Identification of a Clade-Specific HLA-C*03:02 CTL Epitope GY9 Derived from the HIV-1 p17 Matrix Protein
Abstract
:1. Introduction
2. Results
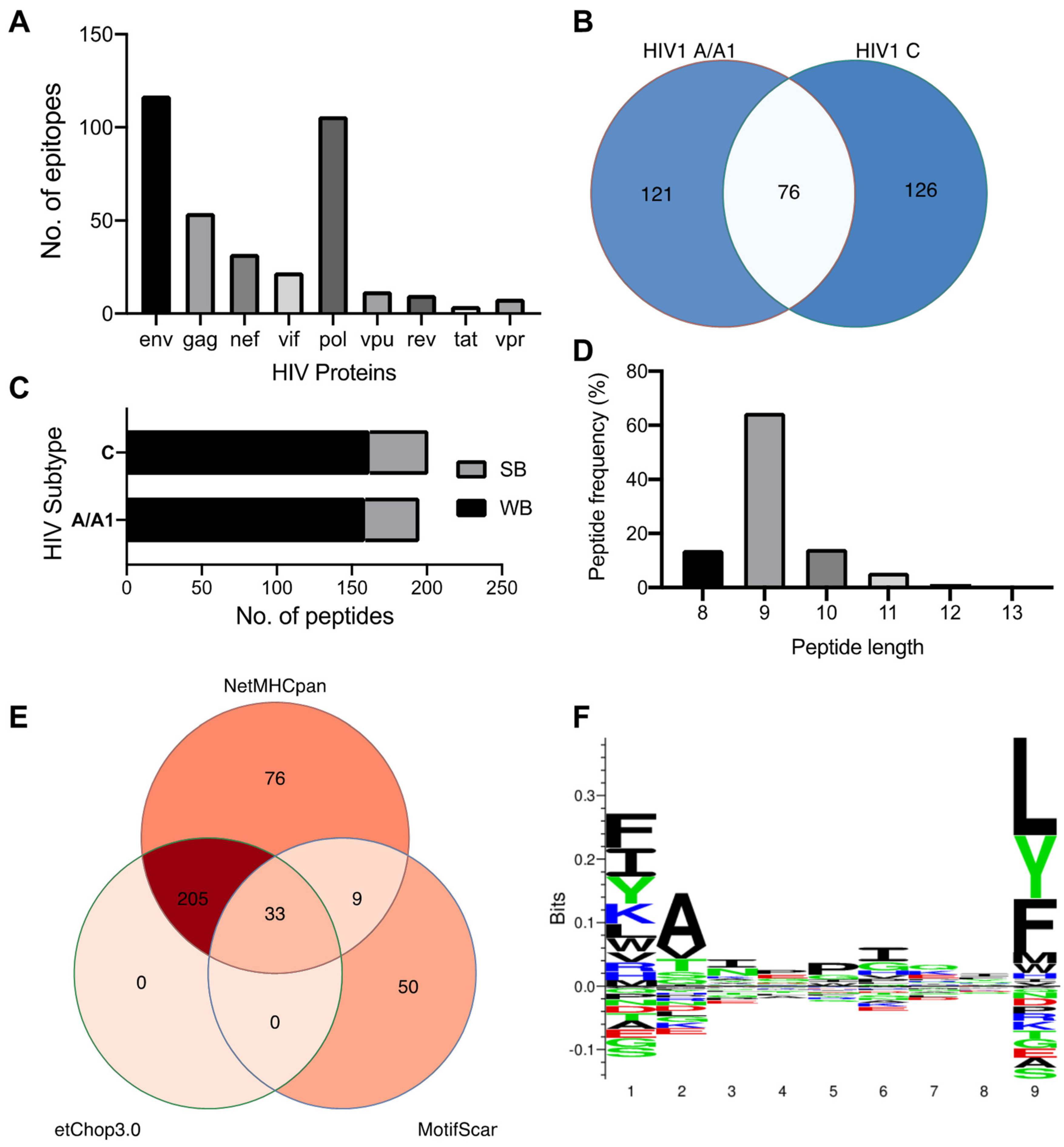
2.1. HIV-1 Clades C and A Have Private and Shared HLA-C*03:02-Epitopes and Preferentially Accommodate Hydrophobic Residues in the Distal Pocket
2.2. C*03:02-Restricted Stable Epitopes Are Mainly Derived from Structural Proteins of HIV
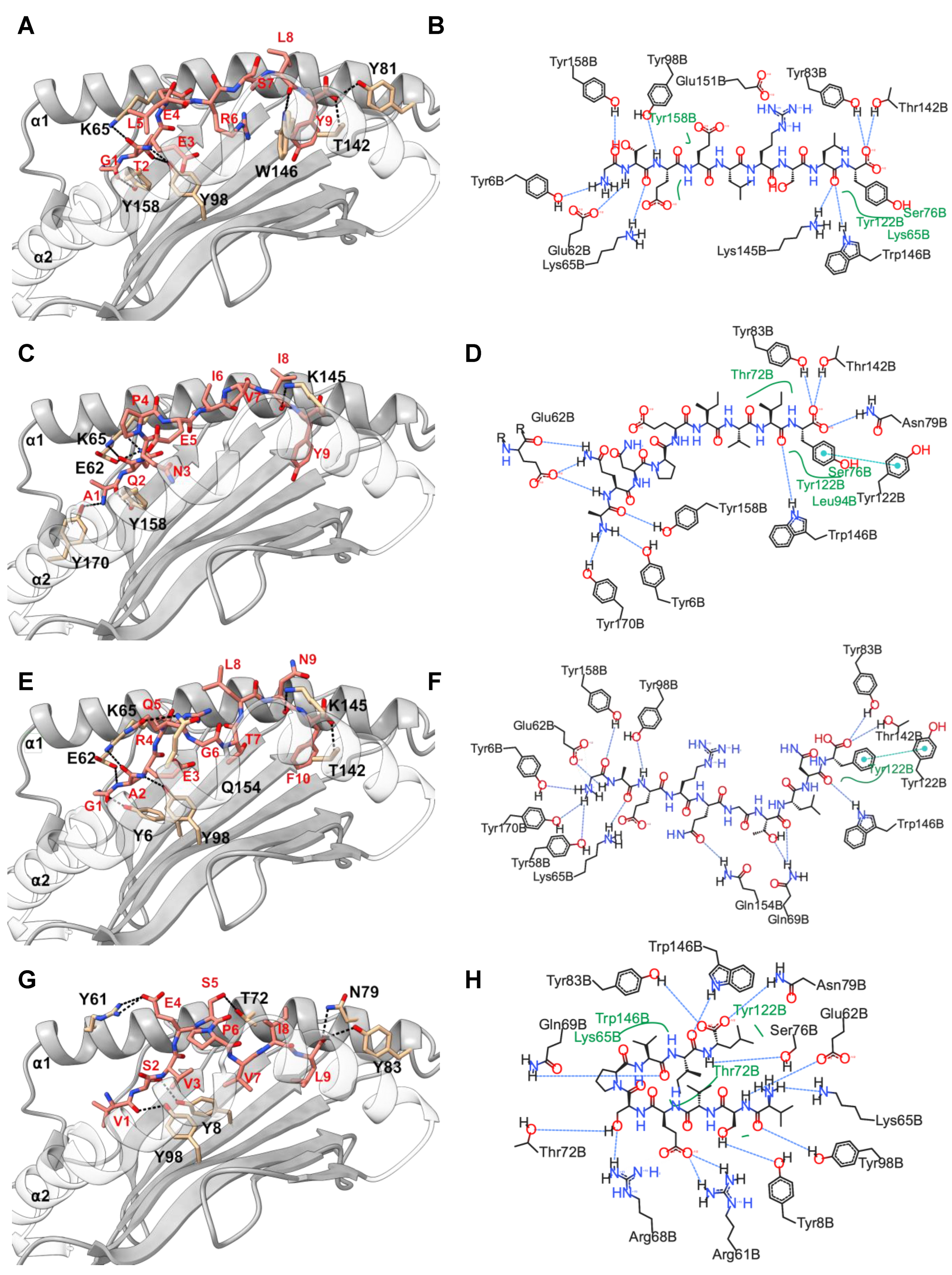
2.3. Positions 62, 142, and 151 in HLA-C*03:02 and P6 in the GY9 Epitope Provide the Structural Basis for the Preferential Binding of GY9
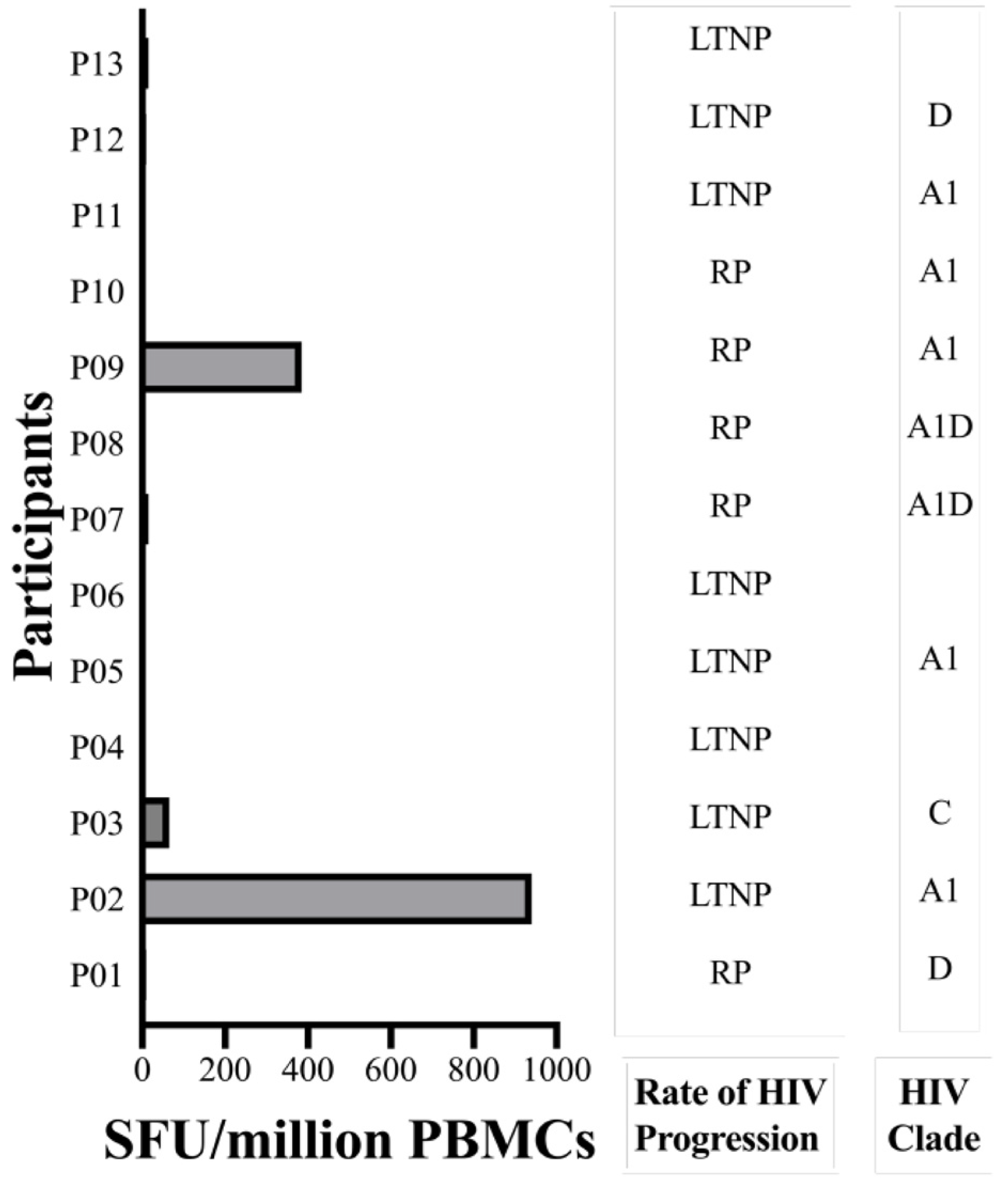
2.4. The GY9 Epitope Elicits a Clade-Specific HLA-C*03:02 IFN-γ Response
3. Discussion
Limitations of the Study
4. Materials and Methods
4.1. Patient Recruitment
4.2. HLA-C*03:02 Homology Modeling and Validation
4.3. HIV-1 Ligand Prediction and Preparation for Docking
4.4. Molecular Docking Protocol and Analysis
4.5. Molecular Dynamics Simulation Protocol and Analysis
4.6. HIV-1 Epitope Conservancy Analysis
4.7. HIV-1 Genotyping
4.8. HIV-1-Specific IFN-γ and IL-2 Dual ELISpot Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McLaren, P.J.; Carrington, M. The Impact of Host Genetic Variation on Infection with HIV-1. Nat. Immunol. 2015, 16, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Cohen, O.J.; Paolucci, S.; Bende, S.M.; Daucher, M.; Moriuchi, H.; Moriuchi, M.; Cicala, C.; Davey, R.T.; Baird, B.; Fauci, A.S. CXCR4 and CCR5 Genetic Polymorphisms in Long-Term Nonprogressive Human Immunodeficiency Virus Infection: Lack of Association with Mutations Other than CCR5-Δ32. J. Virol. 1998, 72, 6215–6217. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.D.; Pereyra, F.; Jia, X.; McLaren, P.J.; Telenti, A.; de Bakker, P.I.W.; De Bakker, P.I.W.; Walker, B.D. The Major Genetic Determinants of HIV-1 Control Affect HLA Class I Peptide Presentation. Science 2010, 330, 1551–1557. [Google Scholar] [CrossRef]
- Boudreau, J.E.; Hsu, K.C. Natural Killer Cell Education in Human Health and Disease. Curr. Opin. Immunol. 2018, 50, 102–111. [Google Scholar] [CrossRef]
- Kaseke, C.; Tano-Menka, R.; Senjobe, F.; Gaiha, G.D. The Emerging Role for CTL Epitope Specificity in HIV Cure Efforts. J. Infect. Dis. 2021, 223, S32–S37. [Google Scholar] [CrossRef]
- Pymm, P.; Tenzer, S.; Wee, E.; Weimershaus, M.; Burgevin, A.; Kollnberger, S.; Gerstoft, J.; Josephs, T.M.; Ladell, K.; McLaren, J.E.; et al. Epitope Length Variants Balance Protective Immune Responses and Viral Escape in HIV-1 Infection. Cell Rep. 2022, 38, 110449. [Google Scholar] [CrossRef] [PubMed]
- Kutsch, O.; Vey, T.; Kerkau, T.; Hünig, T.; Schimpl, A. HIV Type 1 Abrogates TAP-Mediated Transport of Antigenic Peptides Presented by MHC Class I. AIDS Res. Hum. Retroviruses 2002, 18, 1319–1325. [Google Scholar] [CrossRef]
- Li, L.; Bouvier, M. Structures of HLA-A*1101 Complexed with Immunodominant Nonamer and Decamer HIV-1 Epitopes Clearly Reveal the Presence of a Middle, Secondary Anchor Residue. J. Immunol. 2004, 172, 6175–6184. [Google Scholar] [CrossRef]
