Wastewater Treatment with Bacterial Representatives of the Thiothrix Morphotype
Abstract
1. Introduction
2. Types of Wastewater Treatment Plants Where Thiothrix Morphotype Can Occur
2.1. Activated Sludge
2.2. Aerobic Granular Sludge
2.3. Membrane Bioreactors
2.4. Latest Inventions for Membrane Reactors
2.5. Biofilms
3. Methods for Identification of Thiothrix in Wastewater Treatment Plants
4. Metabolic Processes in Wastewater Treatment Involving Thiothrix and Prospects for Thiothrix’s Further Application
4.1. Sulfide Removal
4.2. Enhanced Biological Removal of Phosphorus

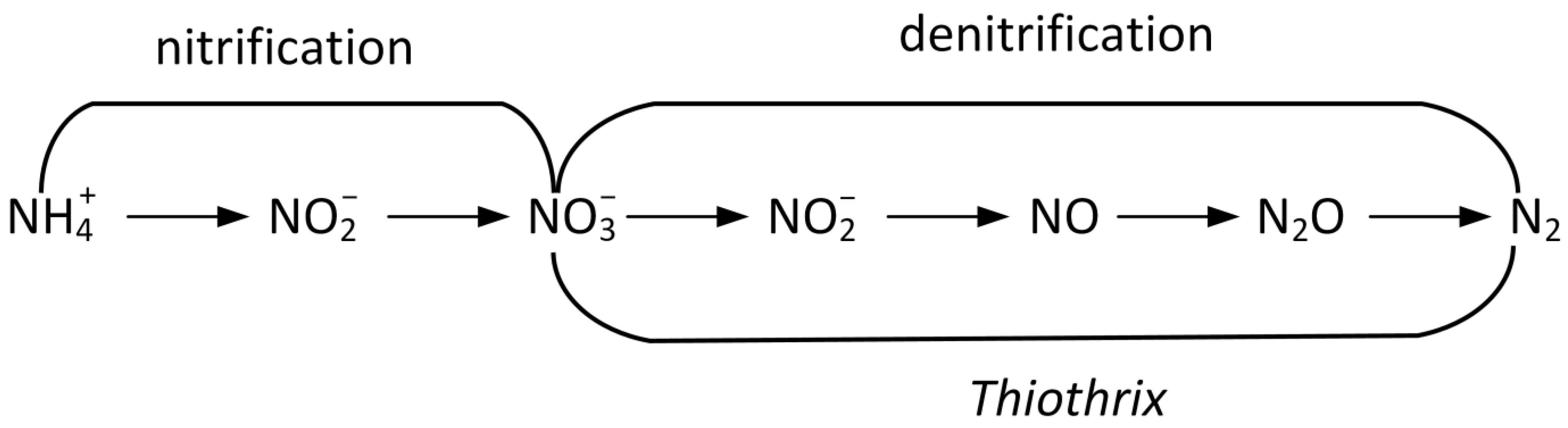
4.3. Nitrogen Removal
4.4. Denitrifying Removal of Phosphorus
5. Factors Affecting Thiothrix Proliferation
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Larkin, J.M. Isolation of Thiothrix in pure culture and observation of a filamentous epiphyte on Thiothrix. Curr. Microbiol. 1980, 4, 155–158. [Google Scholar] [CrossRef]
- Larkin, J.M.; Strohl, W.R. Beggiatoa, Thiothrix, and Thioploca. Annu. Rev. Microbiol. 1983, 37, 341–367. [Google Scholar] [CrossRef] [PubMed]
- Unz, R.F.; Head, I.M.; Genus, I. Thiothrix Winogradsky 1888, 39AL. In Bergey’s Manual of Systematic Bacteriology: Part B: Gammaproteobacteria, 2nd ed.; Garrity, G., Brenner, D.J., Krieg, N.R., Staley, J.T.E., Eds.; Springer: Boston, MA, USA, 2005; Volume 2, pp. 131–142. [Google Scholar]
- Ravin, N.V.; Rudenko, T.S.; Smolyakov, D.D.; Beletsky, A.V.; Gureeva, M.V.; Samylina, O.S.; Grabovich, M.Y. History of the study of the genus Thiothrix: From the first enrichment cultures to pangenomic analysis. Int. J. Mol. Sci. 2022, 23, 9531. [Google Scholar] [CrossRef]
- Ravin, N.V.; Muntyan, M.S.; Smolyakov, D.D.; Rudenko, T.S.; Beletsky, A.V.; Mardanov, A.V.; Grabovich, M.Y. Metagenomics revealed a new genus ‘Candidatus Thiocaldithrix dubininis’ gen. nov., sp. nov. and a new species ‘Candidatus Thiothrix putei’ sp. nov. in the family Thiotrichaceae, some members of which have traits of both Na+- and H+-motive energetics. Int. J. Mol. Sci. 2023, 24, 14199. [Google Scholar] [CrossRef]
- Jenkins, D.; Richard, M.G.; Daigger, G.T. Manual on the Causes and Control of Activated Sludge Bulking and Other Solids Separation Problems, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2003; 236p. [Google Scholar] [CrossRef]
- Eikelboom, D.H. Process Control of Activated Sludge Plants by Microscopic Investigation; IWA Publishing: London, UK, 2000; 156p. [Google Scholar]
- Howarth, R.; Unz, R.F.; Seviour, E.M.; Seviour, R.J.; Blackall, L.L.; Pickup, R.W.; Jones, J.G.; Yaguchi, J.; Head, I.M. Phylogenetic relationships of filamentous sulfur bacteria (Thiothrix spp. and Eikelboom type 021N bacteria) isolated from wastewater-treatment plants and description of Thiothrix eikelboomii sp. nov., Thiothrix unzii sp. nov., Thiothrix fructosivorans sp. nov. and Thiothrix defluvii sp. nov. Int. J. Syst. Bacteriol. 1999, 49, 1817–1827. [Google Scholar] [CrossRef]
- Aruga, S.; Kamagata, Y.; Kohno, T.; Hanada, S.; Nakamura, K.; Kanagawa, T. Characterization of filamentous Eikelboom type 021N bacteria and description of Thiothrix disciformis sp. nov. and Thiothrix flexilis sp. nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 1309–1316. [Google Scholar] [CrossRef]
- Boden, R.; Scott, K.M. Evaluation of the genus Thiothrix Winogradsky 1888 (Approved Lists 1980) emend. Aruga et al. 2002: Reclassification of Thiothrix disciformis to Thiolinea disciformis gen. nov., comb. nov., and of Thiothrix flexilis to Thiofilum flexile gen. nov., comb nov., with emended description of Thiothrix. Int. J. Syst. Evol. Microbiol. 2018, 68, 2226–2239. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, N.; Masakke, Y.; Karthikeyan, S.; Kanazawa, S.; Honda, R.; Yamamoto-Ikemoto, R.; Konstantinidis, K.T. Metagenomic insights into the effect of sulfate on enhanced biological phosphorus removal. Appl. Microbiol. Biotechnol. 2021, 105, 2181–2193. [Google Scholar] [CrossRef]
- Wu, X.; Huang, J.; Lu, Z.; Chen, G.; Wang, J.; Liu, G. Thiothrix eikelboomii interferes oxygen transfer in activated sludge. Water Res. 2019, 151, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Peng, Y.; Yang, X.; Wang, Z.; Zhu, A. Changes in the microbial community structure of filaments and floc formers in response to various carbon sources and feeding patterns. Appl. Microbiol. Biotechnol. 2014, 98, 7633–7644. [Google Scholar] [CrossRef]
- Asvapathanagul, P.; Olson, B.H.; Gedalanga, P.B.; Hashemi, A.; Huang, Z.; La, J. Identification and quantification of Thiothrix eikelboomii using qPCR for early detection of bulking incidents in a full-scale water reclamation plant. Appl. Microbiol. Biotechnol. 2015, 99, 4045–4057. [Google Scholar] [CrossRef]
- Guo, J.; Peng, Y.; Wang, Z.; Yuan, Z.; Yang, X.; Wang, S. Control filamentous bulking caused by chlorine-resistant Type 021N bacteria through adding a biocide CTAB. Water Res. 2012, 46, 6531–6542. [Google Scholar] [CrossRef]
- Kocerba-Soroka, W.; Fiałkowska, E.; Pajdak-Stós, A.; Klimek, B.; Kowalska, E.; Drzewicki, A.; Salvadó, H.; Fyda, J. The use of rotifers for limiting filamentous bacteria Type 021N, a bacteria causing activated sludge bulking. Water Sci. Technol. 2013, 67, 1557–1563. [Google Scholar] [CrossRef] [PubMed]
- Gnida, A.; Żabczyński, S.; Surmacz-Górska, J. Filamentous bacteria in the nitrifying activated sludge. Water Sci. Technol. 2018, 77, 2709–2713. [Google Scholar] [CrossRef]
- Deepnarain, N.; Nasr, M.; Kumari, S.; Stenström, T.A.; Reddy, P.; Pillay, K.; Bux, F. Decision tree for identification and prediction of filamentous bulking at full-scale activated sludge wastewater treatment plant. Process Saf. Environ. Prot. 2019, 126, 25–34. [Google Scholar] [CrossRef]
- Ravin, N.V.; Smolyakov, D.D.; Markov, N.D.; Beletsky, A.V.; Mardanov, A.V.; Rudenko, T.S.; Grabovich, M.Y. tilS and rpoB: New molecular markers for phylogenetic and biodiversity studies of the genus Thiothrix. Microorganisms 2023, 11, 2521. [Google Scholar] [CrossRef]
