_Cytokinins_and_Plant_Development_Cytokinins_Chemistry_Activity_and_Function (accessed on 1 August 2024).
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Schmülling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef]
- Dervinis, C.; Frost, C.J.; Lawrence, S.D.; Novak, N.G.; Davis, J.M. Cytokinin primes plant responses to wounding and reduces insect performance. J. Plant Growth Regul. 2010, 29, 289–296. [Google Scholar] [CrossRef]
- Choi, J.; Choi, D.; Lee, S.; Ryu, C.-M.; Hwang, I. Cytokinins and plant immunity: Old foes or new friends? Trends Plant Sci. 2011, 16, 388–394. [Google Scholar] [CrossRef]
- Denancé, N.; Sánchez-Vallet, A.; Goffner, D.; Molina, A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013, 4, 155. [Google Scholar] [CrossRef]
- Grant, M.R.; Jones, J.D.G. Hormone (dis)harmony moulds plant health and disease. Science 2009, 324, 750–752. [Google Scholar] [CrossRef] [PubMed]
- De Vleesschauwer, D.; Xu, J.; Höfte, M. Making sense of hormone-mediated defense networking: From rice to arabidopsis. Front. Plant Sci. 2014, 5, 611. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.W.; Efetova, M.; Engelmann, J.C.; Kramell, R.; Wasternack, C.; Ludwig-Müller, J.; Hedrich, R.; Deeken, R. Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell 2009, 21, 2948–2962. [Google Scholar] [CrossRef]
- Sardesai, N.; Lee, L.-Y.; Chen, H.; Yi, H.; Olbricht, G.R.; Stirnberg, A.; Jeffries, J.; Xiong, K.; Doerge, R.W.; Gelvin, S.B. Cytokinins secreted by agrobacterium promote transformation by repressing a plant MYB transcription factor. Sci. Signal. 2013, 19, 100. [Google Scholar]
- Giron, D.; Frago, E.; Glevarec, G.; Pieterse, C.M.J.; Dicke, M. Cytokinins as key regulators in plant-microbe-insect interactions: Connecting plant growth and defence. Funct. Ecol. 2013, 27, 599–609. [Google Scholar] [CrossRef]
- Naseem, M.; Wölfling, M.; Dandekar, T. Cytokinins for immunity beyond growth, galls and green islands. Trends Plant Sci. 2014, 19, 481–484. [Google Scholar] [CrossRef]
- Walters, D.R.; McRoberts, N. Plants and biotrophs: A pivotal role for cytokinins? Trends Plant Sci. 2006, 11, 581–586. [Google Scholar] [CrossRef]
- Ludwig-Müller, J.; Schuller, A. What can we learn from clubroots: Alterations in host roots and hormone homeostasis caused by Plasmodiophora brassicae. Eur. J. Plant Pathol. 2008, 121, 291–302. [Google Scholar] [CrossRef]
- Siemens, J.; Keller, I.; Sarx, J.; Kunz, S.; Schuller, A.; Nagel, W.; Schmülling, T.; Parniske, M.; Ludwig-Müller, J. Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Mol. Plant-Microbe Interact. 2006, 19, 480–494. [Google Scholar] [CrossRef]
- Malinowski, R.; Novák, O.; Borhan, M.H.; Spíchal, L.; Strnad, M.; Rolfe, S.A. The role of cytokinins in clubroot disease. Eur. J. Plant Pathol. 2016, 145, 543–557. [Google Scholar] [CrossRef]
- Prerostova, S.; Dobrev, P.I.; Konradyova, V.; Knirsch, V.; Gaudinova, A.; Kramna, B.; Kazda, J.; Ludwig-Müller, J.; Vankova, R. Hormonal responses to Plasmodiophora brassicae infection in brassica napus cultivars differing in their pathogen resistance. Int. J. Mol. Sci. 2018, 19, 4024. [Google Scholar] [CrossRef]
- Malinowski, R.; Smith, J.A.; Fleming, A.J.; Scholes, J.D.; Rolfe, S.A. Gall formation in clubroot-infected Arabidopsis results from an increase in existing meristematic activities of the host but is not essential for the completion of the pathogen life cycle. Plant J. 2012, 71, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Boivin, S.; Fonouni-Farde, C.; Frugier, F. How auxin and cytokinin phytohormones modulate root microbe interactions. Front. Plant Sci. 2016, 7, 1240. [Google Scholar] [CrossRef]