- Carlson, J.M.; Brumme, C.J.; Martin, E.; Listgarten, J.; Brockman, M.A.; Le, A.Q.; Chui, C.K.S.; Cotton, L.A.; Knapp, D.J.H.F.; Riddler, S.A.; et al. Correlates of Protective Cellular Immunity Revealed by Analysis of Population-Level Immune Escape Pathways in HIV-1. J. Virol. 2012, 86, 13202–13216. [Google Scholar] [CrossRef]
- Stewart-Jones, G.B.E.; Gillespie, G.; Overton, I.M.; Kaul, R.; Roche, P.; McMichael, A.J.; Rowland-Jones, S.; Jones, E.Y. Structures of Three HIV-1 HLA-B*5703-Peptide Complexes and Identification of Related HLAs Potentially Associated with Long-Term Nonprogression. J. Immunol. 2005, 175, 2459–2468. [Google Scholar] [CrossRef]
- Kyobe, S.; Mwesigwa, S.; Kisitu, G.P.; Farirai, J.; Katagirya, E.; Mirembe, A.N.; Ketumile, L.; Wayengera, M.; Katabazi, F.A.; Kigozi, E.; et al. Exome Sequencing Reveals a Putative Role for HLA-C*03:02 in Control of HIV-1 in African Pediatric Populations. Front. Genet. 2021, 12, 720213. [Google Scholar] [CrossRef] [PubMed]
- Leslie, A.; Matthews, P.C.; Listgarten, J.; Carlson, J.M.; Kadie, C.; Ndung’U, T.; Brander, C.; Coovadia, H.; Walker, B.D.; Heckerman, D.; et al. Additive Contribution of HLA Class I Alleles in the Immune Control of HIV-1 Infection. J. Virol. 2010, 84, 9879–9888. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Galarza, F.F.; McCabe, A.; dos Santos, E.J.M.; Jones, J.; Takeshita, L.; Ortega-Rivera, N.D.; Del Cid-Pavon, G.M.; Ramsbottom, K.; Ghattaoraya, G.; Alfirevic, A.; et al. Allele Frequency Net Database (AFND) 2020 Update: Gold-Standard Data Classification, Open Access Genotype Data and New Query Tools. Nucleic Acids Res. 2020, 48, D783–D788. [Google Scholar] [CrossRef] [PubMed]
- Gijsbers, E.F.; Anton Feenstra, K.; van Nuenen, A.C.; Navis, M.; Heringa, J.; Schuitemaker, H.; Kootstra, N.A. HIV-1 Replication Fitness of HLA-B*57/58:01 CTL Escape Variants Is Restored by the Accumulation of Compensatory Mutations in Gag. PLoS ONE 2013, 8, e81235. [Google Scholar] [CrossRef] [PubMed]
- Borghans, J.A.M.; Mølgaard, A.; de Boer, R.J.; Keşmir, C. HLA Alleles Associated with Slow Progression to AIDS Truly Prefer to Present HIV-1 p24. PLoS ONE 2007, 2, e920. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Piechocka-Trocha, A.; Miura, T.; Brockman, M.A.; Julg, B.D.; Baker, B.M.; Rothchild, A.C.; Block, B.L.; Schneidewind, A.; Koibuchi, T.; et al. Differential Neutralization of Human Immunodeficiency Virus (HIV) Replication in Autologous CD4 T Cells by HIV-Specific Cytotoxic T Lymphocytes. J. Virol. 2009, 83, 3138–3149. [Google Scholar] [CrossRef]
- Murakoshi, H.; Zou, C.; Kuse, N.; Akahoshi, T.; Chikata, T.; Gatanaga, H.; Oka, S.; Hanke, T.; Takiguchi, M. CD8+ T Cells Specific for Conserved, Cross-Reactive Gag Epitopes with Strong Ability to Suppress HIV-1 Replication. Retrovirology 2018, 15, 46. [Google Scholar] [CrossRef]