- Zhao, X.; Xie, Y.; Sun, B.; Liu, Y.; Zhu, S.; Li, W.; Zhao, M.; Liu, D. Unraveling microbial characteristics of simultaneous nitrification, denitrification and phosphorus removal in a membrane-aerated biofilm reactor. Environ. Res. 2023, 239, 117402. [Google Scholar] [CrossRef]
- Lee, S.; Basu, S.; Tyler, C.W.; Pitt, P.A. A survey of filamentous organisms at the Deer Island treatment plant. Environ. Technol. 2003, 24, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Choi, E.; Yun, Z.; Park, Y.K. Microbial structure and community of RBC biofilm removing nitrate and phosphorus from domestic wastewater. J. Microbiol. Biotechnol. 2008, 18, 1459–1469. [Google Scholar]
- Feng, Y.; Guo, M.; Jia, X.; Liu, N.; Li, X.; Li, X.; Song, L.; Wang, X.; Qiu, L.; Yu, Y. Combined effects of electrical current and nonsteroidal antiinflammatory drugs (NSAIDs) on microbial community in a three-dimensional electrode biological aerated filter (3DE-BAF). Bioresour. Technol. 2020, 309, 123346. [Google Scholar] [CrossRef]
- Seguel Suazo, K.; Dobbeleers, T.; Dries, J. Bacterial community and filamentous population of industrial wastewater treatment plants in Belgium. Appl. Microbiol. Biotechnol. 2024, 108, 43. [Google Scholar] [CrossRef]
- Onetto, C.A.; Eales, K.L.; Grbin, P.R. Remediation of Thiothrix spp. associated bulking problems by raw wastewater feeding: A full-scale experience. Syst. Appl. Microbiol. 2017, 40, 396–399. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.H.; de Muro, M.A.; Nielsen, J.L. Studies on the in situ physiology of Thiothrix spp. present in activated sludge. Environ. Microbiol. 2000, 2, 389–398. [Google Scholar] [CrossRef]
- Wu, G.; Yin, Q. Microbial niche nexus sustaining biological wastewater treatment. NPJ Clean Water 2020, 3, 33. [Google Scholar] [CrossRef]
- Fourest, E.; Craperi, D.; Deschamps-Roupert, C.; Pisicchio, J.-L.; Lenon, G. Occurrence and control of filamentous bulking in aerated wastewater treatment plants of the French paper industry. Water Sci. Technol. 2004, 50, 29–37. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pelletier, C.; Fitzsimmons, M.A.; Deschênes, S.; Paice, M. Impact of septic compounds and operational conditions on the microbiology of an activated sludge system. Water Sci. Technol. 2007, 55, 135–142. [Google Scholar] [CrossRef]
- Dumonceaux, T.J.; Hill, J.E.; Pelletier, C.P.; Paice, M.G.; Van Kessel, A.G.; Hemmingsen, S.M. Molecular characterization of microbial communities in Canadian pulp and paper activated sludge and quantification of a novel Thiothrix eikelboomii-like bulking filament. Can. J. Microbiol. 2006, 52, 494–500. [Google Scholar] [CrossRef]
- Kim, S.B.; Goodfellow, M.; Kelly, J.; Saddler, G.S.; Ward, A.C. Application of oligonucleotide probes for the detection of Thiothrix spp. in activated sludge plants treating paper and board mill wastes. Water Sci. Technol. 2002, 46, 559–564. [Google Scholar] [CrossRef][Green Version]
- Zou, J.; Yang, J.; He, H.; Wang, X.; Mei, R.; Cai, L.; Li, J. Effect of seed sludge type on aerobic granulation, pollutant removal and microbial community in a sequencing batch reactor treating real textile wastewater. Int. J. Environ. Res. Public Health 2022, 19, 10940. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Polonio, E.; Alvim, C.B.; Fernández-Navarro, J.; Mompó-Curell, R.; Mendoza-Roca, J.A.; Bes-Piá, A.; Alonso-Molina, J.L.; Amorós-Muñoz, I. Influence of bisphenol A occurrence in wastewaters on biomass characteristics and activated sludge process performance. Sci. Total Environ. 2021, 778, 146355. [Google Scholar] [CrossRef]
- Henriet, O.; Meunier, C.; Henry, P.; Mahillon, J. Filamentous bulking caused by Thiothrix species is efficiently controlled in full-scale wastewater treatment plants by implementing a sludge densification strategy. Sci. Rep. 2017, 7, 1430. [Google Scholar] [CrossRef]
- Figueroa, M.; Val del Río, A.; Campos, J.L.; Méndez, R.; Mosquera-Corral, A. Filamentous bacteria existence in aerobic granular reactors. Bioprocess Biosyst. Eng. 2015, 38, 841–851. [Google Scholar] [CrossRef] [PubMed]
- van der Waarde, J.; Krooneman, J.; Geurkink, B.; van der Werf, A.; Eikelboom, D.; Beimfohr, C.; Snaidr, J.; Levantesi, C.; Tandoi, V. Molecular monitoring of bulking sludge in industrial wastewater treatment plants. Water Sci. Technol. 2002, 46, 551–558. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, J.; Gong, B.; Wang, Y.; He, Q. Cometabolic degradation of lincomycin in a Sequencing Batch Biofilm Reactor (SBBR) and its microbial community. Bioresour. Technol. 2016, 214, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.F.; Wang, Q.; Zhang, H.; Qi, R. [Investigation for filamentous bacteria community diversity in activated sludge under various kinds and concentration conditions of antibiotics]. Huan Jing Ke Xue 2015, 36, 2239–2244. (In Chinese) [Google Scholar] [PubMed]
- Du, B.; Yang, Q.; Li, X.; Yuan, W.; Chen, Y.; Wang, R. Impacts of long-term exposure to tetracycline and sulfamethoxazole on the sludge granules in an anoxic-aerobic wastewater treatment system. Sci. Total Environ. 2019, 684, 67–77. [Google Scholar] [CrossRef]
- Zhang, B.; He, Y.; Shi, W.; Liu, L.; Li, L.; Liu, C.; Lens, P.N.L. Biotransformation of sulfamethoxazole (SMX) by aerobic granular sludge: Removal performance, degradation mechanism and microbial response. Sci. Total Environ. 2023, 858, 159771. [Google Scholar] [CrossRef]
- Ma, Z.; Wen, X.; Zhao, F.; Xia, Y.; Huang, X.; Waite, D.; Guan, J. Effect of temperature variation on membrane fouling and microbial community structure in membrane bioreactor. Bioresour. Technol. 2013, 133, 462–468. [Google Scholar] [CrossRef]
- Richard, M.G.; Shimizu, G.P.; Jenkins, D. The growth physiology of the filamentous organism Type-021N and its significance to activated sludge bulking. J. WPCF 1985, 57, 1152–1162. Available online: https://www.jstor.org/stable/25042817 (accessed on 19 August 2024).