- Robin, A.H.K.; Hossain, M.R.; Kim, H.T.; Nou, I.S.; Park, J.I. Role of cytokinins in clubroot disease development. Plant Breed. Biotechnol. 2019, 7, 73–82. [Google Scholar] [CrossRef]
- Grsic-Rausch, S.; Kobelt, P.; Siemens, J.M.; Bischoff, M.; Ludwig-Müller, J. Expression and localization of nitrilase during symptom development of the clubroot disease in Arabidopsis. Plant Physiol. 2000, 122, 369–378. [Google Scholar] [CrossRef]
- Miiller, P.; Muller, W.H.; Hiigenberg, P.; Muller, P.; Hiigenberg, W. Isomers of zeatin and zeatin riboside in clubroot tissue: Evidence for trans-zeatin biosynthesis by Plasmodiophora brassicae. Physiol. Plant. 1986, 66, 245–250. [Google Scholar] [CrossRef]
- Siemens, J.; González, M.C.; Wolf, S.; Hofmann, C.; Greiner, S.; Du, Y.; Rausch, T.; Roitsch, T.; Ludwig-Müller, J. Extracellular invertase is involved in the regulation of clubroot disease in Arabidopsis thaliana. Mol. Plant Pathol. 2011, 12, 247–262. [Google Scholar] [CrossRef]
- Devos, S.; Laukens, K.; Deckers, P.; Van Der Straeten, D.; Beeckman, T.; Inzé, D.; Van Onckelen, H.; Witters, E.; Prinsen, E. A hormone and proteome approach to picturing the initial metabolic events during Plasmodiophora brassicae infection on Arabidopsis. Mol. Plant Microbe Interact. 2006, 19, 1431–1443. [Google Scholar] [CrossRef]
- Dekhuijzen, H.M. The occurrence of free and bound cytokinins in dubroots and Plasmodiophora brassicae infected turnip tissue cultures. Plant Cell Rep. 1980, 49, 169–176. [Google Scholar]
- Takei, K.; Sakakibara, H.; Taniguchi, M.; Sugiyama, T. Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: Implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol. 2001, 42, 85–93. [Google Scholar] [CrossRef]
- Miyawaki, K.; Matsumoto-Kitano, M.; Kakimoto, T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: Tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 2004, 37, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Sheen, J.; Müller, B. Cytokinin signaling networks. Annu. Rev. Plant Biol. 2012, 63, 353–380. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmülling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lv, Y.; Zhang, M.; Liu, Y.; Kong, L.; Zou, M.; Lu, G.; Cao, J.; Yu, X. Identification, expression, and comparative genomic analysis of the IPT and CKX gene families in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 14, 594. [Google Scholar] [CrossRef] [PubMed]
- Laila, R.; Robin, A.H.K.; Park, J.I.; Saha, G.; Kim, H.T.; Kayum, M.A.; Nou, I.S. Expression and role of response regulating, biosynthetic and degrading genes for cytokinin signaling during clubroot disease development. Int. J. Mol. Sci. 2020, 21, 3896. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, Y.; Lu, S.; Yang, L.; Zhuang, M.; Zhang, Y.; Lv, H.; Fang, Z.; Hou, X. Genome-wide identification and analysis of cytokinin dehydrogenase/oxidase (CKX) family genes in Brassica oleracea L. reveals their involvement in response to Plasmodiophora brassicae infections. Hortic. Plant J. 2022, 8, 68–80. [Google Scholar] [CrossRef]
- Chen, J.; Wan, H.; Zhu, W.; Dai, X.; Yu, Y.; Zeng, C. Identification and expression analysis of the isopentenyl transferase (IPT) gene family under lack of nitrogen stress in oilseed (Brassica napus L.). Plants 2023, 12, 2166. [Google Scholar] [CrossRef] [PubMed]