- Adland, E.; Carlson, J.M.; Paioni, P.; Kløverpris, H.; Shapiro, R.; Ogwu, A.; Riddell, L.; Luzzi, G.; Chen, F.; Balachandran, T.; et al. Nef-Specific CD8+ T Cell Responses Contribute to HIV-1 Immune Control. PLoS ONE 2013, 8, e73117. [Google Scholar] [CrossRef]
- Altfeld, M.; Addo, M.M.; Eldridge, R.L.; Yu, X.G.; Thomas, S.; Khatri, A.; Strick, D.; Phillips, M.N.; Cohen, G.B.; Islam, S.A.; et al. Vpr Is Preferentially Targeted by CTL During HIV-1 Infection. J. Immunol. 2001, 167, 2743–2752. [Google Scholar] [CrossRef]
- Ferrando-Martínez, S.; Casazza, J.P.; Leal, M.; Machmach, K.; Muñoz-Fernández, M.; Viciana, P.; Koup, R.A.; Ruiz-Mateos, E. Differential Gag-Specific Polyfunctional T Cell Maturation Patterns in HIV-1 Elite Controllers. J. Virol. 2012, 86, 3667–3674. [Google Scholar] [CrossRef]
- Falivene, J.; Ghiglione, Y.; Laufer, N.; Socías, M.E.; Holgado, M.P.; Ruiz, M.J.; Maeto, C.; Figueroa, M.I.; Giavedoni, L.D.; Cahn, P.; et al. Th17 and Th17/Treg Ratio at Early HIV Infection Associate with Protective HIV-Specific CD8+ T-Cell Responses and Disease Progression. Sci. Rep. 2015, 5, 11511. [Google Scholar] [CrossRef]
- Boulet, S.; Song, R.; Kamya, P.; Bruneau, J.; Shoukry, N.H.; Tsoukas, C.M.; Bernard, N.F. HIV Protective KIR3DL1 and HLA-B Genotypes Influence NK Cell Function Following Stimulation with HLA-Devoid Cells. J. Immunol. 2010, 184, 2057–2064. [Google Scholar] [CrossRef] [PubMed]
- Zappacosta, F.; Borrego, F.; Brooks, A.G.; Parker, K.C.; Coligan, J.E. Peptides Isolated from HLA-Cw*0304 Confer Different Degrees of Protection from Natural Killer Cell-Mediated Lysis. Proc. Natl. Acad. Sci. USA 1997, 94, 6313–6318. [Google Scholar] [CrossRef] [PubMed]
- del Moral-Sánchez, I.; Sliepen, K. Strategies for Inducing Effective Neutralizing Antibody Responses against HIV-1. Expert Rev. Vaccines 2019, 18, 1127–1143. [Google Scholar] [CrossRef] [PubMed]
- Arnold, G.F.; Velasco, P.K.; Holmes, A.K.; Wrin, T.; Geisler, S.C.; Phung, P.; Tian, Y.; Resnick, D.A.; Ma, X.; Mariano, T.M.; et al. Broad Neutralization of Human Immunodeficiency Virus Type 1 (HIV-1) Elicited from Human Rhinoviruses That Display the HIV-1 gp41 ELDKWA Epitope. J. Virol. 2009, 83, 5087–5100. [Google Scholar] [CrossRef] [PubMed]
- Payne, R.P.; Branch, S.; Kløverpris, H.; Matthews, P.C.; Koofhethile, C.K.; Strong, T.; Adland, E.; Leitman, E.; Frater, J.; Ndung’U, T.; et al. Differential Escape Patterns within the Dominant HLA-B*57:03-Restricted HIV Gag Epitope Reflect Distinct Clade-Specific Functional Constraints. J. Virol. 2014, 88, 4668–4678. [Google Scholar] [CrossRef] [PubMed]
- Brackenridge, S.; Evans, E.J.; Toebes, M.; Goonetilleke, N.; Liu, M.K.P.; di Gleria, K.; Schumacher, T.N.; Davis, S.J.; McMichael, A.J.; Gillespie, G.M. An Early HIV Mutation within an HLA-B*57-Restricted T Cell Epitope Abrogates Binding to the Killer Inhibitory Receptor 3DL1. J. Virol. 2011, 85, 5415–5422. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sun, W.; Guo, J.; Zhao, G.; Sun, S.; Yu, H.; Guo, Y.; Li, J.; Jin, X.; Du, L.; et al. In Silico Design of a DNA-Based HIV-1 Multi-Epitope Vaccine for Chinese Populations. Hum. Vaccines Immunother. 2015, 11, 795–805. [Google Scholar] [CrossRef]
