The Spiral Model of Evolution: Stable Life Forms of Organisms and Unstable Life Forms of Cancers
Abstract
:1. Introduction
2. The Key Features of Spiral Model of Accumulated Life Functions
- (a)
- The formation of new functionalities coupling with new organic codes via macroevolution;
- (b)
- The extension and modification of existing spiral functionalities via microevolution;
- (c)
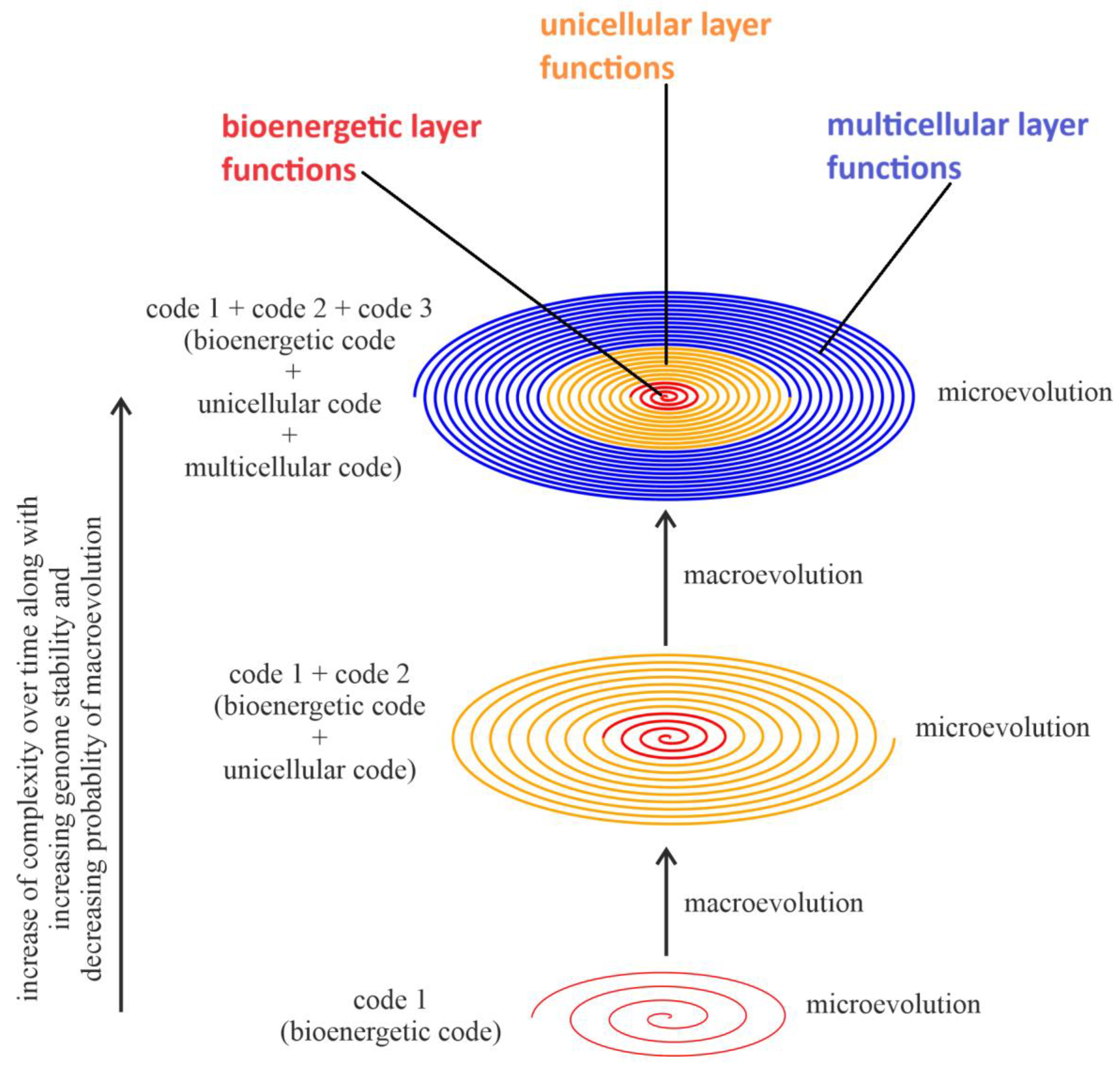
- The improvement of constraint (control) over the already existing spiral functionalities and functions, i.e., improvement of control over intra-spiral functionalities/functions, which is schematically presented in this article by an increase in the squeeze of the spiral of life functions;
- (d)
- The improvement of genome stability by improving control over the functionalities of the bioenergetic layer, resulting in a decreased probability of the occurrence of macroevolution.
3. The Spiral Model of Cellular Evolution
3.1. The Spiral Model Can Integrate Key Life Functions
3.2. Cell Bioenergetic Problems as a Cause among Many of Cancer Transformation
3.3. A Phenomenon of Cancer Transformation as a Result of Disturbances in the Spiral of Life Functions
3.4. Chromosome Instability as a Common Cause of Cancer Transformation
3.5. The Cancer Supersystems Represent New Cellular Ecosystems
4. Information Management in the Spiral Model
5. Selected Aspects of Two-Phased Cancer Evolution
5.1. The Physical Restructuring of the Existing Genome Can Result from Either the Death or the Emergence of a New Life Form
5.2. Scenarios of Competition between Clones
5.3. Cancerous Microenvironment as a Stimulator of Macroevolution
5.4. Cancerous Tissue and Elevated Risk of Metastases Caused by Macroevolution
5.5. Cancer as a Phenomenon with Many Faces—Cancer Progression Is Not Simply Atavism
6. Conclusions and Future Directions
- (a)
- A series of two-phased evolutions are linked by the spiral model. With the establishment of major macroevolutionary innovations, more organic codes form, coupled with increased complexity. The preservation and further modification of these codes provides the mechanisms for long and stable microevolution, allowing populations to grow. Therefore, maintaining the integrity of the chromosome sets or genome is important for given species. In contrast, CIN can harm a species, potentially leading to the emergence of conditions such as cancer.
- (b)
- Macroevolution occurs rapidly, as indicated by the sudden color change, while microevolution is a much longer process, represented by the spiral maintaining the same color. The continuity of the spiral among different species is achieved by different organisms, which function as carriers of information. Specifically, for species with sexual reproduction, the sex function acts as a filter to preserve the karyotype coding, which, in turn, also preserves most of the organic codes [47,111,112]. In cancer, the continuity of the spiral is achieved by different populations of cells with different karyotypes. Comparing the phenotype and genotype profiles of cancer with those of the ancestor (normal cell), cancer cells often lose the constraints on normal cell regulation and collaboration, as well as genome integrity.
