Acidocin A and Acidocin 8912 Belong to a Distinct Subfamily of Class II Bacteriocins with a Broad Spectrum of Antimicrobial Activity
Abstract
1. Introduction
2. Results
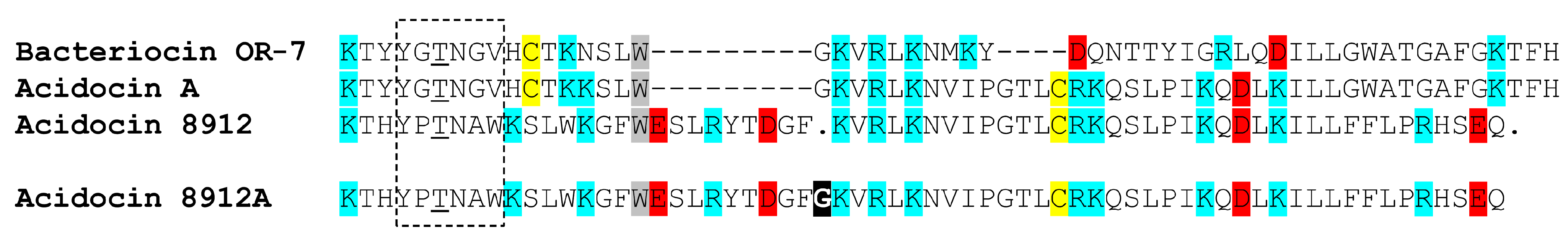
2.1. Acidocin 8912 as a Homolog of Acidocin A
2.2. Expression and Purification of the Recombinant Peptides
2.3. Secondary Structure of Acidodin 8912 and Acidocin 8912A
2.4. Antibacterial Activity
2.5. Antifungal Activity
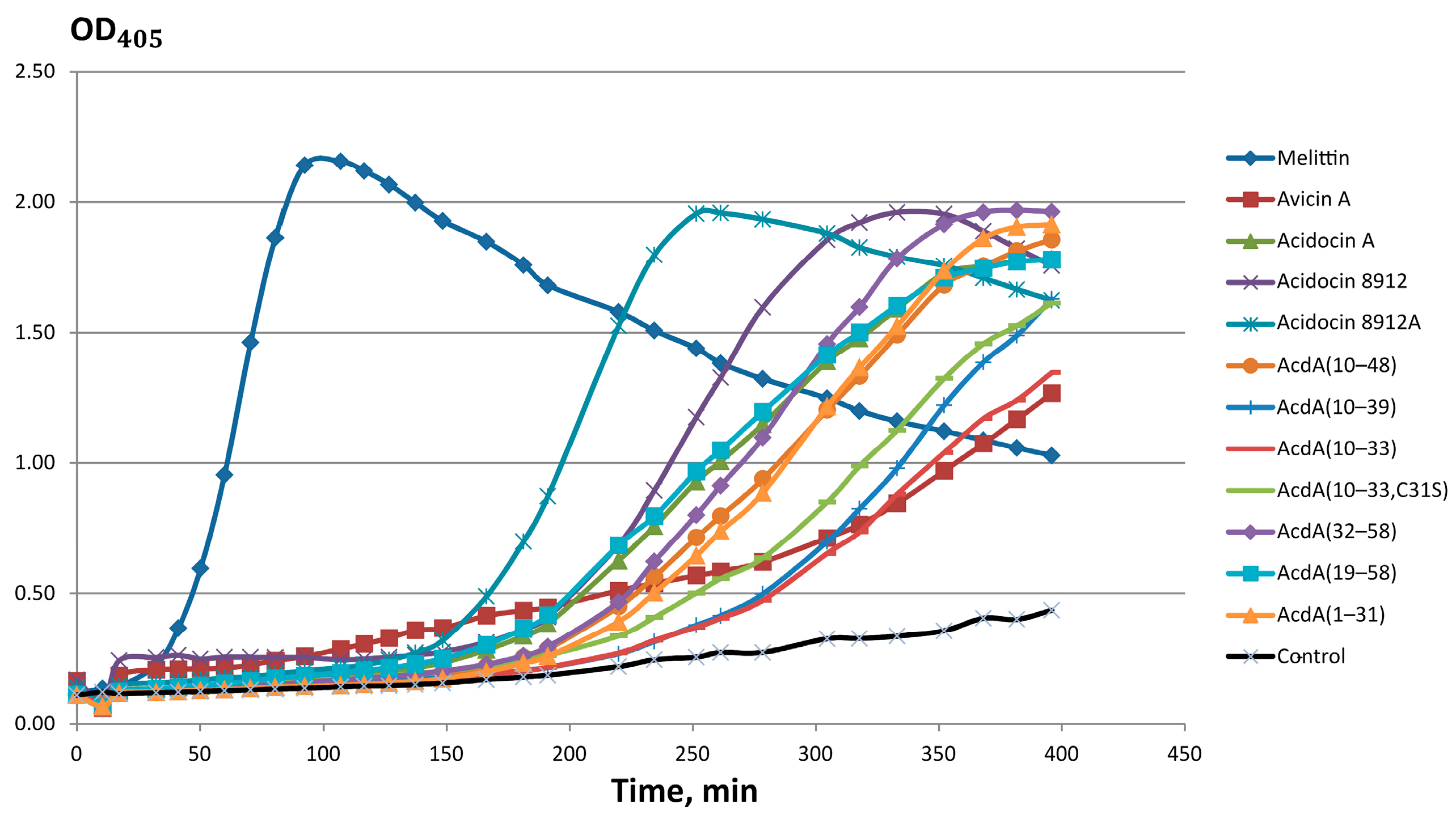
2.6. E. coli ML-35p Membranes Disrupting Activity of Acidocins 8912/8912A
2.7. C. albicans ATCC 18804 Membrane Disrupting Activity of Acidocin A
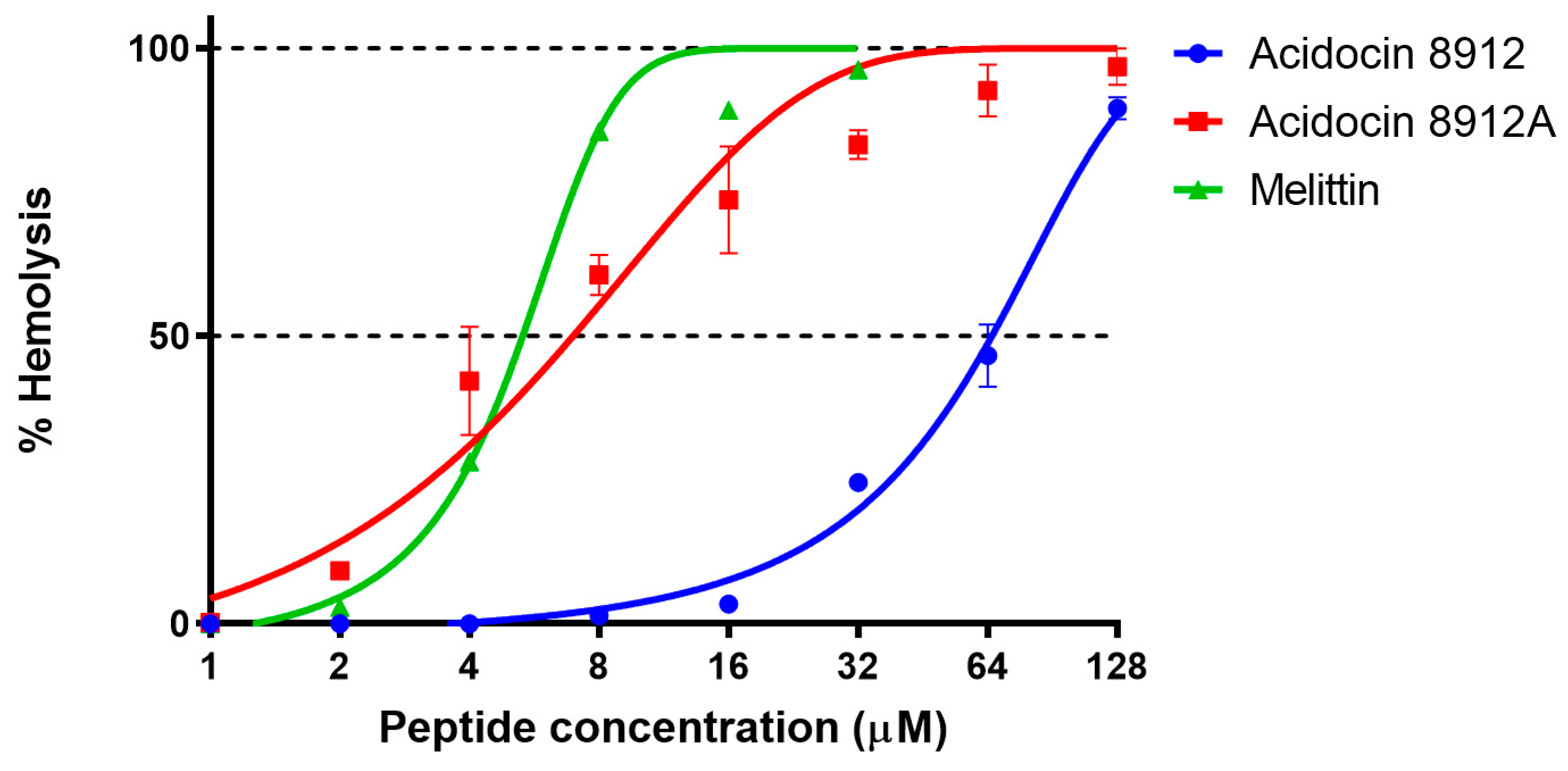
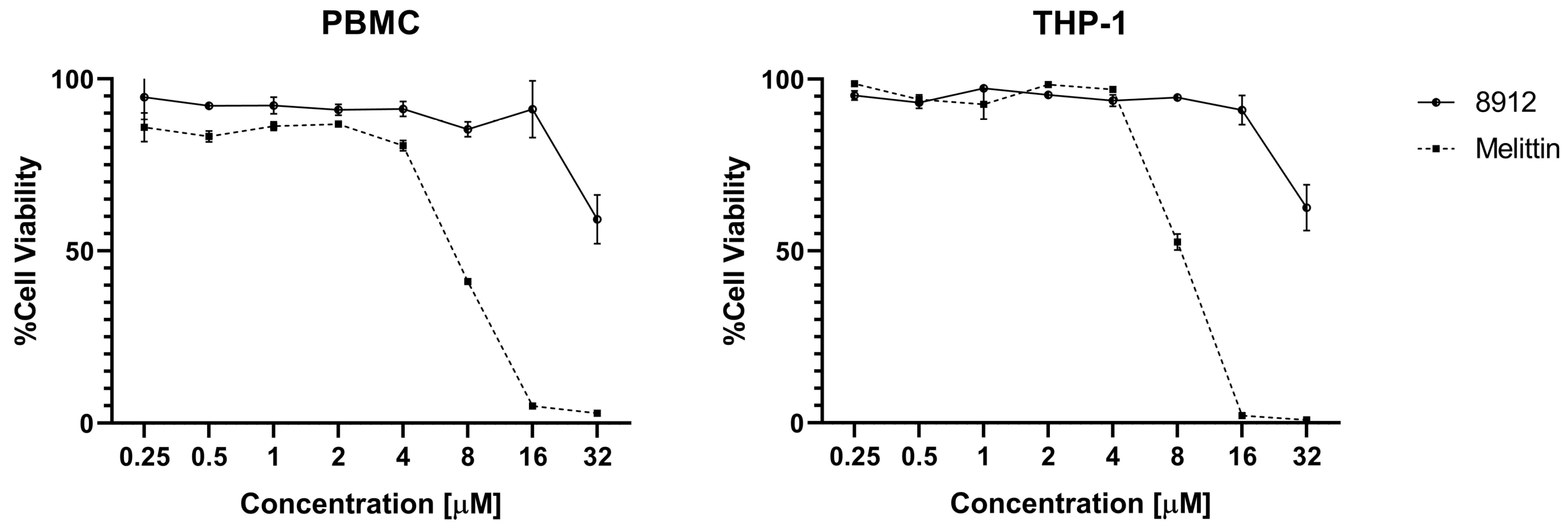
2.8. Hemolytic Activity and Cytotoxicity of Acidocins 8912/8912A
2.9. Proteolytic Stability
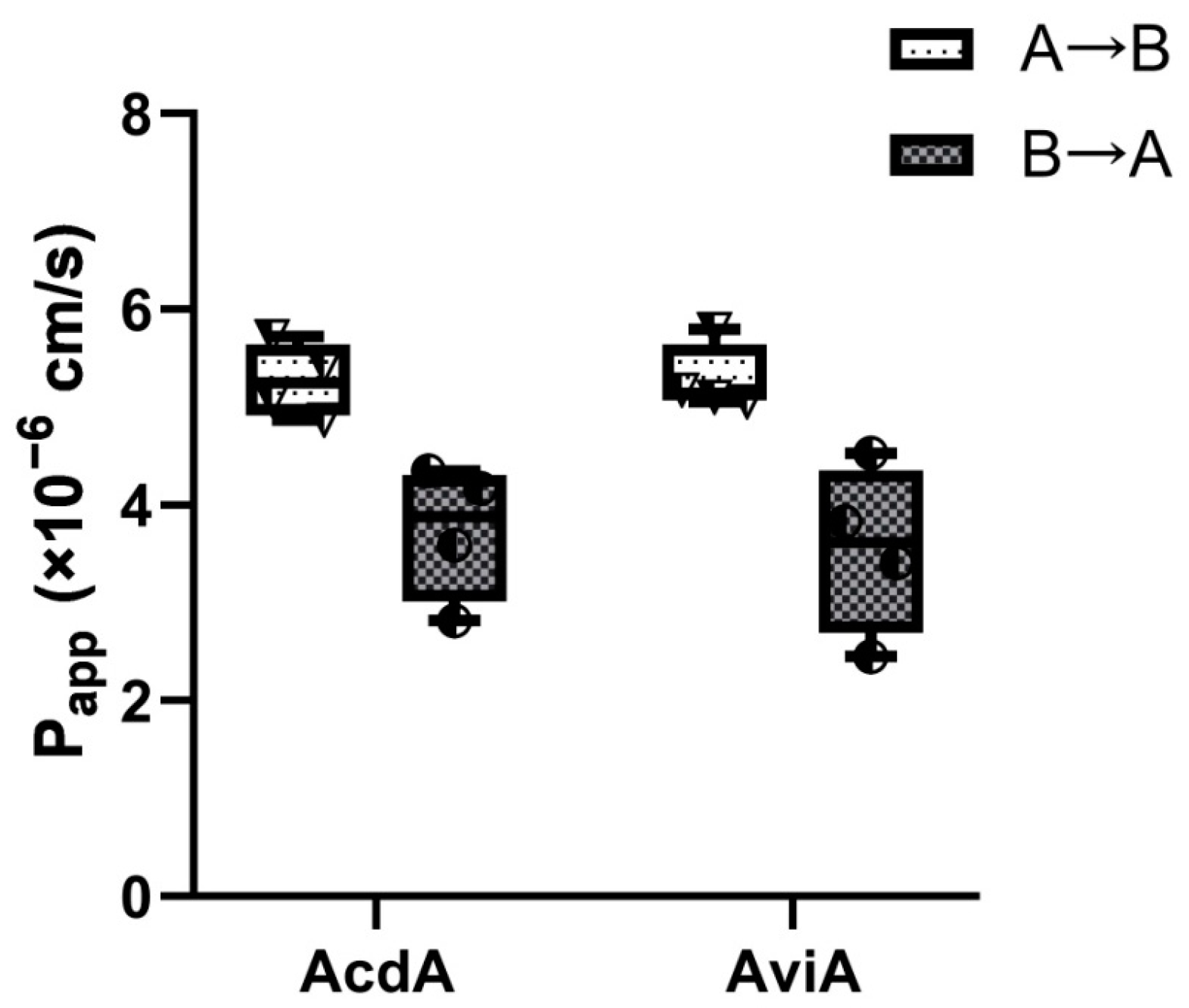
2.10. Transepithelial Transport of Acidocin A and Avicin A
3. Discussion
4. Materials and Methods
4.1. Search for Acidocin A Homologs
4.2. Antimicrobial Peptide Preparations
4.3. Circular Dichroism Spectroscopy
4.4. Antibacterial Activity Assay
4.5. Antifungal Activity Assay
4.6. Membrane Permeability Assay
4.7. Hemolysis Assay
4.8. Cytotoxicity Assay
4.9. Proteolytic Stability Assay
4.10. Caco-2 Monolayer Permeability Assay
4.11. Propidium Iodide Uptake Assay by Flow Cytometry