- Haynes, B.F.; McElrath, M.J. Progress in HIV-1 Vaccine Development. Curr. Opin. HIV AIDS 2013, 8, 326–332. [Google Scholar] [CrossRef]
- Song, H.; Giorgi, E.E.; Ganusov, V.V.; Cai, F.; Athreya, G.; Yoon, H.; Carja, O.; Hora, B.; Hraber, P.; Romero-Severson, E.; et al. Tracking HIV-1 Recombination to Resolve Its Contribution to HIV-1 Evolution in Natural Infection. Nat. Commun. 2018, 9, 1928. [Google Scholar] [CrossRef]
- Hraber, P.; Korber, B.T.; Lapedes, A.S.; Bailer, R.T.; Seaman, M.S.; Gao, H.; Greene, K.M.; McCutchan, F.; Williamson, C.; Kim, J.H.; et al. Impact of Clade, Geography, and Age of the Epidemic on HIV-1 Neutralization by Antibodies. J. Virol. 2014, 88, 12623–12643. [Google Scholar] [CrossRef]
- Chikata, T.; Paes, W.; Akahoshi, T.; Partridge, T.; Murakoshi, H.; Gatanaga, H.; Ternette, N.; Oka, S.; Borrow, P.; Takiguchi, M. Identification of Immunodominant HIV-1 Epitopes Presented by HLA-C*12:02, a Protective Allele, Using an Immunopeptidomics Approach. J. Virol. 2019, 93, e00634-19. [Google Scholar] [CrossRef]
- Bugembe, D.L.; Ekii, A.O.; Ndembi, N.; Serwanga, J.; Kaleebu, P.; Pala, P. Computational MHC-I Epitope Predictor Identifies 95% of Experimentally Mapped HIV-1 Clade A and D Epitopes in a Ugandan Cohort. BMC Infect. Dis. 2020, 20, 172. [Google Scholar] [CrossRef]
- Hsin, J.; Arkhipov, A.; Yin, Y.; Stone, J.E.; Schulten, K. Using VMD: An Introductory Tutorial. Curr. Protoc. Bioinform. 2008, 24, 5.7.1–5.7.48. [Google Scholar] [CrossRef]
- Knapp, B.; van der Merwe, P.A.; Dushek, O.; Deane, C.M. MHC Binding Affects the Dynamics of Different T-Cell Receptors in Different Ways. PLoS Comput. Biol. 2019, 15, e1007338. [Google Scholar] [CrossRef]
- Li, X.; Singh, N.K.; Collins, D.R.; Ng, R.; Zhang, A.; Lamothe-Molina, P.A.; Shahinian, P.; Xu, S.; Tan, K.; Piechocka-Trocha, A.; et al. Molecular Basis of Differential HLA Class I-Restricted T Cell Recognition of a Highly Networked HIV Peptide. Nat. Commun. 2023, 14, 2929. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. Gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef] [PubMed]
- Ndongala, M.L.; Kamya, P.; Boulet, S.; Peretz, Y.; Rouleau, D.; Tremblay, C.; Leblanc, R.; Côté, P.; Baril, J.; Thomas, R.; et al. Changes in Function of HIV-Specific T-Cell Responses with Increasing Time from Infection. Viral Immunol. 2010, 23, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Excler, J.-L.; Robb, M.L.; Kim, J.H. Prospects for a Globally Effective HIV-1 Vaccine. Vaccine 2015, 33, D4–D12. [Google Scholar] [CrossRef] [PubMed]
- Cohen, G.B.; Gandhi, R.T.; Davis, D.M.; Mandelboim, O.; Chen, B.K.; Strominger, J.L.; Baltimore, D. The Selective Downregulation of Class I Major Histocompatibility Complex Proteins by HIV-1 Protects HIV-Infected Cells from NK Cells. Immunity 1999, 10, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Mann, J.K.; Byakwaga, H.; Kuang, X.T.; Le, A.Q.; Brumme, C.J.; Mwimanzi, P.; Omarjee, S.; Martin, E.; Lee, G.Q.; Baraki, B.; et al. Ability of HIV-1 Nef to Downregulate CD4 and HLA Class I Differs among Viral Subtypes. Retrovirology 2013, 10, 100. [Google Scholar] [CrossRef]