- (c)
- For all biological entities, whether independent organisms like humans or cellular organisms like cancers, existence is achieved through mechanisms of information creation, preservation, and amplification during the evolutionary process. For example, the success of organismal evolution depends on an individual’s physiological capability to organize and constrain cellular components for the individual’s function, while the success of cancer evolution depends on breaking down these constraints to form new cellular systems that can compete with the host. CIN is a key player in the conflicting relationship between the host and cancer. When the genome is stable, cancer has a lower chance of success, although the host also reduces its capability for cellular adaptation. When CIN is dominant, new cellular systems can more easily overcome constraints and become cancerous. This is why CIN is a common driver of most cancers. The discussion of bioenergetic disturbances fully supports this view, as many molecular mechanisms are linked to CIN. Indeed, the evolutionary mechanism of cancer, which encompasses the collection and combination of all individual molecular mechanisms, is proposed to unify the field.
- (d)
- During the normal evolution of organisms, when new cellular functionalities are added, the dominance of previously existing functionalities is often adjusted. For example, before typical chromosome formation, individual genes in bacteria might have been more dominant. After chromosome formation, the system-level inheritance takes precedence over the part-level inheritance, becoming an organizer to regulate and constrain the functions of individual genes. The increased organic codes also require genome stability to ensure their maintenance. During cancer evolution, CIN breaks outside the spiral of life functions, removing many constraints and allowing the abnormal activity of unicellular genes, for example. The newly formed genomes no longer follow the host’s system control.
- (e)
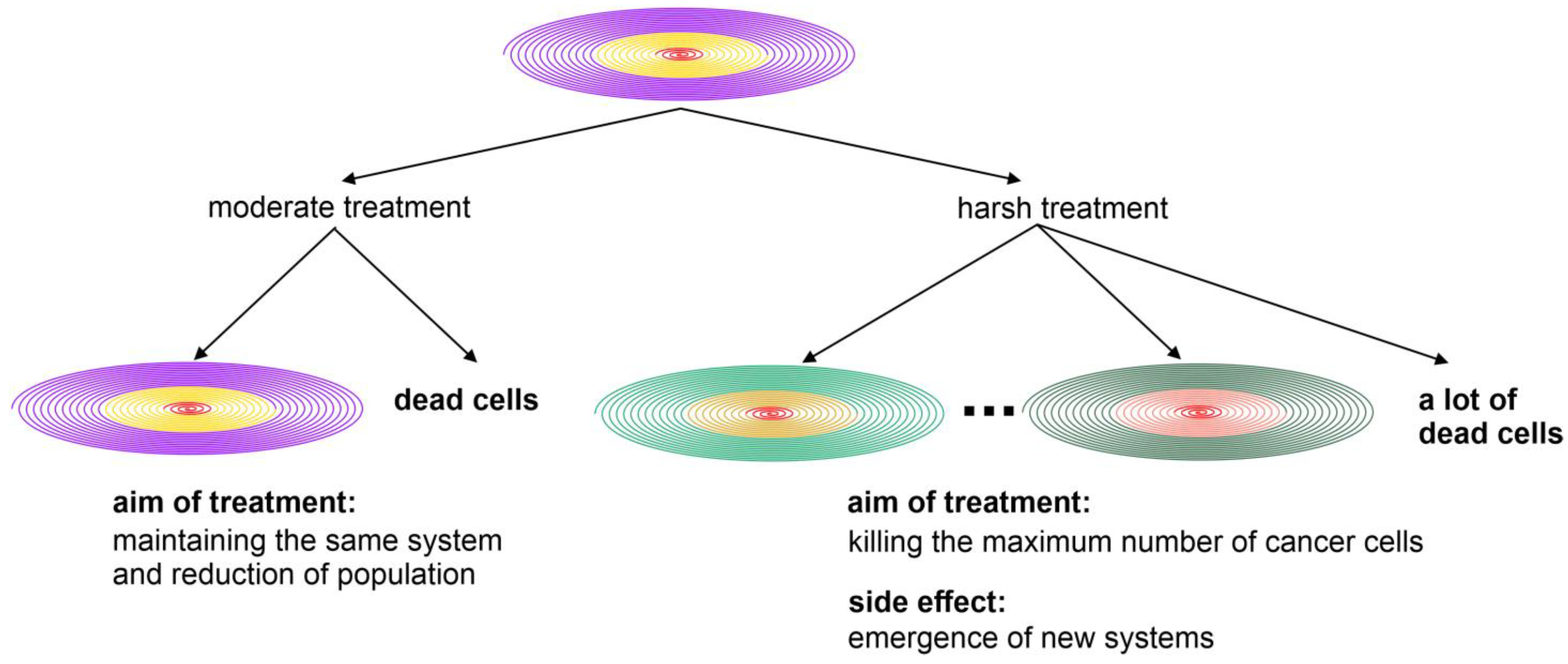
- The battle between the stable microevolution of hosts and the unstable life form of cancer is the very reason why eradicating cancer using powerful drugs proves to be so challenging. This underscores why a singular focus on eradication may be less effective, as treatment-induced crises can actually promote cancer by increasing the probability of macroevolution and, therefore, the opportunity for these unstable life forms to become more aggressive. Novel approaches to combat cancer should be grounded in the understanding of the two-phased evolution and effective information management, because inappropriate treatment may inadvertently harm the host by creating highly unstable genomes. These pharmaceutically created, more aggressive supersystems consist of a mixture of cells, both clonal and nonclonal, representing different stages of evolution. They become extremely challenging to fight, as each of these genetically distanced systems may require different pharmacological agents to eliminate them. Incorrectly established pharmaceutical agents may extinguish some systems, but can also lead to the stimulation of new cancer populations with altered dynamics of both macroevolution and microevolution.
- (f)
- Information management in cancer and host organisms offers new insights into cancer evolution. Cancers possess numerous abilities to generate new information during their evolution, without the constraints of sexual reproduction. This puts them on different competitive playing fields. Given the inherently chaotic nature of cancer evolution, determining the types of data to collect, how to prioritize research, at which phase, and on what scale of systems are still matters of ultimate importance and necessary to debate. For example, one alternative explanation for the mechanism of adaptive therapy [113,114] could be the reduction in genome chaos events. We anticipate that using moderate treatment to slow down microevolution without triggering massive genome chaos could be a new strategy for managing cancers in the future [3,4,31,84].
