Digital Whole Slide Image Analysis of Elevated Stromal Content and Extracellular Matrix Protein Expression Predicts Adverse Prognosis in Triple-Negative Breast Cancer
Abstract
:1. Introduction
2. Results
2.1. Clinicopathological Characteristics of Patients
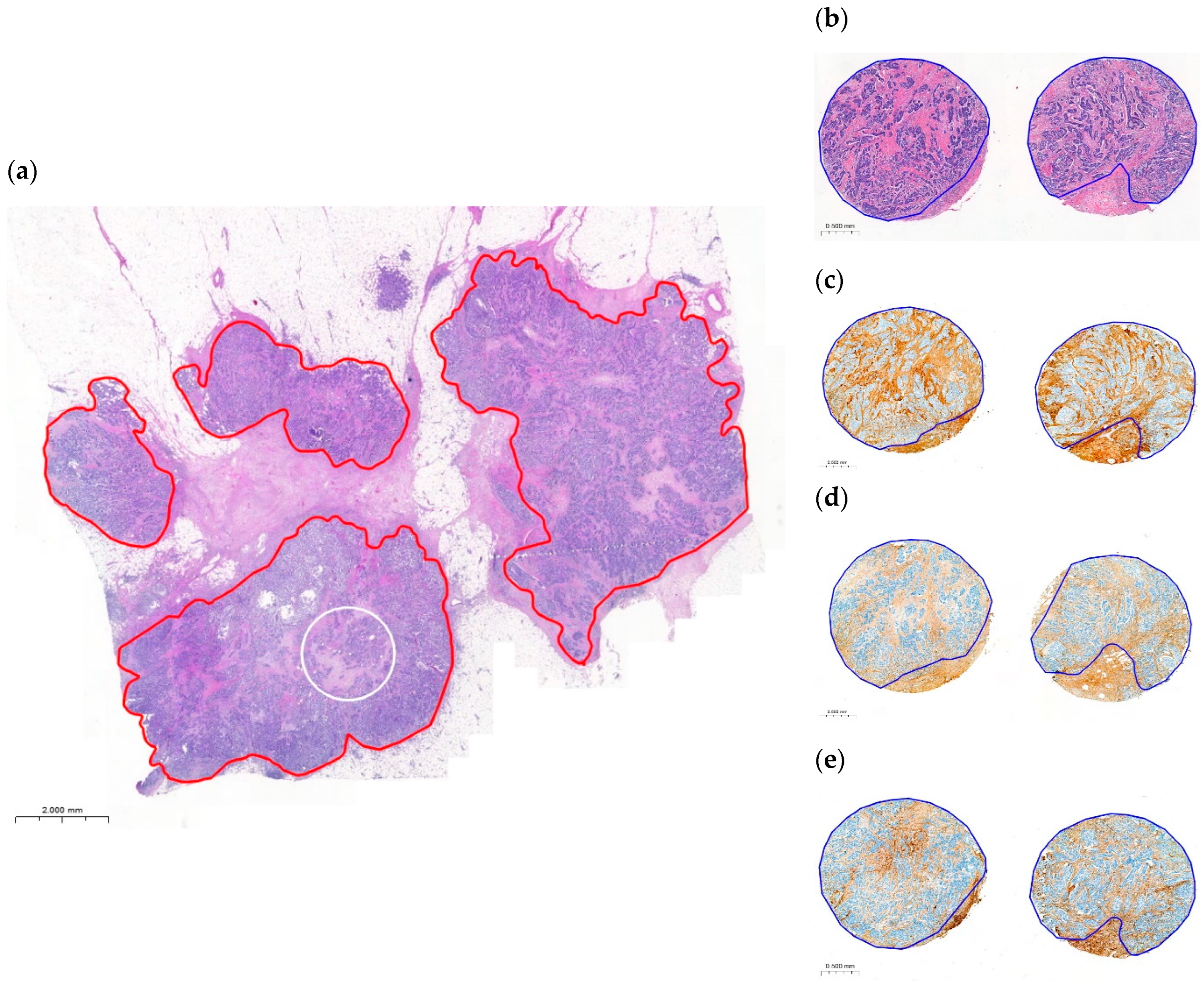
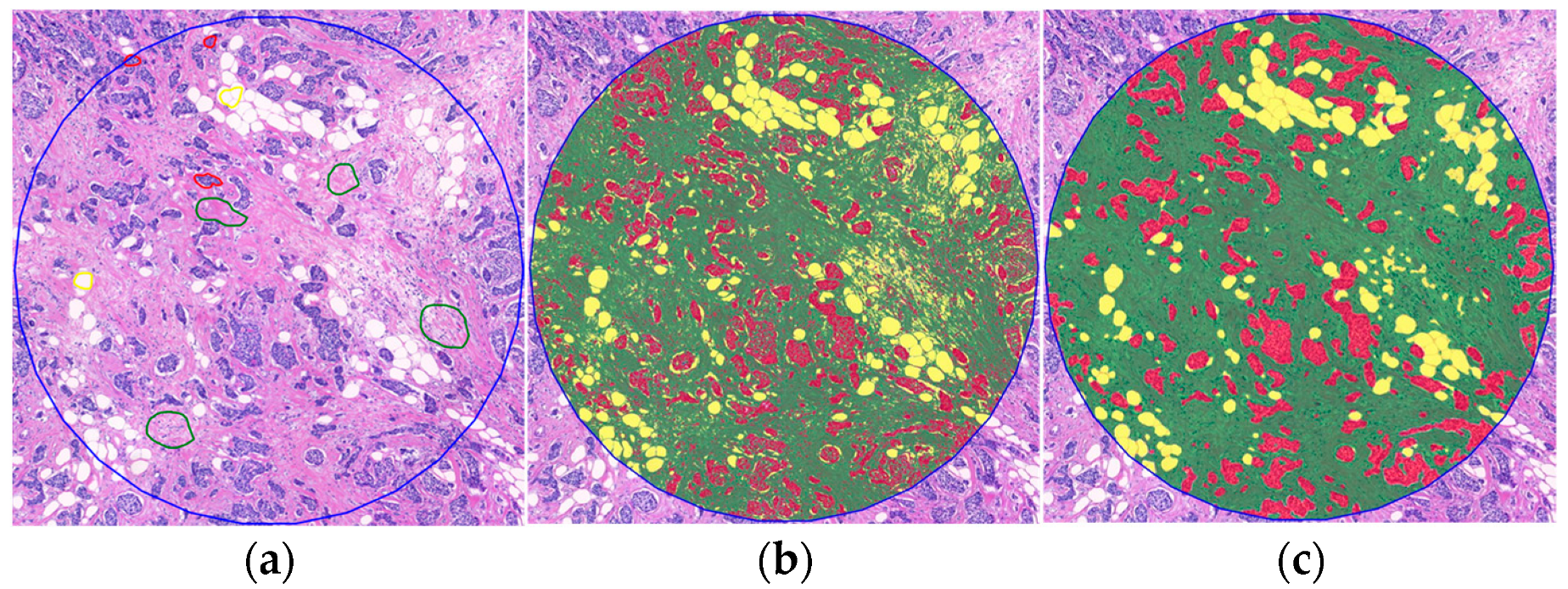
2.2. Variability and Categorization of TSR and OSR on H&E-Stained WSIs
2.3. Association of the TSR and the OSR with the Clinicopathological Factors
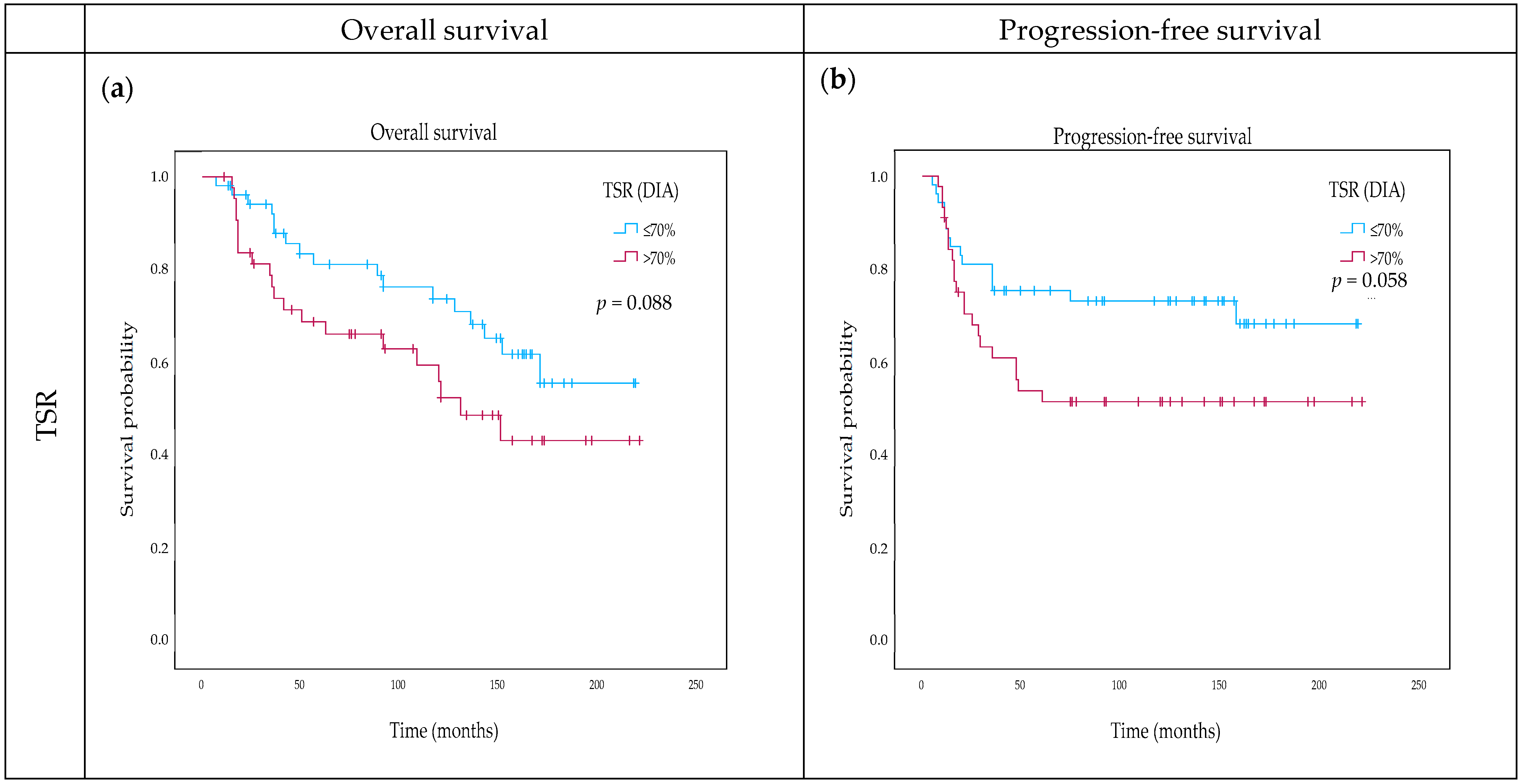
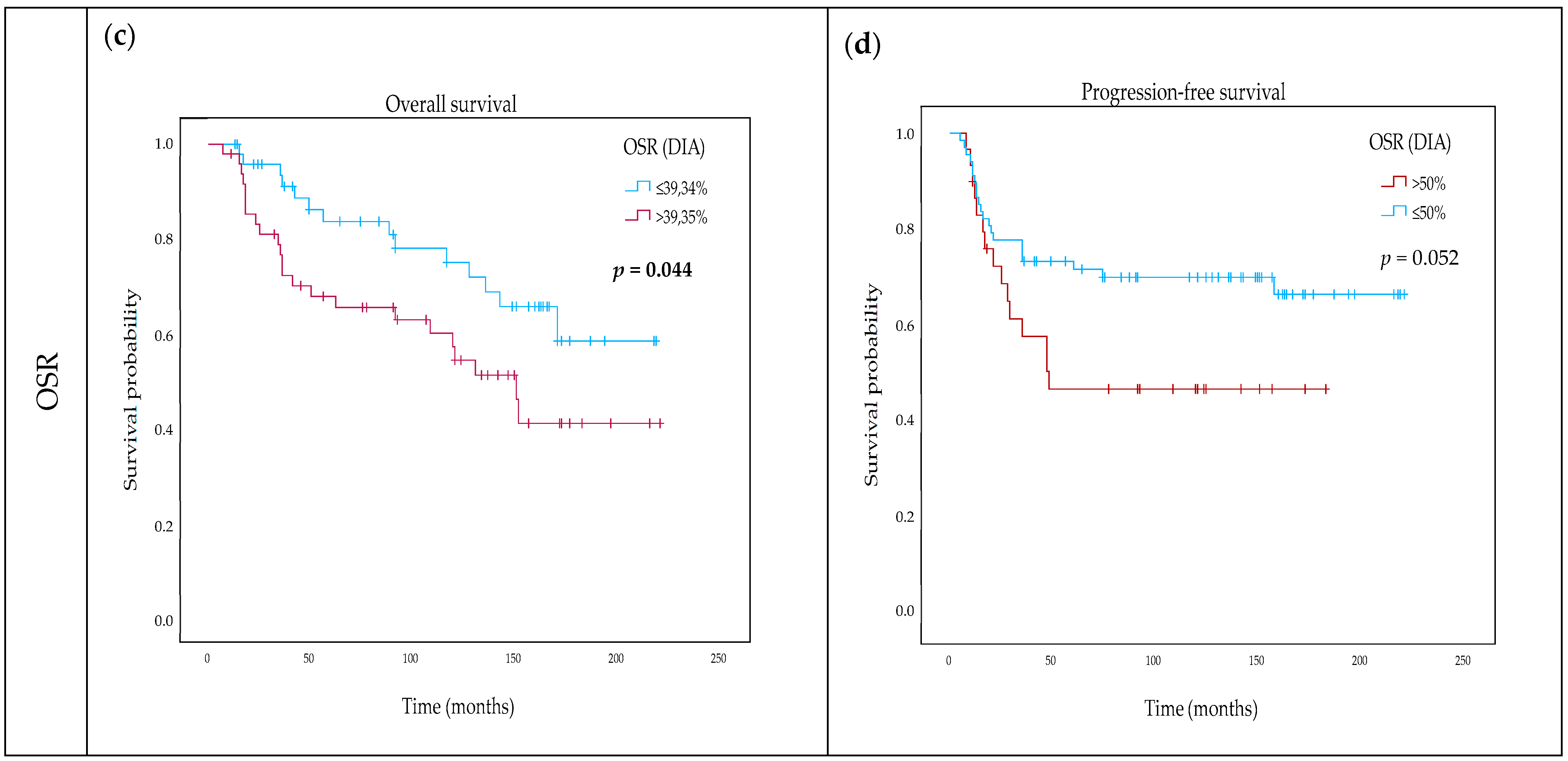
2.4. Impact of TSR and OSR on Survival
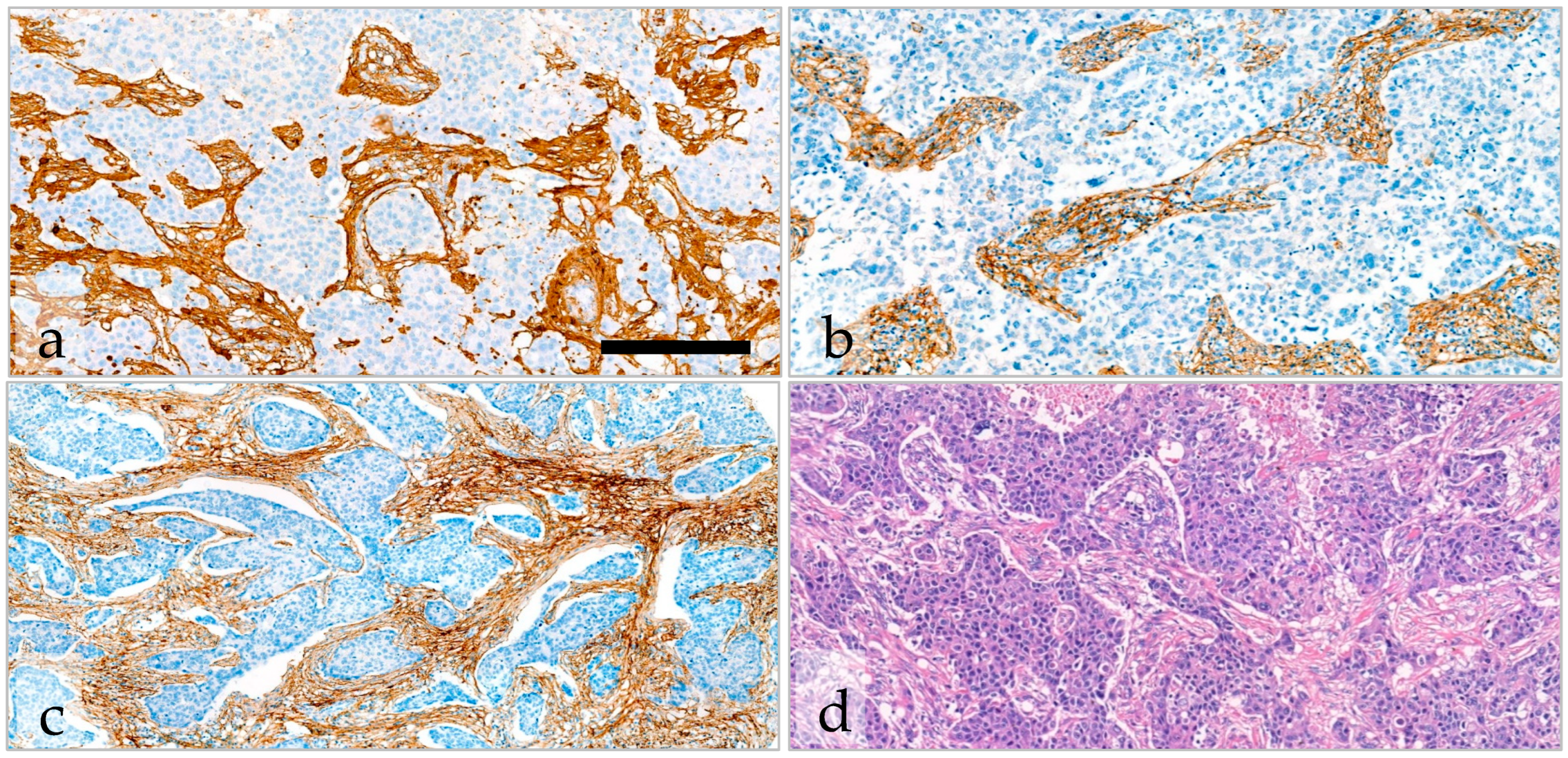
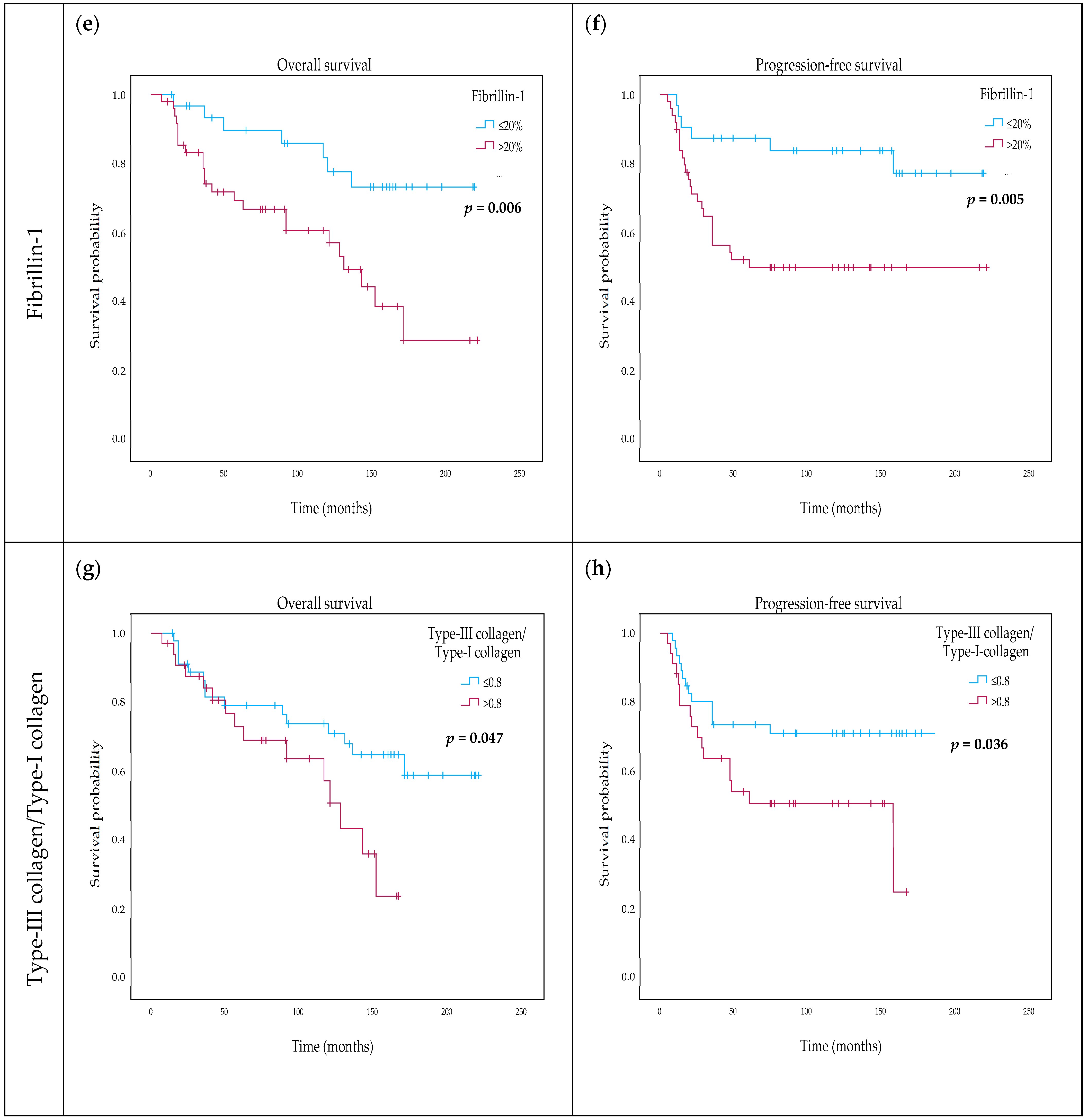
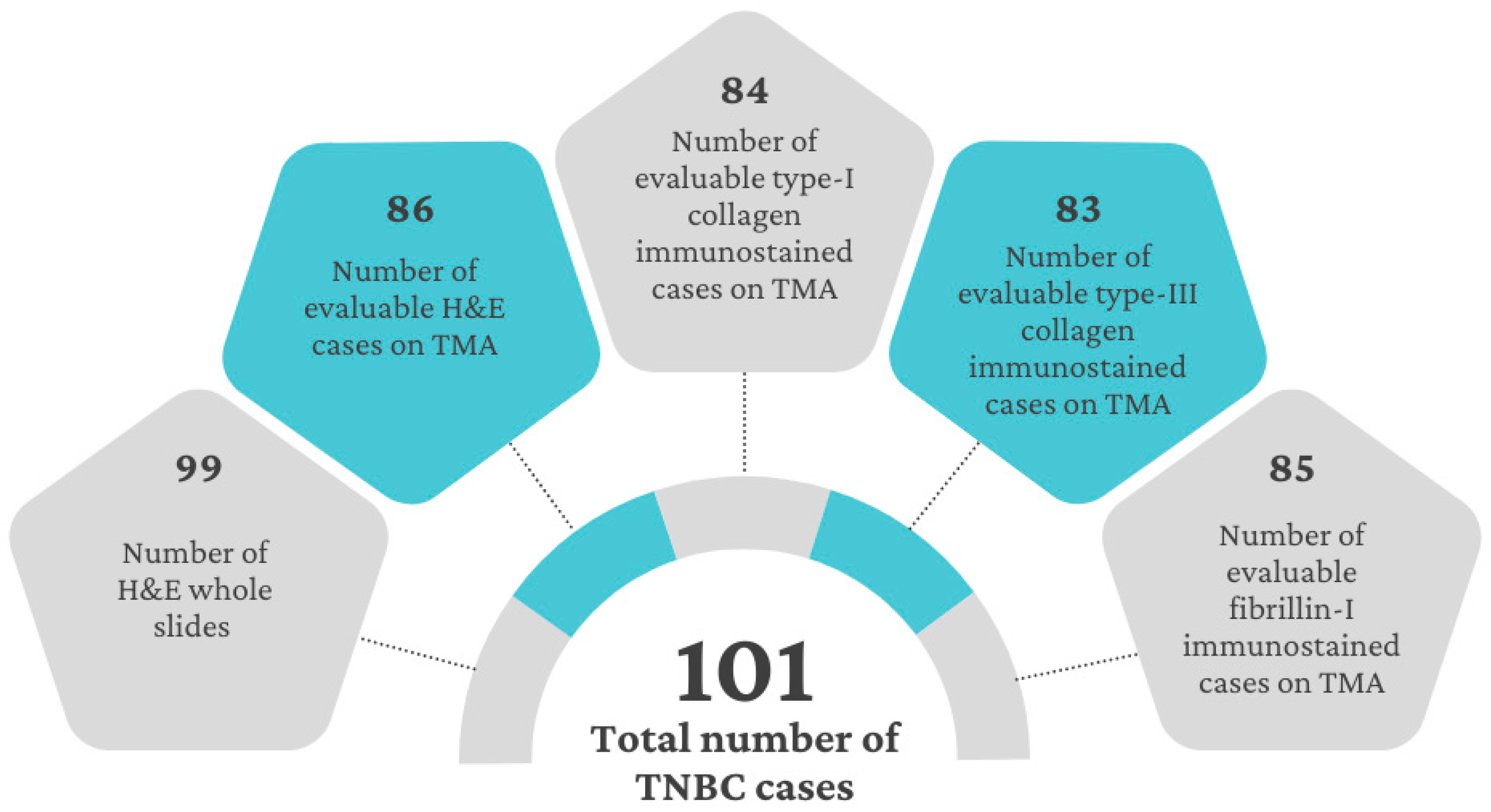
2.5. Variability and Categorization of Type-I Collagen, Type-III Collagen, and Fibrillin-1 Expression and Stromal Content on TMAs
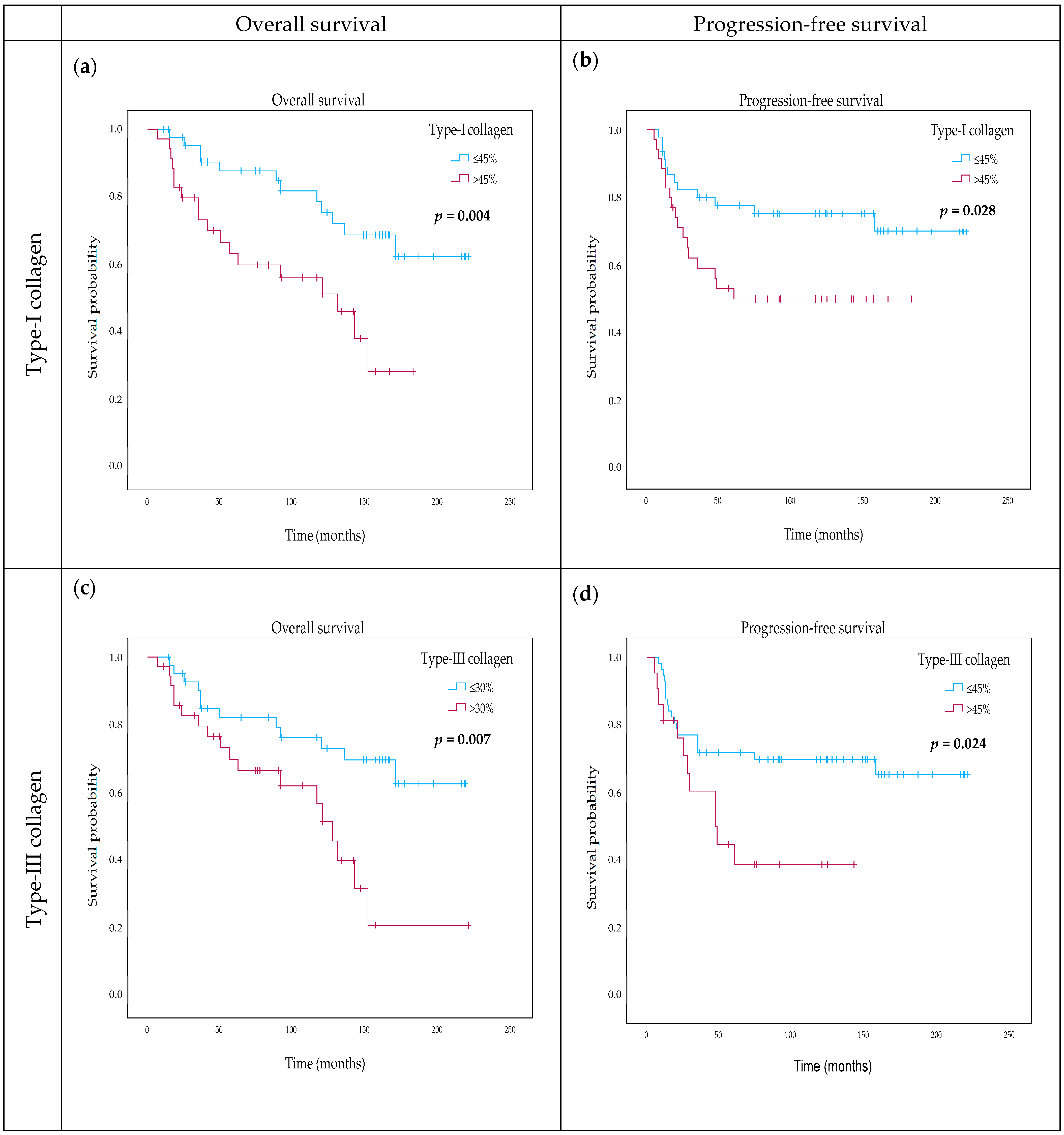
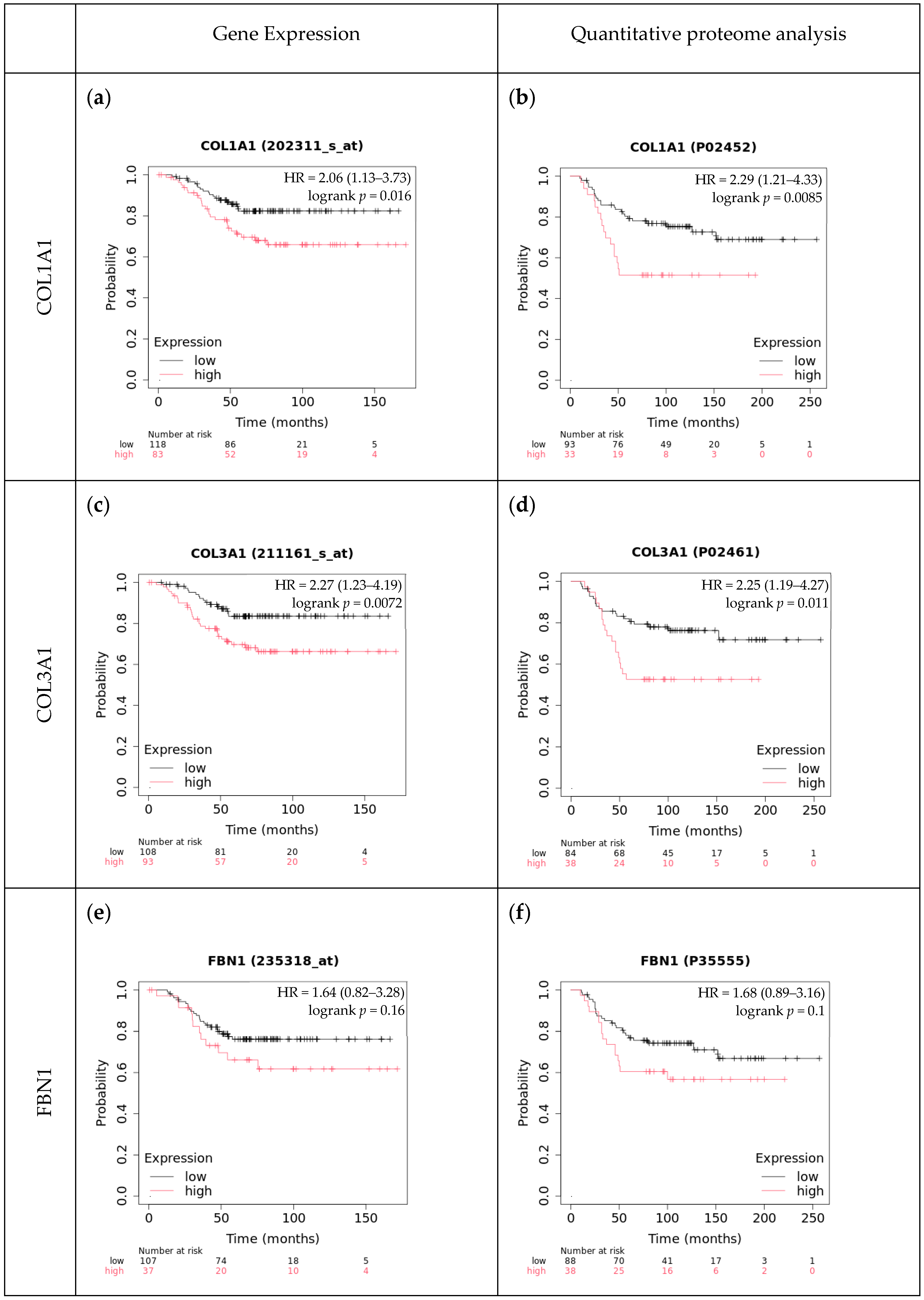
2.6. The Effect of Type-I Collagen, Type-III Collagen, and Fibrillin-1 Expression on Survival
3. Discussion
3.1. Discussion of Study Results
3.2. Limitations of This Study
4. Materials and Methods
4.1. Patients
4.2. Study Design
4.3. Evaluation Methods
4.4. Comparison with Independent Cohorts
4.5. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lukasiewicz, S.; Czeczelewski, M.; Forma, A.; Baj, J.; Sitarz, R.; Stanislawek, A. Breast Cancer-Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies-An Updated Review. Cancers 2021, 13, 4287. [Google Scholar] [CrossRef]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2018; Available online: https://gco.iarc.who.int/today (accessed on 26 June 2024).
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thurlimann, B.; Senn, H.J. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef]
- Mehanna, J.; Haddad, F.G.; Eid, R.; Lambertini, M.; Kourie, H.R. Triple-negative breast cancer: Current perspective on the evolving therapeutic landscape. Int. J. Womens Health 2019, 11, 431–437. [Google Scholar] [CrossRef]
- Mesker, W.E.; Junggeburt, J.M.; Szuhai, K.; de Heer, P.; Morreau, H.; Tanke, H.J.; Tollenaar, R.A. The carcinoma-stromal ratio of colon carcinoma is an independent factor for survival compared to lymph node status and tumor stage. Cell. Oncol. 2007, 29, 387–398. [Google Scholar] [CrossRef]
