Effect of High Glucose on Embryological Development of Zebrafish, Brachyodanio, Rerio through Wnt Pathway
Abstract
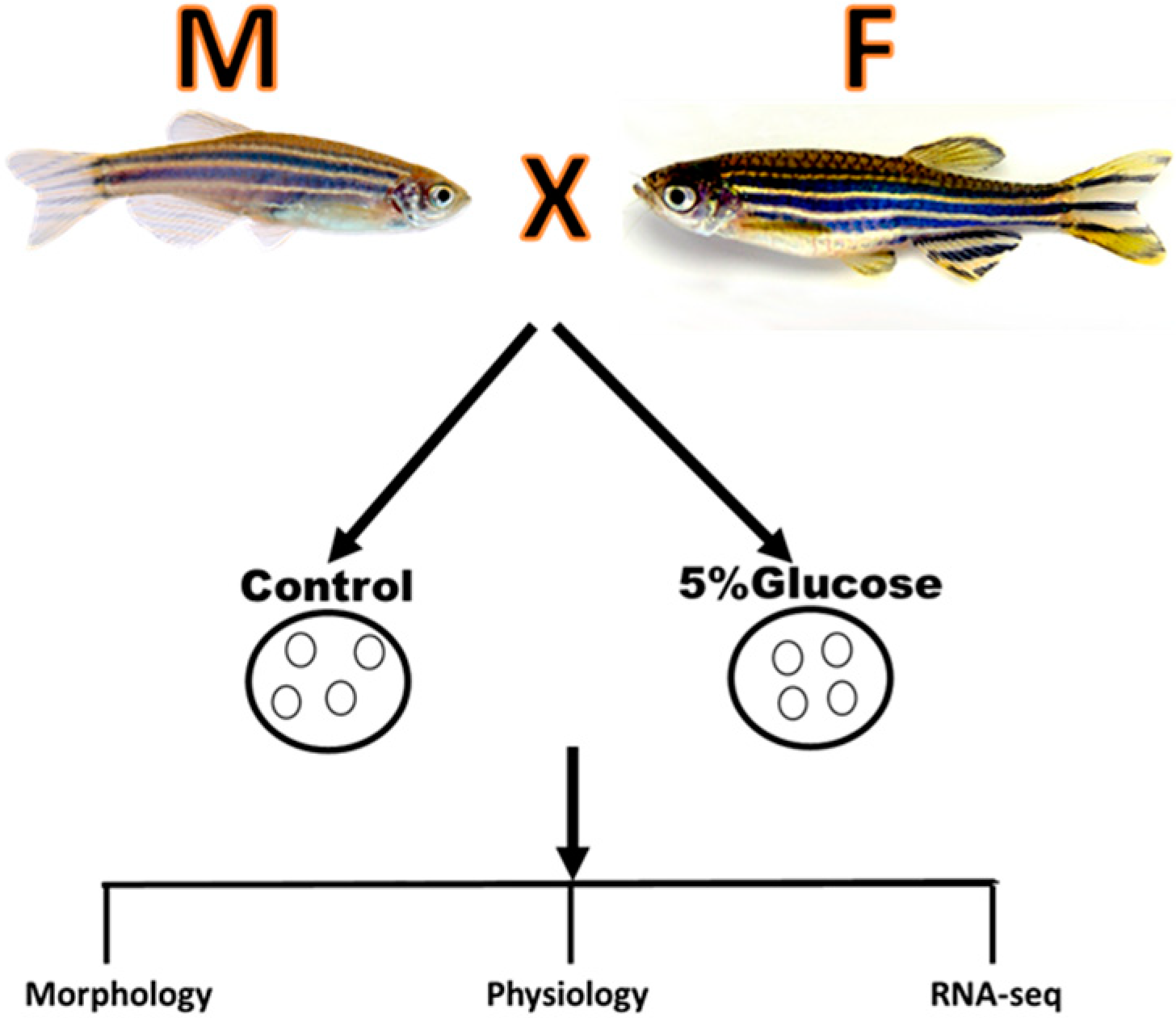
:1. Introduction
2. Results
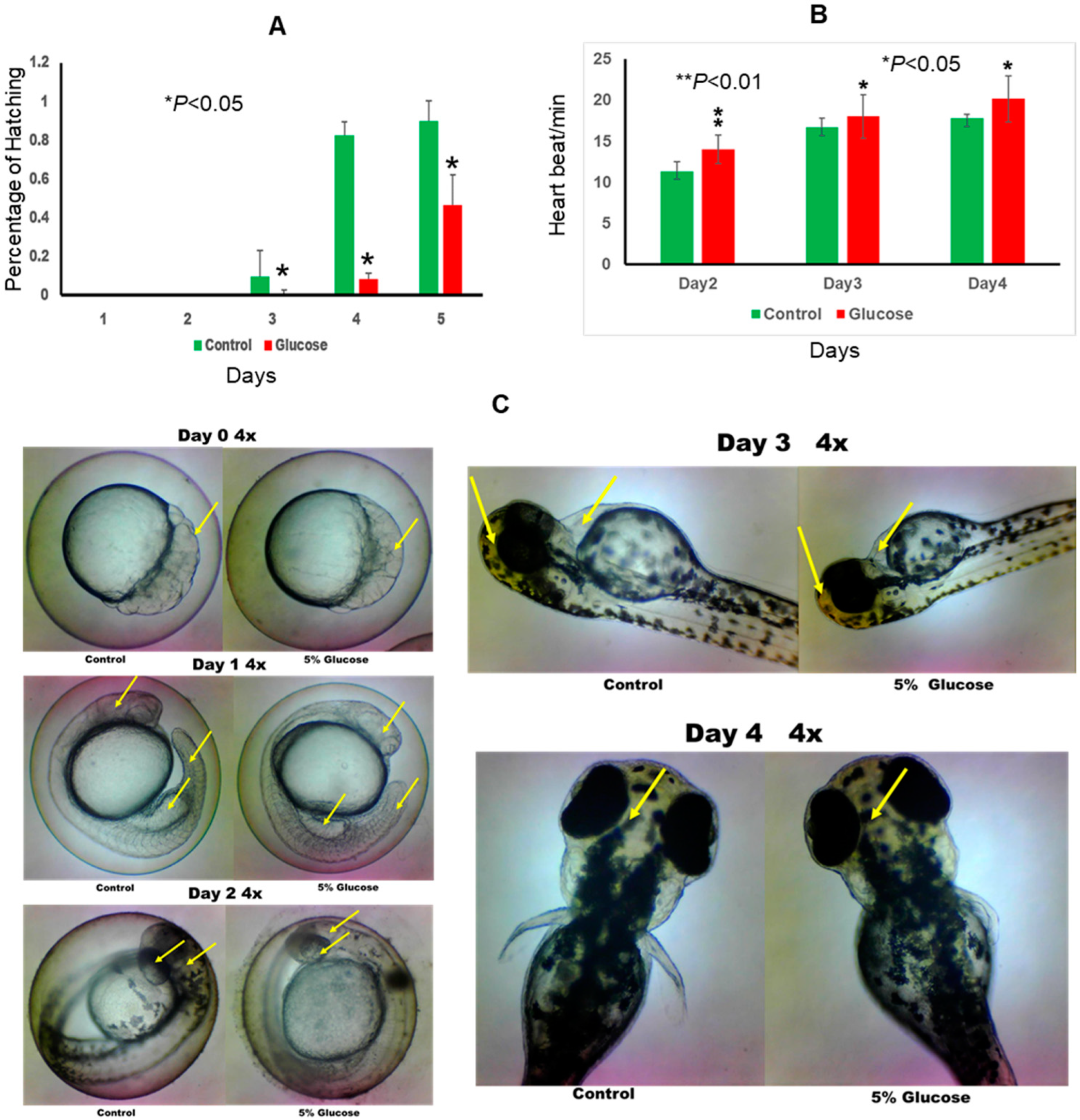
2.1. Effect of Glucose on Zebrafish Embryo Development
2.2. RNA-Seq Analysis Displayed Significant Gene Dysregulation in the High-Glucose-Treated Embryos
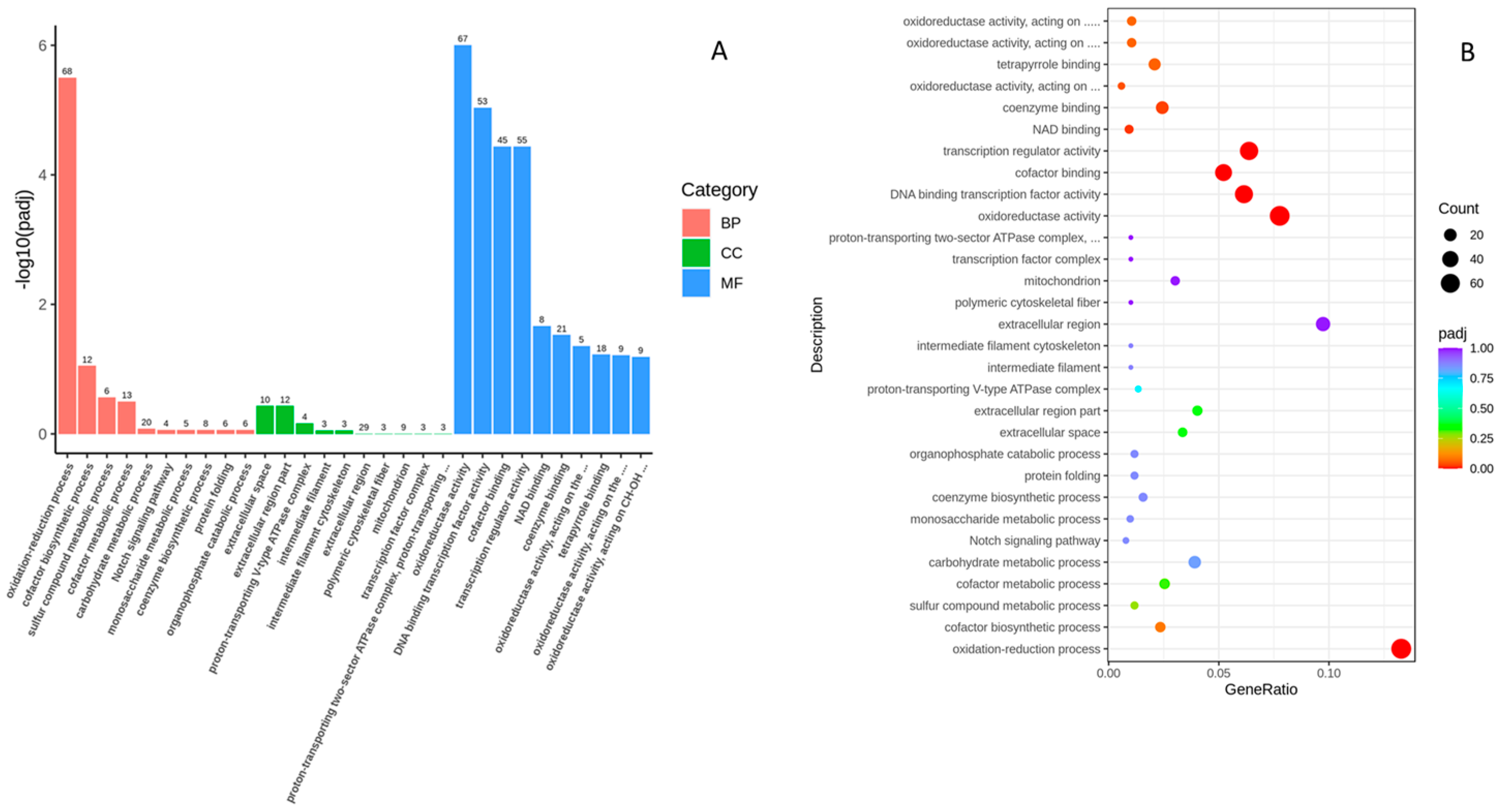
2.3. Functional Analysis of DEGs Using Gene Ontology (GO)Analysis
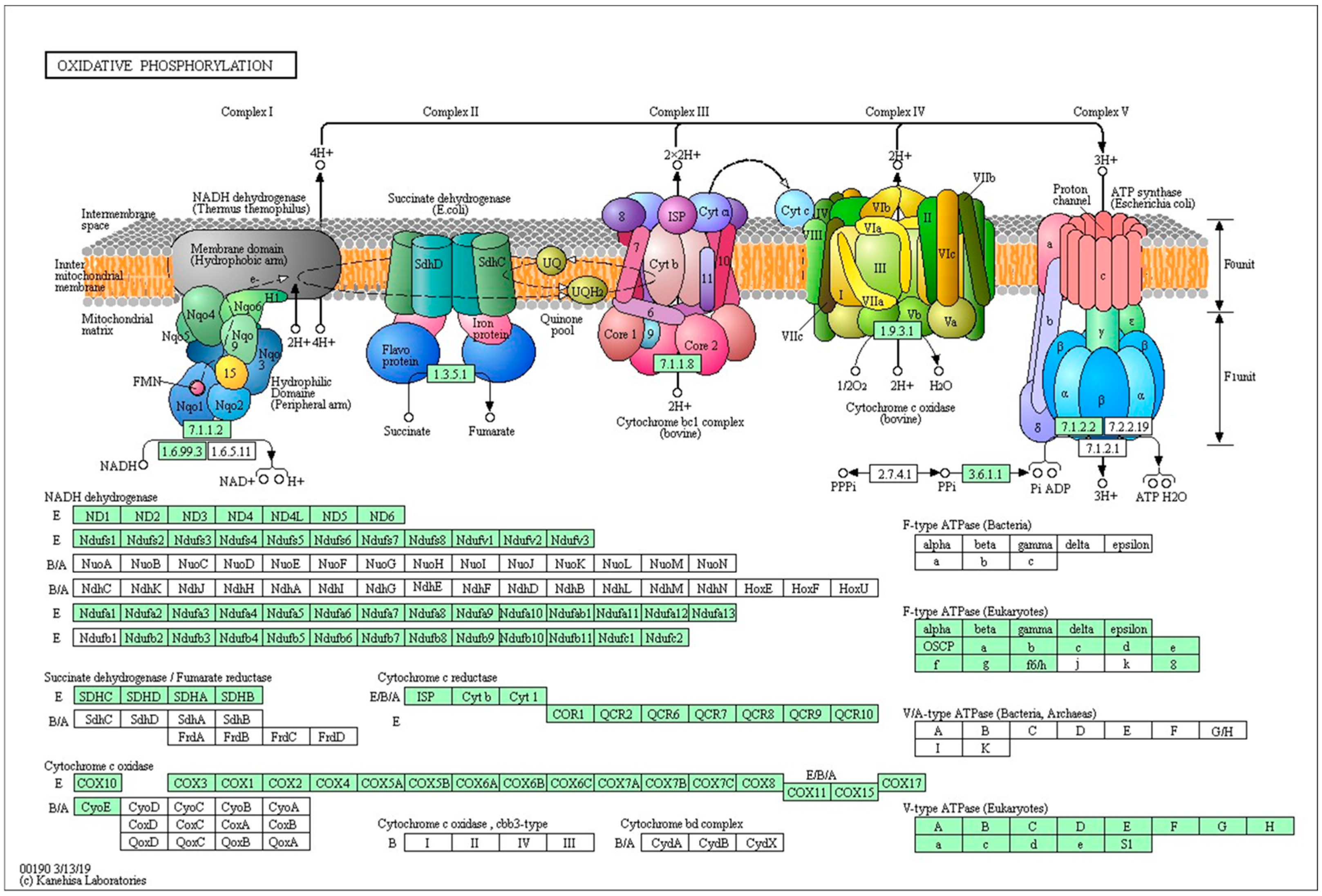
2.4. KEGG Pathway Analysis
3. Discussion
3.1. Limitations in This Study
3.2. Implications and Future Directions
3.3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Animal Husbandry
4.2.1. Preparation of D-Glucose Application
4.2.2. Addition of D-Glucose and Collection of Embryos
4.2.3. Hatch Rate Calculation
4.3. Total RNA Extraction
4.4. RNA Seq
4.5. Reads Mapping to the Reference Genome
4.6. Novel Transcript Prediction
4.7. Quantification of Gene Expression Level
4.8. Differential Expression Analysis
4.9. GO and KEGG Enrichment Analysis of Differentially Expressed Genes
4.10. Gene Set Enrichment Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moon, J.H.; Jang, H.C. Gestational Diabetes Mellitus: Diagnostic Approaches and Maternal-Offspring Complications. Diabetes Metab. J. 2022, 46, 3–14. [Google Scholar] [CrossRef]
- Juan, J.; Yang, H. Prevalence, Prevention, and Lifestyle Intervention of Gestational Diabetes Mellitus in China. Int. J. Environ. Res. Public Health 2020, 18, 9517. [Google Scholar] [CrossRef]
- Hannah, W.; Pradeepa, R.; Anjana, R.M.; Uma, R.; Tiwaskar, M.; Mohan, V. Early Gestational Diabetes Mellitus: An Update. J. Assoc. Physicians India 2023, 71, 101–103. [Google Scholar] [PubMed]