- Fellay, J.; Shianna, K.V.; Ge, D.; Colombo, S.; Ledergerber, B.; Weale, M.; Zhang, K.; Gumbs, C.; Castagna, A.; Cossarizza, A.; et al. A Whole-Genome Association Study of Major Determinants for Host Control of HIV-1. Science 2007, 317, 944–947. [Google Scholar] [CrossRef]
- Kulkarni, S.; Savan, R.; Qi, Y.; Gao, X.; Yuki, Y.; Bass, S.E.; Martin, M.P.; Hunt, P.; Deeks, S.G.; Telenti, A.; et al. Differential microRNA Regulation of HLA-C Expression and Its Association with HIV Control. Nature 2011, 472, 495–498. [Google Scholar] [CrossRef]
- Thomas, R.; Apps, R.; Qi, Y.; Gao, X.; Male, V.; O’Huigin, C.; O’Connor, G.; Ge, D.; Fellay, J.; Martin, J.N.; et al. HLA-C cell Surface Expression and Control of HIV/AIDS Correlate with a Variant Upstream of HLA-C. Nat. Genet. 2009, 41, 1290–1294. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W. T Cell-Based Strategies for HIV-1 Vaccines. Hum. Vaccines Immunother. 2020, 16, 713–722. [Google Scholar] [CrossRef]
- Shin, S.Y. Recent Update in HIV Vaccine Development. Clin. Exp. Vaccine Res. 2016, 5, 6–11. [Google Scholar] [CrossRef]
- Zipeto, D.; Beretta, A. HLA-C and HIV-1: Friends or Foes? Retrovirology 2012, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Buranapraditkun, S.; Hempel, U.; Pitakpolrat, P.; Allgaier, R.L.; Thantivorasit, P.; Lorenzen, S.-I.; Sirivichayakul, S.; Hildebrand, W.H.; Altfeld, M.; Brander, C.; et al. A Novel Immunodominant CD8+ T Cell Response Restricted by a Common HLA-C Allele Targets a Conserved Region of Gag HIV-1 Clade CRF01_AE Infected Thais. PLoS ONE 2011, 6, e23603. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Akahoshi, T.; Murakoshi, H.; Ishizuka, N.; Oka, S.; Takiguchi, M. CTL Recognition of HIV-1-Infected Cells Via Cross-Recognition of Multiple Overlapping Peptides from a Single 11-Mer Pol Sequence. Eur. J. Immunol. 2012, 42, 2621–2631. [Google Scholar] [CrossRef]
- Perdomo-Celis, F.; Taborda, N.A.; Rugeles, M.T. CD8+ T-Cell Response to HIV Infection in the Era of Antiretroviral Therapy. Front. Immunol. 2019, 10, 1896. [Google Scholar] [CrossRef]
- Addo, M.M.; Yu, X.G.; Rathod, A.; Cohen, D.; Eldridge, R.L.; Strick, D.; Johnston, M.N.; Corcoran, C.; Wurcel, A.G.; Fitzpatrick, C.A.; et al. Comprehensive Epitope Analysis of Human Immunodeficiency Virus Type 1 (HIV-1)-Specific T-Cell Responses Directed against the Entire Expressed HIV-1 Genome Demonstrate Broadly Directed Responses, but No Correlation to Viral Load. J. Virol. 2003, 77, 2081–2092. [Google Scholar] [CrossRef]
- Rosás-Umbert, M.; Gunst, J.D.; Pahus, M.H.; Olesen, R.; Schleimann, M.; Denton, P.W.; Ramos, V.; Ward, A.; Kinloch, N.N.; Copertino, D.C.; et al. Administration of Broadly Neutralizing Anti-HIV-1 Antibodies at ART Initiation Maintains Long-Term CD8+ T Cell Immunity. Nat. Commun. 2022, 13, 6473. [Google Scholar] [CrossRef]