- Hagenaars, S.C.; Vangangelt, K.M.H.; Van Pelt, G.W.; Karancsi, Z.; Tollenaar, R.; Green, A.R.; Rakha, E.A.; Kulka, J.; Mesker, W.E. Standardization of the tumor-stroma ratio scoring method for breast cancer research. Breast Cancer Res. Treat. 2022, 193, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Jiang, C.; Zhang, Z.X.; Liu, F.; Zhang, F.; Cheng, Y.F. The tumor-stroma ratio is an independent predictor for survival in nasopharyngeal cancer. Oncol. Res. Treat. 2014, 37, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Ma, W.; Wang, J.; Yu, L.; Zhang, X.; Wang, Z.; Tan, B.; Wang, N.; Bai, B.; Yang, S.; et al. Tumor-stroma ratio is an independent predictor for survival in esophageal squamous cell carcinoma. J. Thorac. Oncol. 2012, 7, 1457–1461. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, J.; Li, J.; Chen, Y.; Guan, X.; Wu, X.; Hao, C.; Sun, Y.; Wang, Y.; Wang, X. Tumor-stroma ratio is an independent predictor for survival in early cervical carcinoma. Gynecol. Oncol. 2014, 132, 81–86. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, L.; Liu, W.; Liu, X. Prognostic Significance of the Tumor-Stroma Ratio in Epithelial Ovarian Cancer. BioMed Res. Int. 2015, 2015, 589301. [Google Scholar] [CrossRef]
- Lv, Z.; Cai, X.; Weng, X.; Xiao, H.; Du, C.; Cheng, J.; Zhou, L.; Xie, H.; Sun, K.; Wu, J.; et al. Tumor-stroma ratio is a prognostic factor for survival in hepatocellular carcinoma patients after liver resection or transplantation. Surgery 2015, 158, 142–150. [Google Scholar] [CrossRef]
- Wu, J.; Liang, C.; Chen, M.; Su, W. Association between tumor-stroma ratio and prognosis in solid tumor patients: A systematic review and meta-analysis. Oncotarget 2016, 7, 68954–68965. [Google Scholar] [CrossRef]
- Karancsi, Z.; Hagenaars, S.C.; Nemeth, K.; Mesker, W.E.; Tokes, A.M.; Kulka, J. Tumour-stroma ratio (TSR) in breast cancer: Comparison of scoring core biopsies versus resection specimens. Virchows Arch. 2023. [Google Scholar] [CrossRef] [PubMed]
- Kramer, C.J.H.; Vangangelt, K.M.H.; van Pelt, G.W.; Dekker, T.J.A.; Tollenaar, R.; Mesker, W.E. The prognostic value of tumour-stroma ratio in primary breast cancer with special attention to triple-negative tumours: A review. Breast Cancer Res. Treat. 2019, 173, 55–64. [Google Scholar] [CrossRef]
- Vangangelt, K.M.H.; Green, A.R.; Heemskerk, I.M.F.; Cohen, D.; van Pelt, G.W.; Sobral-Leite, M.; Schmidt, M.K.; Putter, H.; Rakha, E.A.; Tollenaar, R.; et al. The prognostic value of the tumor-stroma ratio is most discriminative in patients with grade III or triple-negative breast cancer. Int. J. Cancer 2020, 146, 2296–2304. [Google Scholar] [CrossRef] [PubMed]
- Moorman, A.M.; Vink, R.; Heijmans, H.J.; van der Palen, J.; Kouwenhoven, E.A. The prognostic value of tumour-stroma ratio in triple-negative breast cancer. Eur. J. Surg. Oncol. 2012, 38, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Fisher, T.B.; Saini, G.; Rekha, T.S.; Krishnamurthy, J.; Bhattarai, S.; Callagy, G.; Webber, M.; Janssen, E.A.M.; Kong, J.; Aneja, R. Digital image analysis and machine learning-assisted prediction of neoadjuvant chemotherapy response in triple-negative breast cancer. Breast Cancer Res. 2024, 26, 12. [Google Scholar] [CrossRef] [PubMed]
