Integrative Analyses Reveal the Physiological and Molecular Role of Prohexadione Calcium in Regulating Salt Tolerance in Rice
Abstract
1. Introduction
2. Result
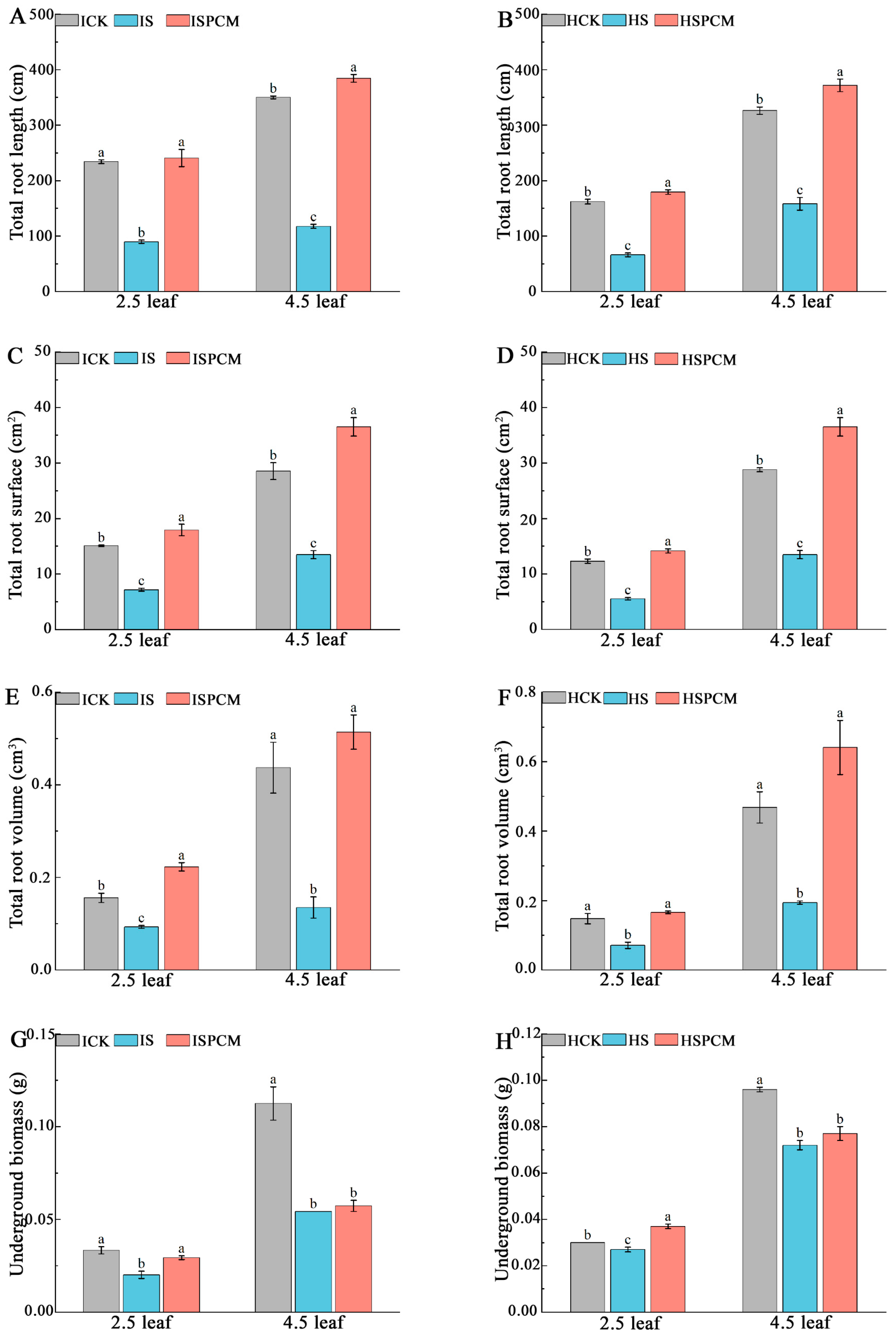
2.1. Effect of Pro-Ca on the Phenotype of Rice Seedlings under Salt Stress
2.2. Effect of Pro-Ca on the Homeostasis of K+ and Na+ in Rice under Salt Stress
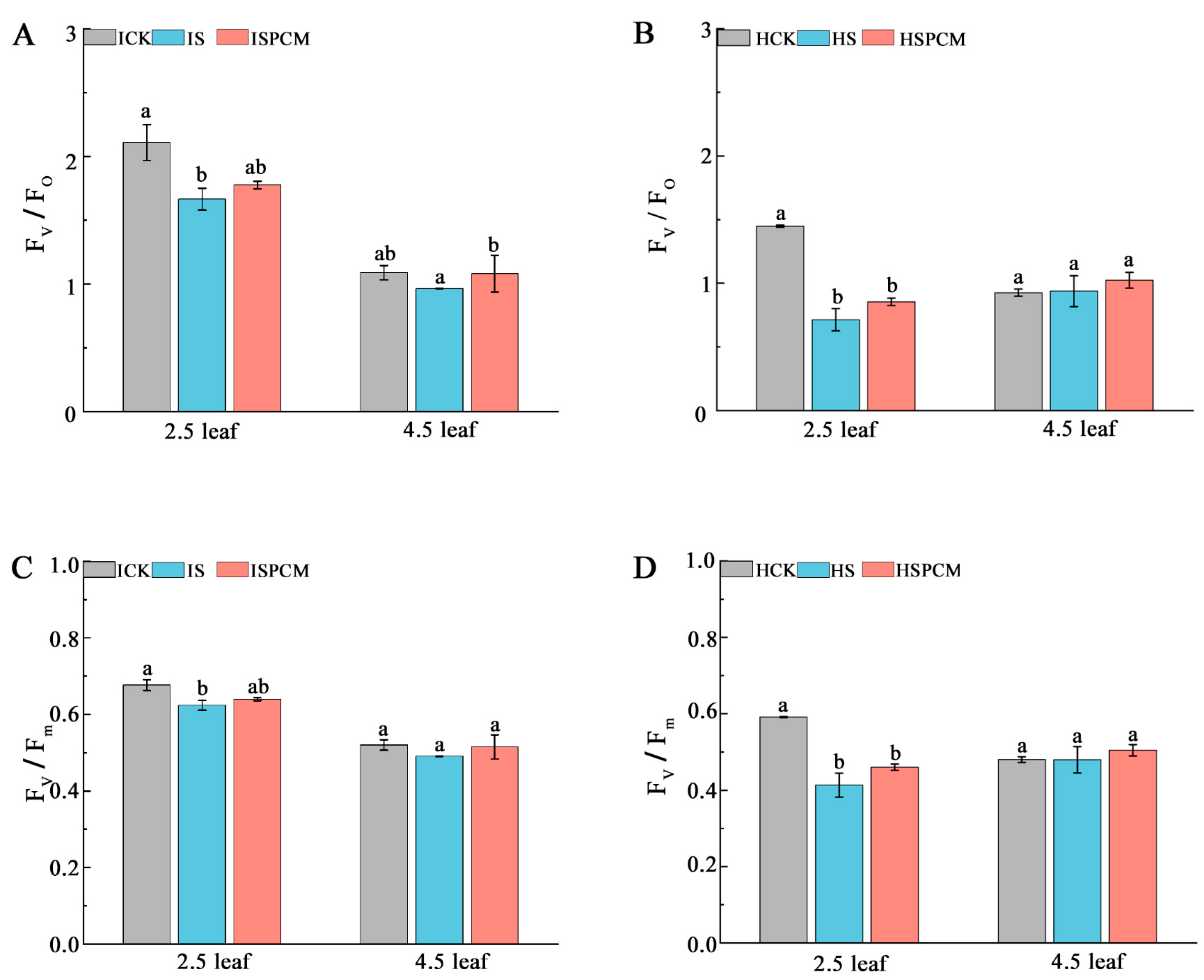
2.3. Effects of Pro-Ca on Photosynthesis- and Fluorescence-Related Parameters of Rice Leaves under Salt Stress
2.4. Analysis of Endogenous Plant Hormone Content
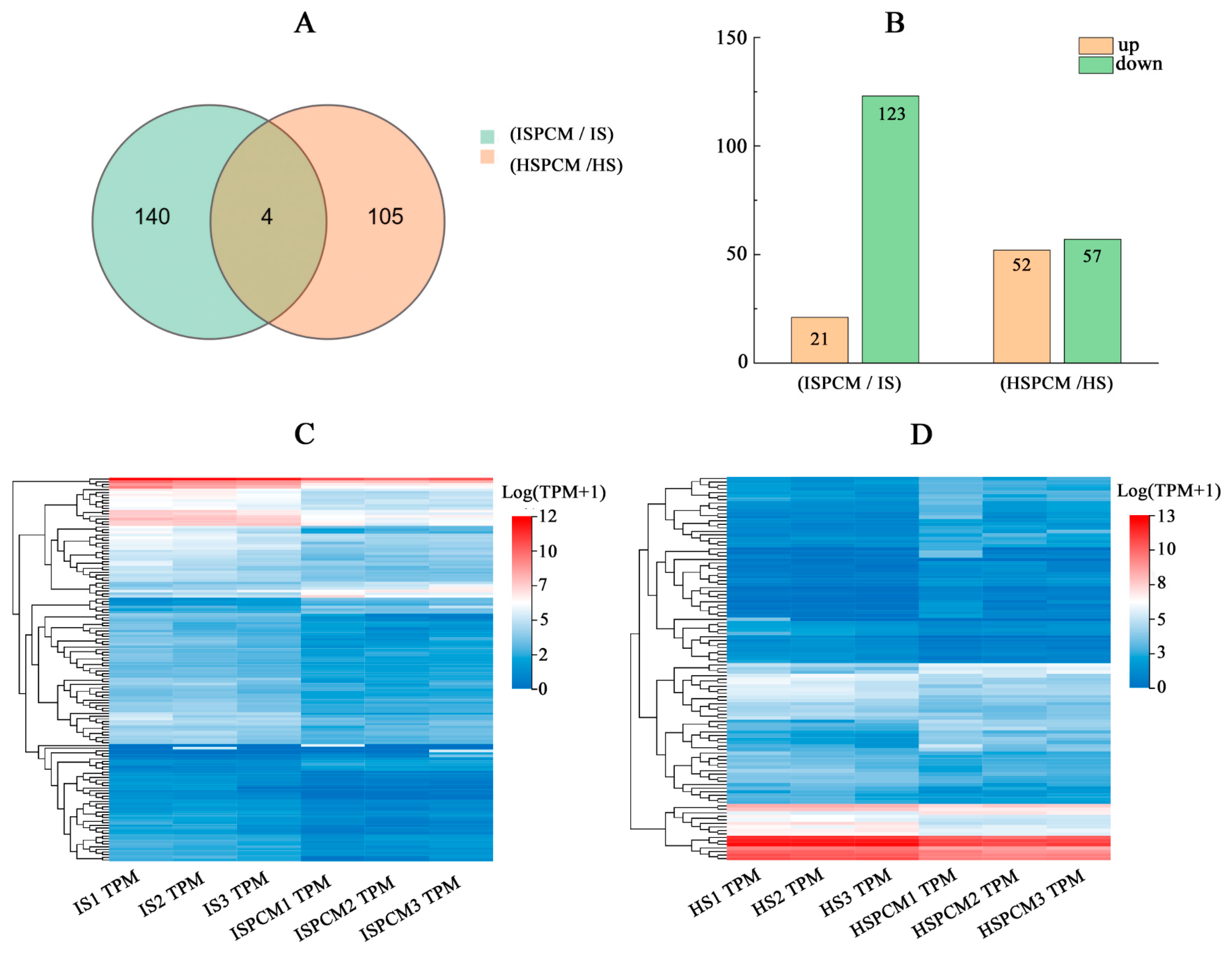
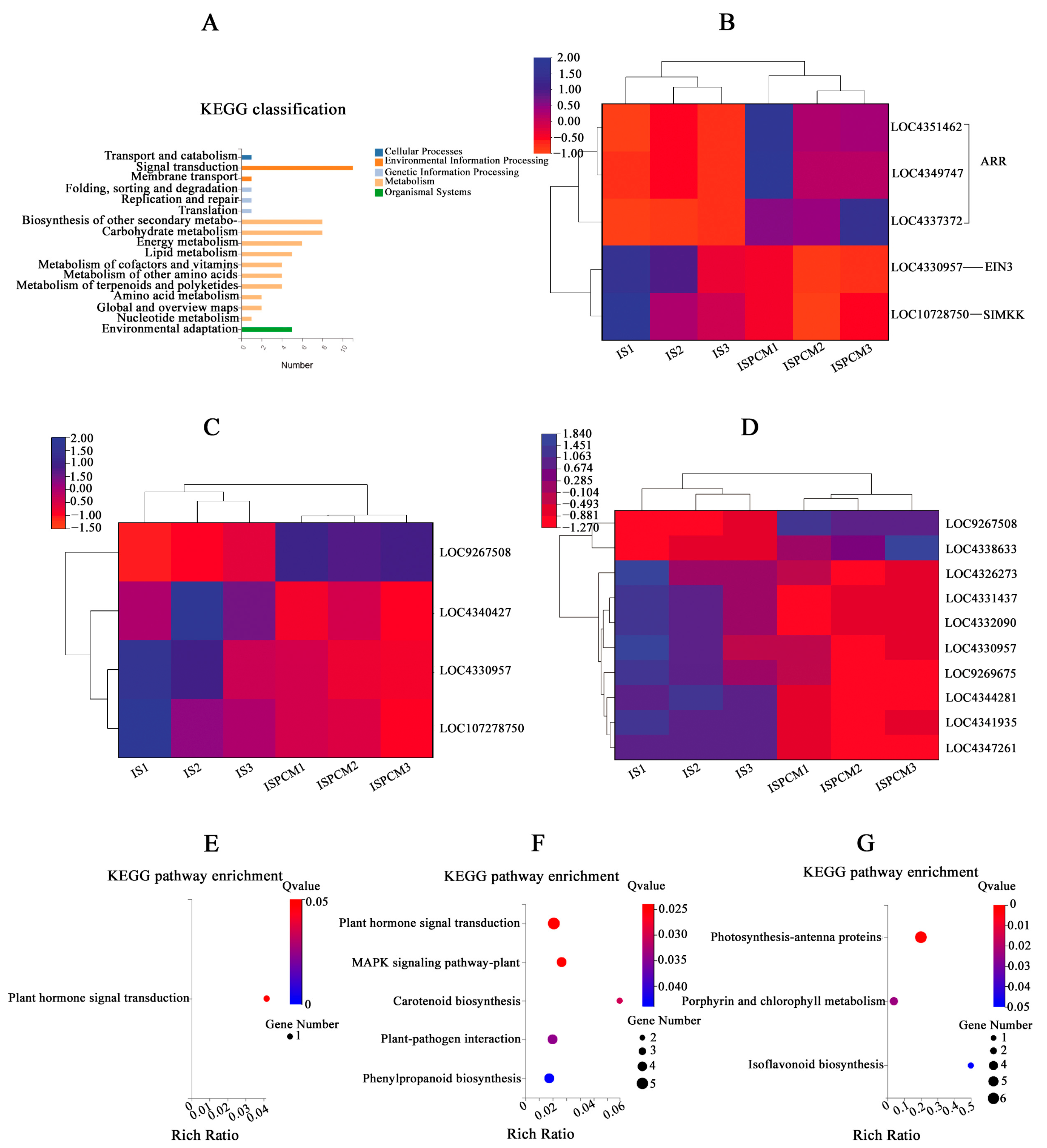
2.5. Effect of Pro-Ca Treatment on Rice Transcriptome under Salt Stress
2.5.1. Reliability Analysis
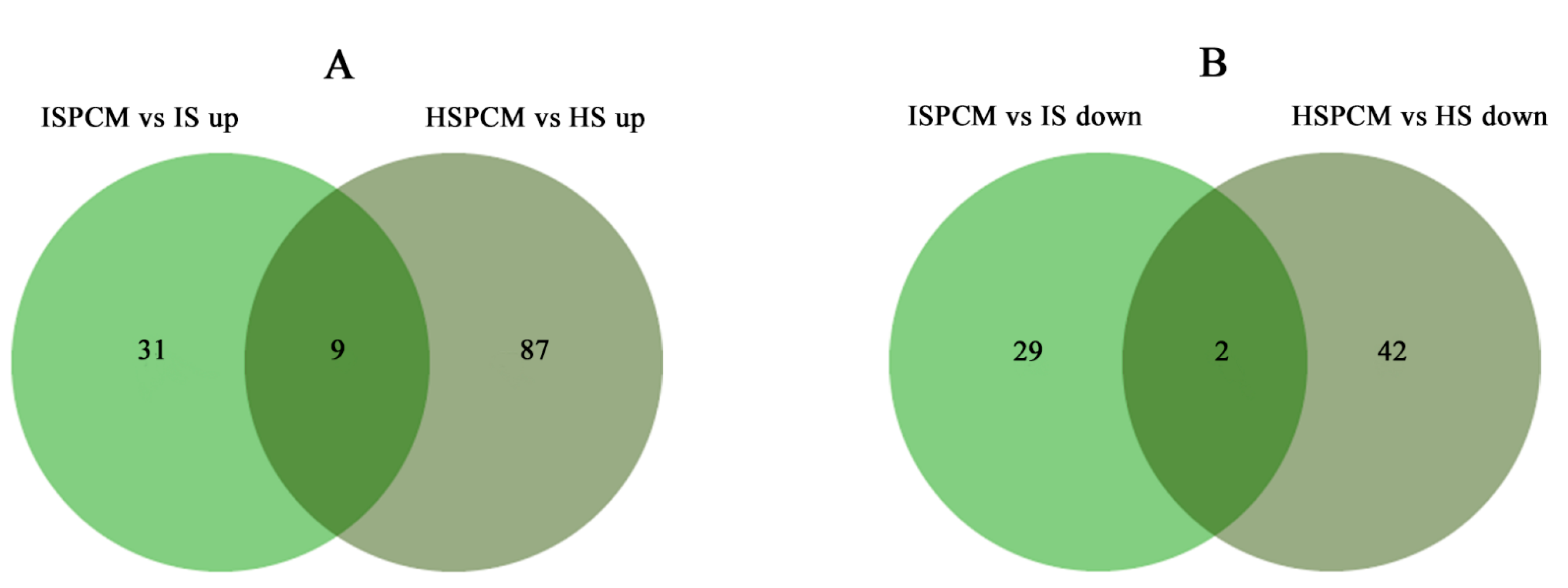
2.5.2. Comparative Analysis of Differentially Expressed Genes under Salt Stress
2.6. Effect of Pro-Ca Treatment on Rice Metabolome under Salt Stress
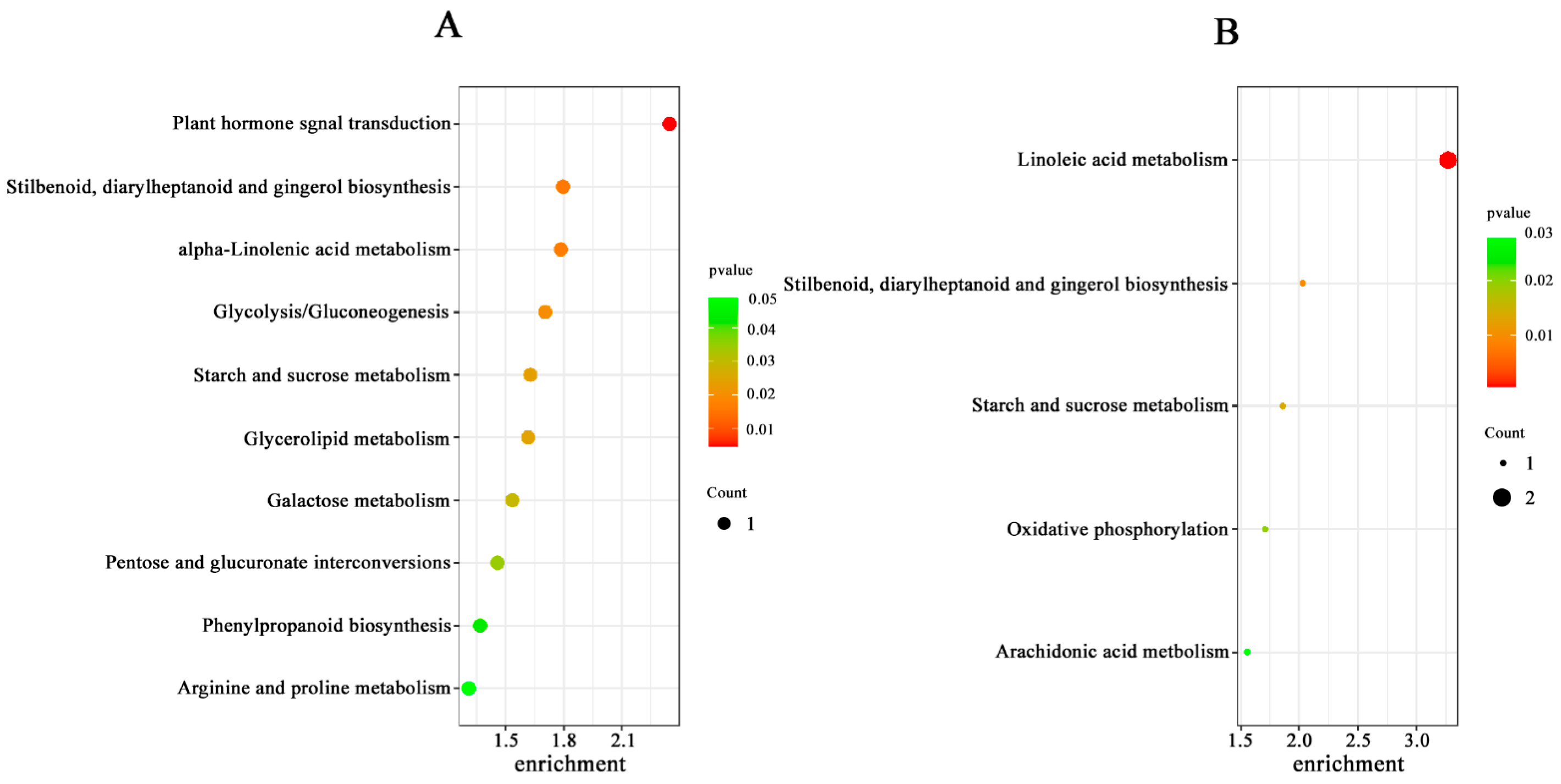
Key Biological Pathways Involved in Transcriptomic and Metabolomic Changes
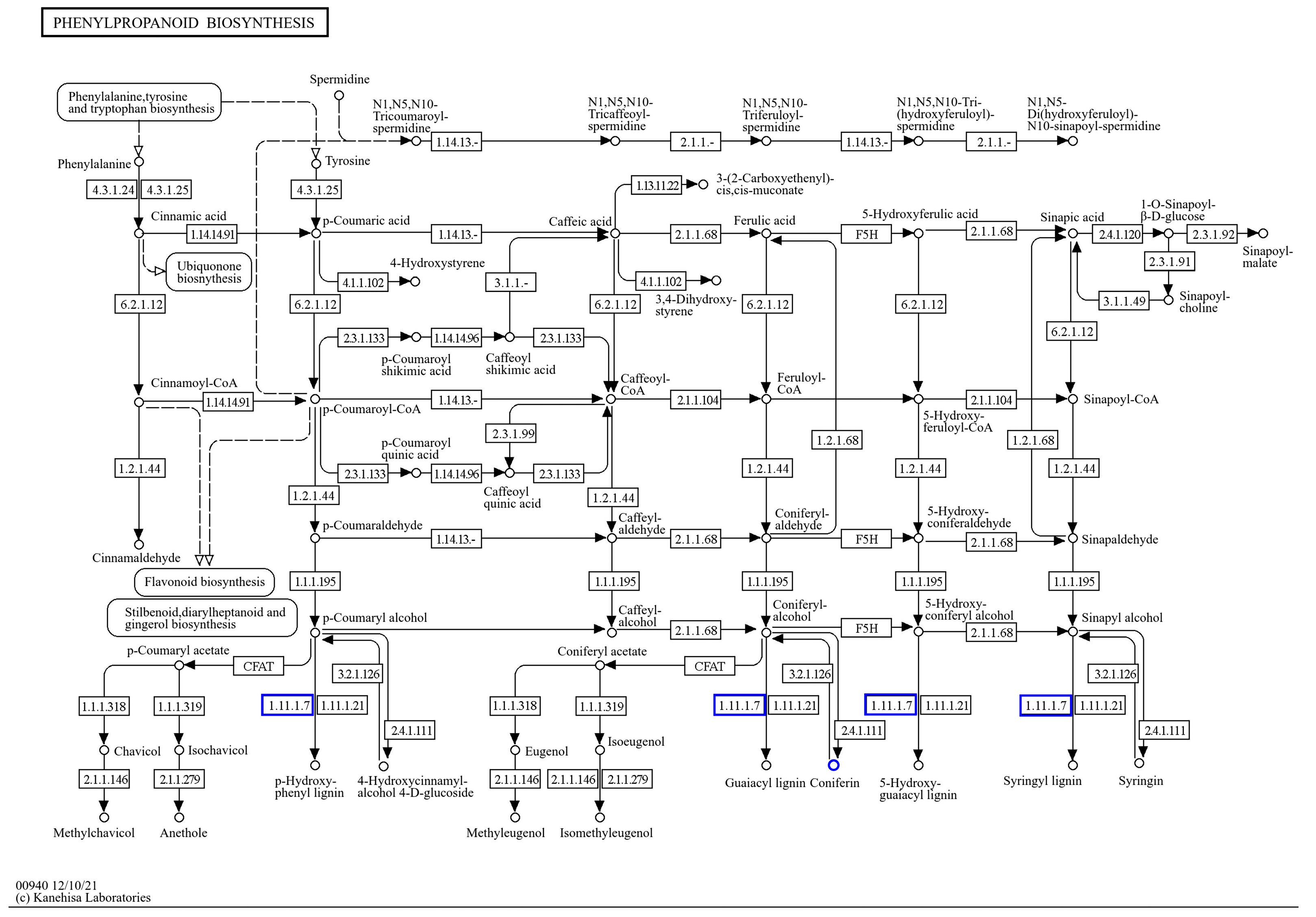
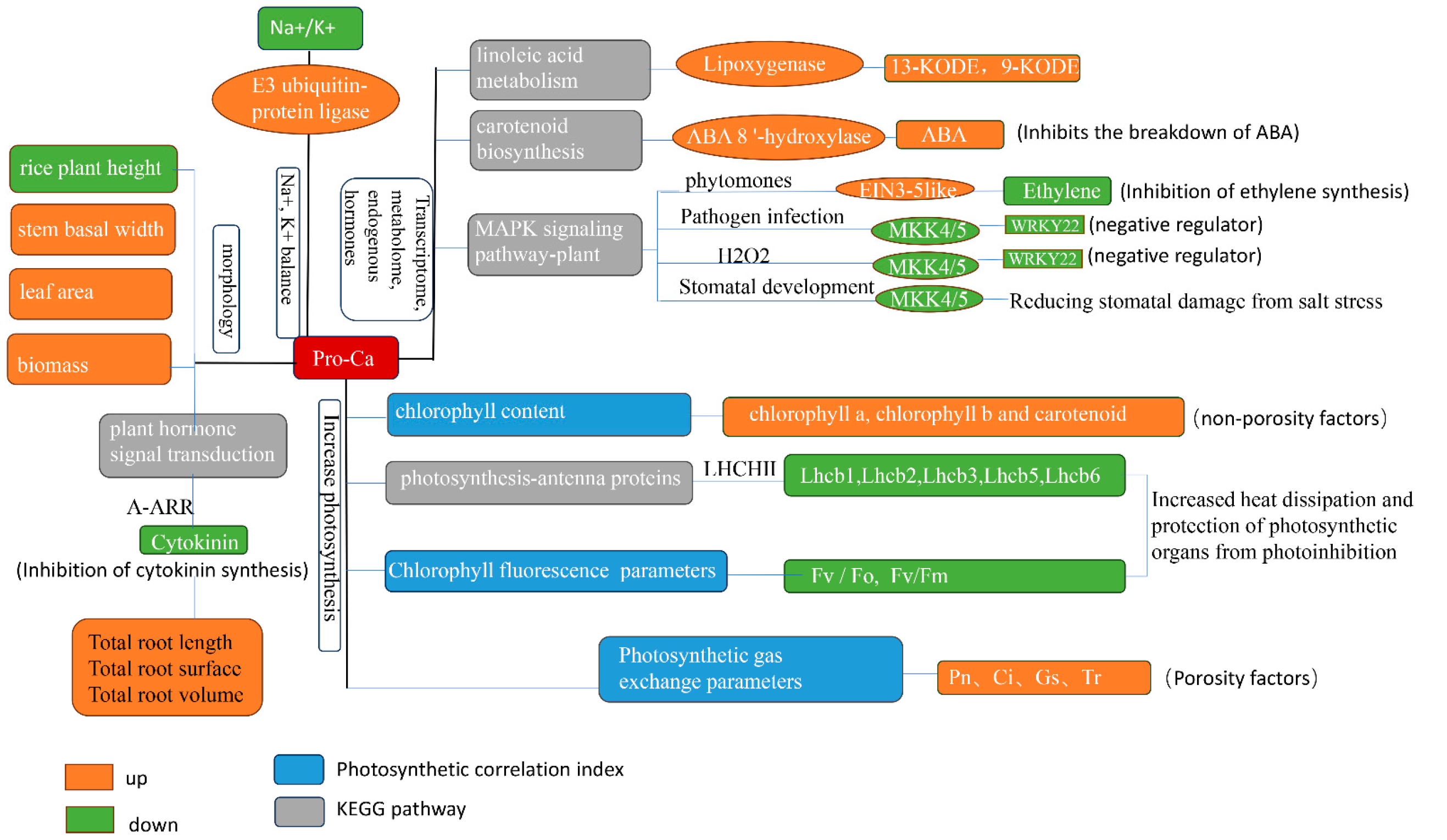
3. Discussion
3.1. Pro-Ca Alleviates the Effects of Salt Stress on Rice by Regulating Morphological Traits
3.2. Pro-Ca Alleviates the Effects of Salt Stress on Rice by Regulating Ion Content
3.3. Effect of Pro-Ca on Photosynthesis in Rice under Salt Stress
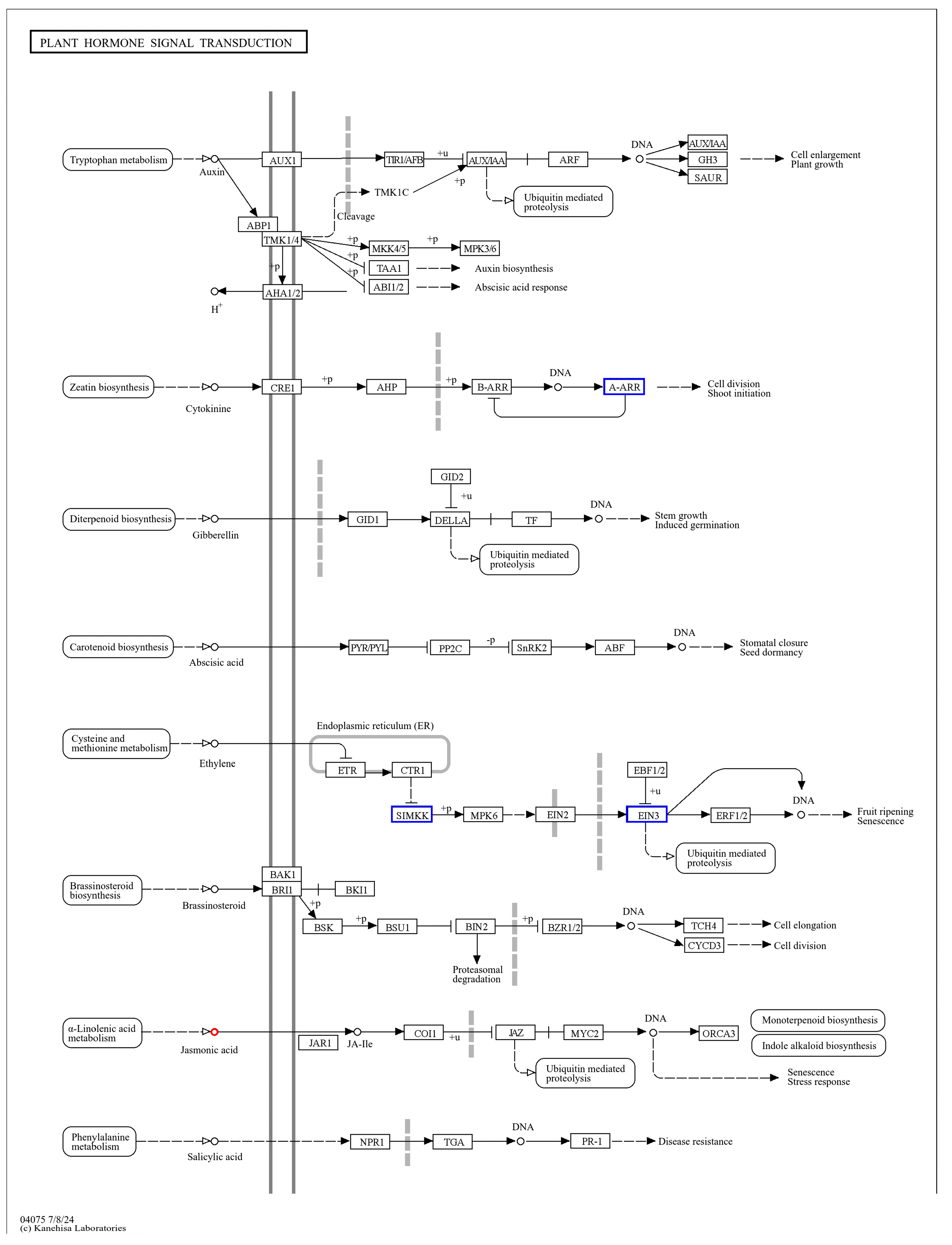
3.4. Pro-Ca Activates Plant Hormone Signal Transduction to Alleviate the Effects of Salt Stress on Rice
3.5. Effect of Pro-Ca on MAPK Signaling Pathway in Rice under Salt Stress
3.6. Effect of Pro-Ca on Carotenoid Biosynthesis in Rice under Salt Stress
3.7. Effect of Pro-Ca on Linoleic Acid Metabolism of Rice under Salt Stress in Rice
4. Conclusions
5. Materials and Methods
5.1. Experimental Materials
5.2. Plant Materials and Growth Conditions
5.3. Measurement of Growth Index
5.4. Measurement of Photosynthetic Gas Exchange Parameters
5.5. Measurement of Chlorophyll Fluorescence Parameters
5.6. Measurement of the Photosynthetic Pigments
5.7. Measurement of Ion-Related Indicators
5.8. Endogenous Hormone Extraction and Determination
5.9. Total RNA Isolation and Transcriptome Analysis
5.10. Real-Time Fluorescence Quantitative PCR (qRT-PCR) Validation
5.11. Metabolite Extraction and Metabolomic Analysis
5.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Landi, S.; Hausman, J.-F.; Guerriero, G.; Esposito, S. Poaceae vs. abiotic stress: Focus on drought and salt stress, recent insights and perspectives. Front. Plant Sci. 2017, 8, 1214. [Google Scholar] [CrossRef] [PubMed]