- Karkia, R.; Giacchino, T.; Shah, S.; Gough, A.; Ramadan, G.; Akolekar, R. Gestational Diabetes Mellitus: Association with Maternal and Neonatal Complications. Medicina 2023, 59, 2096. [Google Scholar] [CrossRef]
- Nakshine, V.S.; Jogdand, S.D. A Comprehensive Review of Gestational Diabetes Mellitus: Impacts on Maternal Health, Fetal Development, Childhood Outcomes, and Long-Term Treatment Strategies. Cureus 2023, 15, e47500. [Google Scholar] [CrossRef]
- Pandaram, A.; Paul, J.; Wankhar, W.; Thakur, A.; Verma, S.; Vasudevan, K.; Wankhar, D.; Kammala, A.K.; Sharma, P.; Jaganathan, R.; et al. Aspartame Causes Developmental Defects and Teratogenicity in Zebra Fish Embryo: Role of Impaired SIRT1/FOXO3a Axis in Neuron Cells. Biomedicines 2024, 12, 855. [Google Scholar] [CrossRef] [PubMed]
- Angom, R.S.; Nakka, N.M.R. Zebrafish as a Model for Cardiovascular and Metabolic Disease: The Future of Precision Medicine. Biomedicines 2024, 12, 693. [Google Scholar] [CrossRef]
- Kudoh, T.; Tsang, M.; Hukriede, N.A.; Chen, X.; Dedekian, M.; Clarke, C.J.; Kiang, A.; Schultz, S.; Epstein, J.A.; Toyama, R.; et al. A gene expression screen in zebrafish embryogenesis. Genome Res. 2001, 11, 1979–1987. [Google Scholar] [CrossRef] [PubMed]
- Amsterdam, A.; Nissen, R.M.; Sun, Z.; Swindell, E.C.; Farrington, S.; Hopkins, N. Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. USA 2004, 101, 12792–12797. [Google Scholar] [CrossRef]