- Jakab, A.; Patai, A.V.; Micsik, T. Digital image analysis provides robust tissue microenvironment-based prognosticators in patients with stage I-IV colorectal cancer. Hum. Pathol. 2022, 128, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Millar, E.K.; Browne, L.H.; Beretov, J.; Lee, K.; Lynch, J.; Swarbrick, A.; Graham, P.H. Tumour Stroma Ratio Assessment Using Digital Image Analysis Predicts Survival in Triple Negative and Luminal Breast Cancer. Cancers 2020, 12, 3749. [Google Scholar] [CrossRef]
- Atallah, N.M.; Wahab, N.; Toss, M.S.; Makhlouf, S.; Ibrahim, A.Y.; Lashen, A.G.; Ghannam, S.; Mongan, N.P.; Jahanifar, M.; Graham, S.; et al. Deciphering the Morphology of Tumor-Stromal Features in Invasive Breast Cancer Using Artificial Intelligence. Mod. Pathol. 2023, 36, 100254. [Google Scholar] [CrossRef]
- Micke, P.; Strell, C.; Mattsson, J.; Martin-Bernabe, A.; Brunnstrom, H.; Huvila, J.; Sund, M.; Warnberg, F.; Ponten, F.; Glimelius, B.; et al. The prognostic impact of the tumour stroma fraction: A machine learning-based analysis in 16 human solid tumour types. eBioMedicine 2021, 65, 103269. [Google Scholar] [CrossRef]
- Yan, D.; Ju, X.; Luo, B.; Guan, F.; He, H.; Yan, H.; Yuan, J. Tumour stroma ratio is a potential predictor for 5-year disease-free survival in breast cancer. BMC Cancer 2022, 22, 1082. [Google Scholar] [CrossRef]
- Albusayli, R.; Graham, J.D.; Pathmanathan, N.; Shaban, M.; Raza, S.E.A.; Minhas, F.; Armes, J.E.; Rajpoot, N. Artificial intelligence-based digital scores of stromal tumour-infiltrating lymphocytes and tumour-associated stroma predict disease-specific survival in triple-negative breast cancer. J. Pathol. 2023, 260, 32–42. [Google Scholar] [CrossRef] [PubMed]
- de Visser, K.E.; Joyce, J.A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef] [PubMed]
- Oskarsson, T. Extracellular matrix components in breast cancer progression and metastasis. Breast 2013, 22 (Suppl. S2), S66–S72. [Google Scholar] [CrossRef]
- Halper, J. Basic Components of Connective Tissues and Extracellular Matrix: Fibronectin, Fibrinogen, Laminin, Elastin, Fibrillins, Fibulins, Matrilins, Tenascins and Thrombospondins. Adv. Exp. Med. Biol. 2021, 1348, 105–126. [Google Scholar] [PubMed]
- Popova, N.V.; Jucker, M. The Functional Role of Extracellular Matrix Proteins in Cancer. Cancers 2022, 14, 238. [Google Scholar] [CrossRef] [PubMed]
- Furler, R.L.; Nixon, D.F.; Brantner, C.A.; Popratiloff, A.; Uittenbogaart, C.H. TGF-beta Sustains Tumor Progression through Biochemical and Mechanical Signal Transduction. Cancers 2018, 10, 199. [Google Scholar] [CrossRef]
