Identification and Functional Exploration of BraGASA Genes Reveal Their Potential Roles in Drought Stress Tolerance and Sexual Reproduction in Brassica rapa L. ssp. pekinensis
Abstract
:1. Introduction
2. Results
2.1. Identification and Physicochemical Characterization of BraGASA Family Genes
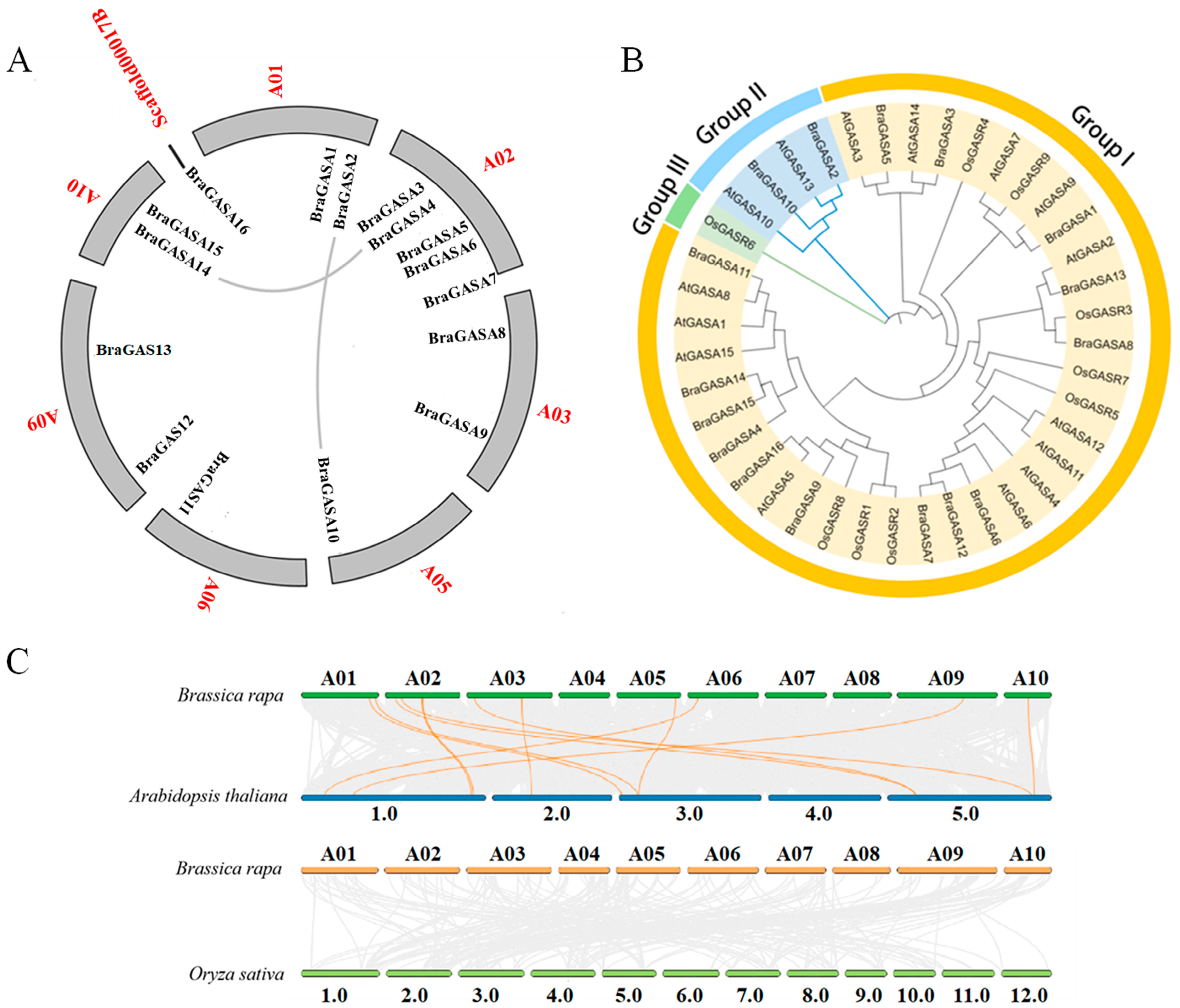
2.2. Phylogenetic Relationships and Synteny
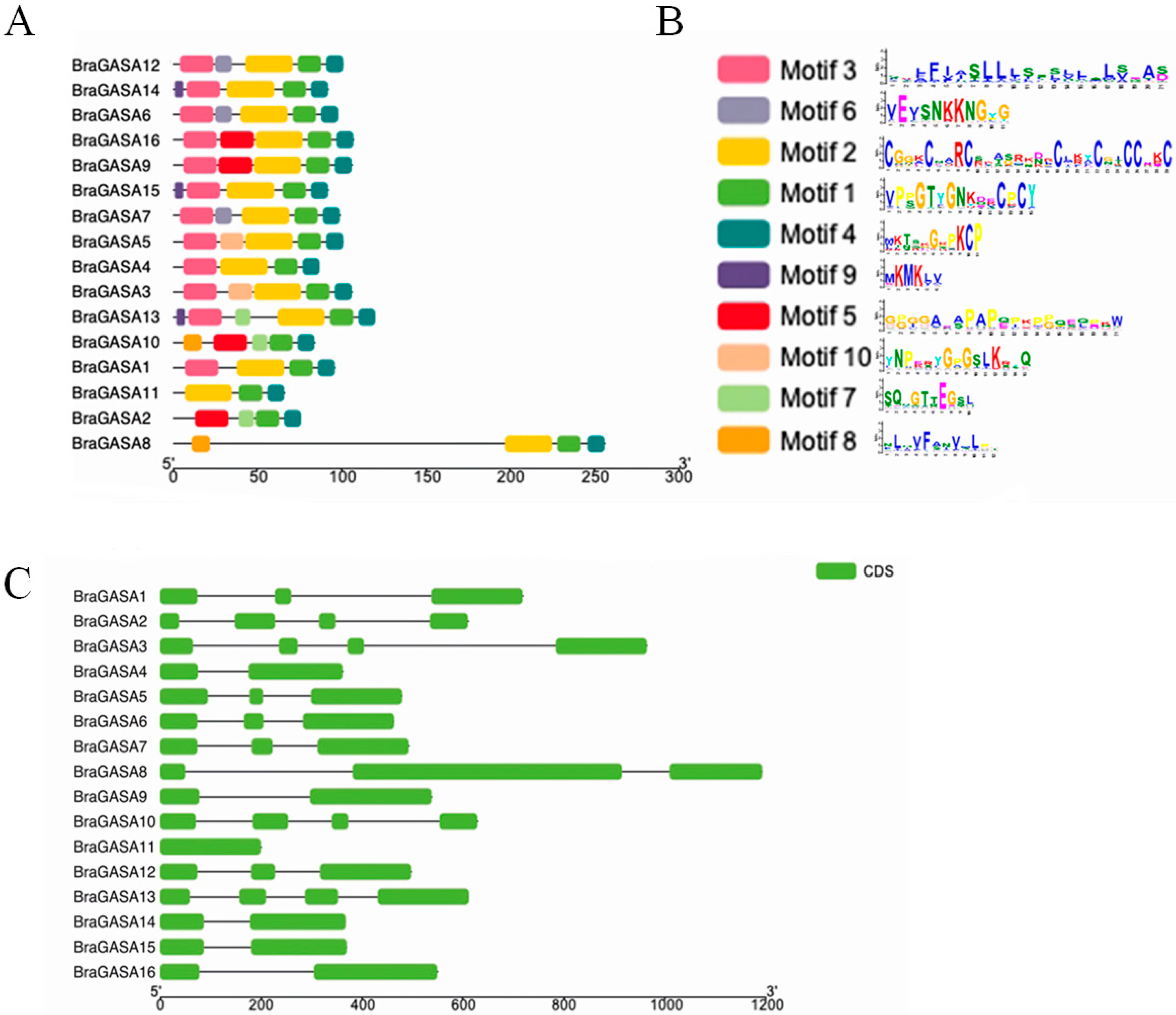
2.3. Gene Structure and Protein Domain
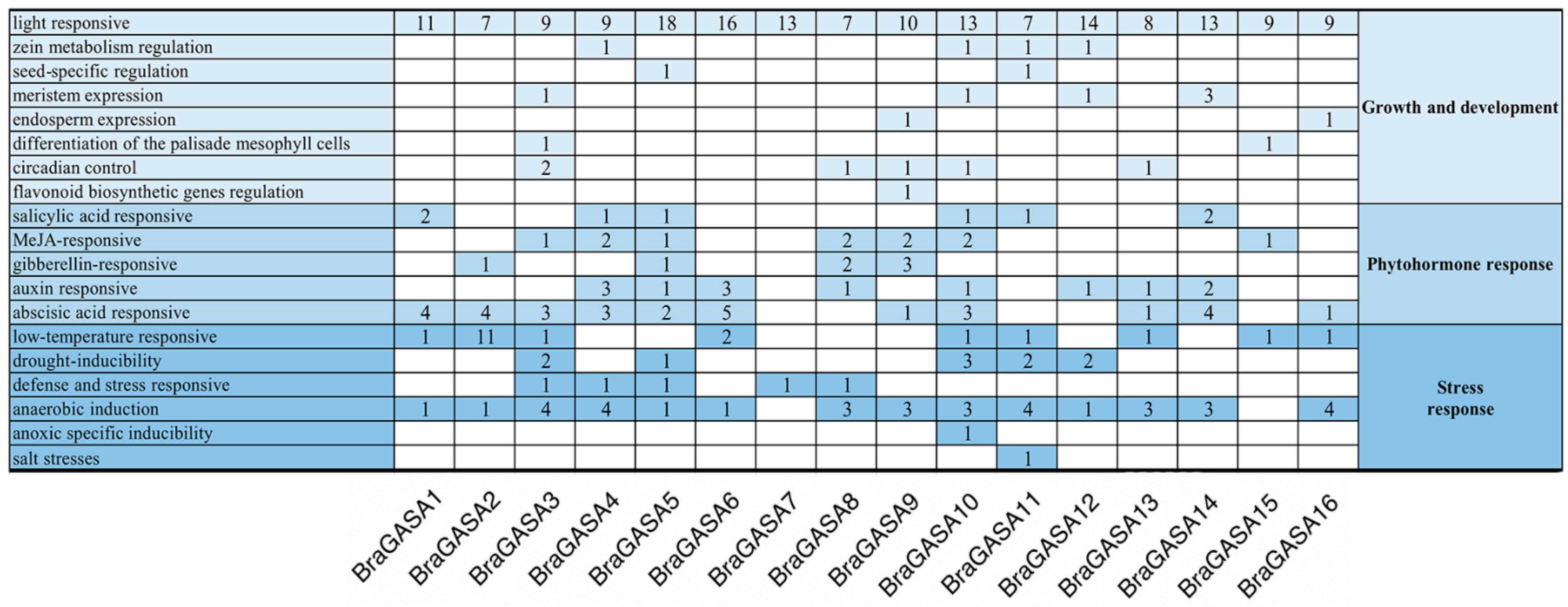
2.4. Analysis of Promoter Cis-Regulatory Elements of BraGASAs
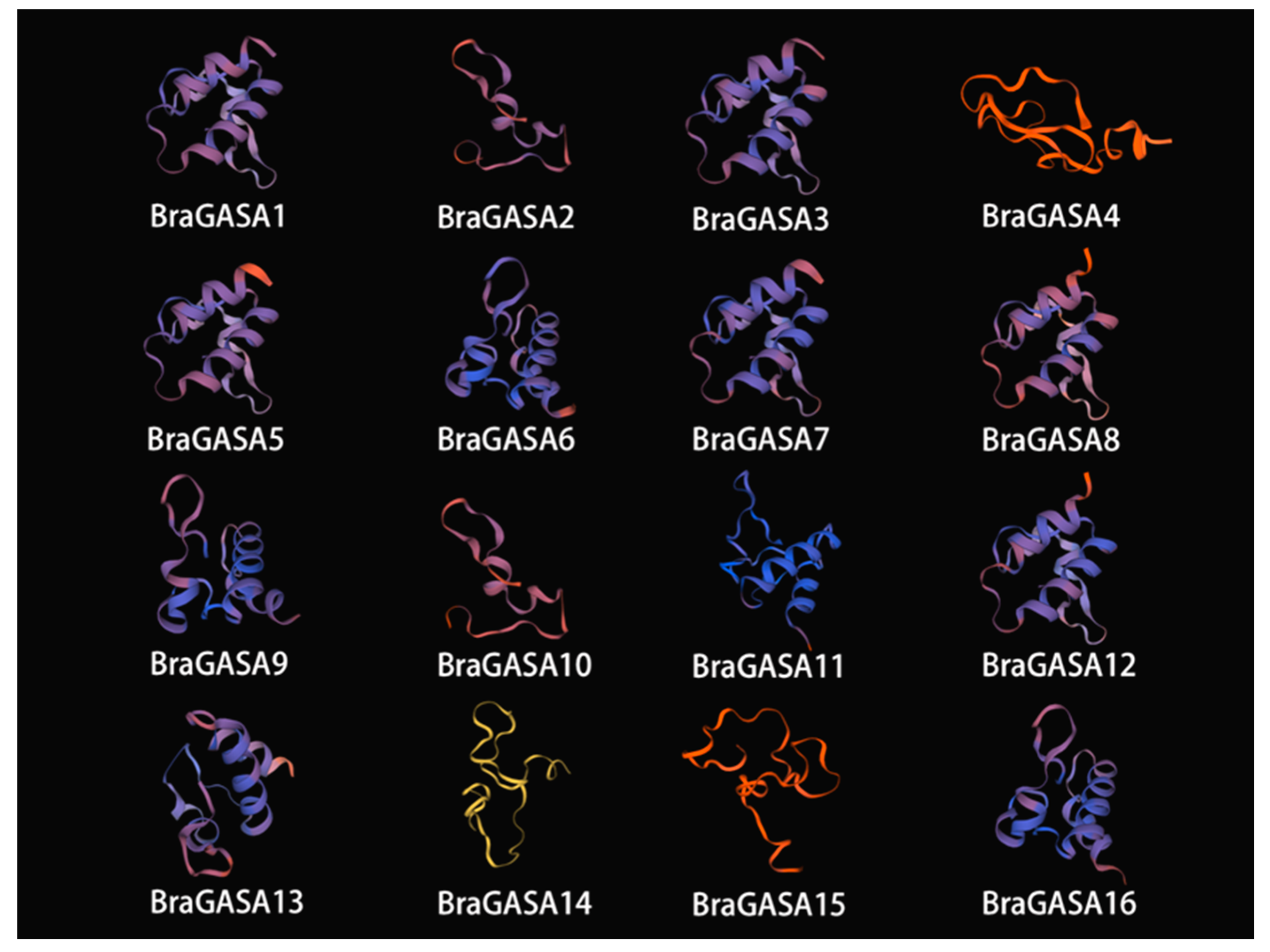
2.5. Analysis of BraGASA Protein Tertiary Structure
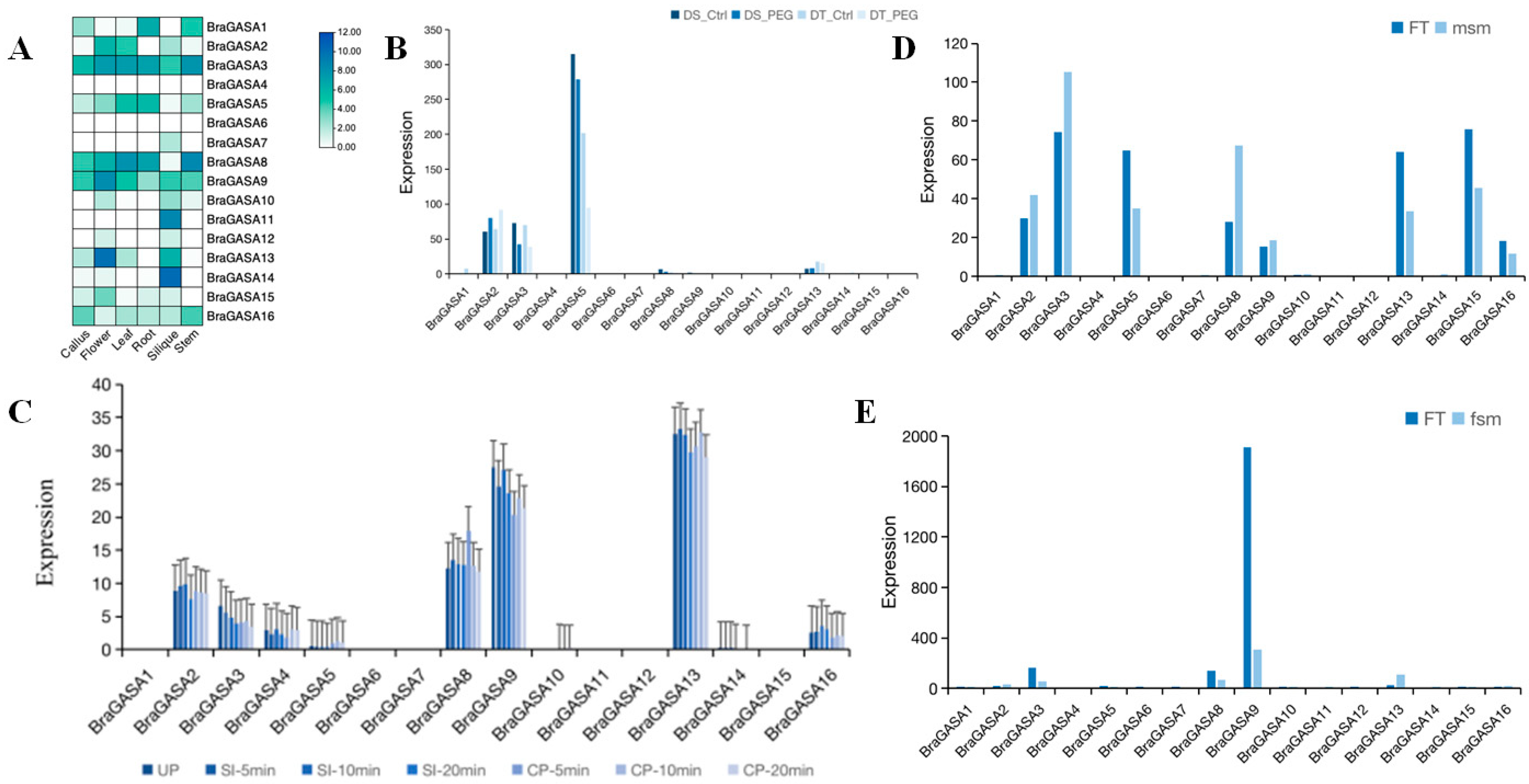
2.6. Gene Expression Analysis of the BraGASAs
2.7. Analysis of Transcriptional Expression under Drought Stress
2.8. Analysis of Sexual Reproduction-Related Expression Profiling of BraGASAs
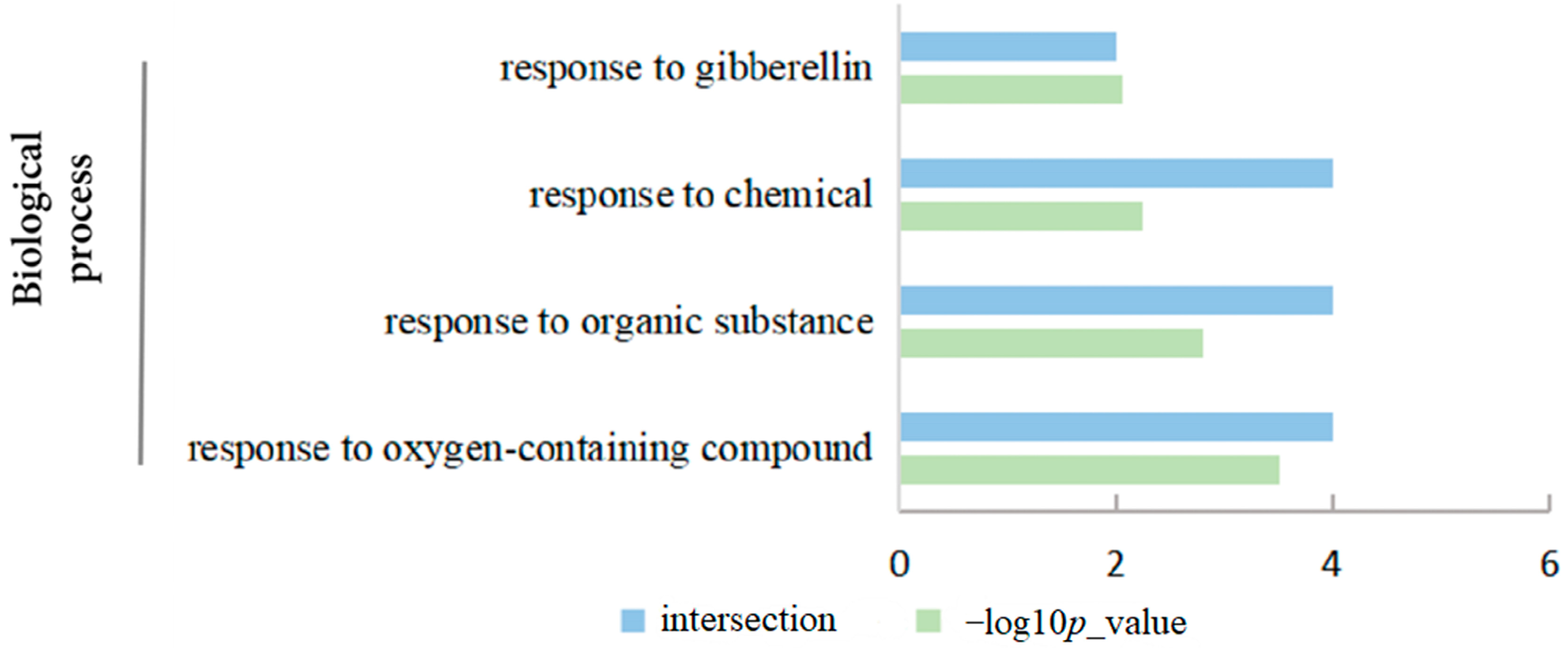
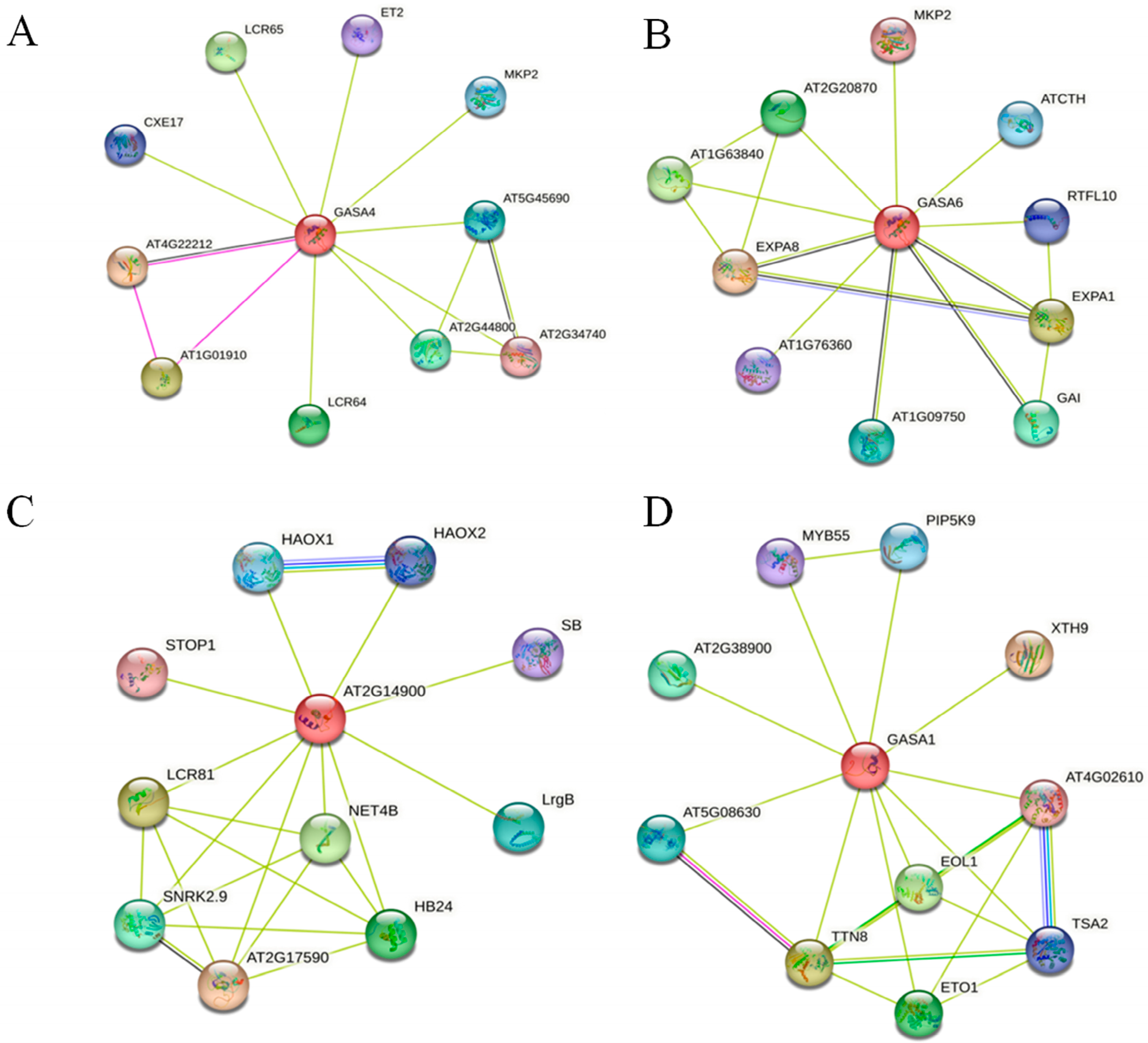
2.9. Functional Annotation and Protein Interaction Analysis
3. Discussion
4. Materials and Methods
4.1. Analysis of GASA Family Proteins: Gene Sequences and Physical and Chemical Properties
4.2. Chromosomal Location, Phylogenetic Tree Construction, and Analysis of Collinearity Relationship
4.3. Prediction of Gene Structure and Conserved Motifs
4.4. Analysis of Cis-Acting Elements
4.5. Prediction of Three-Dimensional Structures of the Proteins