- Fahad, S.; Noor, M.; Adnan, M.; Khan, M.A.; Rahman, I.U.; Alam, M.; Khan, I.A.; Ullah, H.; Mian, I.A.; Hassan, S.; et al. Chapter 28—Abiotic Stress and Rice Grain Quality. In Advances in Rice Research for Abiotic Stress Tolerance; Hasanuzzaman, M., Fujita, M., Nahar, K., Biswas, J.K., Eds.; Woodhead Publishing: Cambridge, UK, 2019; pp. 571–583. [Google Scholar]
- Zhao, J.; Meng, X.; Zhang, Z.; Wang, M.; Nie, F.; Liu, Q. OsLPR5 encoding ferroxidase positively regulates the tolerance to salt stress in rice. Int. J. Mol. Sci. 2023, 24, 8115. [Google Scholar] [CrossRef]
- Ye, S.; Huang, Z.; Zhao, G.; Zhai, R.; Ye, J.; Wu, M.; Yu, F.; Zhu, G.; Zhang, X. Differential physiological responses to salt stress between salt-sensitive and salt-tolerant japonica rice cultivars at the post-germination and seedling stages. Plants 2021, 10, 2433. [Google Scholar] [CrossRef]
- Rajkumari, N.; Chowrasia, S.; Nishad, J.; Ganie, S.A.; Mondal, T.K. Metabolomics-mediated elucidation of rice responses to salt stress. Planta 2023, 258, 111. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Li, Z.; Wang, Y.; Wang, J.; Xiao, M.; Liu, H.; Quan, R.; Zhang, H.; Huang, R.; Zhu, L. Cellulose synthase-like protein OsCSLD4 plays an important role in the response of rice to salt stress by mediating abscisic acid biosynthesis to regulate osmotic stress tolerance. Plant Biotechnol. J. 2022, 20, 468–484. [Google Scholar] [CrossRef]
- Zhang, H.; Irving, L.J.; McGill, C.; Matthew, C.; Zhou, D.; Kemp, P. The effects of salinity and osmotic stress on barley germination rate: Sodium as an osmotic regulator. Ann. Bot. 2010, 106, 1027–1035. [Google Scholar] [CrossRef]
- Yadav, S.; Irfan, M.; Ahmad, A.; Hayat, S. Causes of salinity and plant manifestations to salt stress: A review. J. Environ. Biol. 2011, 32, 667. [Google Scholar]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Zhan, H.; Nie, X.; Zhang, T.; Li, S.; Wang, X.; Du, X.; Tong, W.; Song, W. Melatonin: A small molecule but important for salt stress tolerance in plants. Int. J. Mol. Sci. 2019, 20, 709. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of plant responses to salt stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Jiang, W.; Wang, X.; Wang, Y.; Du, Y.; Zhang, S.; Zhou, H.; Feng, N.; Zheng, D.; Ma, G.; Zhao, L. S-ABA Enhances Rice Salt Tolerance by Regulating Na+/K+ Balance and Hormone Homeostasis. Metabolites 2024, 14, 181. [Google Scholar] [CrossRef] [PubMed]
