Implementing Massive Parallel Sequencing into Biliary Samples Obtained through Endoscopic Retrograde Cholangiopancreatography for Diagnosing Malignant Bile Duct Strictures
Abstract
1. Introduction
2. Results
2.1. Clinicopathological Findings of the Study Population
2.2. Mutation Analysis of Biliary Brush Cytology and Bile Fluid
2.3. Mutations in Biliary Brush Cytology Compared with Bile Fluid Samples
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Massively Parallel Sequencing with Oncomine Comprehensive Assay
4.3. Massively Parallel Sequencing with Oncomine Pan-Cancer Cell-Free Assay
4.4. Classification of Genomic Alterations
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Javle, M.; Bekaii-Saab, T.; Jain, A.; Wang, Y.; Kelley, R.K.; Wang, K.; Kang, H.C.; Catenacci, D.; Ali, S.; Krishnan, S.; et al. Biliary cancer: Utility of next-generation sequencing for clinical management. Cancer 2016, 122, 3838–3847. [Google Scholar] [CrossRef] [PubMed]
- DeOliveira, M.L.; Cunningham, S.C.; Cameron, J.L.; Kamangar, F.; Winter, J.M.; Lillemoe, K.D.; Choti, M.A.; Yeo, C.J.; Schulick, R.D. Cholangiocarcinoma: Thirty-one-year experience with 564 patients at a single institution. Ann. Surg. 2007, 245, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Gelrud, A.; Agarwal, B. Biliary strictures: Diagnostic considerations and approach. Gastroenterol. Rep. 2015, 3, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Nguyen Canh, H.; Harada, K. Adult bile duct strictures: Differentiating benign biliary stenosis from cholangiocarcinoma. Med. Mol. Morphol. 2016, 49, 189–202. [Google Scholar] [CrossRef]
- NIH state-of-the-science statement on endoscopic retrograde cholangiopancreatography (ERCP) for diagnosis and therapy. NIH Consens. State Sci. Statements 2002, 19, 1–26.
- Korc, P.; Sherman, S. ERCP tissue sampling. Gastrointest. Endosc. 2016, 84, 557–571. [Google Scholar] [CrossRef]
- Farrell, R.J.; Jain, A.K.; Brandwein, S.L.; Wang, H.; Chuttani, R.; Pleskow, D.K. The combination of stricture dilation, endoscopic needle aspiration, and biliary brushings significantly improves diagnostic yield from malignant bile duct strictures. Gastrointest. Endosc. 2001, 54, 587–594. [Google Scholar] [CrossRef]
- De Bellis, M.; Fogel, E.L.; Sherman, S.; Watkins, J.L.; Chappo, J.; Younger, C.; Cramer, H.; Lehman, G.A. Influence of stricture dilation and repeat brushing on the cancer detection rate of brush cytology in the evaluation of malignant biliary obstruction. Gastrointest. Endosc. 2003, 58, 176–182. [Google Scholar] [CrossRef]