- (g)
- Genome chaos needs more mathematical understanding (Appendix A): it is challenging to describe genome chaos using current mathematical tools. Biological chaos, with its unique specificity, is much more complex than most physical systems. However, due to the involvement of developmental, physiological, and evolutionary constraints, its predictability can be greater than in physical systems under certain conditions. The question of how to incorporate these key differences into classical chaos theory requires further research. Nevertheless, an increasing number of papers discussing genome chaos from this perspective are being published [115].
- (h)
- The spiral model helps us to understand the mechanisms of both cancer and organismal evolution. Due to the constraints of the genome landscape, evolutionary selection aims to reduce changes in the host genome. In contrast, for successful cancer evolution, favoring system instability leads to the emergence of new systems, in which the supersystem or cancer ecosystem becomes more aggressive. This information is valuable for designing strategies to combat cancer without triggering genome chaos. An example of this would be paying more attention to treatment-induced drug resistance [72,84]. This approach is likely to explain the success of adaptive therapy as well [113,114], providing new prospects for more effective cancer treatments in the near future. One important mechanism of cancer-drug-treatment-induced resistance can be illustrated using the spiral model. The treatment can focus on either microevolution or macroevolution. Using moderate treatment, the cancer population can be reduced without triggering genome chaos. Even though the initial killing is less remarkable, the long-term benefit can be better. Harsh treatments aimed at eliminating all cancer cells can often trigger genome chaos, leading to short-term massive cell death. However, this aggressive treatment can simultaneously result in the emergence of new cancer systems, via rapid genome reorganization, which are drug-resistant and more aggressive. In a sense, treatment aimed at killing cancer cells can promote the success of cancer macroevolution, leading to drug resistance and aggressiveness. This mechanism can explain why aggressive treatment often has good short-term outcomes, but lacks significant long-term survival benefits (Figure 2).
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
- Genome as an Integrated System Rather than a Bag of Genes: The overall genomic architecture, defined by the karyotype (the complete set of chromosomes), is crucial for maintaining system identity, stability, and functionality.
- Karyotype Coding: Responsible for system inheritance, it organizes gene interactions by defining the genetic network structure. Alterations in karyotype coding play a key role in organismal and cancer evolution.
- Two-phased Evolution Unifies Macro- and Microevolution: Unlike the traditional assumption that macroevolution equals the accumulation of microevolution over time, the mechanisms of macro and microevolution differ. Macroevolution involves genome alterations, while microevolution involves gene mutations. This two-phased evolution model reconciles different molecular mechanisms for both genomics and evolution [109,134,135,136].
- Information Management: Traditional information research has overlooked information creation at the system level. Information preservation is crucial for ensuring self-organization into biological routines. Previously, research focused mainly on genetic code usage. The concept of information management reconciles genome and gene functions, providing a new perspective on evolution as information flow, which also drives evolution [31,32,38].
- New Perspective of Inheritance: The genome defines the bioinformation package, rather than genes, representing a holistic view of genetic information. There is a complex and dynamic relationship between multiple scales of genomic and non-genomic inheritance, with the environment serving as an important information context. All inheritance is fuzzy, coding a spectrum of potential phenotypes rather than a defined phenotype, explaining the gaps between genotype and phenotype. Fuzzy inheritance and environmental dynamics form the basis of biological heterogeneity [4].
- New Evolutionary Perspective: Evolutionary constraint is the major function of microevolution. The function of sex is the preservation of system inheritance [56,112]. Developmental and physiological mechanisms also serve as constraints. Genome constraint and gene dynamics are key to understanding the process of somatic change without germline change under “normal” environments, until new genomes form under crisis. This perspective explains the importance of massive extinction events that drive rapid evolution on Earth and aggressive cancer following maximal treatment [32,84,137,138,139,140].
- Redefined Functions of Evolution: In the macroevolutionary phase, chromosomal instability (CIN), either inherited or induced (often arising from environmental crises, including hybridization and accidental access to foreign genomes), promotes genome reorganization via chaos followed by macroevolution. Once new systems or species are formed and survive macro-selection, the function of microevolution is mainly to preserve these systems through biological mechanisms, such as copying or amplifying individuals, where stability is key. Many factors, including selection, as well as physical and developmental constraints, are responsible for such stability. Without it, there is no biological evolution. Therefore, by and large, the evolutionary process is about the conservation of information and its carriers, the organisms, unlike the traditional assumption that evolution is mainly about evolving. With increased complexity in the Earth’s history, more layers of regulation will be required. This is why the spiral can gain more layers with increased complexity and biofunctions. For cancer to be successful within stable organisms, a high level of CIN is constantly needed to break the multiple system constraints. Under stress, more factors can contribute to cancer. Increased reports support this idea [47,141].
- Clinical Implications: Karyotype-defined genome architecture has significant implications for disease diagnosis, treatment, and prevention. Strategies targeting genomic stability and the overall karyotype may be more effective than focusing solely on individual gene mutations. For both cancer research and medical use, prioritizing treatment options based on scales of genomics and phases of evolution is crucial [33,34,35,84,142,143,144,145].
- Information Creation: This mainly refers to system-level information creation through genome-chaos-mediated karyotype reorganization.
- Information Maintenance or Preservation: Karyotype information is primarily preserved through the functions of sex, the developmental process, and genome integrity.
- Information Modification and Transmission: This involves the accurate replication and distribution of genetic information during cell division and reproduction.
- Information Utilization: This encompasses the processes by which genetic information is accessed and used to produce functional molecules and carry out cellular processes.