- Meng, C.; He, Y.; Wei, Z.; Lu, Y.; Du, F.; Ou, G.; Wang, N.; Luo, X.G.; Ma, W.; Zhang, T.C.; et al. MRTF-A mediates the activation of COL1A1 expression stimulated by multiple signaling pathways in human breast cancer cells. Biomed. Pharmacother. 2018, 104, 718–728. [Google Scholar] [CrossRef]
- Hsu, K.S.; Dunleavey, J.M.; Szot, C.; Yang, L.; Hilton, M.B.; Morris, K.; Seaman, S.; Feng, Y.; Lutz, E.M.; Koogle, R.; et al. Cancer cell survival depends on collagen uptake into tumor-associated stroma. Nat. Commun. 2022, 13, 7078. [Google Scholar] [CrossRef]
- Liu, J.; Shen, J.X.; Wu, H.T.; Li, X.L.; Wen, X.F.; Du, C.W.; Zhang, G.J. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov. Med. 2018, 25, 211–223. [Google Scholar]
- Kauppila, S.; Stenback, F.; Risteli, J.; Jukkola, A.; Risteli, L. Aberrant type I and type III collagen gene expression in human breast cancer in vivo. J. Pathol. 1998, 186, 262–268. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, H.Y.; Jung, S.P.; Kim, S.; Lee, J.E.; Nam, S.J.; Bae, J.W. Role of secreted type I collagen derived from stromal cells in two breast cancer cell lines. Oncol. Lett. 2014, 8, 507–512. [Google Scholar] [CrossRef]
- Singh, D.; Rai, V.; Agrawal, D.K. Regulation of Collagen I and Collagen III in Tissue Injury and Regeneration. Cardiol. Cardiovasc. Med. 2023, 7, 5–16. [Google Scholar] [CrossRef]
- Brisson, B.K.; Dekky, B.; Berger, A.C.; Mauldin, E.A.; Loebel, C.; Yen, W.; Stewart, D.C.; Gillette, D.; Assenmacher, C.A.; Cukierman, E.; et al. Tumor-restrictive type III collagen in the breast cancer microenvironment: Prognostic and therapeutic implications. Res. Sq. 2023. [Google Scholar] [CrossRef]
- Brisson, B.K.; Mauldin, E.A.; Lei, W.; Vogel, L.K.; Power, A.M.; Lo, A.; Dopkin, D.; Khanna, C.; Wells, R.G.; Pure, E.; et al. Type III Collagen Directs Stromal Organization and Limits Metastasis in a Murine Model of Breast Cancer. Am. J. Pathol. 2015, 185, 1471–1486. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, L.; Yang, H.; Li, C.; Fang, C. Identification of potential genes related to breast cancer brain metastasis in breast cancer patients. Biosci. Rep. 2021, 41, BSR20211615. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Lin, L.; Li, X.; Wen, R.; Zhang, X. Silencing of COL3A1 represses proliferation, migration, invasion, and immune escape of triple negative breast cancer cells via down-regulating PD-L1 expression. Cell Biol. Int. 2022, 46, 1959–1969. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.; Singh, M.; Eckersley, A.; Cain, S.A.; Sherratt, M.J.; Baldock, C. Fibrillin microfibrils and elastic fibre proteins: Functional interactions and extracellular regulation of growth factors. Semin. Cell Dev. Biol. 2019, 89, 109–117. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Lu, L.; Yang, L.; Yin, S.; Wang, Y.; Qi, Z.; Meng, J.; Zang, R.; Yang, G. Fibrillin-1, induced by Aurora-A but inhibited by BRCA2, promotes ovarian cancer metastasis. Oncotarget 2015, 6, 6670–6683. [Google Scholar] [CrossRef] [PubMed]
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. TNM Classification of Malignant Tumours, 8th ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2016. [Google Scholar]
- Gyorffy, B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput. Struct. Biotechnol. J. 2021, 19, 4101–4109. [Google Scholar] [CrossRef]
- de Kruijf, E.M.; van Nes, J.G.; van de Velde, C.J.; Putter, H.; Smit, V.T.; Liefers, G.J.; Kuppen, P.J.; Tollenaar, R.A.; Mesker, W.E. Tumor-stroma ratio in the primary tumor is a prognostic factor in early breast cancer patients, especially in triple-negative carcinoma patients. Breast Cancer Res. Treat. 2011, 125, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Li, Y.; Wang, Y.; Zhang, S.; Huang, Y.; Yao, S.; Han, C.; Pan, X.; Shi, Z.; Mao, Y.; et al. A deep learning quantified stroma-immune score to predict survival of patients with stage II-III colorectal cancer. Cancer Cell Int. 2021, 21, 585. [Google Scholar] [CrossRef]
- Zhao, K.; Li, Z.; Yao, S.; Wang, Y.; Wu, X.; Xu, Z.; Wu, L.; Huang, Y.; Liang, C.; Liu, Z. Artificial intelligence quantified tumour-stroma ratio is an independent predictor for overall survival in resectable colorectal cancer. eBioMedicine 2020, 61, 103054. [Google Scholar] [CrossRef]
- Acerbi, I.; Cassereau, L.; Dean, I.; Shi, Q.; Au, A.; Park, C.; Chen, Y.Y.; Liphardt, J.; Hwang, E.S.; Weaver, V.M. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. 2015, 7, 1120–1134. [Google Scholar] [CrossRef]
- Kaushik, S.; Pickup, M.W.; Weaver, V.M. From transformation to metastasis: Deconstructing the extracellular matrix in breast cancer. Cancer Metastasis Rev. 2016, 35, 655–667. [Google Scholar] [CrossRef]
- Li, X.; Jin, Y.; Xue, J. Unveiling Collagen’s Role in Breast Cancer: Insights into Expression Patterns, Functions and Clinical Implications. Int. J. Gen. Med. 2024, 17, 1773–1787. [Google Scholar] [CrossRef] [PubMed]
- Jansson, M.; Lindberg, J.; Rask, G.; Svensson, J.; Billing, O.; Nazemroaya, A.; Berglund, A.; Warnberg, F.; Sund, M. Stromal Type I Collagen in Breast Cancer: Correlation to Prognostic Biomarkers and Prediction of Chemotherapy Response. Clin. Breast Cancer 2024, 24, e360–e369.e4. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhang, L.; Wan, D.; Zhou, L.; Zheng, S.; Lin, S.; Qiao, Y. Extracellular matrix and its therapeutic potential for cancer treatment. Signal Transduct. Target. Ther. 2021, 6, 153. [Google Scholar] [CrossRef]