- Navaneethan, U.; Njei, B.; Lourdusamy, V.; Konjeti, R.; Vargo, J.J.; Parsi, M.A. Comparative effectiveness of biliary brush cytology and intraductal biopsy for detection of malignant biliary strictures: A systematic review and meta-analysis. Gastrointest. Endosc. 2015, 81, 168–176. [Google Scholar] [CrossRef]
- Helmy, A.; Saad Eldien, H.M.; Seifeldein, G.S.; Abu-Elfatth, A.M.; Mohammed, A.A. Digital Image Analysis has an Additive Beneficial Role to Conventional Cytology in Diagnosing the Nature of Biliary Ducts Stricture. J. Clin. Exp. Hepatol. 2021, 11, 209–218. [Google Scholar] [CrossRef]
- Dudley, J.C.; Zheng, Z.; McDonald, T.; Le, L.P.; Dias-Santagata, D.; Borger, D.; Batten, J.; Vernovsky, K.; Sweeney, B.; Arpin, R.N.; et al. Next-Generation Sequencing and Fluorescence in Situ Hybridization Have Comparable Performance Characteristics in the Analysis of Pancreaticobiliary Brushings for Malignancy. J. Mol. Diagn. 2016, 18, 124–130. [Google Scholar] [CrossRef]
- Kamp, E.; Dinjens, W.N.M.; van Velthuysen, M.F.; de Jonge, P.J.F.; Bruno, M.J.; Peppelenbosch, M.P.; de Vries, A.C. Next-generation sequencing mutation analysis on biliary brush cytology for differentiation of benign and malignant strictures in primary sclerosing cholangitis. Gastrointest. Endosc. 2023, 97, 456–465.e6. [Google Scholar] [CrossRef] [PubMed]
- Boyd, S.; Mustamäki, T.; Sjöblom, N.; Nordin, A.; Tenca, A.; Jokelainen, K.; Rantapero, T.; Liuksiala, T.; Lahtinen, L.; Kuopio, T.; et al. NGS of brush cytology samples improves the detection of high-grade dysplasia and cholangiocarcinoma in patients with primary sclerosing cholangitis: A retrospective and prospective study. Hepatol. Commun. 2024, 8, e0415. [Google Scholar] [CrossRef] [PubMed]
- Moris, D.; Kostakis, I.D.; Machairas, N.; Prodromidou, A.; Tsilimigras, D.I.; Ravindra, K.V.; Sudan, D.L.; Knechtle, S.J.; Barbas, A.S. Comparison between liver transplantation and resection for hilar cholangiocarcinoma: A systematic review and meta-analysis. PLoS ONE 2019, 14, e0220527. [Google Scholar] [CrossRef]
- Wakai, T.; Shirai, Y.; Sakata, J.; Maruyama, T.; Ohashi, T.; Korira, P.V.; Ajioka, Y.; Hatakeyama, K. Clinicopathological features of benign biliary strictures masquerading as biliary malignancy. Am. Surg. 2012, 78, 1388–1391. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef]
- Nikanjam, M.; Kato, S.; Kurzrock, R. Liquid biopsy: Current technology and clinical applications. J. Hematol. Oncol. 2022, 15, 131. [Google Scholar] [CrossRef]
- Lu, L.; Bi, J.; Bao, L. Genetic profiling of cancer with circulating tumor DNA analysis. J. Genet. Genom. 2018, 45, 79–85. [Google Scholar] [CrossRef]