- Cheng, W.; Zhang, L.; Xu, W.; Sun, Y.; Wan, J.; Li, H.; Wang, Y. Formation and characteristics of filamentous granular sludge. Water Sci. Technol. 2020, 82, 364–372. [Google Scholar] [CrossRef]
- de Graaff, D.R.; van Loosdrecht, M.C.M.; Pronk, M. Stable granulation of seawater-adapted aerobic granular sludge with filamentous Thiothrix bacteria. Water Res. 2020, 175, 115683. [Google Scholar] [CrossRef]
- Garcia, G.P.; Souza, C.L.; Glória, R.M.; Silva, S.Q.; Chernicharo, C.A. Biological oxidation of sulphides by microorganisms present in the scum layer of UASB reactors treating domestic wastewater. Water Sci. Technol. 2012, 66, 1871–1878. [Google Scholar] [CrossRef]
- Cytryn, E.; Minz, D.; Gelfand, I.; Neori, A.; Gieseke, A.; De Beer, D.; Van Rijn, J. Sulfide-oxidizing activity and bacterial community structure in a fluidized bed reactor from a zero-discharge mariculture system. Environ. Sci. Technol. 2005, 39, 1802–1810. [Google Scholar] [CrossRef]
- Rubio-Rincón, F.J.; Welles, L.; Lopez-Vazquez, C.M.; Nierychlo, M.; Abbas, B.; Geleijnse, M.; Nielsen, P.H.; van Loosdrecht, M.C.M.; Brdjanovic, D. Long-term effects of sulphide on the enhanced biological removal of phosphorus: The symbiotic role of Thiothrix caldifontis. Water Res. 2017, 116, 53–64. [Google Scholar] [CrossRef]
- Miao, X.N.; Wang, Q.; Guo, K.C.; Liu, W.R.; Shen, Y.L. [Start-up and optimization of denitrifying phosphorus removal in ABR-MBR coupling process]. Huan Jing Ke Xue 2020, 41, 4150–4160. (In Chinese) [Google Scholar] [CrossRef]
- Dong, K.; Qiu, Y.; Wang, X.; Yu, D.; Yu, Z.; Feng, J.; Wang, J.; Gu, R.; Zhao, J. Towards low carbon demand and highly efficient nutrient removal: Establishing denitrifying phosphorus removal in a biofilm-based system. Bioresour. Technol. 2023, 372, 128658. [Google Scholar] [CrossRef]
- Waqas, S.; Bilad, M.R.; Man, Z.; Wibisono, Y.; Jaafar, J.; Indra Mahlia, T.M.; Khan, A.L.; Aslam, M. Recent progress in integrated fixed-film activated sludge process for wastewater treatment: A review. J. Environ. Manag. 2020, 268, 110718. [Google Scholar] [CrossRef]
- Bakar, S.N.H.A.; Hasan, H.A.; Mohammad, A.W.; Abdullah, S.R.S.; Haan, T.Y.; Ngteni, R.; Yusof, K.M.M. A review of moving-bed biofilm reactor technology for palm oil mill effluent treatment. J. Clean. Prod. 2018, 171, 1532–1545. [Google Scholar] [CrossRef]
- Hong, Y.; Yao, J.Q.; Ma, B.; Xu, S.; Zhang, Y.J. [Filamentous sludge microbial community of a SBR reactor based on high-throughput sequencing]. Huan Jing Ke Xue 2018, 39, 3279–3285. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Williams, T.M.; Unz, R.F. Filamentous sulfur bacteria of activated sludge: Characterization of Thiothrix, Beggiatoa, and Eikelboom type 021N strains. Appl. Environ. Microbiol. 1985, 49, 887–898. [Google Scholar] [CrossRef]
- Tandoi, V.; Caravaglio, N.; Di Dio Balsamo, D.; Majone, M.; Tomei, M.C. Isolation and physiological characterization of Thiothrix sp. Water Sci. Technol. 1994, 29, 261–269. [Google Scholar] [CrossRef]
- Ravin, N.V.; Rossetti, S.; Beletsky, A.V.; Kadnikov, V.V.; Rudenko, T.S.; Smolyakov, D.D.; Moskvitina, M.I.; Gureeva, M.V.; Mardanov, A.V.; Grabovich, M.Y. Two new species of filamentous sulfur bacteria of the genus Thiothrix, Thiothrix winogradskyi sp. nov. and ‘Candidatus Thiothrix sulfatifontis’ sp. nov. Microorganisms 2022, 10, 1300. [Google Scholar] [CrossRef]
- Mardanov, A.V.; Gruzdev, E.V.; Smolyakov, D.D.; Rudenko, T.S.; Beletsky, A.V.; Gureeva, M.V.; Markov, N.D.; Berestovskaya, Y.Y.; Pimenov, N.V.; Ravin, N.V.; et al. Genomic and metabolic insights into two novel Thiothrix species from enhanced biological phosphorus removal systems. Microorganisms 2020, 8, 2030. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, E.J.H.; Pronk, M.; van Loosdrecht, M.C.M. A settling model for full-scale aerobic granular sludge. Water Res. 2020, 186, 116135. [Google Scholar] [CrossRef]
- Wang, G.; Huang, X.; Wang, S.; Yang, F.; Sun, S.; Yan, P.; Chen, Y.; Fang, F.; Guo, J. Effect of food-to-microorganisms ratio on aerobic granular sludge settleability: Microbial community, potential roles and sequential responses of extracellular proteins and polysaccharides. J. Environ. Manag. 2023, 345, 118814. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, J.; Liu, J.; Gao, X.; Wang, X. Effect of C/N on the microbial interactions of aerobic granular sludge system. J. Environ. Manag. 2024, 349, 119505. [Google Scholar] [CrossRef]