4.6. Protein–Protein Interaction Analysis and Gene Ontology Enrichment Analysis
4.7. Analysis of Gene Expression of GASA in B. Rapa
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roxrud, I.; Lid, S.E.; Fletcher, J.C.; Schmidt, E.D.L.; Opsahl-Sorteberg, H.G. GASA4, One of the 14-Member Arabidopsis GASA Family of Small Polypeptides, Regulates Flowering and Seed Development. Plant Cell Physiol. 2007, 48, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Gast, R.T.; Gopalraj, M.; Olszewski, N.E. Characterization of a shoot-specific, GA3- and ABA-regulated gene from tomato. Plant J. 2010, 2, 153–159. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, C.; Peng, J.; Sun, S.; Wang, X. GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana. Plant Mol. Biol. 2009, 69, 745–759. [Google Scholar] [CrossRef]
- Rubinovich, L.; Weiss, D. The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J. 2010, 64, 1018–1027. [Google Scholar] [CrossRef]
- Sheng, F.; Dong, Z.; Lizhi, Z.; Cai, G.; Mingzhi, X.; Mobeen, T.M.; Youmei, L.; Juanjuan, M.; Mingyu, H. Comprehensive analysis of GASA family members in the Malus domestica genome: Identification, characterization, and their expressions in response to apple flower induction. Bmc Genom. 2017, 18, 827. [Google Scholar]
- Bang, A.; Qiannan, W.; Xiaodong, Z.; Bei, Z.; Hongli, L.; Chaozu, H. Comprehensive transcriptional and functional analyses of HbGASA genes reveal their roles in fungal pathogen resistance in Hevea brasiliensis. Tree Genet. Genomes 2018, 14, 41. [Google Scholar]