- Hadano, N.; Murakami, Y.; Uemura, K.; Hashimoto, Y.; Kondo, N.; Nakagawa, N.; Sueda, T.; Hiyama, E. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br. J. Cancer 2016, 115, 59–65. [Google Scholar] [CrossRef]
- Valle, J.W.; Lamarca, A.; Goyal, L.; Barriuso, J.; Zhu, A.X. New Horizons for Precision Medicine in Biliary Tract Cancers. Cancer Discov. 2017, 7, 943–962. [Google Scholar] [CrossRef]
- Nakamura, H.; Arai, Y.; Totoki, Y.; Shirota, T.; Elzawahry, A.; Kato, M.; Hama, N.; Hosoda, F.; Urushidate, T.; Ohashi, S.; et al. Genomic spectra of biliary tract cancer. Nat. Genet. 2015, 47, 1003–1010. [Google Scholar] [CrossRef]
- Lowery, M.A.; Ptashkin, R.; Jordan, E.; Berger, M.F.; Zehir, A.; Capanu, M.; Kemeny, N.E.; O’Reilly, E.M.; El-Dika, I.; Jarnagin, W.R.; et al. Comprehensive Molecular Profiling of Intrahepatic and Extrahepatic Cholangiocarcinomas: Potential Targets for Intervention. Clin. Cancer Res. 2018, 24, 4154–4161. [Google Scholar] [CrossRef] [PubMed]
- Chae, H.; Kim, D.; Yoo, C.; Kim, K.P.; Jeong, J.H.; Chang, H.M.; Lee, S.S.; Park, D.H.; Song, T.J.; Hwang, S.; et al. Therapeutic relevance of targeted sequencing in management of patients with advanced biliary tract cancer: DNA damage repair gene mutations as a predictive biomarker. Eur. J. Cancer 2019, 120, 31–39. [Google Scholar] [CrossRef]
- Boerner, T.; Drill, E.; Pak, L.M.; Nguyen, B.; Sigel, C.S.; Doussot, A.; Shin, P.; Goldman, D.A.; Gonen, M.; Allen, P.J.; et al. Genetic Determinants of Outcome in Intrahepatic Cholangiocarcinoma. Hepatology 2021, 74, 1429–1444. [Google Scholar] [CrossRef]
- Lin, J.; Cao, Y.; Yang, X.; Li, G.; Shi, Y.; Wang, D.; Long, J.; Song, Y.; Mao, J.; Xie, F.; et al. Mutational spectrum and precision oncology for biliary tract carcinoma. Theranostics 2021, 11, 4585–4598. [Google Scholar] [CrossRef] [PubMed]
- Singhi, A.D.; McGrath, K.; Brand, R.E.; Khalid, A.; Zeh, H.J.; Chennat, J.S.; Fasanella, K.E.; Papachristou, G.I.; Slivka, A.; Bartlett, D.L.; et al. Preoperative next-generation sequencing of pancreatic cyst fluid is highly accurate in cyst classification and detection of advanced neoplasia. Gut 2018, 67, 2131–2141. [Google Scholar] [CrossRef] [PubMed]
- Nikiforova, M.N.; Wald, A.I.; Melan, M.A.; Roy, S.; Zhong, S.; Hamilton, R.L.; Lieberman, F.S.; Drappatz, J.; Amankulor, N.M.; Pollack, I.F.; et al. Targeted next-generation sequencing panel (GlioSeq) provides comprehensive genetic profiling of central nervous system tumors. Neuro Oncol. 2016, 18, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Singhi, A.D.; Nikiforova, M.N.; Chennat, J.; Papachristou, G.I.; Khalid, A.; Rabinovitz, M.; Das, R.; Sarkaria, S.; Ayasso, M.S.; Wald, A.I.; et al. Integrating next-generation sequencing to endoscopic retrograde cholangiopancreatography (ERCP)-obtained biliary specimens improves the detection and management of patients with malignant bile duct strictures. Gut 2020, 69, 52–61. [Google Scholar] [CrossRef]