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prokaryotic Antimicrobial Peptides: From Genes to Applications; Drider, D., Rebuffat, S., Eds.; Springer: New York, NY, USA, 2011; ISBN 978-1-4419-7691-8. [Google Scholar]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A Viable Alternative to Antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, Antimicrobial Peptides from Bacterial Origin: Overview of Their Biology and Their Impact against Multidrug-Resistant Bacteria. Microorganisms 2020, 8, 639. [Google Scholar] [CrossRef]
- Zouhir, A.; Hammami, R.; Fliss, I.; Hamida, J.B. A New Structure-Based Classification of Gram-Positive Bacteriocins. Protein J. 2010, 29, 432–439. [Google Scholar] [CrossRef]
- Zimina, M.; Babich, O.; Prosekov, A.; Sukhikh, S.; Ivanova, S.; Shevchenko, M.; Noskova, S. Overview of Global Trends in Classification, Methods of Preparation and Application of Bacteriocins. Antibiotics 2020, 9, 553. [Google Scholar] [CrossRef]
- Klaenhammer, T.R. Genetics of Bacteriocins Produced by Lactic Acid Bacteria. FEMS Microbiol. Rev. 1993, 12, 39–85. [Google Scholar] [CrossRef]
- Du, R.; Ping, W.; Ge, J. Purification, Characterization and Mechanism of Action of Enterocin HDX-2, a Novel Class IIa Bacteriocin Produced by Enterococcus Faecium HDX-2. LWT 2022, 153, 112451. [Google Scholar] [CrossRef]
- Song, D.-F.; Zhu, M.-Y.; Gu, Q. Purification and Characterization of Plantaricin ZJ5, a New Bacteriocin Produced by Lactobacillus Plantarum ZJ5. PLoS ONE 2014, 9, e105549. [Google Scholar] [CrossRef]
- Zhao, S.; Han, J.; Bie, X.; Lu, Z.; Zhang, C.; Lv, F. Purification and Characterization of Plantaricin JLA-9: A Novel Bacteriocin against Bacillus spp. Produced by Lactobacillus Plantarum JLA-9 from Suan-Tsai, a Traditional Chinese Fermented Cabbage. J. Agric. Food Chem. 2016, 64, 2754–2764. [Google Scholar] [CrossRef]