- Jini, D.; Joseph, B. Physiological mechanism of salicylic acid for alleviation of salt stress in rice. Rice Sci. 2017, 24, 97–108. [Google Scholar] [CrossRef]
- Kibria, M.G.; Hossain, M.; Murata, Y.; Hoque, M.A. Antioxidant defense mechanisms of salinity tolerance in rice genotypes. Rice Sci. 2017, 24, 155–162. [Google Scholar] [CrossRef]
- Zhao, L.; Yang, Z.; Guo, Q.; Mao, S.; Li, S.; Sun, F.; Wang, H.; Yang, C. Transcriptomic profiling and physiological responses of halophyte Kochia sieversiana provide insights into salt tolerance. Front. Plant Sci. 2017, 8, 1985. [Google Scholar] [CrossRef]
- Solis, C.A.; Yong, M.-T.; Zhou, M.; Venkataraman, G.; Shabala, L.; Holford, P.; Shabala, S.; Chen, Z.-H. Evolutionary significance of NHX family and NHX1 in salinity stress adaptation in the genus oryza. Int. J. Mol. Sci. 2022, 23, 2092. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ma, C.; Hou, L.; Wu, X.; Wang, D.; Zhang, L.; Liu, P. Exogenous SA affects rice seed germination under salt stress by regulating Na+/K+ balance and endogenous GAs and ABA homeostasis. Int. J. Mol. Sci. 2022, 23, 3293. [Google Scholar] [CrossRef]
- Huang, X.; Zheng, D.; Feng, N.; Huang, A.; Zhang, R.; Meng, F.; Jie, Y.; Mu, B.; Mu, D.; Zhou, H. Effects of prohexadione calcium spraying during the booting stage on panicle traits, yield, and related physiological characteristics of rice under salt stress. PeerJ 2023, 11, e14673. [Google Scholar] [CrossRef]
- Zhang, R.; Zheng, D.; Feng, N.; Qiu, Q.-S.; Zhou, H.; Liu, M.; Li, Y.; Meng, F.; Huang, X.; Huang, A. Prohexadione calcium enhances rice growth and tillering under NaCl stress. PeerJ 2023, 11, e14804. [Google Scholar] [CrossRef]
- Feng, N.; Yu, M.; Li, Y.; Jin, D.; Zheng, D. Prohexadione-calcium alleviates saline-alkali stress in soybean seedlings by improving the photosynthesis and up-regulating antioxidant defense. Ecotoxicol. Environ. Saf. 2021, 220, 112369. [Google Scholar] [CrossRef]
- Meloni, D.A.; Oliva, M.A.; Martinez, C.A.; Cambraia, J. Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ. Exp. Bot. 2003, 49, 69–76. [Google Scholar] [CrossRef]
- Yan, K.; Shao, H.; Shao, C.; Chen, P.; Zhao, S.; Brestic, M.; Chen, X. Physiological adaptive mechanisms of plants grown in saline soil and implications for sustainable saline agriculture in coastal zone. Acta Physiol. Plant. 2013, 35, 2867–2878. [Google Scholar] [CrossRef]