- White, R.J.; Collins, J.E.; Sealy, I.M.; Wali, N.; Dooley, C.M.; Digby, Z.; Stemple, D.L.; Murphy, D.N.; Billis, K.; Hourlier, T.; et al. A high-resolution mRNA expression time course of embryonic development in zebrafish. eLife 2017, 16, e30860. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Parichy, D.M.; Elizondo, M.R.; Mills, M.G.; Gordon, T.N.; Engeszer, R.E. Normal table of postembryonic zebrafish development: Staging by externally visible anatomy of the living fish. Dev. Dyn. 2009, 238, 2975–3015. [Google Scholar] [CrossRef]
- Elo, B.; Villano, C.M.; Govorko, D.; White, L.A. Larval zebrafish as a model for glucose metabolism: Expression of phosphoenolpyruvate carboxykinase as a marker for exposure to anti-diabetic compounds. J. Mol. Endocrinol. 2007, 38, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Castillo, H.A.; Brown, J.; Kaslin, J.; Dwyer, K.M.; Gibert, Y. High glucose levels affect retinal patterning during zebrafish embryogenesis. Sci. Rep. 2019, 9, 4121. [Google Scholar] [CrossRef]
- Dhillon, S.S.; Torell, F.; Donten, M.; Lundstedt-Enkel, K.; Bennett, K.; Rännar, S.; Trygg, J.; Lundstedt, T. Metabolic profiling of zebrafish embryo development from blastula period to early larval stages. PLoS ONE 2019, 14, e0213661. [Google Scholar] [CrossRef]
- Bridget, K.; Cox, C.K.; Garrett, M.R.; Gibert, Y. Excess glucose or fat differentially affects metabolism and appetite-related gene expression during zebrafish embryogenesis. iScience 2023, 26, 107063. [Google Scholar]
- Miao, D.; Ren, J.; Jia, Y.; Jia, Y.; Li, Y.; Huang, H.; Gao, R. PAX1 represses canonical Wnt signaling pathway and plays dual roles during endoderm differentiation. Cell Commun. Signal 2024, 22, 242. [Google Scholar] [CrossRef] [PubMed]
- Westphal, M.; Panza, P.; Kastenhuber, E.; Wehrle, J.; Driever, W. Wnt/β-catenin signaling promotes neurogenesis in the diencephalospinal dopaminergic system of embryonic zebrafish. Sci. Rep. 2022, 19, 1030. [Google Scholar] [CrossRef] [PubMed]
