Circular RNA Profile in Atherosclerotic Disease: Regulation during ST-Elevated Myocardial Infarction
Abstract
:1. Introduction
2. Results
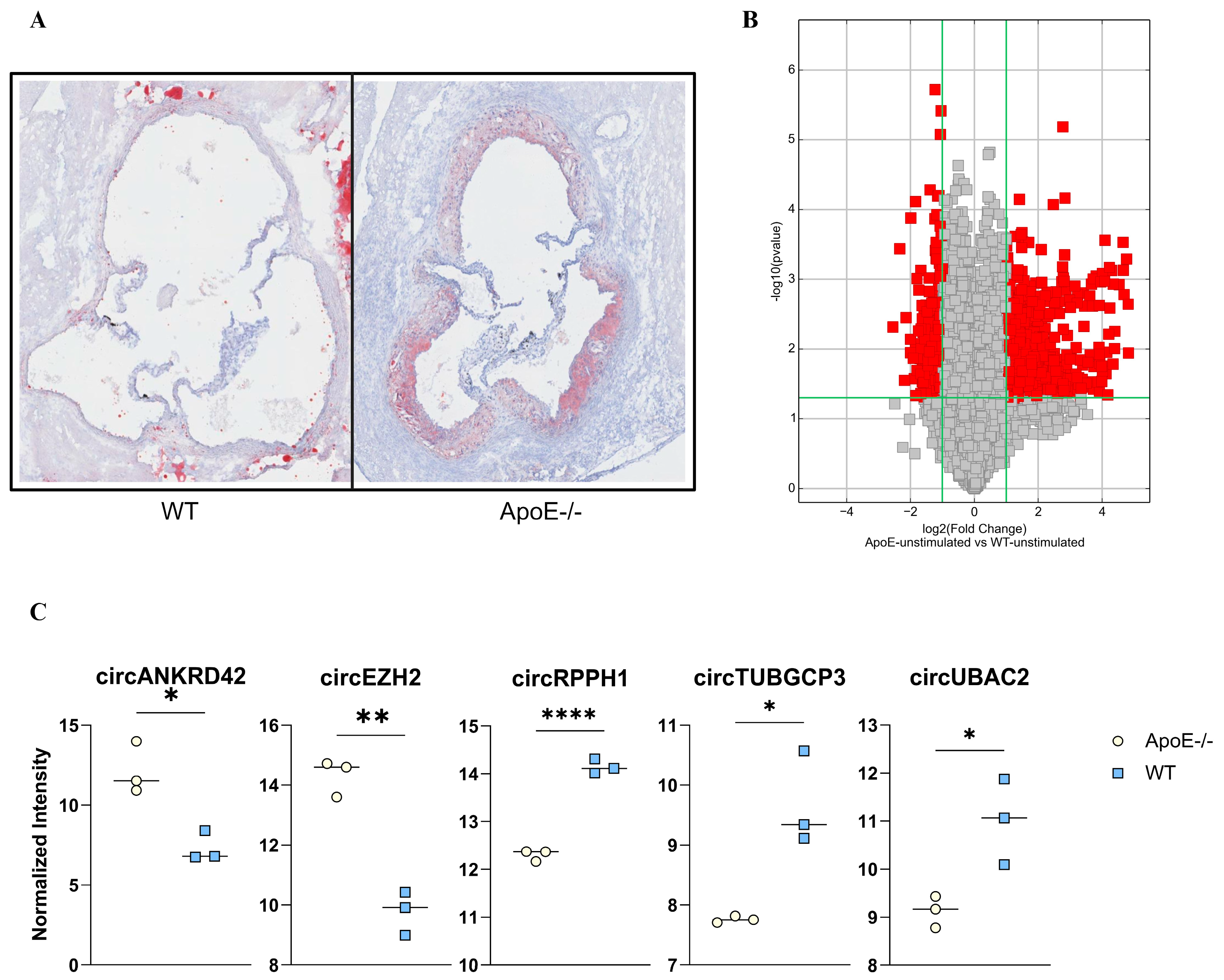
2.1. Atherogenesis Alters the circRNA Expression: Pre-Clinical Explorative Studies
2.2. Atherogenic circRNA Translated to Human Homologs
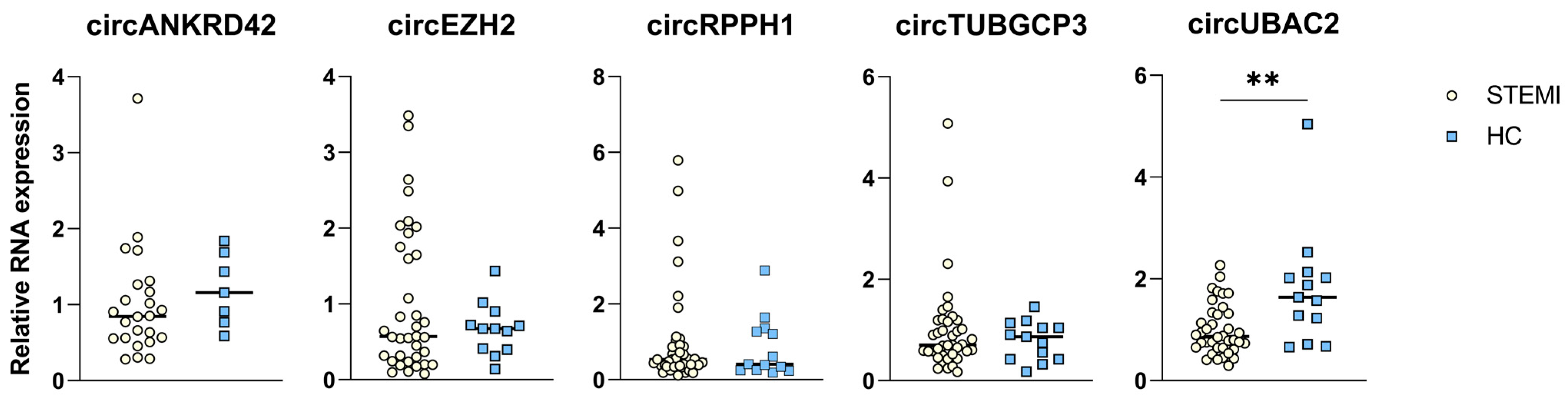
2.3. circRNA Expression in Patients with STEMI
2.4. Negative Correlation between circRNA Levels and Peak TnT Levels and Final Infarct Size
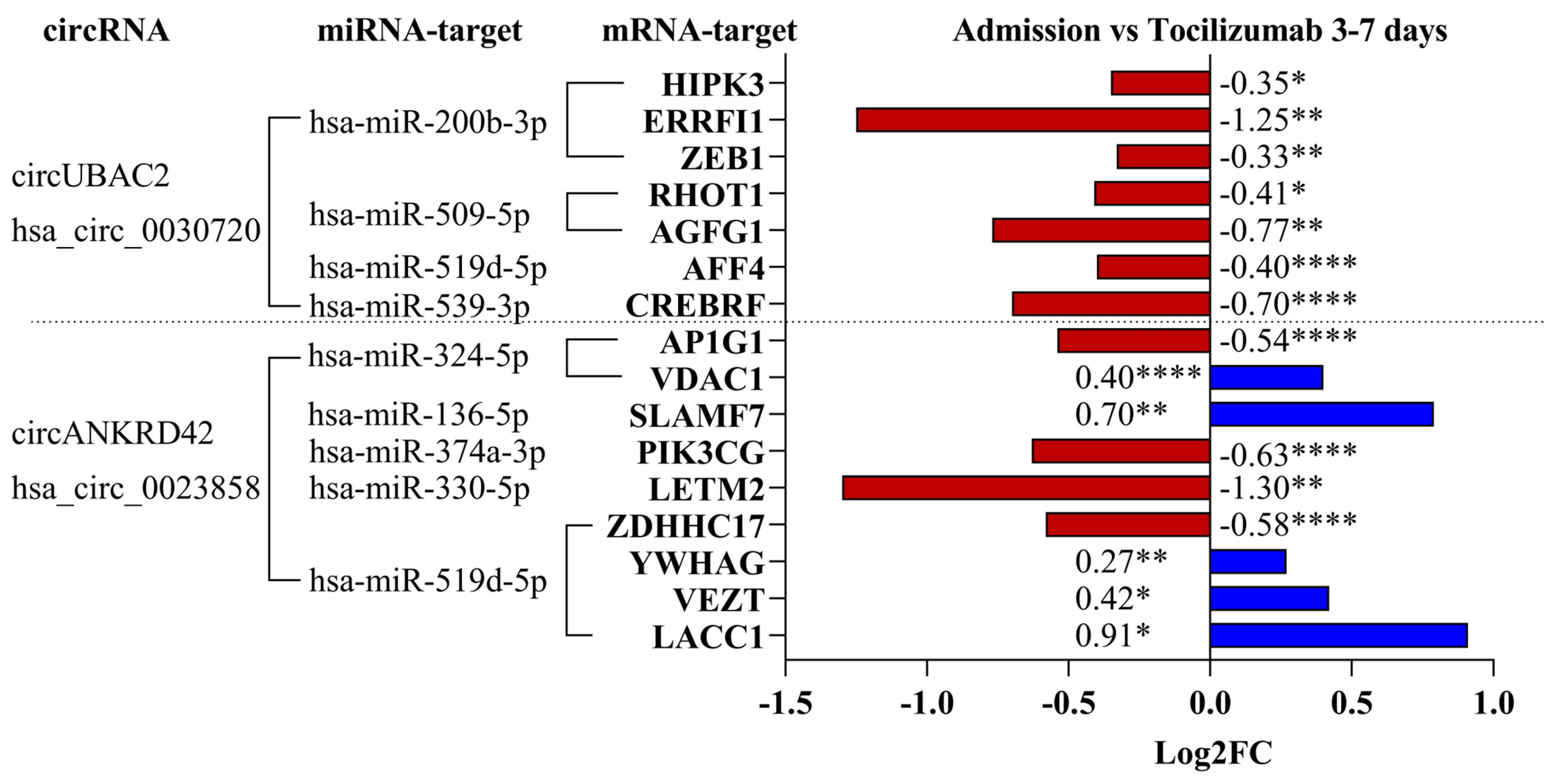
2.5. Regulation of mRNA Targets for the circRNA–miRNA Interaction
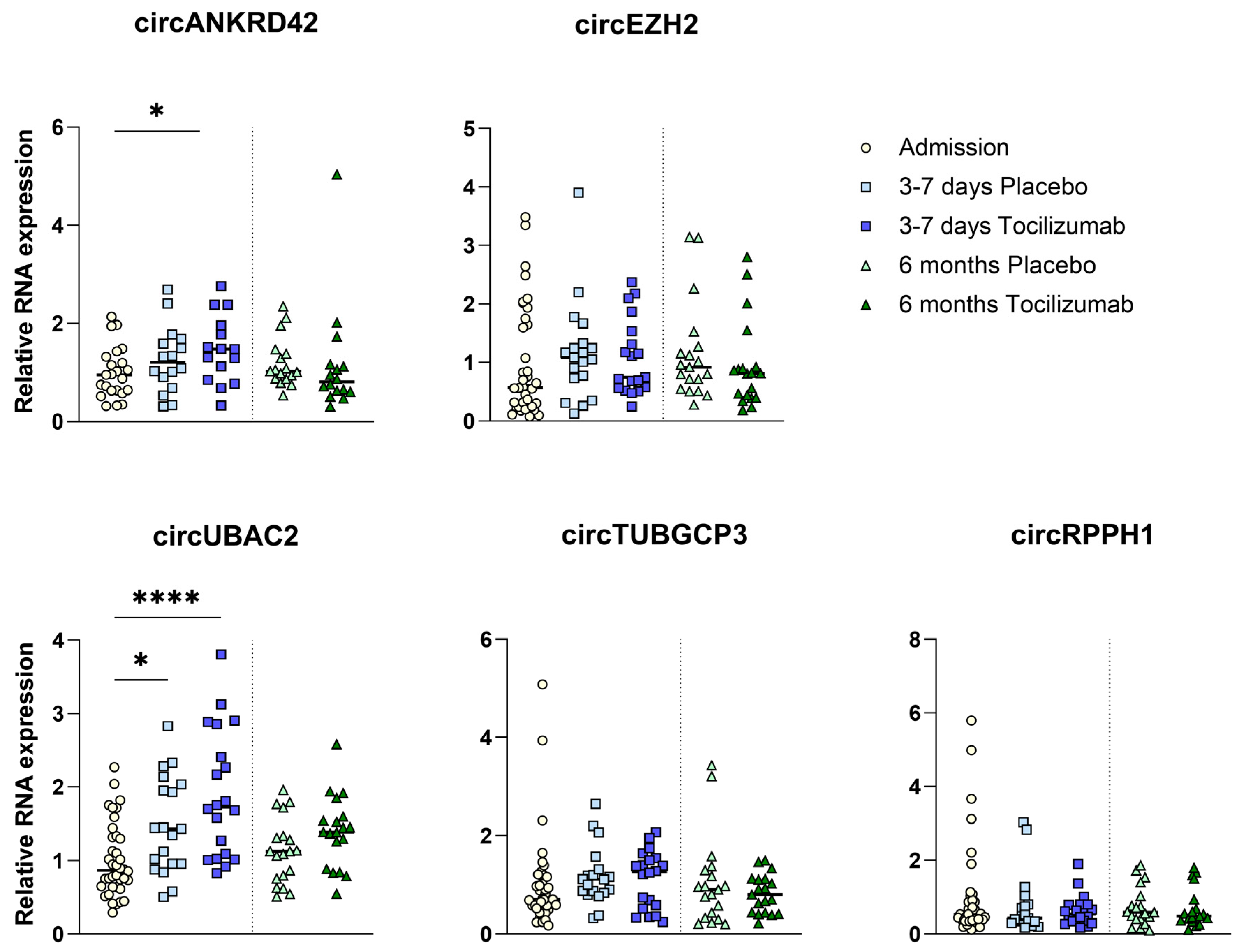
2.6. circRNA Expression with IL-6R Inhibition
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Mouse Models
4.3. Murine Circular RNA Array
4.4. Primer Design
4.5. Human Total RNA-Isolation and cDNA Synthesis
4.6. Quantitative Reverse Transcriptase PCR
4.7. miRNA and mRNA Targets
4.8. RNA Sequencing
4.9. Patients and Study Design of ASSAIL-MI
4.10. Blood Sampling Protocol