- Qian, R.; Ma, X.; Zhang, X.; Hu, Q.; Liu, H.; Zheng, J. Effect of exogenous spermidine on osmotic adjustment, antioxidant enzymes activity, and gene expression of Gladiolus gandavensis seedlings under salt stress. J. Plant Growth Regul. 2021, 40, 1353–1367. [Google Scholar] [CrossRef]
- Biswas, S.; Chatterjee, P.; Biswas, S.; Mazumdar, A.; Biswas, A.K. Salt induced inhibition in photosynthetic parameters and polyamine accumulation in two legume cultivars and its amelioration by pretreatment of seeds with NaCl. Legume Res.-Int. J. 2021, 44, 295–301. [Google Scholar] [CrossRef]
- Liu, H.; Blankenship, R.E. On the interface of light-harvesting antenna complexes and reaction centers in oxygenic photosynthesis. Biochim. Biophys. Acta-Bioenerg. 2019, 1860, 148079. [Google Scholar] [CrossRef] [PubMed]
- Klimmek, F.; Sjödin, A.; Noutsos, C.; Leister, D.; Jansson, S. Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol. 2006, 140, 793–804. [Google Scholar] [CrossRef]
- Xu, Y.-H.; Liu, R.; Yan, L.; Liu, Z.-Q.; Jiang, S.-C.; Shen, Y.-Y.; Wang, X.-F.; Zhang, D.-P. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 2012, 63, 1095–1106. [Google Scholar] [CrossRef]
- Zhao, S.; Gao, H.; Luo, J.; Wang, H.; Dong, Q.; Wang, Y.; Yang, K.; Mao, K.; Ma, F. Genome-wide analysis of the light-harvesting chlorophyll a/b-binding gene family in apple (Malus domestica) and functional characterization of MdLhcb4.3, which confers tolerance to drought and osmotic stress. Plant Physiol. Biochem. 2020, 154, 517–529. [Google Scholar] [CrossRef]
- Cartagena, J.A.; Yao, Y.; Mitsuya, S.; Tsuge, T. Comparative transcriptome analysis of root types in salt tolerant and sensitive rice varieties in response to salinity stress. Physiol. Plant. 2021, 173, 1629–1642. [Google Scholar] [CrossRef]
- Meng, F.; Feng, N.; Zheng, D.; Liu, M.; Zhang, R.; Huang, X.; Huang, A.; Chen, Z. Exogenous Hemin alleviates NaCl stress by promoting photosynthesis and carbon metabolism in rice seedlings. Sci. Rep. 2023, 13, 3497. [Google Scholar] [CrossRef]
- Toma-Fukai, S.; Shimizu, T. Structural diversity of ubiquitin E3 ligase. Molecules 2021, 26, 6682. [Google Scholar] [CrossRef]
- Kim, J.H.; Lim, S.D.; Jung, K.H.; Jang, C.S. Overexpression of a C3HC4-type E3-ubiquitin ligase contributes to salinity tolerance by modulating Na+ homeostasis in rice. Physiol. Plant. 2023, 175, e14075. [Google Scholar] [CrossRef] [PubMed]