References
- Heng, J.; Heng, H.H. Genome Chaos, Information Creation, and Cancer Emergence: Searching for New Frameworks on the 50th Anniversary of the “War on Cancer”. Genes 2021, 13, 101. [Google Scholar] [CrossRef] [PubMed]
- Kasperski, A.; Heng, H.H. Cancer formation as creation and penetration of unknown life spaces. In Cancer Systems Biology and Translational Mathematical Oncology; Salgia, R., Ed.; Oxford University Press: Oxford, UK, 2024. [Google Scholar]
- Heng, H.H. Genome Chaos: Rethinking Genetics, Evolution, and Molecular Medicine; Academic Press: San Diego, CA, USA, 2019. [Google Scholar]
- Heng, H.H. Debating Cancer: The Paradox in Cancer Research; World Scientific Publishing Co.: Singapore, 2015. [Google Scholar]
- Heng, H.H.; Stevens, J.B.; Liu, G.; Bremer, S.W.; Ye, K.J.; Reddy, P.; Wu, G.S.; Wang, Y.A.; Tainsky, M.A.; Ye, C.J. Stochastic cancer progression driven by non-clonal chromosome aberrations. J. Cell. Physiol. 2006, 208, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Gecow, A.; Iantovics, L.B.; Tez, M. Cancer and Chaos and the Complex Network Model of a Multicellular Organism. Biology 2022, 11, 1317. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Nedelcu, A.M. The underexplored links between cancer and the internal body climate: Implications for cancer prevention and treatment. Front. Oncol. 2022, 12, 1040034. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wang, J.; Zellmer, L.; Xu, N.; Liu, M.; Hu, Y.; Ma, H.; Deng, F.; Yang, W.; Liao, D.J. Mutation or not, what directly establishes a neoplastic state, namely cellular immortality and autonomy, still remains unknown and should be prioritized in our research. J. Cancer 2022, 13, 2810–2843. [Google Scholar] [CrossRef]
- Furst, R. The Importance of Henry H. Heng’s Genome Architecture Theory. Prog. Biophys. Mol. Biol. 2021, 165, 153–156. [Google Scholar] [CrossRef]
- Levin, M. Bioelectrical approaches to cancer as a problem of the scaling of the cellular self. Prog. Biophys. Mol. Biol. 2021, 165, 102–113. [Google Scholar] [CrossRef]
- Noble, D. Cellular Darwinism: Regulatory networks, stochasticity, and selection in cancer development. Prog. Biophys. Mol. Biol. 2021, 165, 66–71. [Google Scholar] [CrossRef]
- Shapiro, J.; Noble, D. The value of treating cancer as an evolutionary disease. Prog. Biophys. Mol. Biol. 2021, 165, 1–2. [Google Scholar] [CrossRef]
- Liu, J. The “life code”: A theory that unifies the human life cycle and the origin of human tumors. Semin. Cancer Biol. 2020, 60, 380–397. [Google Scholar] [CrossRef]
- Liu, J. The dualistic origin of human tumors. Semin. Cancer Biol. 2018, 53, 1–16. [Google Scholar] [CrossRef]
- Tian, Y.; Du, W.; Cao, S.; Wu, Y.; Dong, N.; Wang, Y.; Xu, Y. Systematic analyses of glutamine and glutamate metabolisms across different cancer types. Chin. J. Cancer 2017, 36, 88. [Google Scholar] [CrossRef]
- Huang, S. Genetic and non-genetic instability in tumor progression: Link between the fitness landscape and the epigenetic landscape of cancer cells. Cancer Metastasis Rev. 2013, 32, 423–448. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, P.; Shiraishi, T.; Kulkarni, R.V. Cancer: Tilting at windmills? Mol. Cancer 2013, 12, 108. [Google Scholar] [CrossRef] [PubMed]
- Siegel, J.J.; Amon, A. New Insights into the Troubles of Aneuploidy. Annu. Rev. Cell Dev. Biol. 2012, 28, 189–214. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.C.; Lineweaver, C.H. Cancer tumors as Metazoa 1.0: Tapping genes of ancient ancestors. Phys. Biol. 2011, 8, 015001. [Google Scholar] [CrossRef] [PubMed]
- Soto, A.M.; Sonnenschein, C. The tissue organization field theory of cancer: A testable replacement for the somatic mutation theory. BioEssays 2011, 33, 332–340. [Google Scholar] [CrossRef]
- Ao, P.; Galas, D.; Hood, L.; Yin, L.; Zhu, X.M. Towards predictive stochastic dynamical modeling of cancer genesis and progression. Interdiscip. Sci. Comput. Life Sci. 2010, 2, 140–144. [Google Scholar] [CrossRef]
- Pavelka, N.; Rancati, G.; Li, R. Dr Jekyll and Mr Hyde: Role of aneuploidy in cellular adaptation and cancer. Curr. Opin. Cell Biol. 2010, 22, 809–815. [Google Scholar] [CrossRef]
- Vincent, M.D. The Animal within: Carcinogenesis and the Clonal Evolution of Cancer Cells Are Speciation Events Sensu Stricto. Evolution 2010, 64, 1173–1183. [Google Scholar] [CrossRef]
- Wilkins, A.S. The enemy within: An epigenetic role of retrotransposons in cancer initiation. BioEssays 2010, 32, 856–865. [Google Scholar] [CrossRef]
- Ewald, P.W. An evolutionary perspective on parasitism as a cause of cancer. Adv. Parasitol. 2009, 68, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H. The genome-centric concept: Resynthesis of evolutionary theory. BioEssays 2009, 31, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.P.; Ohlsson, R.; Henikoff, S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006, 7, 21–33. [Google Scholar] [CrossRef]
- Duesberg, P.; Rasnick, D. Aneuploidy, the somatic mutation that makes cancer a species of its own. Cell Motil. Cytoskelet. 2000, 47, 81–107. [Google Scholar] [CrossRef]
- Heppner, G.H. Tumor heterogeneity. Cancer Res. 1984, 44, 2259–2265. [Google Scholar] [PubMed]
- Loeb, L.A.; Springgate, C.F.; Battula, N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974, 34, 2311–2321. [Google Scholar]
- Heng, J.; Heng, H.H. Genome chaos: Creating new genomic information essential for cancer macroevolution. Semin. Cancer Biol. 2022, 81, 160–175. [Google Scholar] [CrossRef]
- Heng, H.H. Genome Chaos: Rethinking Genetics, Evolution, and Molecular Medicine, 2nd ed.; Academic Press: Cambridge, MA, USA, 2025. [Google Scholar]
- Ferguson, S.; Jones, A.; Murray, K.; Andrew, R.; Schwessinger, B.; Borevitz, J. Plant genome evolution in the genus Eucalyptus is driven by structural rearrangements that promote sequence divergence. Genome Res. 2024, 34, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Nicolazzo, C.; Francescangeli, F.; Magri, V.; Giuliani, A.; Zeuner, A.; Gazzaniga, P. Is cancer an intelligent species? Cancer Metastasis Rev. 2023, 42, 1201–1218. [Google Scholar] [CrossRef]