4.11. Measurement of Cardiac Markers in the ASSAIL-MI Trial
4.12. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Libby, P. The Changing Landscape of Atherosclerosis. Nature 2021, 592, 524–533. [Google Scholar] [CrossRef]
- Stewart, J.; Manmathan, G.; Wilkinson, P. Primary Prevention of Cardiovascular Disease: A Review of Contemporary Guidance and Literature. JRSM Cardiovasc. Dis. 2017, 6, 2048004016687211. [Google Scholar] [CrossRef]
- The Top 10 Causes of Death. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 21 February 2023).
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef]
- Enuka, Y.; Lauriola, M.; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. Circular RNAs Are Long-Lived and Display Only Minimal Early Alterations in Response to a Growth Factor. Nucleic Acids Res. 2016, 44, 1370–1383. [Google Scholar] [CrossRef]
- Yu, J.; Xie, D.; Huang, N.; Zhou, Q. Circular RNAs as Novel Diagnostic Biomarkers and Therapeutic Targets in Kidney Disease. Front. Med. 2021, 8, 714958. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, J.; Yang, W.; Ye, W.-C. CircRNAs in Colorectal Cancer: Potential Biomarkers and Therapeutic Targets. Cell Death Dis. 2023, 14, 353. [Google Scholar] [CrossRef]
- Wang, X.; Li, H.; Lu, Y.; Cheng, L. Regulatory Effects of Circular RNAs on Host Genes in Human Cancer. Front. Oncol. 2021, 10, 586163. [Google Scholar]
- Sakshi, S.; Jayasuriya, R.; Ganesan, K.; Xu, B.; Ramkumar, K.M. Role of circRNA-miRNA-mRNA Interaction Network in Diabetes and Its Associated Complications. Mol. Ther. Nucleic Acids 2021, 26, 1291–1302. [Google Scholar] [CrossRef] [PubMed]
- Patop, I.L.; Wüst, S.; Kadener, S. Past, Present, and Future of circRNAs. EMBO J. 2019, 38, e100836. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, Y.; Yin, M.; Cheng, Z.; Li, D.; Luo, X.; Liu, X.; Tu, J. Circular RNA circPRDX3 Mediates Neuronal Survival Apoptosis in Ischemic Stroke by Targeting miR-641 and NPR3. Brain Res. 2022, 1797, 148114. [Google Scholar] [CrossRef]