- Li, L.; Huang, J.; Liu, Y. The extracellular matrix glycoprotein fibrillin-1 in health and disease. Front. Cell Dev. Biol. 2023, 11, 1302285. [Google Scholar] [CrossRef]
- Bartha, A.; Gyorffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Cizkova, K.; Foltynkova, T.; Gachechiladze, M.; Tauber, Z. Comparative Analysis of Immunohistochemical Staining Intensity Determined by Light Microscopy, ImageJ and QuPath in Placental Hofbauer Cells. Acta Histochem. Cytochem. 2021, 54, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Szekely, T.; Wichmann, B.; Maros, M.E.; Csizmadia, A.; Bodor, C.; Timar, B.; Krenacs, T. Myelofibrosis progression grading based on type I and type III collagen and fibrillin 1 expression boosted by whole slide image analysis. Histopathology 2023, 82, 622–632. [Google Scholar] [CrossRef] [PubMed]


| All | Visual TSR | Visual OSR | |||||
|---|---|---|---|---|---|---|---|
| n = 99 | Stroma-High (>65) | Stroma-Low (≤65) | t-Test/Log-Rank (p *) | Stroma-High (≥35) | Stroma-Low (<35) | t-Test/Log-Rank (p *) | |
| Age (n = 99) | |||||||
| <48 years | 17 | 5 | 12 | t = 1.8 (0.07) | 3 | 14 | t = 3.5 (<0.001 **) |
| ≥48 years | 82 | 41 | 41 | 48 | 34 | ||
| pT category (n = 97) | |||||||
| pT1b-pT1c | 54 | 21 | 33 | t = 0.83 (0.41) | 27 | 27 | t = 0.03 (0.97) |
| pT2-pT3 | 43 | 23 | 20 | 23 | 20 | ||
| pN category (n = 85) | |||||||
| pN0-pN1mi | 55 | 20 | 35 | t = 3.3 (0.002 **) | 25 | 30 | t = 2.5 (0.013 *) |
| pN1-pN3a | 30 | 19 | 11 | 20 | 10 | ||
| Stromal tumor-infiltrating lymphocyte (n = 99) | |||||||
| ≤20 | 50 | 27 | 23 | t = 1.4 (0.15) | 32 | 18 | t = 3.0 (0.004 **) |
| >20 | 49 | 19 | 30 | 19 | 30 | ||
| Mitotic index (n = 85) | |||||||
| <25 | 36 | 24 | 12 | t = 3.0 (0.003 **) | 25 | 11 | t = 3.7 (0.001 **) |
| ≥25 | 49 | 15 | 34 | 20 | 29 | ||
| Overall survival (n = 99) | |||||||
| Alive | 62 | 28 | 34 | pLR = 0.49 | 27 | 35 | pLR = 0.017 * |
| Dead | 37 | 18 | 19 | 24 | 13 | ||
| Progression-free survival (n = 99) | |||||||
| No progression | 63 | 27 | 36 | pLR = 0.35 | 29 | 34 | pLR = 0.19 |
| Progression | 36 | 19 | 17 | 21 | 15 | ||
| A | ||||||
| Spearman Correlation | Visual TSR Obs. 1/1 | Visual TSR Obs. 1/2 | Visual TSR Obs. 2 | DIA TSR Obs. 1/1 | DIA TSR Obs. 1/2 | DIA TSR Obs. 2 |
| Visual TSR Obs. 1/1 | 1 | 0.979 ** | 0.865 ** | 0.943 ** | 0.946 ** | 0.930 ** |
| Visual TSR Obs. 1/2 | 0.979 ** | 1 | 0.866 ** | 0.928 ** | 0.943 ** | 0.911 ** |
| Visual TSR Obs. 2 | 0.865 ** | 0.866 ** | 1 | 0.876 ** | 0.879 ** | 0.888 ** |