- Shapiro, J.A. What we have learned about evolutionary genome change in the past 7 decades. Biosystems 2022, 215, 104669. [Google Scholar] [CrossRef]
- Pellestor, F. Chromoanagenesis: Cataclysms behind complex chromosomal rearrangements. Mol. Cytogenet. 2019, 12, 6. [Google Scholar] [CrossRef]
- Schubert, I. Macromutations Yielding Karyotype Alterations (and the Process(es) behind Them) Are the Favored Route of Carcinogenesis and Speciation. Cancers 2024, 16, 554. [Google Scholar] [CrossRef] [PubMed]
- Heng, J.; Heng, H.H. Karyotype coding: The creation and maintenance of system information for complexity and biodiversity. BioSystems 2021, 208, 104476. [Google Scholar] [CrossRef] [PubMed]
- Kasperski, A. Life Entrapped in a Network of Atavistic Attractors: How to Find a Rescue. Int. J. Mol. Sci. 2022, 23, 4017. [Google Scholar] [CrossRef]
- Kasperski, A.; Kasperska, R. Study on attractors during organism evolution. Sci. Rep. 2021, 11, 9637. [Google Scholar] [CrossRef] [PubMed]
- Kasperski, A.; Kasperska, R. Bioenergetics of life, disease and death phenomena. Theory Biosci. 2018, 137, 155–168. [Google Scholar] [CrossRef]
- Horne, S.D.; Chowdhury, S.K.; Heng, H.H. Stress, genomic adaptation, and the evolutionary trade-off. Front Genet. 2014, 5, 92. [Google Scholar] [CrossRef]
- Bussey, K.J.; Cisneros, L.H.; Lineweaver, C.H.; Davies, P.C.W. Ancestral gene regulatory networks drive cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 6160–6162. [Google Scholar] [CrossRef]
- Nowell, P.C. The clonal evolution of tumor cell populations. Science 1976, 194, 23–28. [Google Scholar] [CrossRef]
- Cairns, J. Mutation selection and the natural history of cancer. Nature 1975, 255, 197–200. [Google Scholar] [CrossRef]
- Levan, A. Some current problems of cancer cytogenetics. Hereditas 1967, 57, 343–355. [Google Scholar] [CrossRef]
- Heng, H.H. System information creation, preservation, and historical contingencies: How genome chaos unifies macro- and microevolution. In Proceedings of the Potential & Limitations of Evolutionary Processes Conference, Lower Galilee, Israel, 8–12 May 2022. [Google Scholar]
- Barbieri, M. Overview of the third special issue in code biology. BioSystems 2021, 210, 104553. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, M. A general model on the origin of biological codes. BioSystems 2019, 181, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, M. Evolution of the genetic code: The ambiguity-reduction theory. BioSystems 2019, 185, 104024. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, M. The Organic Codes: An Introduction to Semantic Biology; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar] [CrossRef]
- Bayona-Feliu, A.; Herrera-Moyano, E.; Badra-Fajardo, N.; Galván-Femenía, I.; Soler-Oliva, M.E.; Aguilera, A. The chromatin network helps prevent cancer-associated mutagenesis at transcription-replication conflicts. Nat. Commun. 2023, 14, 6890. [Google Scholar] [CrossRef]
- Trigos, A.S.; Pearson, R.B.; Papenfuss, A.T.; Goode, D.L. Altered interactions between unicellular and multicellular genes drive hallmarks of transformation in a diverse range of solid tumors. Proc. Natl. Acad. Sci. USA 2017, 114, 6406–6411. [Google Scholar] [CrossRef]
- Virchow, R. Die Cellularpathologie in Ihrer Begründung auf Physiologische und Pathologische Gewebelehre. Zwanzig Vorlesungen Gehalten Während der Monate Februar, März und Aprilim Pathologischen Institute zu Berlin; A. Hirschwald: Berlin, Germany, 1858. Available online: https://www.loc.gov/item/06041231/ (accessed on 10 December 2022).
- Wilkins, A.S.; Holliday, R. The evolution of meiosis from mitosis. Genetics 2009, 181, 3–12. [Google Scholar] [CrossRef]
- Heng, H.H. Elimination of altered karyotypes by sexual reproduction preserves species identity. Genome 2007, 50, 517–524. [Google Scholar] [CrossRef]
- Yin, S.; Chen, Y.; Yu, C.; Ma, W. From molecular to cellular form: Modeling the first major transition during the arising of life. BMC Ecol. Evol. 2019, 19, 84. [Google Scholar] [CrossRef]
- Kováč, L. Lamarck and Darwin revisited. EMBO Rep. 2019, 20, e47922. [Google Scholar] [CrossRef] [PubMed]
- Mazzocca, A. The Systemic–Evolutionary Theory of the Origin of Cancer (SETOC): A New Interpretative Model of Cancer as a Complex Biological System. Int. J. Mol. Sci. 2019, 20, 4885. [Google Scholar] [CrossRef]
- Kasperski, A. Recognition of Timestamps and Reconstruction of the Line of Organism Development. Processes 2023, 11, 1316. [Google Scholar] [CrossRef]
- Kasperski, A. Genome Attractors as Places of Evolution and Oases of Life. Processes 2021, 9, 1646. [Google Scholar] [CrossRef]
- Giuliani, A. Review of Thomas McCabe (ed.) 2021, Descente and Logic in Biosystematics. Juneau: Perseverant Publishing. Int. J. Biol. Sci. 2021, 5, 87–88. [Google Scholar] [CrossRef]
- Kasperski, A.; Kasperska, R. Selected disease fundamentals based on the unified cell bioenergetics. J. Investig. Biochem. 2013, 2, 93–100. [Google Scholar] [CrossRef]
- Kasperski, A. Modelling of cells bioenergetics. Acta Biotheor. 2008, 56, 233–247. [Google Scholar] [CrossRef]
- Ohnishi, K.; Semi, K.; Yamamoto, T.; Shimizu, M.; Tanaka, A.; Mitsunaga, K.; Okita, K.; Osafune, K.; Arioka, Y.; Maeda, T.; et al. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell 2014, 156, 663–677. [Google Scholar] [CrossRef]
- Shih, I.M. Gestational trophoblastic neoplasia—Pathogenesis and potential therapeutic targets. Lancet Oncol. 2007, 8, 642–650. [Google Scholar] [CrossRef]
- Blagosklonny, M.V. Molecular theory of cancer. Cancer Biol. Ther. 2005, 4, 621–627. [Google Scholar] [CrossRef]
- Sant’ana, D.C.; Pereira, J.P.C.; Cesar, P.H.S.; Trento, M.V.C.; Braga, M.A.; Borges, B.D.B.; Marcussi, S. How do phenolic compounds act in the prevention and treatment of cancer? Rev. Científica Multidiscip. Núcleo Conhecimento 2022, 2, 77–121. [Google Scholar] [CrossRef]