- Singh, P.K.; Sharma, S.; Kumari, A.; Korpole, S. A Non-Pediocin Low Molecular Weight Antimicrobial Peptide Produced by Pediococcus Pentosaceus Strain IE-3 Shows Increased Activity under Reducing Environment. BMC Microbiol. 2014, 14, 226. [Google Scholar] [CrossRef]
- Jiang, Y.-H.; Xin, W.-G.; Yang, L.-Y.; Ying, J.-P.; Zhao, Z.-S.; Lin, L.-B.; Li, X.-Z.; Zhang, Q.-L. A Novel Bacteriocin against Staphylococcus Aureus from Lactobacillus Paracasei Isolated from Yunnan Traditional Fermented Yogurt: Purification, Antibacterial Characterization, and Antibiofilm Activity. J. Dairy Sci. 2022, 105, 2094–2107. [Google Scholar] [CrossRef]
- Rahman, M.S.; Choi, Y.H.; Choi, Y.S.; Yoo, J.C. Glycin-Rich Antimicrobial Peptide YD1 from B. Amyloliquefaciens, Induced Morphological Alteration in and Showed Affinity for Plasmid DNA of E. coli. AMB Express 2017, 7, 8. [Google Scholar] [CrossRef]
- Shan, C.; Wu, H.; Zhu, Y.; Zhou, J.; Yan, W.; Jianhao, Z.; Liu, X. Preservative Effects of a Novel Bacteriocin from Lactobacillus Panis C-M2 Combined with Dielectric Barrier Discharged Cold Plasma (DBD-CP) on Acquatic Foods. Food Sci. Technol. Int. Cienc. Tecnol. Los Aliment. Int. 2023, 29, 406–416. [Google Scholar] [CrossRef]
- Drider, D.; Fimland, G.; Héchard, Y.; McMullen, L.M.; Prévost, H. The Continuing Story of Class IIa Bacteriocins. Microbiol. Mol. Biol. Rev. MMBR 2006, 70, 564–582. [Google Scholar] [CrossRef]