- Yang, Z.; Huang, C.; Wen, X.; Liu, W.; Huang, X.; Li, Y.; Zang, J.; Weng, Z.; Lu, D.; Tsang, C.K.; et al. Circular RNA Circ-FoxO3 Attenuates Blood-Brain Barrier Damage by Inducing Autophagy during Ischemia/Reperfusion. Mol. Ther. 2022, 30, 1275–1287. [Google Scholar] [CrossRef]
- Huang, R.; Zhang, W.; Li, W.; Gao, Y.; Zheng, D.; Bi, G. Overexpressing Circ_0000831 Is Sufficient to Inhibit Neuroinflammation and Vertigo in Cerebral Ischemia through a miR-16-5p-Dependent Mechanism. Exp. Neurol. 2022, 353, 114047. [Google Scholar] [CrossRef]
- Cheng, Q.; Wang, J.; Li, M.; Fang, J.; Ding, H.; Meng, J.; Zhang, J.; Fang, X.; Liu, H.; Ma, C.; et al. CircSV2b Participates in Oxidative Stress Regulation through miR-5107-5p-Foxk1-Akt1 Axis in Parkinson’s Disease. Redox Biol. 2022, 56, 102430. [Google Scholar] [CrossRef]
- Xie, Q.; Ma, Y.; Ren, Z.; Gu, T.; Jiang, Z. Circular RNA: A New Expectation for Cardiovascular Diseases. J. Cell. Biochem. 2024, 125, e30512. [Google Scholar] [CrossRef]