- Assaha, D.V.; Ueda, A.; Saneoka, H.; Al-Yahyai, R.; Yaish, M.W. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front. Physiol. 2017, 8, 509. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Ma, L.; Zhang, C.; Yan, H.; Bao, J.; Gong, M.; Wang, W.; Li, S.; Ma, S.; Chen, B. Grapevine (Vitis vinifera) responses to salt stress and alkali stress: Transcriptional and metabolic profiling. BMC Plant Biol. 2022, 22, 528. [Google Scholar] [CrossRef] [PubMed]
- Bambach, N.; Gilbert, M.E. A dynamic model of RuBP-regeneration limited photosynthesis accounting for photoinhibition, heat and water stress. Agric. For. Meteorol. 2020, 285, 107911. [Google Scholar] [CrossRef]
- Elrad, D.; Niyogi, K.K.; Grossman, A.R. A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 2002, 14, 1801–1816. [Google Scholar] [CrossRef]
- Pascal, A.A.; Liu, Z.; Broess, K.; van Oort, B.; van Amerongen, H.; Wang, C.; Horton, P.; Robert, B.; Chang, W.; Ruban, A. Molecular basis of photoprotection and control of photosynthetic light-harvesting. Nature 2005, 436, 134–137. [Google Scholar] [CrossRef]
- Pour-Aboughadareh, A.; Omidi, M.; Naghavi, M.R.; Etminan, A.; Mehrabi, A.A.; Poczai, P.; Bayat, H. Effect of water deficit stress on seedling biomass and physio-chemical characteristics in different species of wheat possessing the D genome. Agronomy 2019, 9, 522. [Google Scholar] [CrossRef]
- Li, Y.; Gu, X.; Li, Y.; Fang, H.; Chen, P. Ridge-furrow mulching combined with appropriate nitrogen rate for enhancing photosynthetic efficiency, yield and water use efficiency of summer maize in a semi-arid region of China. Agric. Water Manag. 2023, 287, 108450. [Google Scholar] [CrossRef]
- Kiba, T.; Yamada, H.; Sato, S.; Kato, T.; Tabata, S.; Yamashino, T.; Mizuno, T. The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2003, 44, 868–874. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, Q.; Gao, Z.; Wang, Y.; Liu, Y.; Ma, Z.; Chen, Y.; Zhang, Y.; Yan, F.; Li, J. Analysis of phytohormone signal transduction in sophora alopecuroides under salt stress. Int. J. Mol. Sci. 2021, 22, 7313. [Google Scholar] [CrossRef]
- Antoniadi, I.; Mateo-Bonmati, E.; Pernisova, M.; Brunoni, F.; Antoniadi, M.; Villalonga, M.G.-A.; Ament, A.; Karady, M.; Turnbull, C.; Doležal, K. IPT9, a cis-zeatin cytokinin biosynthesis gene, promotes root growth. Front. Plant Sci. 2022, 13, 932008. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.-Y.; Cai, X.; Wan, Y.-T.; Fu, Y.-F.; Yang, X.-Y.; Zhang, Z.-W.; Yuan, S. Relatively Low Light Intensity Promotes Phosphorus Absorption and Enhances the Ethylene Signaling Component EIN3 in Maize, Wheat, and Oilseed Rape. Agronomy 2022, 12, 427. [Google Scholar] [CrossRef]