- Ríos Colombo, N.S.; Chalón, M.C.; Navarro, S.A.; Bellomio, A. Pediocin-like Bacteriocins: New Perspectives on Mechanism of Action and Immunity. Curr. Genet. 2018, 64, 345–351. [Google Scholar] [CrossRef]
- Balandin, S.V.; Sheremeteva, E.V.; Ovchinnikova, T.V. Pediocin-Like Antimicrobial Peptides of Bacteria. Biochem. Biokhimiia 2019, 84, 464–478. [Google Scholar] [CrossRef]
- Yi, Y.; Li, P.; Zhao, F.; Zhang, T.; Shan, Y.; Wang, X.; Liu, B.; Chen, Y.; Zhao, X.; Lü, X. Current Status and Potentiality of Class II Bacteriocins from Lactic Acid Bacteria: Structure, Mode of Action and Applications in the Food Industry. Trends Food Sci. Technol. 2022, 120, 387–401. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, Y.; Li, L.; Jiang, X.; Chen, Z.; Zhao, F.; Yi, Y. Biosynthesis and Production of Class II Bacteriocins of Food-Associated Lactic Acid Bacteria. Fermentation 2022, 8, 217. [Google Scholar] [CrossRef]
- Antoshina, D.V.; Balandin, S.V.; Bogdanov, I.V.; Vershinina, M.A.; Sheremeteva, E.V.; Toropygin, I.Y.; Finkina, E.I.; Ovchinnikova, T.V. Antimicrobial Activity and Immunomodulatory Properties of Acidocin A, the Pediocin-like Bacteriocin with the Non-Canonical Structure. Membranes 2022, 12, 1253. [Google Scholar] [CrossRef]
- Kanatani, K.; Oshimura, M.; Sano, K. Isolation and Characterization of Acidocin A and Cloning of the Bacteriocin Gene from Lactobacillus Acidophilus. Appl. Environ. Microbiol. 1995, 61, 1061–1067. [Google Scholar] [CrossRef]