- Dong, L.; Wang, F.; Liu, T.; Dong, Z.; Wang, D. Natural variation of TaGASR7-A1 affects grain length in common wheat under multiple cultivation conditions. Mol. Breed. 2014, 34, 937–947. [Google Scholar] [CrossRef]
- Segura, A.; Moreno, M.; Madueño, F.; Molina, A.; García-Olmedo, F. Snakin-1, a Peptide from Potato That Is Active Against Plant Pathogens. Mol. Plant-Microbe Interact. MPMI 1999, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Enriqueta, M.C.; Bellido, M.L.; Nicolás, G.-C.; Laura, M.P.; Francisco, A.R.; González-Reyes, J.A.; Caballero, J.L.; Juan, M.B.; Rosario, B.P. FaGAST2, a strawberry ripening-related gene, acts together with FaGAST1 to determine cell size of the fruit receptacle. Plant Cell Physiol. 2013, 54, 218–236. [Google Scholar]
- Li, K.-L.; Bai, X.; Li, Y.; Hua, C. GsGASA1 mediated root growth inhibition in response to chronic cold stress is marked by the accumulation of DELLAs. J. Plant Physiol. 2011, 168, 2153–2160. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, X. Overexpression of GASA5 increases the sensitivity of Arabidopsis to heat stress. J. Plant Physiol. 2011, 168, 2093–2101. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Yu, H.; Zhong, C.; Zhang, X.; Peng, J.; Wang, X. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J. Exp. Bot. 2013, 6, 1637–1647. [Google Scholar] [CrossRef]