- Naing, A.H.; Campol, J.R.; Kang, H.; Xu, J.; Chung, M.Y.; Kim, C.K. Role of ethylene biosynthesis genes in the regulation of salt stress and drought stress tolerance in petunia. Front. Plant Sci. 2022, 13, 844449. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Dai, W.; Zhang, C.; Wang, Y.; Wu, M.; Zhao, Y.; Ma, Q.; Xiang, Y.; Cheng, B. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 2017, 246, 1215–1231. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, N.A. Unraveling the MAPK signaling network in stomatal development. Plant Cell 2009, 21, 3413. [Google Scholar] [CrossRef][Green Version]
- Okazaki, M.; Kittikorn, M.; Ueno, K.; Mizutani, M.; Hirai, N.; Kondo, S.; Ohnishi, T.; Todoroki, Y. Abscinazole-E2B, a practical and selective inhibitor of ABA 8′-hydroxylase CYP707A. Bioorg. Med. Chem. 2012, 20, 3162–3172. [Google Scholar] [CrossRef]
- Okamoto, M.; Kushiro, T.; Jikumaru, Y.; Abrams, S.R.; Kamiya, Y.; Seki, M.; Nambara, E. ABA 9′-hydroxylation is catalyzed by CYP707A in Arabidopsis. Phytochemistry 2011, 72, 717–722. [Google Scholar] [CrossRef]
- Shrestha, K.; Pant, S.; Huang, Y. Genome-wide identification and classification of Lipoxygenase gene family and their roles in sorghum-aphid interaction. Plant Mol. Biol. 2021, 105, 527–541. [Google Scholar] [CrossRef]
- Babenko, L.; Shcherbatiuk, M.M.; Skaterna, T.D.; Kosakivska, I. Lipoxygenases and their metabolites in formation of plant stress tolerance. Ukr. Biochem. J. 2017, 89, 5–21. [Google Scholar] [CrossRef]
- Shaban, M.; Ahmed, M.M.; Sun, H.; Ullah, A.; Zhu, L. Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses. BMC Genom. 2018, 19, 599. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, H.; Feng, N.; Zheng, D.; Ma, G.; Feng, S.; Liu, M.; Yu, M.; Huang, X.; Huang, A. Physiological and transcriptome analysis reveals that prohexadione-calcium promotes rice seedling’s development under salt stress by regulating antioxidant processes and photosynthesis. PLoS ONE 2023, 18, e0286505. [Google Scholar] [CrossRef] [PubMed]
- Sales, C.R.; Molero, G.; Evans, J.R.; Taylor, S.H.; Joynson, R.; Furbank, R.T.; Hall, A.; Carmo-Silva, E. Phenotypic variation in photosynthetic traits in wheat grown under field versus glasshouse conditions. J. Exp. Bot. 2022, 73, 3221–3237. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, F.-H.; Wang, W.-H.; Zheng, C.-J.; Lin, G.-H.; Dong, X.-J.; He, J.-X.; Pei, Z.-M.; Zheng, H.-L. Hydrogen sulphide enhances photosynthesis through promoting chloroplast biogenesis, photosynthetic enzyme expression, and thiol redox modification in Spinacia oleracea seedlings. J. Exp. Bot. 2011, 62, 4481–4493. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.; Johnson, G.N. Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef]