- Xu, G.; Liu, G.; Wang, Z.; Li, Y.; Fang, W. Circular RNAs: Promising Treatment Targets and Biomarkers of Ischemic Stroke. Int. J. Mol. Sci. 2024, 25, 178. [Google Scholar] [CrossRef]
- Broch, K.; Anstensrud, A.K.; Woxholt, S.; Sharma, K.; Tøllefsen, I.M.; Bendz, B.; Aakhus, S.; Ueland, T.; Amundsen, B.H.; Damås, J.K.; et al. Randomized Trial of Interleukin-6 Receptor Inhibition in Patients with Acute ST-Segment Elevation Myocardial Infarction. J. Am. Coll. Cardiol. 2021, 77, 1845–1855. [Google Scholar] [CrossRef] [PubMed]
- Abbate, A.; Trankle, C.R.; Buckley, L.F.; Lipinski, M.J.; Appleton, D.; Kadariya, D.; Canada, J.M.; Carbone, S.; Roberts, C.S.; Abouzaki, N.; et al. Interleukin-1 Blockade Inhibits the Acute Inflammatory Response in Patients with ST-Segment-Elevation Myocardial Infarction. J. Am. Heart Assoc. 2020, 9, e014941. [Google Scholar] [CrossRef]
- Hjerteinfarkt. Available online: https://legehandboka.no/handboken/kliniske-kapitler/hjertekar/tilstander-og-sykdommer/koronarsykdom/hjerteinfarkt/ (accessed on 22 February 2023).