- Büyük, l.; Okay, A.; Gorska, M.; Lhan, E.; Aras, S. Identification and characterization of the Pvul-GASA gene family in the Phaseolus vulgaris and expression patterns under salt stress. Doga Turk. J. Bot. 2021, 45, 655–670. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, F.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef]
- Cai, C.; Wang, X.; Liu, B.; Wu, J.; Liang, J.; Cui, Y.; Cheng, F.; Wang, X. Brassica rapa Genome 2.0: A Reference Upgrade through Sequence Re-assembly and Gene Re-annotation. Mol. Plant 2017, 10, 649–651. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. BRAD V3.0: An upgraded Brassicaceae database. Nucleic Acids Res. 2022, 50, D1432–D1441. [Google Scholar] [CrossRef]
- Zhang, Z.; Guo, J.; Cai, X.; Li, Y.; Xi, X.; Lin, R.; Lin, R.; Liang, J.; Wang, J. Improved Reference Genome Annotation of Brassica rapa by Pacific Biosciences RNA Sequencing. Front. Plant Sci. 2022, 13, 841618. [Google Scholar] [CrossRef]
- Cai, X.; Chang, L.; Zhang, T.; Chen, H.; Zhang, L.; Lin, R.; Liang, J.; Wu, J.; Freeling, M.; Wang, X. Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 2021, 22, 166. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Liu, W.; Xu, J.; Liu, Z.; Feng, H. The SAP function in pistil development was proved by two allelic mutations in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Plant Biol. 2020, 20, 538. [Google Scholar] [CrossRef] [PubMed]