- Joglekar, A.V.; Liu, Z.; Weber, J.K.; Ouyang, Y.; Jeppson, J.D.; Noh, W.J.; Lamothe-Molina, P.A.; Chen, H.; Kang, S.-G.; Bethune, M.T.; et al. T Cell Receptors for the HIV KK10 Epitope from Patients with Differential Immunologic Control Are Functionally Indistinguishable. Proc. Natl. Acad. Sci. USA 2018, 115, 1877–1882. [Google Scholar] [CrossRef]
- Tassiopoulos, K.; Landay, A.; Collier, A.C.; Connick, E.; Deeks, S.G.; Hunt, P.; Lewis, D.E.; Wilson, C.; Bosch, R. CD28-Negative CD4+ and CD8+ T Cells in Antiretroviral Therapy–Naive HIV-Infected Adults Enrolled in Adult Clinical Trials Group Studies. J. Infect. Dis. 2012, 205, 1730–1738. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, M.C.; Nelde, A.; Weber, J.K.; Schreitmüller, C.M.; Martrus, G.; Huynh, T.; Bunders, M.J.; Lunemann, S.; Stevanovic, S.; Zhou, R.; et al. HIV-1 Induced Changes in HLA-C∗03: 04-Presented Peptide Repertoires Lead to Reduced Engagement of Inhibitory Natural Killer Cell Receptors. AIDS 2020, 34, 1713–1723. [Google Scholar] [CrossRef] [PubMed]
- Knapp, B.; Ospina, L.; Deane, C.M. Avoiding False Positive Conclusions in Molecular Simulation: The Importance of Replicas. J. Chem. Theory Comput. 2018, 14, 6127–6138. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Knapp, B.; Wright, D.W.; Deane, C.M.; Coveney, P.V. Rapid, Precise, and Reproducible Prediction of Peptide–MHC Binding Affinities from Molecular Dynamics That Correlate Well with Experiment. J. Chem. Theory Comput. 2015, 11, 3346–3356. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, M.; Destiarani, W.; Widayat, W.; Yosua, Y.; Gumilar, G.; Tanudireja, A.S.; Rohmatulloh, F.G.; Maulana, F.A.; Baroroh, U.; Hardianto, A.; et al. Coarse-Grained Molecular Dynamics-Guided Immunoinformatics to Explain the Binder and Non-Binder Classification of Cytotoxic T-Cell Epitope for SARS-CoV-2 Peptide-Based Vaccine Discovery. PLoS ONE 2023, 18, e0292156. [Google Scholar] [CrossRef] [PubMed]
- Koita, O.A.; Dabitao, D.; Mahamadou, I.; Tall, M.; Dao, S.; Tounkara, A.; Guiteye, H.; Noumsi, C.; Thiero, O.; Kone, M.; et al. Confirmation of Immunogenic Consensus Sequence HIV-1 T-cell Epitopes in Bamako, Mali and Providence, Rhode Island. Hum. Vaccines 2006, 2, 119–128. [Google Scholar] [CrossRef]
- Goepfert, P.A.; Bansal, A.; Edwards, B.H.; Ritter, G.D.; Tellez, I.; McPherson, S.A.; Sabbaj, S.; Mulligan, M.J. A Significant Number of Human Immunodeficiency Virus Epitope-Specific Cytotoxic T Lymphocytes Detected by Tetramer Binding Do Not Produce Gamma Interferon. J. Virol. 2000, 74, 10249–10255. [Google Scholar] [CrossRef]