- Xu, P.; Xie, Z.; Shi, L.; Yan, X.; Fu, Z.; Ma, J.; Zhang, W.; Wang, H.; Xu, B.; He, Q. Distinct responses of aerobic granular sludge sequencing batch reactors to nitrogen and phosphorus deficient conditions. Sci. Total Environ. 2022, 834, 155369. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.H.; Ren, Q.; Xu, X.L.; Fang, J.Y.; Wang, T.; Wang, K.M.; Wang, H.Y. Enhancing stability of aerobic granules by microbial selection pressure using height-adjustable influent strategy. Water Res. 2021, 201, 117356. [Google Scholar] [CrossRef]
- Rahman, T.U.; Roy, H.; Islam, M.R.; Tahmid, M.; Fariha, A.; Mazumder, A.; Tasnim, N.; Pervez, M.N.; Cai, Y.; Naddeo, V.; et al. The advancement in Membrane Bioreactor (MBR) technology toward sustainable industrial wastewater management. Membranes 2023, 13, 181. [Google Scholar] [CrossRef]
- Huang, R.; Geng, M.; Gao, S.; Yin, X.; Tian, J. In-depth insight into improvement of simultaneous nitrification and denitrification/biofouling control by increasing sludge concentration in membrane reactor: Performance, microbial assembly and metagenomic analysis. Bioresour. Technol. 2023, 393, 130013. [Google Scholar] [CrossRef]
- Xiao, X.; Guo, H.; Ma, F.; Zhang, J.; Ma, X.; You, S. New insights into mycelial pellets for aerobic sludge granulation in membrane bioreactor: Bio-functional interactions among metazoans, microbial communities and protein expression. Water Res. 2023, 228, 119361. [Google Scholar] [CrossRef]
- Wang, Z.; Meng, F.; He, X.; Zhou, Z.; Huang, L.N.; Liang, S. Optimisation and performance of NaClO-assisted maintenance cleaning for fouling control in membrane bioreactors. Water Res. 2014, 53, 1–11. [Google Scholar] [CrossRef]
- Yang, L.; Jiao, Y.; Xu, X.; Pan, Y.; Su, C.; Duan, X.; Sun, H.; Liu, S.; Wang, S.; Shao, Z. Superstructures with atomic-level arranged perovskite and oxide layers for advanced oxidation with an enhanced non-free radical pathway. ACS Sustain. Chem. Eng. 2022, 10, 1899–1909. [Google Scholar] [CrossRef]
- Yang, N.; Yu, J.; Zhang, L.; Sun, Y.; Zhang, L.; Jiang, B. Integration of efficient LaSrCoO4 perovskite and polyacrylonitrile membrane to enhance mass transfer for rapid contaminant degradation. J. Water Process. Eng. 2023, 53, 103804. [Google Scholar] [CrossRef]
- Hassard, F.; Biddle, L.; Cartmell, E.; Jefferson, B.; Tyrrel, S.; Stephenson, T. Rotating biological contactors for wastewater treatment—A review. Process Saf. Environ. Prot. 2015, 94, 285–306. [Google Scholar] [CrossRef]
- Galván, A.; de Castro, F. Relationships among filamentous microorganisms in rotating biological contactors. Indian J. Microbiol. 2007, 47, 15–25. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ferrera, I.; Massana, R.; Casamayor, E.O.; Balagué, V.; Sánchez, O.; Pedrós-Alió, C.; Mas, J. High-diversity biofilm for the oxidation of sulfide-containing effluents. Appl. Microbiol. Biotechnol. 2004, 64, 726–734. [Google Scholar] [CrossRef]
- Moissl, C.; Rudolph, C.; Huber, R. Natural communities of novel archaea and bacteria with a string-of-pearls-like morphology: Molecular analysis of the bacterial partners. Appl. Environ. Microbiol. 2002, 68, 933–937. [Google Scholar] [CrossRef]
- Wagner, M.; Amann, R.; Kampfer, P.; Assmus, B.; Hartmann, A.; Hutzler, P.; Springer, N.; Schleifer, K. Identification and In-Situ detection of Gram-negative filamentous bacteria in activated sludge. Syst. Appl. Microbiol. 1994, 17, 405–417. [Google Scholar]
- Kanagawa, T.; Kamagata, Y.; Aruga, S.; Kohno, T.; Horn, M.; Wagner, M. Phylogenetic analysis of and oligonucleotide probe development for eikelboom type 021N filamentous bacteria isolated from bulkingactivated sludge. Appl. Environ. Microbiol. 2000, 66, 5043–5052. [Google Scholar] [CrossRef]
- Maestre, J.P.; Rovira, R.; Alvarez-Hornos, F.J.; Fortuny, M.; Lafuente, J.; Gamisans, X.; Gabriel, D. Bacterial community analysis of a gas-phase biotrickling filter for biogas mimics desulfurization through the rRNA approach. Chemosphere 2010, 80, 872–880. [Google Scholar] [CrossRef]
- Lou, I.; de Los Reyes, F.L., 3rd. Substrate uptake tests and quantitative FISH show differences in kinetic growth of bulking and non-bulking activated sludge. Biotechnol. Bioeng. 2005, 92, 729–739. [Google Scholar] [CrossRef]