- Wu, S.; Zhu, W.; Thompson, P.; Hannun, Y.A. Evaluating intrinsic and non-intrinsic cancer risk factors. Nat. Commun. 2018, 9, 3490. [Google Scholar] [CrossRef]
- Li, X.; Zhong, Y.; Zhang, X.; Sood, A.K.; Liu, J. Spatiotemporal view of malignant histogenesis and macroevolution via formation of polyploid giant cancer cells. Oncogene 2023, 42, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, R.; McManus, K.J. Chromosome Instability; Implications in Cancer Development, Progression, and Clinical Outcomes. Cancers 2020, 12, 824. [Google Scholar] [CrossRef]
- Ye, C.J.; Sharpe, Z.; Alemara, S.; Mackenzie, S.; Liu, G.; Abdallah, B.; Horne, S.; Regan, S.; Heng, H.H. Micronuclei and Genome Chaos: Changing the System Inheritance. Genes 2019, 10, 366. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.J.; Regan, S.; Liu, G.; Alemara, S.; Heng, H.H. Understanding aneuploidy in cancer through the lens of system inheritance, fuzzy inheritance and emergence of new genome systems. Mol. Cytogenet. 2018, 11, 31. [Google Scholar] [CrossRef]
- Thompson, L.L.; Jeusset, L.M.; Lepage, C.C.; McManus, K.J. Evolving Therapeutic Strategies to Exploit Chromosome Instability in Cancer. Cancers 2017, 9, 151. [Google Scholar] [CrossRef]
- Liu, G.; Stevens, J.B.; Horne, S.D.; Abdallah, B.Y.; Ye, K.J.; Bremer, S.W.; Ye, C.J.; Chen, D.J.; Heng, H.H. Genome chaos: Survival strategy during crisis. Cell Cycle 2014, 13, 528–537. [Google Scholar] [CrossRef]
- Heng, H.H.; Bremer, S.W.; Stevens, J.B.; Horne, S.D.; Liu, G.; Abdallah, B.Y.; Ye, K.J.; Ye, C.J. Chromosomal instability (CIN): What it is and why it is crucial to cancer evolution. Cancer Metastasis Rev. 2013, 32, 325–340. [Google Scholar] [CrossRef]
- Zhang, C.Z.; Pellman, D. Cancer Genomic Rearrangements and Copy Number Alterations from Errors in Cell Division. Annu. Rev. Cancer Biol. 2022, 6, 245–268. [Google Scholar] [CrossRef]
- Zhou, X.; Zhou, M.; Zheng, M.; Tian, S.; Yang, X.; Ning, Y.; Li, Y.; Zhang, S. Polyploid giant cancer cells and cancer progression. Front. Cell Dev. Biol. 2022, 10, 1017588. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Ciriano, I.; Lee, J.J.K.; Xi, R.; Jain, D.; Jung, Y.L.; Yang, L.; Gordenin, D.; Klimczak, L.J.; Zhang, C.-Z.; Pellman, D.S.; et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat. Genet. 2020, 52, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Bunting, S.F.; Nussenzweig, A. End-joining, translocations and cancer. Nat. Rev. Cancer 2013, 13, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, A.W.; Collins, C.C. In Brief: Chromothripsis and cancer. J. Pathol. 2013, 231, 1–3. [Google Scholar] [CrossRef]
- Forment, J.V.; Kaidi, A.; Jackson, S.P. Chromothripsis and cancer: Causes and consequences of chromosome shattering. Nat. Rev. Cancer 2012, 12, 663–670. [Google Scholar] [CrossRef]
- Stephens, P.J.; Greenman, C.D.; Fu, B.; Yang, F.; Bignell, G.R.; Mudie, L.J.; Pleasance, E.D.; Lau, K.W.; Beare, D.; Stebbings, L.A.; et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011, 144, 27–40. [Google Scholar] [CrossRef]
- Ye, J.C.; Horne, S.; Zhang, J.Z.; Jackson, L.; Heng, H.H. Therapy Induced Genome Chaos: A Novel Mechanism of Rapid Cancer Drug Resistance. Front. Cell Dev. Biol. 2021, 9, 676344. [Google Scholar] [CrossRef]
- Friedman, R. Drug resistance in cancer: Molecular evolution and compensatory proliferation. Oncotarget 2016, 7, 11746–11755. [Google Scholar] [CrossRef]
- Foo, J.; Michor, F. Evolution of resistance to anti-cancer therapy during general dosing schedules. J. Theor. Biol. 2010, 263, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.W. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 2012, 1, a011403. [Google Scholar] [CrossRef]
- Brandvain, Y.; Wade, M.J. The functional transfer of genes from the mitochondria to the nucleus: The effects of selection, mutation, population size and rate of self-fertilization. Genetics 2009, 182, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef]
- Adams, K.L.; Palmer, J.D. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol. Phylogenetics Evol. 2003, 29, 380–395. [Google Scholar] [CrossRef]
- Gray, M.W. Evolution of organellar genomes. Curr. Opin. Genet. Dev. 1999, 9, 678–687. [Google Scholar] [CrossRef]
- Gabaldón, T.; Huynen, M.A. From endosymbiont to host-controlled organelle: The hijacking of mitochondrial protein synthesis and metabolism. PLoS Comput. Biol. 2007, 3, e219. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, N.; Frazier, A.E.; Pfanner, N. The Protein Import Machinery of Mitochondria. J. Biol. Chem. 2004, 279, 14473–14476. [Google Scholar] [CrossRef]
- Wiedemann, N.; Pfanner, N. Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 2017, 86, 685–714. [Google Scholar] [CrossRef]
- Chacinska, A.; Koehler, C.M.; Milenkovic, D.; Lithgow, T.; Pfanner, N. Importing mitochondrial proteins: Machineries and mechanisms. Cell 2009, 138, 628–644. [Google Scholar] [CrossRef] [PubMed]
- Comaills, V.; Castellano-Pozo, M. Chromosomal Instability in Genome Evolution: From Cancer to Macroevolution. Biology 2023, 12, 671. [Google Scholar] [CrossRef]
- Akagi, K.; Li, J.; Broutian, T.R.; Padilla-Nash, H.; Xiao, W.; Jiang, B.; Rocco, J.W.; Teknos, T.N.; Kumar, B.; Wangsa, D.; et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014, 24, 185–199. [Google Scholar] [CrossRef]
- Alvarez, E.G.; Demeulemeester, J.; Otero, P.; Jolly, C.; Garcia-Souto, D.; Pequeno-Valtierra, A.; Zamora, J.; Tojo, M.; Temes, J.; Baez-Ortega, A.; et al. Aberrant integration of Hepatitis B virus DNA promotes major restructuring of human hepatocellular carcinoma genome architecture. Nat. Commun. 2021, 12, 6910. [Google Scholar] [CrossRef] [PubMed]
- Burn, A. How the Ancient Viral DNA in Our Genome Affects Disease and Development. Scientific American. Published 19 October 2022. Available online: https://www.scientificamerican.com/article/how-the-ancient-viral-dna-in-our-genome-affects-disease-and-development/ (accessed on 10 December 2022).