- Feng, Y.; Ye, D.; Wang, Z.; Pan, H.; Lu, X.; Wang, M.; Xu, Y.; Yu, J.; Zhang, J.; Zhao, M.; et al. The Role of Interleukin-6 Family Members in Cardiovascular Diseases. Front. Cardiovasc. Med. 2022, 9, 818890. [Google Scholar] [CrossRef]
- Huang, M.; Yang, D.; Xiang, M.; Wang, J. Role of Interleukin-6 in Regulation of Immune Responses to Remodeling after Myocardial Infarction. Heart Fail. Rev. 2015, 20, 25–38. [Google Scholar] [CrossRef]
- Ridker, P.M.; Rane, M. Interleukin-6 Signaling and Anti-Interleukin-6 Therapeutics in Cardiovascular Disease. Circ. Res. 2021, 128, 1728–1746. [Google Scholar] [CrossRef] [PubMed]
- Huse, C.; Anstensrud, A.K.; Michelsen, A.E.; Ueland, T.; Broch, K.; Woxholt, S.; Yang, K.; Sharma, K.; Tøllefsen, I.M.; Bendz, B.; et al. Interleukin-6 Inhibition in ST-Elevation Myocardial Infarction: Immune Cell Profile in the Randomised ASSAIL-MI Trial. eBioMedicine 2022, 80, 104013. [Google Scholar] [CrossRef]
- Panda, A.C. Circular RNAs Act as miRNA Sponges. In Circular RNAs: Biogenesis and Functions; Xiao, J., Ed.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2018; pp. 67–79. ISBN 9789811314261. [Google Scholar]
- Li, Q.; Wang, Y.; An, Y.; Wang, J.; Gao, Y. The Particular Expression Profiles of Circular RNA in Peripheral Blood of Myocardial Infarction Patients by RNA Sequencing. Front. Cardiovasc. Med. 2022, 9, 810257. [Google Scholar] [CrossRef]
- Xu, P.; Zhang, J.; Wang, M.; Liu, B.; Li, R.; Li, H.; Zhai, N.; Liu, W.; Lv, C.; Song, X. hnRNPL-Activated circANKRD42 Back-Splicing and circANKRD42-Mediated Crosstalk of Mechanical Stiffness and Biochemical Signal in Lung Fibrosis. Mol. Ther. 2022, 30, 2370–2387. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.B.; Kapoor, P.; Howard, J.K.; Shah, A.M.; Lord, G.M. An Update on the Roles of Immune System-Derived microRNAs in Cardiovascular Diseases. Cardiovasc. Res. 2021, 117, 2434–2449. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An Online Database for Prediction of Functional microRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Liu, W.; Wang, X. Prediction of Functional microRNA Targets by Integrative Modeling of microRNA Binding and Target Expression Data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Chen, B.; Dong, L.; Zhang, J.; Hao, Y.; Chi, W.; Song, D. Exploring Shared Pathways and the Shared Biomarker ERRFI1 in Obstructive Sleep Apnoea and Atherosclerosis Using Integrated Bioinformatics Analysis. Sci. Rep. 2023, 13, 15103. [Google Scholar] [CrossRef]
- Duan, Z.; Yang, M.; Yang, J.; Wu, Z.; Zhu, Y.; Jia, Q.; Ma, X.; Yin, Y.; Zheng, J.; Yang, J.; et al. AGFG1 Increases Cholesterol Biosynthesis by Disrupting Intracellular Cholesterol Homeostasis to Promote PDAC Progression. Cancer Lett. 2024, 598, 217130. [Google Scholar] [CrossRef]
- Fang, Y.; Cao, H.; Gong, X.; Chen, Y.; Zhuang, Y.; Zhou, S.; Chen, Y.; Jiang, Y.; Ji, X.; Peng, H.; et al. AFF4 Predicts the Prognosis of Colorectal Cancer Patients and Suppresses Colorectal Cancer Metastasis via Promoting CDH1 Expression. Front. Oncol. 2022, 12, 797392. [Google Scholar] [CrossRef]
- Camara, A.K.S.; Zhou, Y.; Wen, P.-C.; Tajkhorshid, E.; Kwok, W.-M. Mitochondrial VDAC1: A Key Gatekeeper as Potential Therapeutic Target. Front. Physiol. 2017, 8, 460. [Google Scholar] [CrossRef]
- O’Connell, P.; Blake, M.K.; Godbehere, S.; Amalfitano, A.; Aldhamen, Y.A. SLAMF7 Modulates B Cells and Adaptive Immunity to Regulate Susceptibility to CNS Autoimmunity. J. Neuroinflamm. 2022, 19, 241. [Google Scholar] [CrossRef]