- Tahara, T.; Kanatani, K.; Yoshida, K.; Miura, H.; Sakamoto, M.; Oshimura, M. Purification and Some Properties of Acidocin 8912, a Novel Bacteriocin Produced by Lactobacillus Acidophilus TK8912. Biosci. Biotechnol. Biochem. 1992, 56, 1212–1215. [Google Scholar] [CrossRef][Green Version]
- Kanatani, K.; Tahara, T.; Oshimura, M.; Sano, K.; Umezawa, C. Cloning and Nucleotide Sequence of the Gene for Acidocin 8912, a Bacteriocin from Lactobacillus Acidophilus TK8912. Lett. Appl. Microbiol. 1995, 21, 384–386. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, S.; Qin, Y.; Ding, W.; Tu, Y.; Chen, X.; Wu, Y.; Yanhua, L.; Cai, X. Experimental Evaluation of the Transport Mechanisms of PoIFN-α in Caco-2 Cells. Front. Pharmacol. 2017, 8, 781. [Google Scholar] [CrossRef]
- Press, B. Optimization of the Caco-2 Permeability Assay to Screen Drug Compounds for Intestinal Absorption and Efflux. Methods Mol. Biol. Clifton NJ 2011, 763, 139–154. [Google Scholar] [CrossRef]
- Zhu, L.; Zeng, J.; Wang, C.; Wang, J. Structural Basis of Pore Formation in the Mannose Phosphotransferase System by Pediocin PA-1. Appl. Environ. Microbiol. 2022, 88, e0199221. [Google Scholar] [CrossRef]
- Kjos, M.; Nes, I.F.; Diep, D.B. Class II One-Peptide Bacteriocins Target a Phylogenetically Defined Subgroup of Mannose Phosphotransferase Systems on Sensitive Cells. Microbiol. Read. Engl. 2009, 155, 2949–2961. [Google Scholar] [CrossRef]
- Soltani, S.; Zirah, S.; Rebuffat, S.; Couture, F.; Boutin, Y.; Biron, E.; Subirade, M.; Fliss, I. Gastrointestinal Stability and Cytotoxicity of Bacteriocins From Gram-Positive and Gram-Negative Bacteria: A Comparative In Vitro Study. Front. Microbiol. 2021, 12, 780355. [Google Scholar] [CrossRef]