- Garg, V.; Geurten, B.R.H. Diving deep: Zebrafish models in motor neuron degeneration research. Front. Neurosci. 2024, 18, 1424025. [Google Scholar] [CrossRef]
- Aman, A.J.; Fulbright, A.N.; Parichy, D.M. Wnt/β-catenin regulates an ancient signaling network during zebrafish scale development. eLife 2018, 7, e37001. [Google Scholar] [CrossRef]
- Sutton, G.; Kelsh, R.N.; Scholpp, S. Review: The Role of Wnt/β-Catenin Signalling in Neural Crest Development in Zebrafish. Front. Cell Dev. Biol. 2021, 9, 782445. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Anai, S.; Ota, S.; Ishitani, S.; Oginuma, M.; Ishitani, T. Determining zebrafish dorsal organizer size by a negative feedback loop between canonical/non-canonical Wnts and Tlr4/NFκB. Nat. Commun. 2023, 14, 7194. [Google Scholar] [CrossRef]
- Akieda, Y.; Ogamino, S.; Furuie, H.; Ishitani, S.; Akiyoshi, R.; Nogami, J.; Masuda, T.; Shimizu, N.; Ohkawa, Y.; Ishitani, T. Cell competition corrects noisy Wnt morphogen gradients to achieve robust patterning in the zebrafish embryo. Nat. Commun. 2019, 10, 4710. [Google Scholar] [CrossRef] [PubMed]
- Heisenberg, C.P.; Tada, M.; Rauch, G.J.; Saúde, L.; Concha, M.L.; Geisler, R.; Stemple, D.L.; Smith, J.C.; Wilson, S.W. Silberblick/Wnt11 mediates convergent extension movements during zebrafish gastrulation. Nature 2000, 405, 76–81. [Google Scholar] [CrossRef]
- Wang, X.; Lee, J.E.; Dorsky, R.I. Identification of Wnt-responsive cells in the zebrafish hypothalamus. Zebrafish 2009, 6, 49–58. [Google Scholar] [CrossRef]