- Gulez, G.; de Los Reyes, F.L., 3rd. Multiple approaches to assess filamentous bacterial growth in activated sludge under different carbon source conditions. J. Appl. Microbiol. 2009, 106, 682–691. [Google Scholar] [CrossRef]
- Gaval, G.; Pernelle, J.J. Impact of the repetition of oxygen deficiencies on the filamentous bacteria proliferation in activated sludge. Water Res. 2003, 37, 1991–2000. [Google Scholar] [CrossRef] [PubMed]
- Vervaeren, H.; De Wilde, K.; Matthys, J.; Boon, N.; Raskin, L.; Verstraete, W. Quantification of an Eikelboom type 021N bulking event with fluorescence in situ hybridization and real-time PCR. Appl. Microbiol. Biotechnol. 2005, 68, 695–704. [Google Scholar] [CrossRef]
- Chen, R.; Shuai, J.; Xie, Y.; Wang, B.; Hu, X.; Guo, W.; Lyu, W.; Zhou, D.; Mosa, A.; Wang, H. Aerobic granulation and microbial community succession in sequencing batch reactors treating the low strength wastewater: The dual effects of weak magnetic field and exogenous signal molecule. Chemosphere 2022, 309, 136762. [Google Scholar] [CrossRef] [PubMed]
- He, X.M.; Ding, L.L.; Zhang, L.L.; Gu, Z.J.; Ren, H.Q. [Performance, sludge characteristics, and the microbial community dynamics of bulking sludge under different nitrogen and phosphorus imbalances]. Huan Jing Ke Xue 2018, 39, 1782–1793. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Arumugam, K.; Bessarab, I.; Haryono, M.A.S.; Liu, X.; Zuniga-Montanez, R.E.; Roy, S.; Qiu, G.; Drautz-Moses, D.I.; Law, Y.Y.; Wuertz, S.; et al. Recovery of complete genomes and non-chromosomal replicons from activated sludge enrichment microbial communities with long read metagenome sequencing. npj Biofilms Microbiomes 2021, 7, 23. [Google Scholar] [CrossRef]
- Begmatov, S.; Beletsky, A.V.; Dorofeev, A.G.; Pimenov, N.V.; Mardanov, A.V.; Ravin, N.V. Metagenomic insights into the wastewater resistome before and after purification at large-scale wastewater treatment plants in the Moscow city. Sci. Rep. 2024, 14, 6349. [Google Scholar] [CrossRef]
- Yang, M.; Chen, T.; Liu, Y.X.; Huang, L. Visualizing set relationships: EVenn’s comprehensive approach to Venn diagrams. iMeta 2024, 3, e184. [Google Scholar] [CrossRef]
- Chernitsyna, S.M.; Elovskaya, I.S.; Bukin, S.V.; Bukin, Y.S.; Pogodaeva, T.V.; Kwon, D.A.; Zemskaya, T.I. Genomic and morphological characterization of a new Thiothrix species from a sulfide hot spring of the Zmeinaya bay (Northern Baikal, Russia). Antonie Van Leeuwenhoek 2024, 117, 23. [Google Scholar] [CrossRef]
- Cytryn, E.; Minz, D.; Gieseke, A.; van Rijn, J. Transient development of filamentous Thiothrix species in a marine sulfide oxidizing, denitrifying fluidized bed reactor. FEMS Microbiol. Lett. 2006, 256, 22–29. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maestre, J.P.; Rovira, R.; Gamisans, X.; Kinney, K.A.; Kirisits, M.J.; Lafuente, J.; Gabriel, D. Characterization of the bacterial community in a biotrickling filter treating high loads of H2S by molecular biology tools. Water Sci. Technol. 2009, 59, 1331–1337. [Google Scholar] [CrossRef]
- Wen, D.D.; Yuan, L.J.; Chen, X.; Wang, Y.; Shen, T.T.; Liu, X.B. [Effect of microbial community structure and metabolites on sludge settling ability under three different switching condition processes]. Huan Jing Ke Xue 2018, 39, 4644–4652. (In Chinese) [Google Scholar] [CrossRef]
- Wentzel, M.C.; Comeau, Y.; Ekama, G.A.; van Loosdrecht, M.C.M.; Brdjanovic, D. Enhanced biological phosphorus removal. In Biological Wastewater Treatment. Principles, Modelling and Design; Henze, M., van Loosdrecht, M., Ekama, G., Brdjanovic, D., Eds.; IWA Publishing: London, UK, 2008; pp. 155–220. [Google Scholar]
- Zuthi, M.F.; Guo, W.S.; Ngo, H.H.; Nghiem, L.D.; Hai, F.I. Enhanced biological phosphorus removal and its modeling for the activated sludge and membrane bioreactor processes. Bioresour. Technol. 2013, 139, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Ahn, K.; Kornberg, A. Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J. Biol. Chem. 1990, 265, 11734–11739. [Google Scholar]
- Zhang, H.; Ishige, K.; Kornberg, A. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc. Natl. Acad. Sci. USA 2002, 99, 16678–16683. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Guanglei, Q.; Zuniga-Montanez, R.; Williams, R.B.; Wuertz, S. Recent advances in understanding the ecophysiology of enhanced biological phosphorus removal. Curr. Opin. Biotechnol. 2021, 67, 166–174. [Google Scholar] [CrossRef]