- Bouezzedine, F.; El Baba, R.; Haidar Ahmad, S.; Herbein, G. Polyploid Giant Cancer Cells Generated from Human Cytomegalovirus-Infected Prostate Epithelial Cells. Cancers 2023, 15, 4994. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, A.M.; Peter, D. Artificial Intelligence in Cancer Research: Trends, Challenges and Future Directions. Life 2022, 12, 1991. [Google Scholar] [CrossRef]
- Cross, W.; Kovac, M.; Mustonen, V.; Temko, D.; Davis, H.; Baker, A.M.; Biswas, S.; Arnold, R.; Chegwidden, L.; Gatenbee, C.; et al. The evolutionary landscape of colorectal tumorigenesis. Nat. Ecol. Evol. 2018, 2, 1661–1672. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, L. Rapid and sensitive dot-matrix methods for genome analysis. Bioinformatics 2004, 20, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Ermini, L.; Taurone, S.; Greco, A.; Artico, M. Cancer progression: A single cell perspective. Eur. Rev. Med. Pharmacol. Sci. 2023, 27, 5721–5747. [Google Scholar] [CrossRef] [PubMed]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef]
- Lipsick, J. A History of Cancer Research: Tyrosine Kinases. Cold Spring Harb. Perspect. Biol. 2019, 1, a035592. [Google Scholar] [CrossRef]
- Kasperski, A.; Heng, H.H. Two-Phased Cancer Evolution: The Pattern and Scale of Genomic and Non-Genomic Landscapes. In Cancer through the Lens of Evolution and Ecology; Somarelli, J.A., Johnson, N.A., Eds.; CRC Press: Boca Raton, FL, USA, 2024; ISBN 9781003307921. [Google Scholar] [CrossRef]
- Paul, D. Cancer as a form of life: Musings of the cancer and evolution symposium. Prog. Biophys. Mol. Biol. 2021, 165, 120–139. [Google Scholar] [CrossRef]
- Vendramin, R.; Litchfield, K.; Swanton, C. Cancer evolution: Darwin and beyond. EMBO J. 2021, 40, e108389. [Google Scholar] [CrossRef]
- Heng, H.H. Cancer genome sequencing: The challenges ahead. Bioessays 2007, 29, 783–794. [Google Scholar] [CrossRef]
- Crkvenjakov, R.; Heng, H.H. Further illusions: On key evolutionary mechanisms that could never fit with Modern Synthesis. Prog. Biophys. Mol. Biol. 2022, 169–170, 3–11. [Google Scholar] [CrossRef]
- Gorelick, R.; Heng, H.H. Sex reduces genetic variation: A multidisciplinary review. Evolution 2011, 65, 1088–1098. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Brown, J.S. The Evolution and Ecology of Resistance in Cancer Therapy. Cold Spring Harb. Perspect. Med. 2020, 10, a040972. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Silva, A.S.; Gillies, R.J.; Frieden, B.R. Adaptive therapy. Cancer Res. 2009, 69, 4894–4903. [Google Scholar] [CrossRef]
- Shityakov, S.; Kravtsov, V.; Skorb, E.V.; Nosonovsky, M. Ergodicity Breaking and Self-Destruction of Cancer Cells by Induced Genome Chaos. Entropy 2023, 26, 37. [Google Scholar] [CrossRef]
- Mirzayans, R. Changing the Landscape of Solid Tumor Therapy from Apoptosis-Promoting to Apoptosis-Inhibiting Strategies. Curr. Issues Mol. Biol. 2024, 46, 5379–5396. [Google Scholar] [CrossRef]
- Weinberg, R.A. It took a long, long time: Ras and the race to cure cancer. Cell 2024, 187, 1574–1577. [Google Scholar] [CrossRef] [PubMed]
- Baverstock, K. The gene: An appraisal. Prog. Biophys. Mol. Biol. 2021, 164, 46–62. [Google Scholar] [CrossRef]
- Raza, A. The First Cell: And the Human Costs of Pursuing Cancer to the Last; Basic Books: New York, NY, USA, 2019. [Google Scholar]
- Tez, M.; Tez, S. Cancer is the chaotic search for adaptation to previously unknown environments. Theor. Biol. Forum 2016, 109, 149–154. [Google Scholar] [CrossRef]
- Joyner, M.J. Has Neo-Darwinism failed clinical medicine: Does systems biology have to? Prog. Biophys. Mol. Biol. 2015, 117, 107–112. [Google Scholar] [CrossRef]
- Joyner, M.J.; Prendergast, F.G. Chasing Mendel: Five questions for personalized medicine. J. Physiol. 2014, 592, 2381–2388. [Google Scholar] [CrossRef]
- Weinberg, R.A. Coming full circle-from endless complexity to simplicity and back again. Cell 2014, 157, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, I.; Sasieni, P.; Bodmer, W. How many mutations in a cancer? Am. J. Pathol. 2002, 160, 755–758. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Iourov, I.Y.; Vorsanova, S.G.; Yurov, Y.B. Systems Cytogenomics: Are We Ready Yet? Curr. Genom. 2021, 22, 75–78. [Google Scholar] [CrossRef]
- Zhao, T.; Zwaenepoel, A.; Xue, J.Y. Kao, S.M.; Li, Z.; Schranz, M.E.; Van de Peer, Y. Whole-genome microsynteny-based phylogeny of angiosperms. Nat. Commun. 2021, 12, 3498. [Google Scholar] [CrossRef] [PubMed]
- Van der Mude, A. Structure encoding in DNA. J. Theor. Biol. 2020, 492, 110205. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301. [Google Scholar] [CrossRef]
- Shapiro, J.A. How Chaotic Is Genome Chaos? Cancers 2021, 13, 1358. [Google Scholar] [CrossRef] [PubMed]
- Pellestor, F.; Gatinois, V. Chromoanagenesis: A piece of the macroevolution scenario. Mol. Cytogenet. 2020, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.; Navarrete-Meneses, P.; Molina, B.; Cervantes-Barragán, D.E.; Lozano, V.; Gallardo, E.; Marchetti, F.; Frias, S. Genomic chaos in peripheral blood lymphocytes of Hodgkin’s lymphoma patients one year after ABVD chemotherapy/radiotherapy. Environ. Mol. Mutagen. 2018, 59, 755–768. [Google Scholar] [CrossRef]
- Mojica, E.A.; Kültz, D. Physiological mechanisms of stress-induced evolution. J. Exp. Biol. 2022, 225 (Suppl. S1), jeb243264. [Google Scholar] [CrossRef]