- Zhou, S.; Zhong, Z.; Lu, Y.; Li, Y.; Yao, H.; Zhao, Y.; Guo, T.; Yang, K.; Li, Y.; Chen, S.; et al. A LETM2-Regulated PI3K-Akt Signaling Axis Reveals a Prognostic and Therapeutic Target in Pancreatic Cancer. Cancers 2022, 14, 4722. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Oh, J.; Flavell, R.A.; Crawford, J.M. LACC1 Bridges NOS2 and Polyamine Metabolism in Inflammatory Macrophages. Nature 2022, 609, 348–353. [Google Scholar] [CrossRef]
- Huang, V. Endogenous miRNAa: miRNA-Mediated Gene Upregulation. In RNA Activation; Li, L.-C., Ed.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2017; pp. 65–79. ISBN 978-981-10-4310-9. [Google Scholar]
- Matter, M.A.; Paneni, F.; Libby, P.; Frantz, S.; Stähli, B.E.; Templin, C.; Mengozzi, A.; Wang, Y.-J.; Kündig, T.M.; Räber, L.; et al. Inflammation in Acute Myocardial Infarction: The Good, the Bad and the Ugly. Eur. Heart J. 2024, 45, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Ramachandra, C.J.A.; Hernandez-Resendiz, S.; Crespo-Avilan, G.E.; Lin, Y.-H.; Hausenloy, D.J. Mitochondria in Acute Myocardial Infarction and Cardioprotection. eBioMedicine 2020, 57, 1–9. [Google Scholar] [CrossRef]
- Zhao, H.; Tan, Z.; Zhou, J.; Wu, Y.; Hu, Q.; Ling, Q.; Ling, J.; Liu, M.; Ma, J.; Zhang, D.; et al. The Regulation of circRNA and lncRNAprotein Binding in Cardiovascular Diseases: Emerging Therapeutic Targets. Biomed. Pharmacother. 2023, 165, 115067. [Google Scholar] [CrossRef]
- Kong, X.Y.; Huse, C.; Yang, K.; Øgaard, J.; Berges, N.; Vik, E.S.; Nawaz, M.S.; Quiles-Jiménez, A.; Abbas, A.; Gregersen, I.; et al. Endonuclease V Regulates Atherosclerosis through C-C Motif Chemokine Ligand 2-Mediated Monocyte Infiltration. J. Am. Heart Assoc. 2021, 10, e020656. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Zhang, Q.; Yang, Z.; An, F.; Nie, H.; Wang, H.; Yang, C.; Sun, J.; Chen, K.; Zhou, J.; et al. CircEZH2/miR-133b/IGF2BP2 Aggravates Colorectal Cancer Progression via Enhancing the Stability of m6A-Modified CREB1 mRNA. Mol. Cancer 2022, 21, 140. [Google Scholar] [CrossRef]
- Huang, Y.; Zheng, W.; Ji, C.; Wang, X.; Yu, Y.; Deng, X.; Zhou, X.; Fang, L. Circular RNA circRPPH1 Promotes Breast Cancer Progression via circRPPH1-miR-512-5p-STAT1 Axis. Cell Death Discov. 2021, 7, 376. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Nie, Y.; Liu, D.; Zhu, D.; Wu, K.; Zhang, Y.; Li, W.; Tian, X.; Wang, H.; et al. CircTUBGCP3 Facilitates the Tumorigenesis of Lung Adenocarcinoma by Sponging miR-885-3p. Cancer Cell Int. 2021, 21, 651. [Google Scholar] [CrossRef]
- Panda, A.C.; Gorospe, M. Detection and Analysis of Circular RNAs by RT-PCR. Bio Protocol. 2018, 8, e2775. [Google Scholar] [CrossRef]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA Targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef]
- Rehmsmeier, M.; Steffen, P.; Höchsmann, M.; Giegerich, R. Fast and Effective Prediction of microRNA/Target Duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The Biochemical Basis of microRNA Targeting Efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Frankish, A.; Diekhans, M.; Ferreira, A.-M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE Reference Annotation for the Human and Mouse Genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Love, M.I.; Soneson, C.; Hickey, P.F.; Johnson, L.K.; Pierce, N.T.; Shepherd, L.; Morgan, M.; Patro, R. Tximeta: Reference Sequence Checksums for Provenance Identification in RNA-Seq. PLoS Comput. Biol. 2020, 16, e1007664. [Google Scholar] [CrossRef]