- Gardner, S.G.; Johns, K.D.; Tanner, R.; McCleary, W.R. The PhoU Protein from Escherichia coli interacts with PhoR, PstB, and metals to form a phosphate-signaling complex at the membrane. J. Bacteriol. 2014, 196, 1741–1752. [Google Scholar] [CrossRef]
- Ravin, N.V.; Rudenko, T.S.; Smolyakov, D.D.; Beletsky, A.V.; Rakitin, A.L.; Markov, N.D.; Fomenkov, A.; Sun, L.; Roberts, R.J.; Novikov, A.A.; et al. Comparative genome analysis of the genus Thiothrix involving three novel species, Thiothrix subterranea sp. nov. Ku-5, Thiothrix litoralis sp. nov. AS and “Candidatus Thiothrix anitrata” sp. nov. A52, revealed the conservation of the pathways of dissimilatory sulfur metabolism and variations in the genetic inventory for nitrogen metabolism and autotrophic carbon fixation. Front. Microbiol. 2021, 12, 760289. [Google Scholar] [CrossRef]
- Singleton, C.M.; Petriglieri, F.; Wasmund, K.; Nierychlo, M.; Kondrotaite, Z.; Petersen, J.F.; Peces, M.; Dueholm, M.S.; Wagner, M.; Nielsen, P.H. The novel genus, ‘Candidatus Phosphoribacter’, previously identified as Tetrasphaera, is the dominant polyphosphate accumulating lineage in EBPR wastewater treatment plants worldwide. ISME J. 2022, 16, 1605–1616. [Google Scholar] [CrossRef]
- Tian, Y.; Chen, H.; Chen, L.; Deng, X.; Hu, Z.; Wang, C.; Wei, C.; Qiu, G.; Wuertz, S. Glycine adversely affects enhanced biological phosphorus removal. Water Res. 2022, 209, 117894. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.H.; McIlroy, S.J.; Albertsen, M.; Nierychlo, M. Re-evaluating the microbiology of the enhanced biological phosphorus removal process. Curr. Opin. Biotechnol. 2019, 57, 111–118. [Google Scholar] [CrossRef]
- Coats, E.R.; Deyo, B.; Brower, N.; Brinkman, C.K. Effects of anaerobic HRT and VFA loading on the kinetics and stoichiometry of enhanced biological phosphorus removal. Water Environ. Res. 2021, 93, 1608–1618. [Google Scholar] [CrossRef]
- Rey-Martínez, N.; Badia-Fabregat, M.; Guisasola, A.; Baeza, J.A. Glutamate as sole carbon source for enhanced biological phosphorus removal. Sci. Total Environ. 2019, 657, 1398–1408. [Google Scholar] [CrossRef]
- Molenaar, D.; van der Rest, M.E.; Drysch, A.; Yücel, R. Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Corynebacterium glutamicum. J. Bacteriol. 2000, 182, 6884–6891. [Google Scholar] [CrossRef] [PubMed]
- van der Rest, M.E.; Frank, C.; Molenaar, D. Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Escherichia coli. J. Bacteriol. 2000, 182, 6892–6899. [Google Scholar] [CrossRef] [PubMed]
- Wissenbach, U.; Kröger, A.; Unden, G. The specific functions of menaquinone and demethylmenaquinone in anaerobic respiration with fumarate, dimethylsulfoxide, trimethylamine N-oxide and nitrate by Escherichia coli. Arch. Microbiol. 1990, 154, 60–66. [Google Scholar] [CrossRef]
- Wallace, B.J.; Young, I.G. Role of quinones in electron transport to oxygen and nitrate in Escherichia coli. Studies with a ubiA- menA- double quinone mutant. Biochim. Biophys. Acta 1977, 461, 84–100. [Google Scholar] [CrossRef]
- Trubitsyn, I.V.; Andreevskikh, Z.G.; Yurevich, L.I.; Belousova, E.V.; Tutukina, M.N.; Merkel, A.Y.; Dubinina, G.A.; Grabovich, M.Y. Capacity for nitrate respiration as a new aspect of metabolism of the filamentous sulfur bacteria of the genus Thiothrix. Microbiology 2013, 82, 15–21. [Google Scholar] [CrossRef]
- Trubitsyn, I.V.; Belousova, E.V.; Tutukina, M.N.; Merkel, A.Y.; Dubinina, G.A.; Grabovich, M.Y. Expansion of ability of denitrification within the filamentous colorless sulfur bacteria of the genus Thiothrix. FEMS Microbiol. Lett. 2014, 358, 72–80. [Google Scholar] [CrossRef]
- Martinez-Espinosa, R.M.; Dridge, E.J.; Bonete, M.J.; Butt, J.N.; Butler, C.S.; Sargent, F.; Richardson, D.J. Look on the positive side! The orientation, identification and bioenergetics of ‘Archaeal’ membrane-bound nitrate reductases. FEMS Microbiol. Lett. 2007, 276, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, U.; Rückert, A.; Jeoung, J.H.; Görisch, H. Malate:quinone oxidoreductase is essential for growth on ethanol or acetate in Pseudomonas aeruginosa. Microbiology 2002, 148, 3839–3847. [Google Scholar] [CrossRef] [PubMed]