| DIA TSR Obs. 1/1 | 0.943 ** | 0.928 ** | 0.876 ** | 1 | 0.961 ** | 0.964 ** |
| DIA TSR Obs. 1/2 | 0.946 ** | 0.943 ** | 0.879 ** | 0.961 ** | 1 | 0.962 ** |
| B | ||||||
| Pearson Correlation | Visual OSR | DIA OSR Obs. 1/1 | DIA OSR Obs. 1/2 | DIA OSR Obs. 2 | ||
| Visual OSR | 1 | 0.833 ** | 0.833 ** | 0.843 ** | ||
| DIA OSR Obs. 1/1 | 0.833 ** | 1 | 0.911 ** | 0.920 ** | ||
| DIA OSR 2 Obs. 1/2 | 0.833 ** | 0.911 ** | 1 | 0.918 ** | ||
| PFS | Univariate HR | CI | p | Multivariate HR | CI | p |
| DIA OSR (continuous) | 1.012 | 0.996–1.034 | 0.113 | 1.007 | 0.984–1.031 | 0.540 |
| DIA OSR categorical (≤39.34% vs. above) | 1.907 | 0.979–3.715 | 0.058 | |||
| Number of positive lymph nodes | 1.220 | 1.107–1.345 | <0.001 ** | 1.179 | 1.065–1.306 | 0.002 ** |
| Presence of fibrotic foci | 2.119 | 1.007–4.458 | 0.048 * | 2.043 | 0.894–4.670 | 0.090 |
| sTIL categorical (≤20% vs. above) | 0.498 | 0.252–0.985 | 0.045 * | |||
| OS | Univariate HR | CI | p | Multivariate HR | CI | p |
| DIA OSR (continuous) | 1.023 | 1.003–1.044 | 0.023 * | 1.037 | 1.007–1.041 | 0.028 * |
| DIA OSR categorical (≤39.34% vs. above) | 1.955 | 1.004–3.808 | 0.045 * | |||
| Age | 1.039 | 1.009–1.069 | 0.01 * | 1.023 | 0.989–1.059 | 0.188 |
| Number of positive lymph nodes | 1.14 | 1.039–1.250 | 0.006 ** | 1.083 | 0.96–1.223 | 0.195 |
| Presence of necrosis | 2.739 | 1.043–7.191 | 0.041 * | |||
| sTIL categorical (≤20 vs. above) | 0.499 | 0.256–0.972 | 0.041 * | 0.995 | 0.973–1.017 | 0.631 |
| Mitotic index (MI) | 1.017 | 0.097–1.038 | 0.096 | 1.040 | 1.040–1.015 | 0.002 ** |
| Characteristics of the TMAs (n) | H&E (86) | Type-I Collagen (84) | Type-III Collagen (83) | Fibrillin-1 (85) |
|---|---|---|---|---|
| Median percentage of positivity (range) | 37.5 (5–80) | 41.8 (5.3–85) | 27.8 (1.7–85.4) | 30 (2–71.6) |
| Cut-offs used for further calculations * (number or cases below; above) | OS, PFS: 45% (46; 35) | OS: 30% (37; 43) PFS: 45% (58; 22) | OS, PFS: 20% (32; 50) | |
| Median intensity (range) | 198 (126–270) | 191 (143–261) | ||
| Median DQ * PQ (range) | 5507 (309–18,444) | 5050 (372–18,687) | ||
| Median type-I collagen rate (range) | 0.76 (0.05–2.58) | 0.64 (0.18–1.56) |
| Pearson’s Correlation | Stromal Ratio (H&E TMA) | Type-I Collagen | Type-III Collagen | Fibrillin-1 |
|---|---|---|---|---|
| Stromal ratio (H&E TMA) | 1 | 0.864 ** | 0.718 ** | 0.727 ** |
| Type-I collagen | 0.864 ** | 1 | 0.811 ** | 0.809 ** |
| Type-III collagen | 0.718 ** | 0.811 ** | 1 | 0.793 ** |
| Fibrillin-1 | 0.727 ** | 0.809 ** | 0.793 ** | 1 |
| OSR (WSI) | 0.551 ** | 0.627 ** | 0.603 ** | 0.590 ** |
| OS | Univariate HR | CI | p | Multivariate HR | CI | p |
| DIA type-I collagen categorical (≤45% vs. above) | 2.826 | 1.340–5.961 | 0.006 ** | 3.075 | 1.309–7.228 | 0.01 * |