- Hickey, J.M.; Wike, C.W.; Nie, X.; Guo, Y.; Tan, M.; Murphy, P.J.; Cairns, B.R. Establishment of developmental gene silencing by ordered polycomb complex recruitment in early zebrafish embryos. eLife 2022, 11, e67738. [Google Scholar]
- Abrams, E.W.; Mullins, M.C. Early zebrafish development: It’s in the maternal genes. Curr. Opin. Genet. Dev. 2009, 19, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Lammer, E.; Carr, G.J.; Wendler, K.; Rawlings, J.M.; Belanger, S.E.; Braunbeck, T. Is the fish embryo toxicity test (FET) with the zebrafish (Danio rerio) a potential alternative for the fish acute toxicity test? Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2009, 149, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Curcio, V.; Macirella, R.; Sesti, S.; Ahmed, A.I.M.; Talarico, F.; Tagarelli, A.; Mezzasalma, M.; Brunelli, E. Morphological and Functional Alterations Induced by Two Ecologically Relevant Concentrations of Lead on Danio rerio Gills. Int. J. Mol. Sci. 2022, 23, 9165. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.; Antonescu, C. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]


| Gene | Biological Process | Molecular Function |
|---|---|---|
| Kcnj1a.2 | Transport; ion transport | Enable ATP-activated inward rectifier potassium channel activity |
| Eomesa | Enables DNA binding activity and transcription factor activity in blastoderm, blastodisc, brain, eye, and immune system | DNA-binding and DNA-binding transcription factor activity |
| zgc:77439 | Enables N,N-dimethylaniline monooxygenase activity, NADP binding activity and flavin adenine dinucleotide binding activity | NADP binding activity in response to xenobiotic stimulus in endoplasmic reticulum |