- Retshabile, G.; Mlotshwa, B.C.; Williams, L.; Mwesigwa, S.; Mboowa, G.; Huang, Z.; Rustagi, N.; Swaminathan, S.; Katagirya, E.; Kyobe, S.; et al. Whole-Exome Sequencing Reveals Uncaptured Variation and Distinct Ancestry in the Southern African Population of Botswana. Am. J. Hum. Genet. 2018, 102, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Bbosa, N.; Kaleebu, P.; Ssemwanga, D. HIV Subtype Diversity Worldwide. Curr. Opin. HIV AIDS 2019, 14, 153–160. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera? A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Antunes, D.A.; Moll, M.; Devaurs, D.; Jackson, K.R.; Lizée, G.; Kavraki, L.E. DINC 2.0: A New Protein–Peptide Docking Webserver Using an Incremental Approach. Cancer Res 2017, 77, e55–e57. [Google Scholar] [CrossRef]
- El-Hachem, N.; Haibe-Kains, B.; Khalil, A.; Kobeissy, F.H.; Nemer, G. AutoDock and AutoDockTools for Protein-Ligand Docking: Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1(BACE1) as a Case Study. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2017; pp. 391–403. [Google Scholar]
- Kadukova, M.; Grudinin, S. Convex-PL: A Novel Knowledge-Based Potential for Protein-Ligand Interactions Deduced from Structural Databases Using Convex Optimization. J. Comput. Aided. Mol. Des. 2017, 31, 943–958. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field: A Force Field for Drug-like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Miah, M.M.; Tabassum, N.; Afroj Zinnia, M.; Islam, A.B.M.M.K. Drug and Anti-Viral Peptide Design to Inhibit the Monkeypox Virus by Restricting A36R Protein. Bioinform. Biol. Insights 2022, 16, 11779322221141164. [Google Scholar] [CrossRef]
- Vishnoi, S.; Bhattacharya, S.; Walsh, E.M.; Okoh, G.I.; Thompson, D. Computational Peptide Design Cotargeting Glucagon and Glucagon-like Peptide-1 Receptors. J. Chem. Inf. Model. 2023, 63, 4934–4947. [Google Scholar] [CrossRef] [PubMed]
- Lapointe, H.R.; Dong, W.; Lee, G.Q.; Bangsberg, D.R.; Martin, J.N.; Mocello, A.R.; Boum, Y.; Karakas, A.; Kirkby, D.; Poon, A.F.Y.; et al. HIV Drug Resistance Testing by High-Multiplex “Wide” Sequencing on the MiSeq Instrument. Antimicrob. Agents Chemother. 2015, 59, 6824–6833. [Google Scholar] [CrossRef] [PubMed]
- Tachbele, E.; Kyobe, S.; Katabazi, F.A.; Kigozi, E.; Mwesigwa, S.; Joloba, M.; Messele, A.; Amogne, W.; Legesse, M.; Pieper, R.; et al. Genetic Diversity and Acquired Drug Resistance Mutations Detected by Deep Sequencing in Virologic Failures among Antiretroviral Treatment Experienced Human Immunodeficiency Virus-1 Patients in a Pastoralist Region of Ethiopia. Infect. Drug Resist. 2021, 14, 4833–4847. [Google Scholar] [CrossRef] [PubMed]
- Pineda-Peña, A.-C.; Faria, N.R.; Imbrechts, S.; Libin, P.; Abecasis, A.B.; Deforche, K.; Gómez-López, A.; Camacho, R.J.; de Oliveira, T.; Vandamme, A.-M. Automated Subtyping of HIV-1 Genetic Sequences for Clinical and Surveillance Purposes: Performance Evaluation of the New REGA Version 3 and Seven Other Tools. Infect. Genet. Evol. 2013, 19, 337–348. [Google Scholar] [CrossRef]

| HIV-1 Clade | Protein and Position | Peptide Sequence | No. of H Bonds | RMSD a | Binding Energy (kcal/mol) | Convex-PL Score | Amino Acids Involved in H Bond Interaction | |
|---|---|---|---|---|---|---|---|---|
| Structural | ||||||||