- Buc, E.; Lesurtel, M.; Belghiti, J. Is preoperative histological diagnosis necessary before referral to major surgery for cholangiocarcinoma? HPB 2008, 10, 98–105. [Google Scholar] [CrossRef][Green Version]
- Nagino, M.; Hirano, S.; Yoshitomi, H.; Aoki, T.; Uesaka, K.; Unno, M.; Ebata, T.; Konishi, M.; Sano, K.; Shimada, K.; et al. Clinical practice guidelines for the management of biliary tract cancers 2019: The 3rd English edition. J. Hepatobiliary Pancreat. Sci. 2021, 28, 26–54. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.J.; Mills, P.R.; Carter, R.; O’Donohue, J.; Fullarton, G.; Imrie, C.W.; Murray, W.R. Brush cytology in the assessment of pancreatico-biliary strictures: A review of 406 cases. J. Clin. Pathol. 2001, 54, 449–455. [Google Scholar] [CrossRef]
- Bank, J.S.; Witt, B.L.; Taylor, L.J.; Adler, D.G. Diagnostic yield and accuracy of a new cytology brush design compared to standard brush cytology for evaluation of biliary strictures. Diagn. Cytopathol. 2018, 46, 234–238. [Google Scholar] [CrossRef]
- Ding, S.M.; Lu, A.L.; Xu, B.Q.; Shi, S.H.; Edoo, M.I.A.; Zheng, S.S.; Li, Q.Y. Accuracy of brush cytology in biliopancreatic strictures: A single-center cohort study. J. Int. Med. Res. 2021, 49, 300060520987771. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Hao, X.; Liu, B.; Liu, S.; Yuan, Y. Bile liquid biopsy in biliary tract cancer. Clin. Chim. Acta 2023, 551, 117593. [Google Scholar] [CrossRef]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.N.; Zhang, C.; Li, Z.; Kong, L.C.; Wang, X.H.; Gu, Z.C.; Wang, J.L. MicroRNA expression signatures of atrial fibrillation: The critical systematic review and bioinformatics analysis. Exp. Biol. Med. 2020, 245, 42–53. [Google Scholar] [CrossRef]
- Driescher, C.; Fuchs, K.; Haeberle, L.; Goering, W.; Frohn, L.; Opitz, F.V.; Haeussinger, D.; Knoefel, W.T.; Keitel, V.; Esposito, I. Bile-Based Cell-Free DNA Analysis Is a Reliable Diagnostic Tool in Pancreatobiliary Cancer. Cancers 2020, 13, 39. [Google Scholar] [CrossRef]
- Pishvaian, M.J.; Joseph Bender, R.; Matrisian, L.M.; Rahib, L.; Hendifar, A.; Hoos, W.A.; Mikhail, S.; Chung, V.; Picozzi, V.; Heartwell, C.; et al. A pilot study evaluating concordance between blood-based and patient-matched tumor molecular testing within pancreatic cancer patients participating in the Know Your Tumor (KYT) initiative. Oncotarget 2017, 8, 83446–83456. [Google Scholar] [CrossRef]