| foxg1d | Transcription regulation, DNA binding activity | Enables sequence specific double-stranded DNA binding activity in nucleus |
| zgc:15840 | Transport; urea transmembrane transporter activity | Enables serine-type endopeptidase activity and urea transmembrane transporter activity |
| tbr1b | Commitment of neuronal cells to specific neuron type in forebrain, neuron projection development, transcription regulation | DNA-binding transcription factor activity |
| pcdh1g32 | Calcium ion binding activity and cell adhesion | Homophilic cell adhesion via plasma membrane adhesion molecules in the brain |
| tert | Telomerase activity; erythrocyte maturation in brain, eye, hear, liver, pleuroperitoneal region, intestinal epithelial structure maintenance | Telomere maintenance via telomerase |
| plp1a | myelination | Structural constituent of myelin sheath |
| TIAM2 | GTPase activity, axonogenesis regulation, signal transduction | enables GTPase activator activity and guanyl-nucleotide exchange factor activity |
| Gene | Biological Process | Molecular Function |
|---|---|---|
| apoda.2 | lipid metabolic process, response to reactive oxygen species, aging, lipid transport | Enables lipid binding activity |
| lct | Carbohydrate metabolic processes, hydrolase activity | Hydrolyzes O-glycosyl compounds |
| tnfrsf11b | Response to mechanical stimulus | Tumor necrosis factor receptor; head and scale |
| opn5 | Absorption of visible light | Enables 11-cis retinal binding activity and all-trans retinal binding activity |
| rgra | G protein-coupled receptor signaling, cellular response to light stimulus and phototransduction | Enables G protein-coupled photoreceptor activity |
| zgc:153154 | Small molecule binding in extracellular region | Small molecule binding activity |