- Herzog, M.; Dorne, A.M.; Grellet, F. GASA, a gibberellin-regulated gene family from Arabidopsis thaliana related to the tomato GAST1 gene. Plant Mol. Biol. 1995, 27, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Z.; Xu, Y.; Joo, X.S.; Kim, S.K.; Xue, Z.; Xu, Z.; Wang, Z.; Chong, K. OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J. Cell Mol. Biol. 2009, 57, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Wang, X. Progress in Cysteine-rich Gibberellic Acid-stimulated Arabidopsis Protein. Chin. Bull. Bot. 2016, 51, 1–8. [Google Scholar]
- Aubert, D.; Chevillard, M.; Dorne, A.M.; Arlaud, G.; Herzog, M. Expression patterns of GASA genes in Arabidopsis thaliana: The GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol. Biol. 1998, 36, 871–883. [Google Scholar] [CrossRef]
- Wang, H.; Wei, T.; Wang, X.; Zhang, L.; Yang, M.; Chen, L.; Song, S.; Wang, C.; Chen, C. Transcriptome Analyses from Mutant Salvia miltiorrhiza Reveals Important Roles for SmGASA4 during Plant Development. Int. J. Mol. Sci. 2018, 19, 2088. [Google Scholar] [CrossRef]
- Yang, D.H.; Fenning, T.; Tang, S.; Xia, X.; Yin, W. Genome-wide transcriptional response of Populus euphratica to long-term drought stress. Plant Sci. Int. J. Exp. Plant Biol. 2012, 195, 24–35. [Google Scholar] [CrossRef]