- Harbhajanka, A.; Michael, C.W.; Janaki, N.; Gokozan, H.N.; Wasman, J.; Bomeisl, P.; Yoest, J.; Sadri, N. Tiny but mighty: Use of next generation sequencing on discarded cytocentrifuged bile duct brushing specimens to increase sensitivity of cytological diagnosis. Mod. Pathol. 2020, 33, 2019–2025. [Google Scholar] [CrossRef]
- Han, J.Y.; Ahn, K.S.; Kim, T.S.; Kim, Y.H.; Cho, K.B.; Shin, D.W.; Baek, W.K.; Suh, S.I.; Jang, B.C.; Kang, K.J. Liquid Biopsy from Bile-Circulating Tumor DNA in Patients with Biliary Tract Cancer. Cancers 2021, 13, 4581. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, M.; Rosenblatt, K.P.; Li, L.; Rakhade, M.; Amato, R.J. Validation and Clinical Applications of a Comprehensive Next Generation Sequencing System for Molecular Characterization of Solid Cancer Tissues. Front. Mol. Biosci. 2019, 6, 82. [Google Scholar] [CrossRef] [PubMed]
- Bankov, K.; Döring, C.; Schneider, M.; Hartmann, S.; Winkelmann, R.; Albert, J.G.; Bechstein, W.O.; Zeuzem, S.; Hansmann, M.L.; Peveling-Oberhag, J.; et al. Sequencing of intraductal biopsies is feasible and potentially impacts clinical management of patients with indeterminate biliary stricture and cholangiocarcinoma. Clin. Transl. Gastroenterol. 2018, 9, 151. [Google Scholar] [CrossRef]
- Alvisi, G.; Termanini, A.; Soldani, C.; Portale, F.; Carriero, R.; Pilipow, K.; Costa, G.; Polidoro, M.; Franceschini, B.; Malenica, I.; et al. Multimodal single-cell profiling of intrahepatic cholangiocarcinoma defines hyperactivated Tregs as a potential therapeutic target. J. Hepatol. 2022, 77, 1359–1372. [Google Scholar] [CrossRef]
- Brooks, C.; Gausman, V.; Kokoy-Mondragon, C.; Munot, K.; Amin, S.P.; Desai, A.; Kipp, C.; Poneros, J.; Sethi, A.; Gress, F.G.; et al. Role of Fluorescent In Situ Hybridization, Cholangioscopic Biopsies, and EUS-FNA in the Evaluation of Biliary Strictures. Dig. Dis. Sci. 2018, 63, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Barr Fritcher, E.G.; Voss, J.S.; Brankley, S.M.; Campion, M.B.; Jenkins, S.M.; Keeney, M.E.; Henry, M.R.; Kerr, S.M.; Chaiteerakij, R.; Pestova, E.V.; et al. An Optimized Set of Fluorescence In Situ Hybridization Probes for Detection of Pancreatobiliary Tract Cancer in Cytology Brush Samples. Gastroenterology 2015, 149, 1813–1824.e1. [Google Scholar] [CrossRef]
- Gonda, T.A.; Viterbo, D.; Gausman, V.; Kipp, C.; Sethi, A.; Poneros, J.M.; Gress, F.; Park, T.; Khan, A.; Jackson, S.A.; et al. Mutation Profile and Fluorescence In Situ Hybridization Analyses Increase Detection of Malignancies in Biliary Strictures. Clin. Gastroenterol. Hepatol. 2017, 15, 913–919.e1. [Google Scholar] [CrossRef]