- Harold, L.K.; Jinich, A.; Hards, K.; Cordeiro, A.; Keighley, L.M.; Cross, A.; McNeil, M.B.; Rhee, K.; Cook, G.M. Deciphering functional redundancy and energetics of malate oxidation in mycobacteria. J. Biol. Chem. 2022, 298, 101859. [Google Scholar] [CrossRef]
- Kumar, R.; Sharma, P.; Chauhan, A.; Singh, N.; Prajapati, V.M.; Singh, S.K. Malate:quinone oxidoreductase knockout makes Mycobacterium tuberculosis susceptible to stress and affects its in vivo survival. Microbes Infect. 2024, 26, 105215. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.T.; Chang, H.H.; Egamberdieva, D.; Kamilova, F.; Lugtenberg, B.; Kuo, C.H. Genome analysis of Pseudomonas fluorescens PCL1751: A Rhizobacterium that controls root diseases and alleviates salt stress for its plant host. PLoS ONE 2015, 10, e0140231. [Google Scholar] [CrossRef]
- Guigliarelli, B.; Asso, M.; More, C.; Augier, V.; Blasco, F.; Pommier, J.; Giordano, G.; Bertrand, P. EPR and redox characterization of iron-sulfur centers in nitrate reductases A and Z from Escherichia coli: Evidence for a high-potential and a low-potential class and their relevance in the electron-transfer mechanism. Eur. J. Biochem. 1992, 207, 61–68. [Google Scholar] [CrossRef]
- Zhang, C.; Guisasola, A.; Baeza, J.A. Exploring the stability of an A-stage-EBPR system for simultaneous biological removal of organic matter and phosphorus. Chemosphere 2023, 313, 137576. [Google Scholar] [CrossRef]
- Wang, H.; Lin, L.; Zhang, L.; Han, P.; Ju, F. Microbiome assembly mechanism and functional potential in enhanced biological phosphorus removal system enriched with Tetrasphaera-related polyphosphate accumulating organisms. Environ. Res. 2023, 233, 116494. [Google Scholar] [CrossRef]
- Arumugam, K.; Bessarab, I.; Haryono, M.A.; Liu, X.; Zuniga-Montanez, R.E.; Roy, S.; Qiu, G.; Drautz-Moses, D.I.; Law, Y.Y.; Wuertz, S.; et al. Analysis procedures for assessing recovery of high quality, complete, closed genomes from Nanopore long read metagenome sequencing. BioRxiv 2020. [Google Scholar] [CrossRef]
- Ishige, K.; Zhang, H.; Kornberg, A. Polyphosphate kinase (PPK2), a potent, polyphosphate-driven generator of GTP. Proc. Natl. Acad. Sci. USA 2002, 99, 16684–16688. [Google Scholar] [CrossRef]
- Kosgey, K.; Zungu, P.V.; Bux, F.; Kumari, S. Biological nitrogen removal from low carbon wastewater. Front. Microbiol. 2022, 13, 968812. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, Q.; Chen, X.; Zhu, Y.; Yuan, C.; Zhang, C.; Zhao, T. Efficiency and microbial diversity of aeration solid-phase denitrification process bioaugmented with HN-AD bacteria for the treatment of low C/N wastewater. Environ. Res. 2021, 202, 111786. [Google Scholar] [CrossRef]
- Xia, Z.; Wang, Q.; She, Z.; Gao, M.; Zhao, Y.; Guo, L.; Jin, C. Nitrogen removal pathway and dynamics of microbial community with the increase of salinity in simultaneous nitrification and denitrification process. Sci. Total Environ. 2019, 697, 134047. [Google Scholar] [CrossRef]
- Salcedo Moyano, A.J.; Delforno, T.P.; Subtil, E.L. Simultaneous nitrification-denitrification (SND) using a thermoplastic gel as support: Pollutants removal and microbial community in a pilot-scale biofilm membrane bioreactor. Environ. Technol. 2021, 13, 4411–4425. [Google Scholar] [CrossRef]
- Luan, Y.N.; Yin, Y.; An, Y.; Zhang, F.; Wang, X.; Zhao, F.; Xiao, Y.; Liu, C. Investigation of an intermittently-aerated moving bed biofilm reactor in rural wastewater treatment under low dissolved oxygen and C/N condition. Bioresour. Technol. 2022, 358, 127405. [Google Scholar] [CrossRef]
- Tang, K.; Baskaran, V.; Nemati, M. Bacteria of the sulphur cycle: An overview of microbiology, biokinetics and their role in petroleum and mining industries. Biochem. Eng. J. 2009, 44, 73–94. [Google Scholar] [CrossRef]
- Feng, L.; Jia, R.; Zeng, Z.; Yang, G.; Xu, X. Simultaneous nitrification-denitrification and microbial community profile in an oxygen-limiting intermittent aeration SBBR with biodegradable carriers. Biodegradation 2018, 29, 473–486. [Google Scholar] [CrossRef]