- Huang, D.; Wu, W.; Abrams, S.R.; Cutler, A.J. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 2008, 59, 2991–3007. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shi, S.; Tao, Q.; Tao, Y.; Miao, J.; Peng, X.; Li, C.; Yang, Z.; Zhou, G.; Liang, G. OsGASR9 positively regulates grain size and yield in rice (Oryza sativa). Plant Sci. Int. J. Exp. Plant Biol. 2019, 286, 17–27. [Google Scholar] [CrossRef]
- Nahirñak, V.; Rivarola, M.; Gonzalez de Urreta, M.; Paniego, N.; Hopp, H.E.; Almasia, N.I.; Vazquez-Rovere, C. Genome-wide analysis of the Snakin/GASA gene family in Solanum tuberosum cv. Kennebec. Am. J. Potato Res. 2016, 93, 172–188. [Google Scholar] [CrossRef]
- Ahmad, B.; Yao, J.; Zhang, S.; Li, X.; Zhang, X.; Yadav, V.; Wang, X. Genome-Wide Characterization and Expression Profiling of GASA Genes during Different Stages of Seed Development in Grapevine (Vitis vinifera L.) Predict Their Involvement in Seed Development. Int. J. Mol. Sci. 2020, 21, 1088. [Google Scholar] [CrossRef]
- Masahiko, I.; Masakazu, H.; Nobuko, F.; Tomohiro, K.; Yasujiro, M. Glucosinolate metabolism, functionality and breeding for the improvement of Brassicaceae vegetables. Breed. Sci. 2014, 64, 48–59. [Google Scholar] [CrossRef]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef] [PubMed]
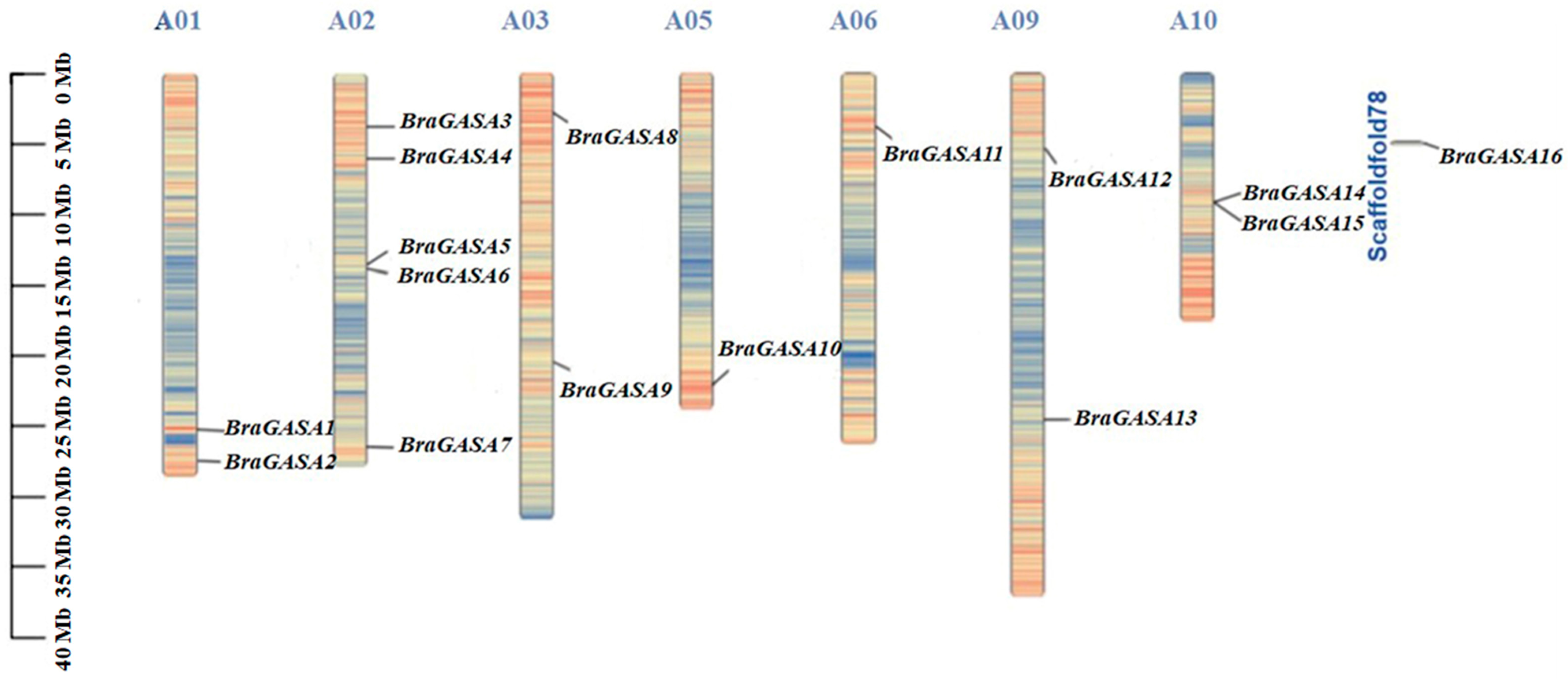

| Gene ID | Gene Name | Chromosome | pI | MW (Da) | Protein Length (aa) | A.thaliana ID | A.thaliana Name |
|---|---|---|---|---|---|---|---|
| Bra021407 | BraGASA1 | A01:25317858:25318576 | 9.46 | 10,760.72 | 291 | AT3G02885 | AtGASA5 |