- Schmülling, T.; Werner, T.; Riefler, M.; Krupková, E.; Manns, I.B.Y. Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J. Plant Res. 2003, 116, 241–252. [Google Scholar] [CrossRef]
- Kakimoto, T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferases. Plant Cell Physiol. 2001, 42, 677–685. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Yang, S.; Kim, E.; Ko, Y.; Hwang, S.; Shin, J.; Shim, J.E.; Shim, H.; Kim, H.; Kim, C.; et al. AraNet v2: An improved database of co-functional gene networks for the study of Arabidopsis thaliana and 27 other nonmodel plant species. Nucleic Acids Res. 2015, 43, 996–1002. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Wei, X.; Gao, R.; Huo, F.; Nie, X.; Tong, W.; Song, W. Genome-wide identification of PYL gene family in wheat: Evolution, expression and 3d structure analysis. Genomics 2021, 113, 854–866. [Google Scholar] [CrossRef]
- Sun, X.; Zhu, L.; Hao, Z.; Wu, W.; Xu, L.; Yang, Y.; Zhang, J.; Lu, Y.; Shi, J.; Chen, J. Genome-wide identification and abiotic-stress-responsive expression of CKX gene family in liriodendron chinense. Plants 2023, 12, 2157. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; Pires, J.C. Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009, 17, 699–717. [Google Scholar] [CrossRef]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Hasegawa, Y.; Saito, M.; Shirasawa, S.; Fukushima, A.; Ito, T.; Fujii, H.; Kishitani, S.; Kitashiba, H.; Nishio, T. Extensive chromosome homoeology among brassiceae species were revealed by comparative genetic mapping with high-density est-based SNP markers in radish (Raphanus Sativus L.). DNA Res. 2011, 18, 401–411. [Google Scholar] [CrossRef]
- Nelson, M.N.; Parkin, I.A.P.; Lydiate, D.J. The mosaic of ancestral karyotype blocks in the Sinapis alba L. Genome 2011, 54, 33–41. [Google Scholar] [CrossRef]
- Cheng, F.; Liang, J.; Cai, C.; Cai, X.; Wu, J.; Wang, X. Genome sequencing supports a multi-vertex model for brassiceae species. Curr. Opin. Plant Biol. 2017, 36, 79–87. [Google Scholar] [CrossRef]
- Paritosh, K.; Gupta, V.; Yadava, S.K.; Singh, P.; Pradhan, A.K.; Pental, D. RNA-Seq based SNPs for mapping in Brassica juncea (AABB): Synteny analysis between the two constituent genomes a (from B. rapa) and b (from B. nigra) shows highly divergent gene block arrangement and unique block fragmentation patterns. BMC Genom. 2014, 15, 396. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jeong, Y.M.; Kim, N.; Ahn, B.O.; Oh, M.; Chung, W.H.; Chung, H.; Jeong, S.; Lim, K.B.; Hwang, Y.J.; Kim, G.B.; et al. Elucidating the triplicated ancestral genome structure of radish based on chromosome-level comparison with the brassica genomes. Theor. Appl. Genet. 2016, 129, 1357–1372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, T.; Wang, J.; Wang, P.; Qiu, Y.; Zhao, W.; Pang, S.; Li, X.; Wang, H.; Song, J.P.; et al. Pan-Genome of Raphanus highlights genetic variation and introgression among domesticated, wild, and weedy radishes. Mol. Plant 2021, 14, 2032–2055. [Google Scholar] [CrossRef] [PubMed]