- Kolomeichuk, L.V.; Efimova, M.V.; Zlobin, I.E.; Kreslavski, V.D.; Murgan, O.G.K.; Kovtun, I.S.; Khripach, V.A.; Kuznetsov, V.V.; Allakhverdiev, S.I. 24-Epibrassinolide alleviates the toxic effects of NaCl on photosynthetic processes in potato plants. Photosynth. Res. 2020, 146, 151–163. [Google Scholar] [CrossRef]
- Pan, X.; Welti, R.; Wang, X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nat. Protoc. 2010, 5, 986–992. [Google Scholar] [CrossRef]
- Fan, M.; Tang, X.; Yang, Z.; Wang, J.; Zhang, X.; Yan, X.; Li, P.; Xu, N.; Liao, Z. Integration of the transcriptome and proteome provides insights into the mechanism calcium regulated of Ulva prolifera in response to high-temperature stress. Aquaculture 2022, 557, 738344. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef]






| Variety | Treatment | Leaf Na+ | Leaf K+ | Leaf Na+/K+ | Root Na+ | Root K+ | Root Na+/K+ |
|---|---|---|---|---|---|---|---|
| IR29 | CK | 0.83 ± 0.00 c | 20.93 ± 0.11 b | 0.04 ± 0.00 c | 1.59 ± 0.00 b | 16.61 ± 0.00 b | 0.10 ± 0.00 b |
| S | 11.48 ± 0.03 a | 20.35 ± 0.00 c | 0.56 ± 0.00 a | 7.94 ± 0.01 a | 14.23 ± 0.00 c | 0.56 ± 0.00 a | |
| SPCM | 1.57 ± 0.00 b | 21.86 ± 0.11 a | 0.07 ± 0.00 b | 1.48 ± 0.00 c | 18.70 ± 0.11 a | 0.08 ± 0.00 c | |
| HD96-1 | CK | 0.85 ± 0.00 c | 29.32 ± 0.11 b | 0.03 ± 0.00 b | 1.60 ± 0.00 b | 16.62 ± 0.00 b | 0.10 ± 0.00 b |
| S | 10.38 ± 0.02 a | 24.83 ± 0.12 c | 0.42 ± 0.00 a | 6.43 ± 0.00 a | 14.10 ± 0.00 c | 0.46 ± 0.00 a | |
| SPCM | 0.90 ± 0.00 b | 29.11 ± 0.11 a | 0.03 ± 0.00 b | 1.84 ± 0.01 c | 17.13 ± 0.00 a | 0.11 ± 0.00 c |
| Sampling Period | Treatment | Chl a (mg·g−1) | Chl b (mg·g−1) | Chl a+b (mg·g−1) | Car (mg·g−1) |
|---|---|---|---|---|---|
| 2.5 leaf | ICK | 23.960 ± 1.070 a | 8.516 ± 0.574 a | 32.472 ± 1.645 a | 51.145 ± 2.188 a |
| IS | 18.602 ± 0.600 b | 6.266 ± 0.273 b | 24.867 ± 0.871 b | 40.445 ± 1.154 b | |
| ISPCM | 19.418 ± 0.434 b | 6.506 ± 0.160 b | 25.924 ± 0.594 b | 44.676 ± 0.634 b | |
| HCK | 16.874 ± 0.288 a | 5.578 ± 0.106 a | 22.452 ± 0.394 a | 35.509 ± 0.562 a | |
| HS | 15.569 ± 0.906 a | 5.117 ± 0.315 a | 20.686 ± 1.220 a | 33.332 ± 1.905 a | |