| Bra034095 | BraGASA2 | A01:27475061:27475671 | 7.81 | 8400.38 | 231 | AT1G74670 | AtGASA6 |
| Bra023513 | BraGASA3 | A02:3715274:3716239 | 9.36 | 11,704.76 | 321 | AT5G15230 | AtGASA4 |
| Bra020281 | Br4GASA4 | A02:6005564:6005926 | 8.97 | 9647.36 | 264 | AT5G59845 | AtGASA10 |
| Bra008162 | BraGASA5 | A02:13527439:13527919 | 8.95 | 11,443.45 | 306 | AT1G74670 | AtGASA6 |
| Bra008222 | BraGASA6 | A02:13905515:13905979 | 9 | 10,578.39 | 297 | AT1G75750 | AtGASA1 |
| Bra029227 | BraGASA7 | A02:26446073:26446566 | 8.79 | 10,558.37 | 300 | AT1G75750 | AtGASA1 |
| Bra006273 | BraGASA8 | A03:2778795:2779988 | 10.14 | 27,191.56 | 771 | AT5G14920 | AtGASA13 |
| Bra013115 | BraGASA9 | A03:20378333:20378871 | 8.86 | 11,232.3 | 321 | AT2G14900 | AtGASA7 |
| Bra029820 | BraGASA10 | A05:22152528:22153157 | 9.61 | 9217.66 | 255 | AT3G10185 | AtGASA15 |
| Bra019917 | BraGASA11 | A06:3722606:3722806 | 8.48 | 7172.29 | 201 | AT2G39540 | AtGASA8 |
| Bra038550 | BraGASA12 | A09:5247947:5248445 | 8.99 | 10,838.64 | 306 | AT1G75750 | AtGASA1 |
| Bra024530 | BraGASA13 | A09:24582390:24583002 | 9.1 | 13,187.23 | 363 | AT1G22690 | AtGASA9 |
| Bra002526 | BraGASA14 | A10:9052766:9053136 | 8.62 | 10,241.93 | 279 | AT5G59845 | AtGASA10 |
| Bra002525 | BraGASA15 | A10:9057487:9057855 | 8.75 | 10,229.9 | 279 | AT5G59845 | AtGASA10 |