- Zhao, J.; Feng, L.; Yang, G.; Dai, J.; Mu, J. Development of simultaneous nitrification-denitrification (SND) in biofilm reactors with partially coupled a novel biodegradable carrier for nitrogen-rich water purification. Bioresour. Technol. 2017, 243, 800–809. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Sun, Y.; Wan, D.; Abbasi, H.N.; Guo, Z.; Geng, H.; Wang, X.; Chen, Y. Simultaneous denitrification and phosphorus removal: A review on the functional strains and activated sludge processes. Sci. Total Environ. 2022, 835, 155409. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Wang, C.; Zhang, M.; Wu, Q.; Chen, M.; Ding, C.; He, T. [Denitrifying phosphate accumulating organisms and its mechanism of nitrogen and phosphorus removal]. Sheng Wu Gong Cheng Xue Bao 2023, 39, 1009–1025. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Gao, C.D.; Zhang, N.; Han, H.; Ren, H.; Li, Y.; Hou, C.Y.; Wang, C.D.; Peng, Y.Z. [Microbial diversity of filamentous sludge bulking at low temperature]. Huan Jing Ke Xue 2020, 41, 3373–3383. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zou, J.; Zhang, L.; Sun, J. Aerobic granular sludge for simultaneous accumulation of mineral phosphorus and removal of nitrogen via nitrite in wastewater. Bioresour. Technol. 2014, 154, 178–184. [Google Scholar] [CrossRef]
- Ai, S.; Du, L.; Nie, Z.; Liu, W.; Kang, H.; Wang, F.; Bian, D. Characterization of a novel micro-pressure double-cycle reactor for low temperature municipal wastewater treatment. Environ. Technol. 2023, 44, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Zhang, Y.; Zhou, X.; Jiang, M. Effect of nitrate concentration on filamentous bulking under low level of dissolved oxygen in an airlift inner circular anoxic-aerobic incorporate reactor. J. Environ. Sci. 2013, 25, 1736–1744. [Google Scholar] [CrossRef]
- Lei, Z.; Xue, J.; Feng, Y.; Li, Y.Y.; Kong, Z.; Chen, R. Sludge granulation in PN/A enhances nitrogen removal from mainstream anaerobically pretreated wastewater. Sci. Total Environ. 2023, 895, 165048. [Google Scholar] [CrossRef]
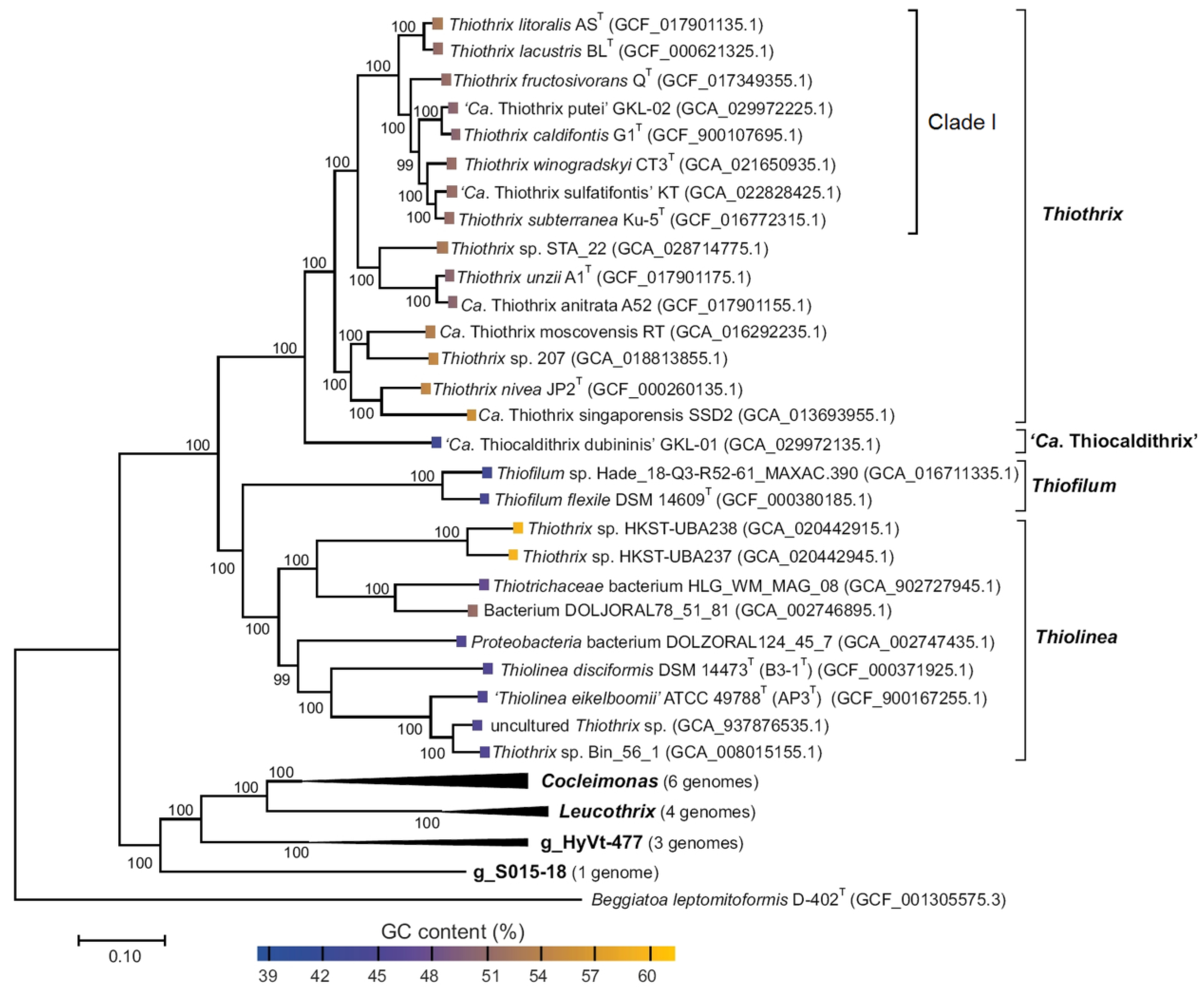
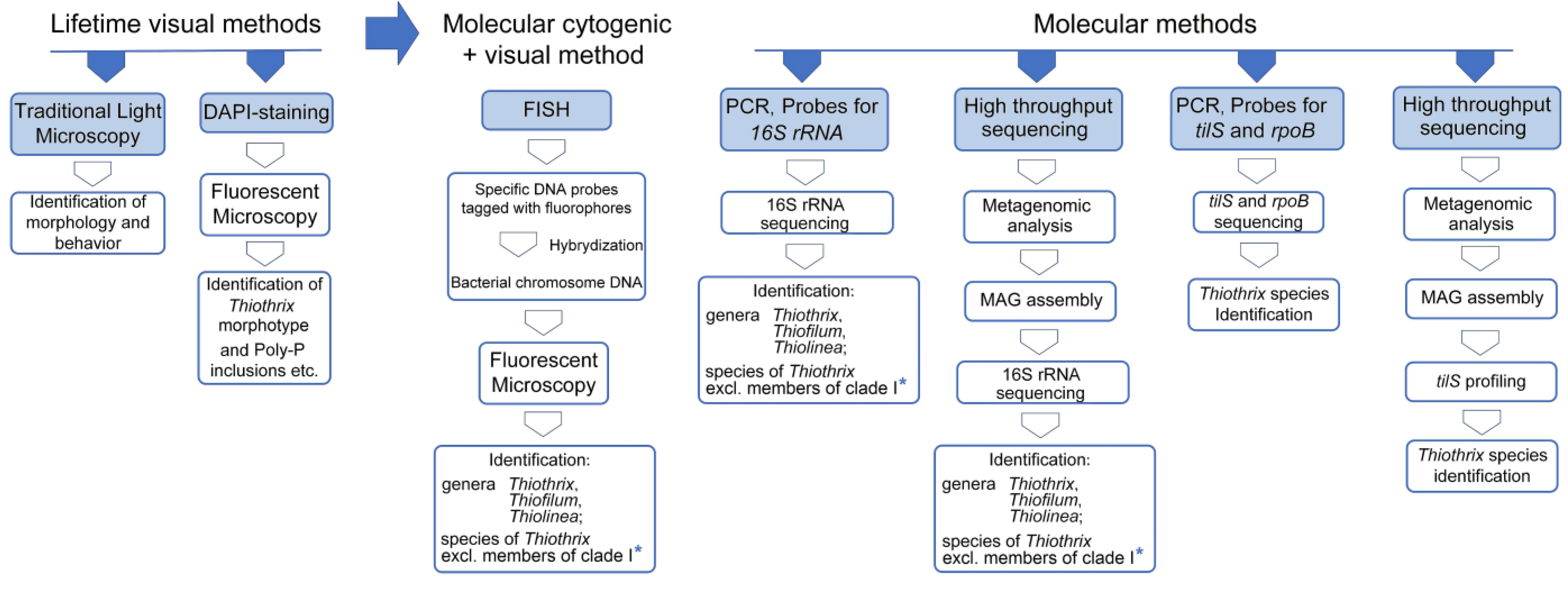
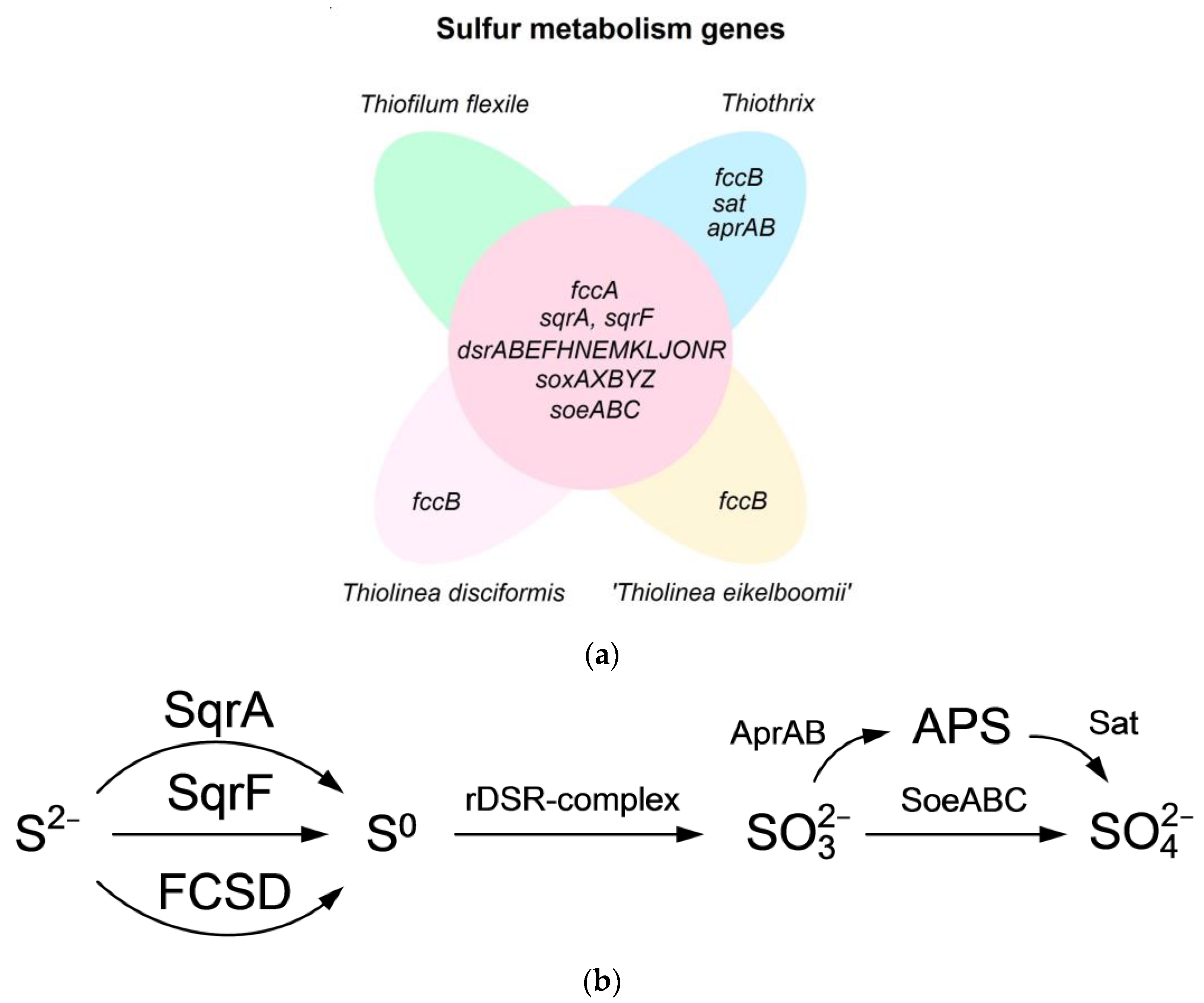
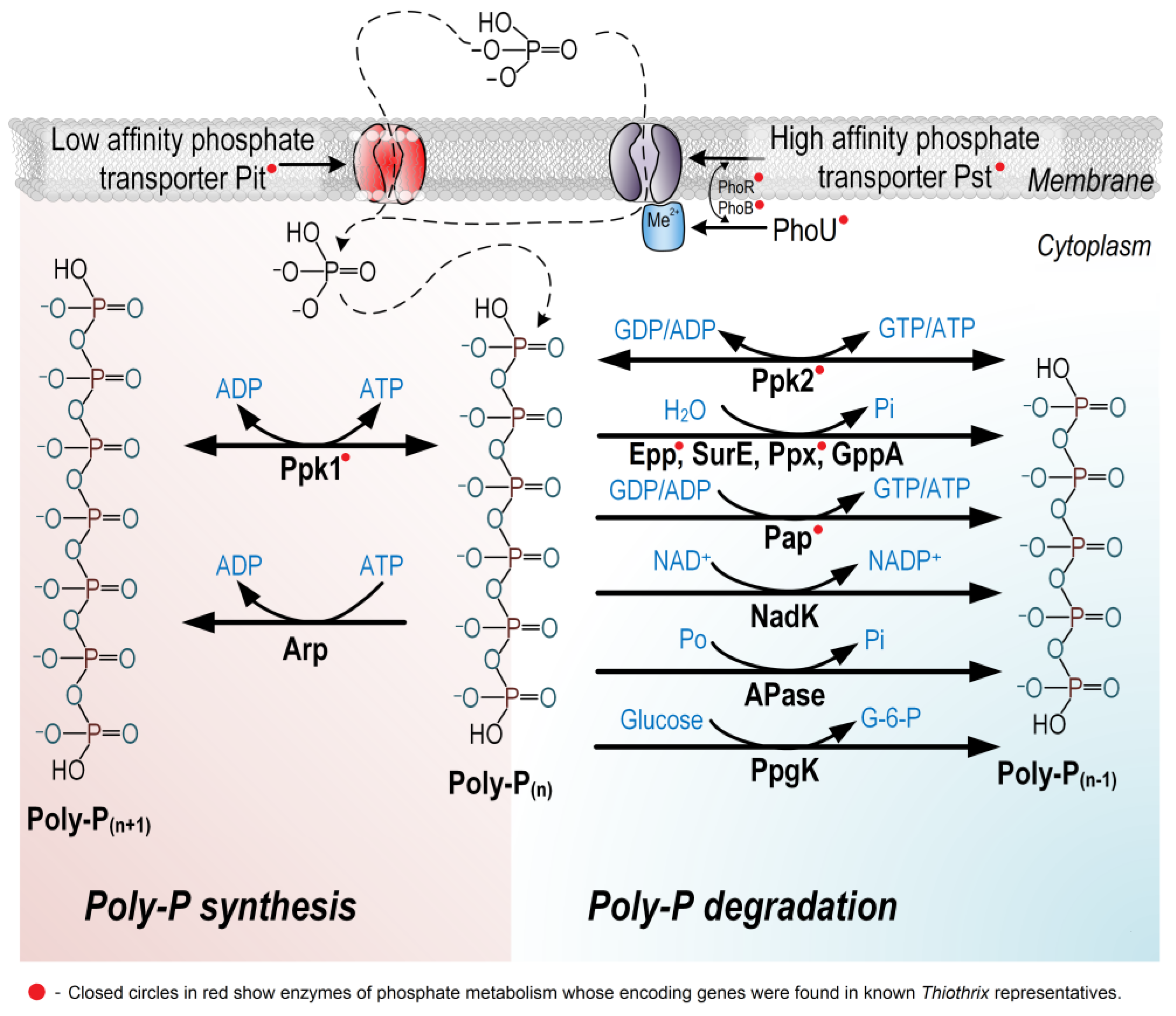

| Strain | Dissimilatory Nitrate Reduction | Assimilatory Nitrate and Nitrite Reduction | ||||
|---|---|---|---|---|---|---|
| NO3− → NO2− | NO2− → NO | NO → N2O | N2O → N2 | NO3− → NO2− | NO2− → NH3 | |
| MAG Thiothrix sp. 207 | narGHIJ, napAB | nirS | norBC | nosZ | nasA, nasD | nirBD |
| MAG Thiothrix sp. STA 22 | narGHIJ | nirS | norBC | nosZ | nasA, nasD | nirBD |
| T. litoralis AST | narGHIJ | nirS | norBC | - | nasA, nasD | nirBD |
| T. fructosivorans QT | narGHIJ | nirS | norBC | - | nasA, nasD | nirBD |
| T. caldifontis G1T | narGHIJ | nirS | norBC | - | nasA, nasD | nirBD |
| T. winogradskyi CT3T | narGHIJ | nirS | norBC | - | nasA, nasD | nirBD |
| T. unzii A1T | narGHIJ | nirS | norBC | - | nasA, nasD | nirBD |
| ‘Ca. Thiothrix singaporensis’ SSD2 | narGHIJ | nirS | norBC | - | nasA, nasD | nirBD |
| ‘Ca. Thiothrix moscovensis’ RT | narGHIJ | - | norBC | - | nasA, nasD | nirBD |
| ‘Ca. Thiothrix putei’ GK-02 | narGHIJ | - | norBC | - | - | - |
| T. subterranea Ku-5T | narGHIJ | - | - | - | nasA, nasD | nirBD |
| T. lacustris BLT | narGHIJ | - | - | - | nasA, nasD | nirBD |
| ‘Ca. Thiothrix sulfatifontis’ KT | - | - | - | - | nasA, nasD | nirBD |
| T. nivea JP2T | napAB | - | - | - | nasA, nasD | nirBD |
| ‘Ca. Thiothrix anitrata’ A52 | - | - | - | - | - | - |
| ‘Ca. Thiothrix namsaraevi’ | narGHIJ | - | - | - | nasA | nirBD |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gureeva, M.V.; Muntyan, M.S.; Ravin, N.V.; Grabovich, M.Y. Wastewater Treatment with Bacterial Representatives of the Thiothrix Morphotype. Int. J. Mol. Sci. 2024, 25, 9093. https://doi.org/10.3390/ijms25169093
Gureeva MV, Muntyan MS, Ravin NV, Grabovich MY. Wastewater Treatment with Bacterial Representatives of the Thiothrix Morphotype. International Journal of Molecular Sciences. 2024; 25(16):9093. https://doi.org/10.3390/ijms25169093
Chicago/Turabian StyleGureeva, Maria V., Maria S. Muntyan, Nikolai V. Ravin, and Margarita Yu. Grabovich. 2024. "Wastewater Treatment with Bacterial Representatives of the Thiothrix Morphotype" International Journal of Molecular Sciences 25, no. 16: 9093. https://doi.org/10.3390/ijms25169093
APA StyleGureeva, M. V., Muntyan, M. S., Ravin, N. V., & Grabovich, M. Y. (2024). Wastewater Treatment with Bacterial Representatives of the Thiothrix Morphotype. International Journal of Molecular Sciences, 25(16), 9093. https://doi.org/10.3390/ijms25169093