| Placebo (n = 19) A | Tocilizumab (n = 21) B | |
|---|---|---|
| Demographics | ||
| Age, years | 64 ± 6 | 65 ± 6 |
| Body mass index, kg/m2 | 24.4 ± 2.8 | 25.2 ± 3.3 |
| Male | 9 (47) | 11 (52) |
| White | 19 (100) | 21 (100) |
| Smoking status | ||
| Never smokers | 6 (32) | 7 (33) |
| Previous smokers | 4 (21) | 7 (33) |
| Current smokers | 9 (47) | 7 (33) |
| Prior conditions | ||
| Angina pectoris | 1 (5) | 0 |
| Cerebrovascular disease | 1 (5) | 1 (5) |
| Hypertension | 7 (37) | 7 (33) |
| Treatment | ||
| ACE inhibitor or ARB | 4 (21) | 1 (5) |
| Oral anticoagulants | 0 | 1 (5) |
| Platelet inhibitor | 1 (5) | 3 (14) |
| Beta-blocker | 1 (5) | 4 (19) |
| Calcium antagonist | 2 (10) | 3 (14) |
| Diuretic | 1 (5) | 0 |
| Statin | 3 (16) | 5 (24) |
| Up-front DAPT | 19 (100) | 21 (100) |
| Clinical characteristics | ||
| Blood pressure at admission, mm Hg | ||
| Systolic | 124 ± 22 | 130 ± 23 |
| Diastolic | 77 ± 14 | 77 ± 21 |
| Heart rate at admission, beats/min | 70 ± 16 | 70 ± 15 |
| Time from symptom onset to arrival at PCI center, min | 184 ± 80 | 144 ± 64 |
| Door-to-balloon time, min | 27 ± 15 | 21 ± 8 |
| Killip class | ||
| I | 18 (95) | 20 (95) |
| II | 1 (5) | 1 (5) |
| GRACE risk score | 147 ± 20 | 147 ± 24 |
| Infarct location | ||
| Left anterior descending branch | 3 (16) | 5 (24) |
| Circumflex or marginal | 5 (26) | 2 (10) |
| Right coronary artery | 11 (58) | 13 (62) |
| Laboratory values | ||
| Hemoglobin, g/dL | 14.1 ± 1.3 | 13.9 ± 1.5 |
| Platelet count, 109/L | 251 ± 52 | 261 ± 64 |
| Total white blood cell count, 109/L | 10.5 ± 3.4 | 11.7 ± 4.0 |
| Aspartate transaminase, U/L | 29 (24–39) | 29 (23–38) |
| Troponin T, ng/L | 58 (28–95) | 33 (19–62) |
| CK-MB, μg/L | 6.0 (2.8–23.8) | 4.4 (2.7–6.3) |
| NT-proBNP, ng/L | 155 (58–347) | 107 (52–212) |
| Creatinine, mmol/L | 64 ± 13 | 70 ± 13 |
| Glucose, mmol/L | 8.1 ± 2.0 | 8.2 ± 1.3 |
| HbA1c, mmol/mol | 36 (33–48) | 38 (36–39) |
| Total cholesterol, mmol/L | 5.5 ± 1.1 | 5.4 ± 1.1 |
| HDL cholesterol, mmol/L | 1.3 (1.1–1.7) | 1.2 (1.0–1.3) |
| LDL cholesterol, mmol/L | 3.8 ± 0.9 | 3.9 ± 1.0 |
| C-reactive protein, mg/L | 4.1 (2.1–5.0) | 1.7 (0.9–5.0) |
| Albumin, g/L | 42 ± 6 | 40 ± 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holme, F.A.; Huse, C.; Kong, X.Y.; Broch, K.; Gullestad, L.; Anstensrud, A.K.; Andersen, G.Ø.; Amundsen, B.H.; Kleveland, O.; Quiles-Jimenez, A.; et al. Circular RNA Profile in Atherosclerotic Disease: Regulation during ST-Elevated Myocardial Infarction. Int. J. Mol. Sci. 2024, 25, 9014. https://doi.org/10.3390/ijms25169014
Holme FA, Huse C, Kong XY, Broch K, Gullestad L, Anstensrud AK, Andersen GØ, Amundsen BH, Kleveland O, Quiles-Jimenez A, et al. Circular RNA Profile in Atherosclerotic Disease: Regulation during ST-Elevated Myocardial Infarction. International Journal of Molecular Sciences. 2024; 25(16):9014. https://doi.org/10.3390/ijms25169014
Chicago/Turabian StyleHolme, Fredric A., Camilla Huse, Xiang Yi Kong, Kaspar Broch, Lars Gullestad, Anne Kristine Anstensrud, Geir Ø. Andersen, Brage H. Amundsen, Ola Kleveland, Ana Quiles-Jimenez, and et al. 2024. "Circular RNA Profile in Atherosclerotic Disease: Regulation during ST-Elevated Myocardial Infarction" International Journal of Molecular Sciences 25, no. 16: 9014. https://doi.org/10.3390/ijms25169014