| Bra039830 | BraGASA16 | Scaffold000178:118569:119118 | 8.6 | 11,378.4 | 324 | AT2G14900 | AtGASA7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Sun, X.; Zhou, J.; Liu, L.; Huang, L.; Hu, Q. Identification and Functional Exploration of BraGASA Genes Reveal Their Potential Roles in Drought Stress Tolerance and Sexual Reproduction in Brassica rapa L. ssp. pekinensis. Int. J. Mol. Sci. 2024, 25, 9643. https://doi.org/10.3390/ijms25179643
Zhao Y, Sun X, Zhou J, Liu L, Huang L, Hu Q. Identification and Functional Exploration of BraGASA Genes Reveal Their Potential Roles in Drought Stress Tolerance and Sexual Reproduction in Brassica rapa L. ssp. pekinensis. International Journal of Molecular Sciences. 2024; 25(17):9643. https://doi.org/10.3390/ijms25179643
Chicago/Turabian StyleZhao, Yanting, Xinjie Sun, Jingyuan Zhou, Lixuan Liu, Li Huang, and Qizan Hu. 2024. "Identification and Functional Exploration of BraGASA Genes Reveal Their Potential Roles in Drought Stress Tolerance and Sexual Reproduction in Brassica rapa L. ssp. pekinensis" International Journal of Molecular Sciences 25, no. 17: 9643. https://doi.org/10.3390/ijms25179643
APA StyleZhao, Y., Sun, X., Zhou, J., Liu, L., Huang, L., & Hu, Q. (2024). Identification and Functional Exploration of BraGASA Genes Reveal Their Potential Roles in Drought Stress Tolerance and Sexual Reproduction in Brassica rapa L. ssp. pekinensis. International Journal of Molecular Sciences, 25(17), 9643. https://doi.org/10.3390/ijms25179643