- Astier, C.; Ngo, C.; Colmet-Daage, L.; Marty, V.; Bawa, O.; Nicotra, C.; Ngo-Camus, M.; Italiano, A.; Massard, C.; Scoazec, J.Y.; et al. Molecular profiling of biliary tract cancers reveals distinct genomic landscapes between circulating and tissue tumor DNA. Exp. Hematol. Oncol. 2024, 13, 2. [Google Scholar] [CrossRef]
- Goyal, L.; Saha, S.K.; Liu, L.Y.; Siravegna, G.; Leshchiner, I.; Ahronian, L.G.; Lennerz, J.K.; Vu, P.; Deshpande, V.; Kambadakone, A.; et al. Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion-Positive Cholangiocarcinoma. Cancer Discov. 2017, 7, 252–263. [Google Scholar] [CrossRef]
- Nagai, K.; Kuwatani, M.; Hirata, K.; Suda, G.; Hirata, H.; Takishin, Y.; Furukawa, R.; Kishi, K.; Yonemura, H.; Nozawa, S.; et al. Genetic Analyses of Cell-Free DNA in Pancreatic Juice or Bile for Diagnosing Pancreatic Duct and Biliary Tract Strictures. Diagnostics 2022, 12, 2704. [Google Scholar] [CrossRef]
- Grasso, C.; Butler, T.; Rhodes, K.; Quist, M.; Neff, T.L.; Moore, S.; Tomlins, S.A.; Reinig, E.; Beadling, C.; Andersen, M.; et al. Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J. Mol. Diagn. 2015, 17, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Takayasu, T.; Zorofchian Moghadamtousi, S.; Arevalo, O.; Chen, M.; Lan, C.; Duose, D.; Hu, P.; Zhu, J.J.; Roy-Chowdhuri, S.; et al. Evaluation of the Oncomine Pan-Cancer Cell-Free Assay for Analyzing Circulating Tumor DNA in the Cerebrospinal Fluid in Patients with Central Nervous System Malignancies. J. Mol. Diagn. 2021, 23, 171–180. [Google Scholar] [CrossRef]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef] [PubMed]
- Javle, M.; Churi, C.; Kang, H.C.; Shroff, R.; Janku, F.; Surapaneni, R.; Zuo, M.; Barrera, C.; Alshamsi, H.; Krishnan, S.; et al. HER2/neu-directed therapy for biliary tract cancer. J. Hematol. Oncol. 2015, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Zhang, Y.; Van Horn, R.D.; Yin, T.; Buchanan, S.; Yadav, V.; Mochalkin, I.; Wong, S.S.; Yue, Y.G.; Huber, L.; et al. Oncogenic BRAF Deletions That Function as Homodimers and Are Sensitive to Inhibition by RAF Dimer Inhibitor LY3009120. Cancer Discov. 2016, 6, 300–315. [Google Scholar] [CrossRef] [PubMed]
- Singhi, A.D.; Ali, S.M.; Lacy, J.; Hendifar, A.; Nguyen, K.; Koo, J.; Chung, J.H.; Greenbowe, J.; Ross, J.S.; Nikiforova, M.N.; et al. Identification of Targetable ALK Rearrangements in Pancreatic Ductal Adenocarcinoma. J. Natl. Compr. Cancer Netw. 2017, 15, 555–562. [Google Scholar] [CrossRef]