- Kültz, D. Evolution of cellular stress response mechanisms. J. Exp. Zool. Part A Ecol. Integr. Physiol. 2020, 333, 359–378. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.J.; Eldredge, N. Punctuated equilibrium comes of age. Nature 1993, 366, 223–227. [Google Scholar] [CrossRef]
- Shapiro, J.A. Evolution: A View from the 21st Century, Fortified. In Why Evolution Works as Well as It Does; Cognition Press: Chicago, IL, USA, 2022. [Google Scholar]
- Mudd, A.B.; Bredeson, J.V.; Baum, R.; Hockemeyer, D.; Rokhsar, D.S. Analysis of muntjac deer genome and chromatin architecture reveals rapid karyotype evolution. Commun. Biol. 2020, 3, 480. [Google Scholar] [CrossRef]
- Simakov, O.; Marlétaz, F.; Yue, J.X.; O’Connell, B.; Jenkins, J.; Brandt, A.; Calef, R.; Tung, C.-H.; Huang, T.-K.; Schmutz, J.; et al. Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol. 2020, 4, 820–830. [Google Scholar] [CrossRef]
- Murat, F.; Armero, A.; Pont, C.; Klopp, C.; Salse, J. Reconstructing the genome of the most recent common ancestor of flowering plants. Nat. Genet. 2017, 49, 490–496. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Vainshelbaum, N.M.; Lazovska, M.; Karklins, R.; Salmina, K.; Zayakin, P.; Rumnieks, F.; Inashkina, I.; Pjanova, D.; Erenpreiss, J. The Price of Human Evolution: Cancer-Testis Antigens, the Decline in Male Fertility and the Increase in Cancer. Int. J. Mol. Sci. 2023, 24, 11660. [Google Scholar] [CrossRef]
- Pienta, K.J.; Hammarlund, E.U.; Austin, R.H.; Axelrod, R.; Brown, J.S.; Amend, S.R. Cancer cells employ an evolutionarily conserved polyploidization program to resist therapy. Semin Cancer Biol. 2022, 81, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Davoli, T.; Uno, H.; Wooten, E.C.; Elledge, S.J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017, 355, eaaf8399. [Google Scholar] [CrossRef] [PubMed]
- Jamal-Hanjani, M.; Wilson, G.A.; McGranahan, N.; Birkbak, N.J.; Watkins, T.B.K.; Veeriah, S.; Shafi, S.; Johnson, D.H.; Mitter, R.; Rosenthal, R.; et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 376, 2109–2121. [Google Scholar] [CrossRef]
- Park, S.Y.; Gönen, M.; Kim, H.J.; Michor, F.; Polyak, K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J. Clin Investig. 2010, 120, 636–644. [Google Scholar] [CrossRef]
- Pellestor, F.; Gaillard, J.B.; Schneider, A.; Puechberty, J.; Gatinois, V. Chromoanagenesis, the mechanisms of a genomic chaos. Semin. Cell Dev. Biol. 2022, 123, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Maciejowski, J.; Li, Y.; Bosco, N.; Campbell, P.J.; De Lange, T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell 2015, 163, 1641–1654. [Google Scholar] [CrossRef]
- Baca, S.C.; Prandi, D.; Lawrence, M.S.; Mosquera, J.M.; Romanel, A.; Drier, Y.; Park, K.; Kitabayashi, N.; MacDonald, T.Y.; Ghandi, M.; et al. Punctuated evolution of prostate cancer genomes. Cell 2013, 153, 666–677. [Google Scholar] [CrossRef]
- Holland, A.J.; Cleveland, D.W. Chromoanagenesis and cancer: Mechanisms and consequences of localized, complex chromosomal rearrangements. Nat. Med. 2012, 18, 1630–1638. [Google Scholar] [CrossRef]
- Inaki, K.; Liu, E.T. Structural mutations in cancer: Mechanistic and functional insights. Trends Genet. 2012, 28, 550–559. [Google Scholar] [CrossRef]
- Righolt, C.; Mai, S. Shattered and stitched chromosomes-chromothripsis and chromoanasynthesis-manifestations of a new chromosome crisis? Genes Chromosomes Cancer 2012, 51, 975–981. [Google Scholar] [CrossRef]
- Setlur, S.R.; Lee, C. Tumor archaeology reveals that mutations love company. Cell 2012, 149, 959–961. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Erez, A.; Nagamani, S.C.; Dhar, S.U.; Kołodziejska, K.E.; Dharmadhikari, A.V.; Cooper, M.L.; Wiszniewska, J.; Zhang, F.; Withers, M.; et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 2011, 146, 889–903. [Google Scholar] [CrossRef] [PubMed]
- Tubio, J.M.; Estivill, X. Cancer: When catastrophe strikes a cell. Nature 2011, 470, 476–477. [Google Scholar] [CrossRef]
- Weihua, Z.; Lin, Q.; Ramoth, A.J.; Fan, D.; Fidler, I.J. Formation of solid tumors by a single multinucleated cancer cell. Cancer 2011, 117, 4092–4099. [Google Scholar] [CrossRef] [PubMed]
- Duesberg, P. Chromosomal chaos and cancer. Sci. Am. 2007, 296, 52–59. [Google Scholar] [CrossRef]
- Prinz, R. Meaning Relies on Codes but Depends on Agents. In Pathways to the Origin and Evolution of Meanings in the Universe; Sharov, A., Mikhailovsky, G.E., Eds.; Scrivener Publishing LLC: Beverly Hills, MA, USA, 2024; pp. 245–264. [Google Scholar] [CrossRef]
- Miller, W.B., Jr.; Baluška, F.; Reber, A.S.; Slijepčević, P. Biology in the 21st century: Natural selection is cognitive selection. Prog. Biophys. Mol. Biol. 2024, 190, 170–184. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasperski, A.; Heng, H.H. The Spiral Model of Evolution: Stable Life Forms of Organisms and Unstable Life Forms of Cancers. Int. J. Mol. Sci. 2024, 25, 9163. https://doi.org/10.3390/ijms25179163
Kasperski A, Heng HH. The Spiral Model of Evolution: Stable Life Forms of Organisms and Unstable Life Forms of Cancers. International Journal of Molecular Sciences. 2024; 25(17):9163. https://doi.org/10.3390/ijms25179163
Chicago/Turabian StyleKasperski, Andrzej, and Henry H. Heng. 2024. "The Spiral Model of Evolution: Stable Life Forms of Organisms and Unstable Life Forms of Cancers" International Journal of Molecular Sciences 25, no. 17: 9163. https://doi.org/10.3390/ijms25179163