| HSPCM | 16.487 ± 1.380 a | 5.497 ± 0.478 a | 21.983 ± 18.858 a | 33.921 ± 2.721 a | |
| 4.5 leaf | ICK | 23.102 ± 0.358 a | 7.989 ± 0.136 a | 31.091 ± 0.493 a | 59.904 ± 0.844 a |
| IS | 18.853 ± 1.408 b | 6.177 ± 0.516 b | 25.029 ± 1.924 b | 48.741 ± 4.045 b | |
| ISPCM | 23.249 ± 0.751 a | 8.453 ± 0.345 a | 31.702 ± 1.924 b | 60.319 ± 1.969 a | |
| HCK | 29.979 ± 0.152 a | 14.166 ± 0.360 a | 44.145 ± 0.489 a | 65.902 ± 0.199 a | |
| HS | 24.074 ± 0.575 c | 8.518 ± 0.298 c | 32.592 ± 0.873 c | 59.974 ± 1.195 b | |
| HSPCM | 27.090 ± 0.531 b | 11.284 ± 0.496 b | 38.374 ± 1.027 b | 61.815 ± 0.784 b |
| Variety | Index | Compounds | Class | Material Category | Pvalue | Log2FC | Type |
|---|---|---|---|---|---|---|---|
| IR29 | mT9G | meta-Topolin-9-glucoside | CK | cytokinin | 0.191 | −Inf | down |
| DHZROG | Dihydrozeatin-O-glucoside riboside | CK | cytokinin | 0.074 | −2.011 | down | |
| BAP | 6-Benzyladenine | CK | cytokinin | 0.015 | −Inf | down | |
| HD96-1 | cZ | cis-Zeatin | CK | cytokinin | 1.403 | −1.140 | down |
| K9G | Kinetin-9-glucoside | CK | cytokinin | 0.007 | −Inf | down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, R.; Li, Y.; Feng, N.-J.; Zheng, D.-F.; Du, Y.-W.; Khan, A.; Xue, Y.-B.; Zhang, J.-Q.; Feng, Y.-N. Integrative Analyses Reveal the Physiological and Molecular Role of Prohexadione Calcium in Regulating Salt Tolerance in Rice. Int. J. Mol. Sci. 2024, 25, 9124. https://doi.org/10.3390/ijms25169124
Deng R, Li Y, Feng N-J, Zheng D-F, Du Y-W, Khan A, Xue Y-B, Zhang J-Q, Feng Y-N. Integrative Analyses Reveal the Physiological and Molecular Role of Prohexadione Calcium in Regulating Salt Tolerance in Rice. International Journal of Molecular Sciences. 2024; 25(16):9124. https://doi.org/10.3390/ijms25169124
Chicago/Turabian StyleDeng, Rui, Yao Li, Nai-Jie Feng, Dian-Feng Zheng, You-Wei Du, Aaqil Khan, Ying-Bin Xue, Jian-Qin Zhang, and Ya-Nan Feng. 2024. "Integrative Analyses Reveal the Physiological and Molecular Role of Prohexadione Calcium in Regulating Salt Tolerance in Rice" International Journal of Molecular Sciences 25, no. 16: 9124. https://doi.org/10.3390/ijms25169124
APA StyleDeng, R., Li, Y., Feng, N.-J., Zheng, D.-F., Du, Y.-W., Khan, A., Xue, Y.-B., Zhang, J.-Q., & Feng, Y.-N. (2024). Integrative Analyses Reveal the Physiological and Molecular Role of Prohexadione Calcium in Regulating Salt Tolerance in Rice. International Journal of Molecular Sciences, 25(16), 9124. https://doi.org/10.3390/ijms25169124