- Javle, M.; Lowery, M.; Shroff, R.T.; Weiss, K.H.; Springfeld, C.; Borad, M.J.; Ramanathan, R.K.; Goyal, L.; Sadeghi, S.; Macarulla, T.; et al. Phase II Study of BGJ398 in Patients With FGFR-Altered Advanced Cholangiocarcinoma. J. Clin. Oncol. 2018, 36, 276–282. [Google Scholar] [CrossRef]
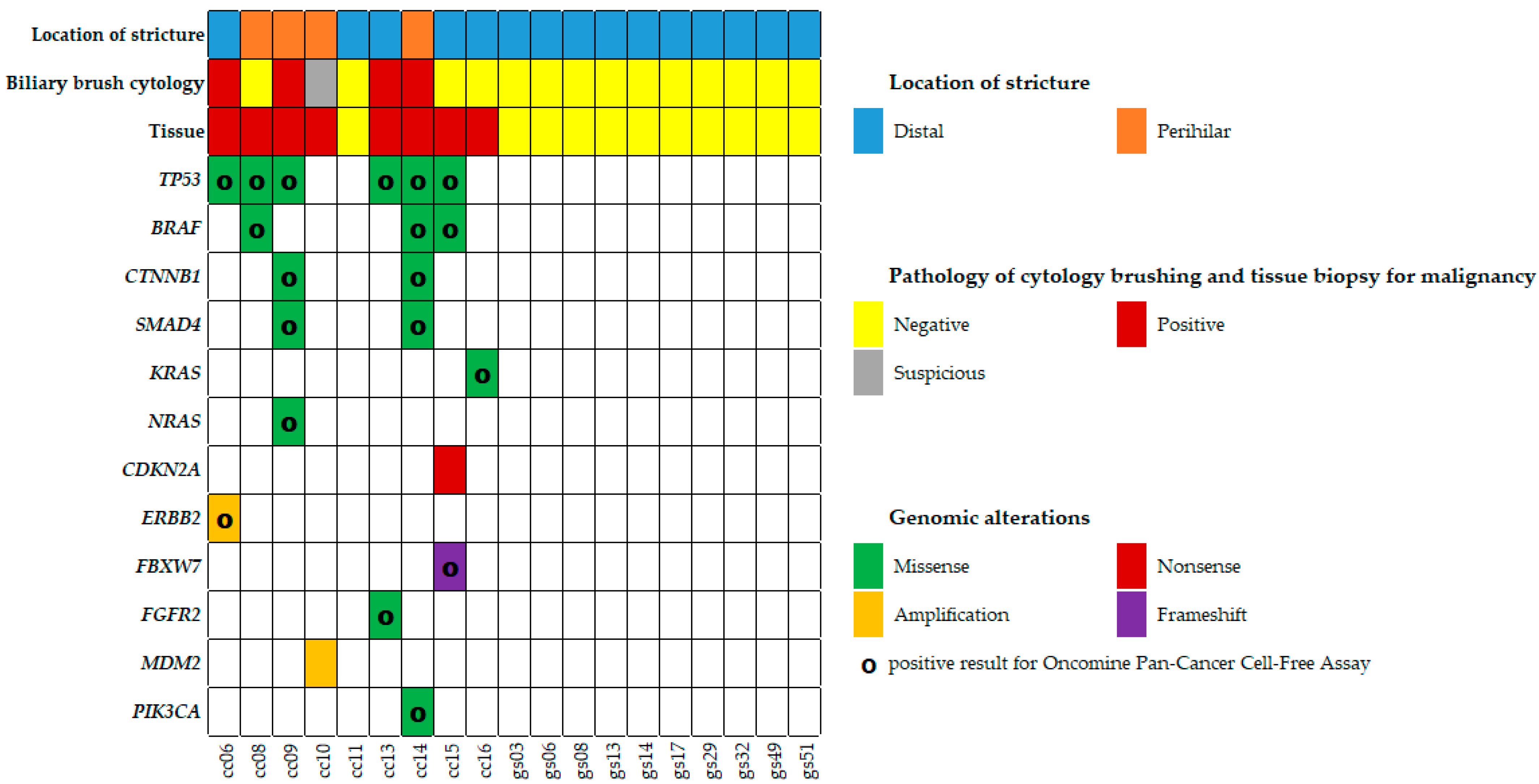
| Case | Diagnosis | Pathology | Cytology | Treatment | Prognosis | OS (Day) | |
|---|---|---|---|---|---|---|---|
| Initial | Confirmed | ||||||
| cc06 | eCCA | dBDC | AC | Positive * | PPPD, Adjuvant ChT | death | 622 |
| cc08 | eCCA | pCCA | AC | Negative † | ConserTx | death | 239 |
| dc09 | eCCA | pCCA | AC | Positive | ConserTx | F/U loss | 325 |
| cc10 | eCCA | pCCA | AC | Suspicious ‡ | ConserTx | F/U loss | 120 |
| cc11 | eCCA | SC | SC, IgG4-related | Negative | ConserTx | alive | 2284 |
| cc13 | eCCA | dBDC | AC | Positive | ConserTx | death | 267 |
| cc14 | eCCA | pCCA | AC | Positive | PPPD | F/U loss | 408 |
| cc15 | eCCA | dBDC | AC | Negative | PPPD | alive | 2001 |
| cc16 | eCCA | dBDC | AC with SRC | Negative | ConserTx | death | 303 |
| Case | S/A (y) | Gene | Mutation | Mutation Change | Depth | VAF | rsID | Class | OPCCFA |
|---|---|---|---|---|---|---|---|---|---|
| Type | (×) | (%) | (%) | ||||||
| cc06 | M/61 | TP53 | missense | c.832C>G/p.P278A | 1965 | 64 | rs17849781 | tier 2 | 3.7 |
| ERBB2 | amplification (8 copies) | tier 2 | 1.8 | ||||||
| cc08 | M/72 | TP53 | missense | c.1039G>A/p.A347T | 2000 | 46 | rs1597349147 | tier 2 | 3.7 |
| BRAF | missense | c.1397G>A/p.G466E | 1999 | 6 | rs121913351 | tier 2 | 0.8 | ||
| cc09 | F/79 | TP53 | missense | c.535C>T/p.H179Y | 1694 | 70 | rs587780070 | tier 2 | 3.1 |
| CTNNB1 | missense | c.134C>G/p.S45C | 2000 | 14 | rs121913409 | tier 2 | 0.6 | ||
| SMAD4 | missense | c.1051G>A/p.D351N | 2000 | 15 | rs1057519739 | tier 2 | 0.3 | ||
| NRAS | missense | c.182A>T/p.Q61L | 2000 | 43 | rs11554290 | tier 2 | 2.6 | ||
| cc10 | M/58 | MDM2 | amplification (6 copies) | tier 2 | N.A. | ||||
| cc11 | M/64 | Not Detected | N.D. | ||||||
| cc13 | M/76 | TP53 | missense | c.707A>G/p.236C | 1970 | 62 | rs730882026 | tier 2 | 2.9 |
| FGFR2 | missense | c.1144T>C/p.C382R | 1885 | 36 | rs121913474 | tier 1 | 1.4 | ||
| cc14 | F/76 | TP53 | missense | c.523C>T/p.R175C | 1222 | 45 | rs138729528 | tier 2 | 3.9 |
| BRAF | missense | c.1781A>G/p.D594G | 1022 | 28 | rs121913338 | tier 2 | 2.6 | ||
| CTNNB1 | missense | c.134C>G/p.S45C | 1999 | 67 | rs121913409 | tier 2 | 2.5 | ||
| SMAD4 | missense | c.1081C>T/p.R361C | 2000 | 30 | rs80338963 | tier 2 | 0.7 | ||