| cd74b | Ear development and neuromast development | MHC class II protein binding activity, cytokine receptor activity, macrophage migration inhibitory factor binding activity |
| zgc:153911 | T cell receptor signaling, cytokine production regulation | Signaling receptor binding activity in membrane |
| pah | Cellular response to estrogen stimulus in structures incl. Digestive system, epidermis, eye, optic vesicle, and periderm | Phenylalanine 4-monooxygenase activity |
| rasef | GDP and GTP binding in cytosol and perinuclear region of cytoplasm | Enables GDP and GTP binding activity |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thompson, E.; Hensley, J.; Taylor, R.S. Effect of High Glucose on Embryological Development of Zebrafish, Brachyodanio, Rerio through Wnt Pathway. Int. J. Mol. Sci. 2024, 25, 9443. https://doi.org/10.3390/ijms25179443
Thompson E, Hensley J, Taylor RS. Effect of High Glucose on Embryological Development of Zebrafish, Brachyodanio, Rerio through Wnt Pathway. International Journal of Molecular Sciences. 2024; 25(17):9443. https://doi.org/10.3390/ijms25179443
Chicago/Turabian StyleThompson, Ebony, Justin Hensley, and Renfang Song Taylor. 2024. "Effect of High Glucose on Embryological Development of Zebrafish, Brachyodanio, Rerio through Wnt Pathway" International Journal of Molecular Sciences 25, no. 17: 9443. https://doi.org/10.3390/ijms25179443
APA StyleThompson, E., Hensley, J., & Taylor, R. S. (2024). Effect of High Glucose on Embryological Development of Zebrafish, Brachyodanio, Rerio through Wnt Pathway. International Journal of Molecular Sciences, 25(17), 9443. https://doi.org/10.3390/ijms25179443