- Baindara, P.; Chaudhry, V.; Mittal, G.; Liao, L.M.; Matos, C.O.; Khatri, N.; Franco, O.L.; Patil, P.B.; Korpole, S. Characterization of the Antimicrobial Peptide Penisin, a Class Ia Novel Lantibiotic from Paenibacillus sp. Strain A3. Antimicrob. Agents Chemother. 2016, 60, 580–591. [Google Scholar] [CrossRef]
- Ehmann, D.; Wendler, J.; Koeninger, L.; Larsen, I.S.; Klag, T.; Berger, J.; Marette, A.; Schaller, M.; Stange, E.F.; Malek, N.P.; et al. Paneth Cell α-Defensins HD-5 and HD-6 Display Differential Degradation into Active Antimicrobial Fragments. Proc. Natl. Acad. Sci. USA 2019, 116, 3746–3751. [Google Scholar] [CrossRef]
- Arapidi, G.P.; Urban, A.S.; Osetrova, M.S.; Shender, V.O.; Butenko, I.O.; Bukato, O.N.; Kuznetsov, A.A.; Saveleva, T.M.; Nos, G.A.; Ivanova, O.M.; et al. Non-Human Peptides Revealed in Blood Reflect the Composition of Small Intestine Microbiota. BMC Biol. 2023, 22, 178. [Google Scholar]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Puigbò, P.; Guzmán, E.; Romeu, A.; Garcia-Vallvé, S. OPTIMIZER: A Web Server for Optimizing the Codon Usage of DNA Sequences. Nucleic Acids Res. 2007, 35, W126–W131. [Google Scholar] [CrossRef]