| PIK3CA | missense | c.1624G>A/p.E542K | 1963 | 37 | rs121913273 | tier 2 | 0.3 | ||
| cc15 | M/71 | TP53 | missense | c.817C>T/p.R273C | 2000 | 31 | rs121913343 | tier 2 | 3.4 |
| BRAF | missense | c.1780G>A/p.D594N | 2000 | 16 | rs397516896 | tier 2 | 0.3 | ||
| CDKN2A | nonsense | c.205G>T/p.E69* | 1434 | 47 | rs121913383 | tier 2 | N.A. | ||
| FBXW7 | frameshift | c.58delA/p.R20Efs*9 | 1981 | 15 | N.A. | tier 2 | 2.2 | ||
| FBXW7 | frameshift | c.1218delG/p.W406Cfs*9 | 1994 | 13 | N.A. | tier 2 | 3.1 | ||
| cc16 | F/90 | KRAS | missense | c.182A>T/p.Q61L | 2000 | 26 | rs121913240 | tier 2 | 4.3 |
| Case | S/A (y) | Gene | Mutation | Mutation Change | Depth | VAF | rsID | Class | OPCCFA |
|---|---|---|---|---|---|---|---|---|---|
| Type | (×) | (%) | |||||||
| gs03 | F/34 | Not Detected | N.D. | ||||||
| gs06 | F/62 | TET2 | missense | c.4076G>A/p.R1359H | 1999 | 29 | rs775677220 | tier 3 | N.A. |
| gs08 | M/57 | Not Detected | N.D. | ||||||
| gs13 | F/59 | Not Detected | N.D. | ||||||
| gs14 | F/64 | Not Detected | N.D. | ||||||
| gs17 | M/47 | Not Detected | N.D. | ||||||
| gs29 | M/55 | ATM | missense | c.7328G>A/p.R2443 | 2000 | 13 | rs587782310 | tier 3 | N.A. |
| gs32 | M/42 | Not Detected | N.D. | ||||||
| gs49 | F/43 | Not Detected | N.D. | ||||||
| gs51 | M/56 | Not Detected | N.D. | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, W.; Gwack, J.; Park, J. Implementing Massive Parallel Sequencing into Biliary Samples Obtained through Endoscopic Retrograde Cholangiopancreatography for Diagnosing Malignant Bile Duct Strictures. Int. J. Mol. Sci. 2024, 25, 9461. https://doi.org/10.3390/ijms25179461
Park W, Gwack J, Park J. Implementing Massive Parallel Sequencing into Biliary Samples Obtained through Endoscopic Retrograde Cholangiopancreatography for Diagnosing Malignant Bile Duct Strictures. International Journal of Molecular Sciences. 2024; 25(17):9461. https://doi.org/10.3390/ijms25179461
Chicago/Turabian StylePark, Wonsuk, Jin Gwack, and Joonhong Park. 2024. "Implementing Massive Parallel Sequencing into Biliary Samples Obtained through Endoscopic Retrograde Cholangiopancreatography for Diagnosing Malignant Bile Duct Strictures" International Journal of Molecular Sciences 25, no. 17: 9461. https://doi.org/10.3390/ijms25179461
APA StylePark, W., Gwack, J., & Park, J. (2024). Implementing Massive Parallel Sequencing into Biliary Samples Obtained through Endoscopic Retrograde Cholangiopancreatography for Diagnosing Malignant Bile Duct Strictures. International Journal of Molecular Sciences, 25(17), 9461. https://doi.org/10.3390/ijms25179461