- Mun, J.H.; Kwon, S.J.; Yang, T.J.; Seol, Y.J.; Jin, M.; Kim, J.A.; Lim, M.H.; Kim, J.S.; Baek, S.; Choi, B.S.; et al. Genome-wide comparative analysis of the brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009, 10, R111. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Barton, N.H. Chromosomal speciation and molecular divergence-accelerated evolution in rearranged chromosomes. Science 2003, 300, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.M.; Chung, W.H.; Chung, H.; Kim, N.; Park, B.S.; Lim, K.B.; Yu, H.J.; Mun, J.H. Comparative analysis of the radish genome based on a conserved ortholog set (COS) of brassica. Theor. Appl. Genet. 2014, 127, 1975–1989. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.M.; Mun, J.H.; Lee, I.; Woo, J.C.; Hong, C.B.; Kim, S.G. Distinct roles of the first introns on the expression of Arabidopsis profilin gene family members. Plant Physiol. 2006, 140, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Fedorova, L.; Fedorov, A. Introns in gene evolution. Genetica 2003, 118, 123–131. [Google Scholar] [CrossRef]
- Ren, X.Y.; Vorst, O.; Fiers, M.W.E.J.; Stiekema, W.J.; Nap, J.P. In plants, highly expressed genes are the least compact. Trends Genet. 2006, 22, 528–532. [Google Scholar] [CrossRef]
- Tan, M.; Li, G.; Qi, S.; Liu, X.; Chen, X.; Ma, J.; Zhang, D.; Han, M. Identification and expression analysis of the IPT and CKX gene families during axillary bud outgrowth in apple (Malus domestica Borkh.). Gene 2018, 651, 106–117. [Google Scholar] [CrossRef]
- Feng, Y.; Lv, J.; Peng, M.; Li, J.; Wu, Y.; Gao, M.; Wu, X.; Wang, Y.; Wu, T.; Zhang, X.; et al. Genome-wide identification and characterization of the IPT family members in nine rosaceae species and a functional analysis of MdIPT5b in cold resistance. Hortic. Plant J. 2023, 9, 616–630. [Google Scholar] [CrossRef]
- García-Gutiérrez, Á.; Cánovas, F.M.; Ávila, C. Glutamate synthases from conifers: Gene structure and phylogenetic studies. BMC Genom. 2018, 19, 65. [Google Scholar] [CrossRef] [PubMed]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Wang, D.; Wang, R.; Kong, N.; Zhang, C.; Yang, C.; Wu, W.; Ma, H.; Chen, Q. Genome-wide analysis of the potato Hsp20 gene family: Identification, genomic organization and expression profiles in response to heat stress. BMC Genom. 2018, 19, 61. [Google Scholar] [CrossRef]
- Zhou, F.; Guo, Y.; Qiu, L.J. Genome-wide identification and evolutionary analysis of leucine-rich repeat receptor-like protein kinase genes in soybean. BMC Plant Biol. 2016, 16, 58. [Google Scholar] [CrossRef]
- Chen, J.; Piao, Y.; Liu, Y.; Li, X.; Piao, Z. Genome-wide identification and expression analysis of chitinase gene family in brassica rapa reveals its role in clubroot resistance. Plant Sci. 2018, 270, 257–267. [Google Scholar] [CrossRef]
- Sham, A.; Moustafa, K.; Al-Ameri, S.; Al-Azzawi, A.; Iratni, R.; AbuQamar, S. Identification of Arabidopsis candidate genes in response to biotic and abiotic stresses using comparative microarrays. PLoS ONE 2015, 10, e0125666. [Google Scholar] [CrossRef]
- Yu, T.F.; Zhao, W.Y.; Fu, J.D.; Liu, Y.W.; Chen, M.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S.; Xi, Y.J. Genome-wide analysis of CDPK family in foxtail millet and determination of SiCDPK24 functions in drought stress. Front. Plant Sci. 2018, 9, 651. [Google Scholar] [CrossRef]
- Song, S.; Xu, Y.; Huang, D.; Miao, H.; Liu, J.; Jia, C.; Hu, W.; Valarezo, A.V.; Xu, B.; Jin, Z. Identification of a novel promoter from banana aquaporin family gene (MaTIP1;2) which responses to drought and salt-stress in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2018, 128, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Idrovo Espín, F.M.; Peraza-Echeverria, S.; Fuentes, G.; Santamaría, J.M. In silico cloning and characterization of the TGA (TGACG MOTIF-BINDING FACTOR) transcription factors subfamily in Carica papaya. Plant Physiol. Biochem. 2012, 54, 113–122. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, R.; Wang, D.; Zhang, J.; Zang, S.; Zou, W.; Feng, A.; You, C.; Su, Y.; Wu, Q.; et al. Dissecting the features of TGA gene family in Saccharum and the functions of ScTGA1 under biotic stresses. Plant Physiol. Biochem. 2023, 200, 107760. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Huh, S.U.; Kojima, M.; Sakakibara, H.; Paek, K.H.; Hwang, I. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 2010, 19, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Dai, Y.; Noman, M.; Li, R.; Li, X.; Wu, X.; Wang, H.; Song, F.; Li, D. Genome-wide characterization and functional analysis of the melon TGA gene family in disease resistance through ectopic overexpression in Arabidopsis thaliana. Plant Physiol. Biochem. 2024, 212, 108784. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Mao, W.; Tang, W.; Soares, M.A.; Li, H. Wild rosa endophyte M7SB41-mediated host plant’s powdery mildew resistance. J. Fungi 2023, 9, 620. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wu, M.; Wang, X.; Li, M.; Gao, X.; Xu, X.; Zhang, Y.; Liu, X.; Yu, L.; Zhang, Y. Common Bean (Phaseolus vulgaris L.) NAC transcriptional factor PvNAC52 enhances transgenic Arabidopsis resistance to salt, alkali, osmotic, and ABA stress by upregulating stress-responsive genes. Int. J. Mol. Sci. 2024, 25, 5818. [Google Scholar] [CrossRef]
- Yu, K.; Yu, Y.; Bian, L.; Ni, P.; Ji, X.; Guo, D.; Zhang, G.; Yang, Y. Genome-wide identification of cytokinin oxidases/dehydrogenase (CKXs) in grape and expression during berry set. Sci. Hortic. 2021, 280, 109917. [Google Scholar] [CrossRef]
- Zamioudis, C.; Hanson, J.; Pieterse, C.M. β-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 2014, 204, 368–379. [Google Scholar] [CrossRef]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. BRAD V3.0: An upgraded Brassicaceae database. Nucleic Acids Res. 2022, 7, 1432–1441. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, d255–d259. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Hereditas 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Machanick, P.; Bailey, T.L. MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 2011, 27, 1696–1697. [Google Scholar] [CrossRef]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.A.; Haubold, B.; Mitchell-Olds, T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 2000, 17, 1483–1498. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qiu, Y.; Wang, X.; Yue, Z.; Yang, X.; Chen, X.; Zhang, X.; Shen, D.; Wang, H.; Song, J.; et al. Insights into the species-specific metabolic engineering of glucosinolates in radish (Raphanus sativus L.) based on comparative genomic analysis. Sci. Rep. 2017, 22, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yue, Z.; Mei, S.; Qiu, Y.; Yang, X.; Chen, X.; Cheng, F.; Wu, Z.; Sun, Y.; Jing, Y.; et al. A de novo genome of a Chinese radish cultivar. Hortic. Plant J. 2015, 1, 155–164. [Google Scholar]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Bowie, J.U.; Lüthy, R.; Eisenberg, D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Pontius, J.; Richelle, J.; Wodak, S.J. Deviations from standard atomic volumes as a quality measure for protein crystal structures. J. Mol. Biol. 1996, 264, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Maiti, R.; Van Domselaar, G.H.; Zhang, H.; Wishart, D.S. SuperPose: A simple server for sophisticated structural superposition. Nucleic Acids Res. 2004, 32 (Suppl. 2), W590–W594. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, 605–612. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).