| C/A1 | Gag71 * | GTEELRSLY | GY9 | 7 | 0.811 | –8.6 | 7.23 | Lys65, Tyr83, Tyr98, Thr142, Trp146 and Tyr158 |
| A1 | Env58 | KAYETEMHN | KN9 | 11 | 0.202 | –7.4 | 7.36 | Tyr6, Arg61, Glu62, Lys65, Tyr66, Ser76, Glu151 and Tyr158 |
| C | Pol43 * | GAERQGTLNF | GF10 | 7 | 1.165 | –8.7 | 7.22 | Tyr6, Glu62, Lys65, Tyr98, Thr142, Lys145 and Gln154 |
| C | Pol324 * | AQNPEIVIY | AY9 | 6 | 1.547 | –7.7 | 7.16 | Glu62, Lys65, Lys145, Tyr158 and Tyr170 |
| Regulatory | ||||||||
| C/A1 | Nef84 | GAFDLSFFL | GL9 | 7 | 1.030 | –9.1 | 7.24 | Tyr8, Glu62, Lys65, Thr72, Tyr98, Lys145 and Tyr158 |
| C/A1 | Nef114 | WVYNTQGYF | WF9 | 7 | 0.873 | –9.7 | 7.53 | Tyr6, Arg68, Tyr98, Thr142, Lys145, Trp146 and Glu151 |
| A1 | Vif128 | VVSPRCEY | VY8 | 8 | 1.473 | –8.3 | 7.10 | Gln69, Thr72, Ser76, Tyr83, Tyr98, Thr142, Ly145 and Glu151 |
| A1 | Rev109 * | VSVESPVIL | VL9 | 7 | 1.638 | –8.0 | 7.27 | Tyr8, Arg61, Thr72, Asn79, Tyr83 and Tyr98 |
| HIV-1 Subtype | Peptide Sequence | Energy Components (kcal/mol) | ||||
|---|---|---|---|---|---|---|
| Van der Waals | Electrostatics | Polar Solvation | ΔG Binding Energy | |||
| Structural | ||||||
| C/A1 | GTEELRSLY | GY9 | −53.01 | −547.44 | 521.66 | −88.41 |
| C | GAERQGTLNF | GF10 | −53.45 | −335.96 | 347.44 | −50.59 |
| C | AQNPEIVIY | AY9 | −68.75 | −258.85 | 285.79 | −51.24 |
| Regulatory | ||||||
| A1 | VSVESPVIL | VL9 | −67.29 | −367.42 | 394.36 | −49.73 |
| HIV1 Clade a | Amino Acid Residue and Position | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| G1 | T2 | E3 | E4 | L5 | R6 | S7 | L8 | Y9 | |
| A1 | . | . | . | . | . | R/K | . | . | Y/F |
| C | . | . | . | . | . | K | . | . | Y/F/H |
| D | . | . | . | . | I | K | . | . | Y/F |
| K | . | . | . | . | I | K | . | . | Y/F |
| ΔΔG GY9 b | NA | −8.18 | −10.3 | −4.82 | −2.83 | −27.00 | −5.85 | −2.82 | −7.33 |
| Conservancy score c | 5 | 4 | 2 | 3 | 3 | 2 | 5 | 5 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kyobe, S.; Mwesigwa, S.; Nkurunungi, G.; Retshabile, G.; Egesa, M.; Katagirya, E.; Amujal, M.; Mlotshwa, B.C.; Williams, L.; Sendagire, H.; et al. Identification of a Clade-Specific HLA-C*03:02 CTL Epitope GY9 Derived from the HIV-1 p17 Matrix Protein. Int. J. Mol. Sci. 2024, 25, 9683. https://doi.org/10.3390/ijms25179683
Kyobe S, Mwesigwa S, Nkurunungi G, Retshabile G, Egesa M, Katagirya E, Amujal M, Mlotshwa BC, Williams L, Sendagire H, et al. Identification of a Clade-Specific HLA-C*03:02 CTL Epitope GY9 Derived from the HIV-1 p17 Matrix Protein. International Journal of Molecular Sciences. 2024; 25(17):9683. https://doi.org/10.3390/ijms25179683
Chicago/Turabian StyleKyobe, Samuel, Savannah Mwesigwa, Gyaviira Nkurunungi, Gaone Retshabile, Moses Egesa, Eric Katagirya, Marion Amujal, Busisiwe C. Mlotshwa, Lesedi Williams, Hakim Sendagire, and et al. 2024. "Identification of a Clade-Specific HLA-C*03:02 CTL Epitope GY9 Derived from the HIV-1 p17 Matrix Protein" International Journal of Molecular Sciences 25, no. 17: 9683. https://doi.org/10.3390/ijms25179683