| DIA type-III collagen (continuous) | 1.022 | 1.004–1.040 | 0.017 * | 1.030 | 1.008–1.054 | 0.008 ** |
| DIA type-III collagen categorical (≤30% vs. above) | 2.697 | 1.269–5.729 | 0.01 * | 3.467 | 1.411–8.522 | 0.007 ** |
| DIA fibrillin-1 (continuous) | 1.031 | 1.007–1.056 | 0.011 * | 1.040 | 1.009–1.071 | 0.01 * |
| DIA fibrillin-1 categorical (≤20% vs. above) | 3.129 | 1.331–7.354 | 0.009 ** | 3.439 | 1.248–9.479 | 0.017 * |
| PFS | Univariate HR | CI | p | Multivariate HR | CI | p |
| DIA type-I collagen categorical (≤45% vs. above) | 2.255 | 1.068–4.760 | 0.033 * | 2.725 | 1.197–2.725 | 0.017 * |
| DIA type-III collagen (continuous) | 1.018 | 1.001–1.036 | 0.037 * | 1.033 | 1.009–1.058 | 0.008 ** |
| DIA type-III collagen categorical (≤30% vs. above) | 2.280 | 1.087–4.785 | 0.029 * | 3.825 | 1.364–10.731 | 0.011 * |
| DIA fibrillin-1 (continuous) | 1.030 | 1.007–1.054 | 0.012 * | 1.045 | 1.013–1.077 | 0.005 ** |
| DIA fibrillin-1 categorical (≤20% vs. above) | 3.388 | 1.372–8.368 | 0.008 ** | 3.815 | 1.275–11.574 | 0.018 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karancsi, Z.; Gregus, B.; Krenács, T.; Cserni, G.; Nagy, Á.; Szőcs-Trinfa, K.F.; Kulka, J.; Tőkés, A.M. Digital Whole Slide Image Analysis of Elevated Stromal Content and Extracellular Matrix Protein Expression Predicts Adverse Prognosis in Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2024, 25, 9445. https://doi.org/10.3390/ijms25179445
Karancsi Z, Gregus B, Krenács T, Cserni G, Nagy Á, Szőcs-Trinfa KF, Kulka J, Tőkés AM. Digital Whole Slide Image Analysis of Elevated Stromal Content and Extracellular Matrix Protein Expression Predicts Adverse Prognosis in Triple-Negative Breast Cancer. International Journal of Molecular Sciences. 2024; 25(17):9445. https://doi.org/10.3390/ijms25179445
Chicago/Turabian StyleKarancsi, Zsófia, Barbara Gregus, Tibor Krenács, Gábor Cserni, Ágnes Nagy, Klementina Fruzsina Szőcs-Trinfa, Janina Kulka, and Anna Mária Tőkés. 2024. "Digital Whole Slide Image Analysis of Elevated Stromal Content and Extracellular Matrix Protein Expression Predicts Adverse Prognosis in Triple-Negative Breast Cancer" International Journal of Molecular Sciences 25, no. 17: 9445. https://doi.org/10.3390/ijms25179445
APA StyleKarancsi, Z., Gregus, B., Krenács, T., Cserni, G., Nagy, Á., Szőcs-Trinfa, K. F., Kulka, J., & Tőkés, A. M. (2024). Digital Whole Slide Image Analysis of Elevated Stromal Content and Extracellular Matrix Protein Expression Predicts Adverse Prognosis in Triple-Negative Breast Cancer. International Journal of Molecular Sciences, 25(17), 9445. https://doi.org/10.3390/ijms25179445