- Shenkarev, Z.O.; Panteleev, P.V.; Balandin, S.V.; Gizatullina, A.K.; Altukhov, D.A.; Finkina, E.I.; Kokryakov, V.N.; Arseniev, A.S.; Ovchinnikova, T.V. Recombinant Expression and Solution Structure of Antimicrobial Peptide Aurelin from Jellyfish Aurelia Aurita. Biochem. Biophys. Res. Commun. 2012, 429, 63–69. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and Broth Dilution Methods to Determine the Minimal Inhibitory Concentration (MIC) of Antimicrobial Substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Shamova, O.V.; Orlov, D.S.; Zharkova, M.S.; Balandin, S.V.; Yamschikova, E.V.; Knappe, D.; Hoffmann, R.; Kokryakov, V.N.; Ovchinnikova, T.V. Minibactenecins ChBac7.Nα and ChBac7. Nβ—Antimicrobial Peptides from Leukocytes of the Goat Capra Hircus. Acta Naturae 2016, 8, 136–146. [Google Scholar] [CrossRef]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (Resazurin) Fluorescent Dye for the Assessment of Mammalian Cell Cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar] [CrossRef]
- Rampersad, S.N. Multiple Applications of Alamar Blue as an Indicator of Metabolic Function and Cellular Health in Cell Viability Bioassays. Sensors 2012, 12, 12347–12360. [Google Scholar] [CrossRef]
- Bogdanov, I.V.; Fateeva, S.I.; Voropaev, A.D.; Ovchinnikova, T.V.; Finkina, E.I. Immunomodulatory Effects of the Pea Defensin Psd1 in the Caco-2/Immune Cells Co-Culture upon Candida Albicans Infection. Int. J. Mol. Sci. 2023, 24, 7712. [Google Scholar] [CrossRef]
- UCAST Definitive Document E.DEF 7.3.2 Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Yeasts. Available online: https://www.eucast.org/astoffungi/methodsinantifungalsusceptibilitytesting/susceptibility_testing_of_yeasts/ (accessed on 17 June 2024).

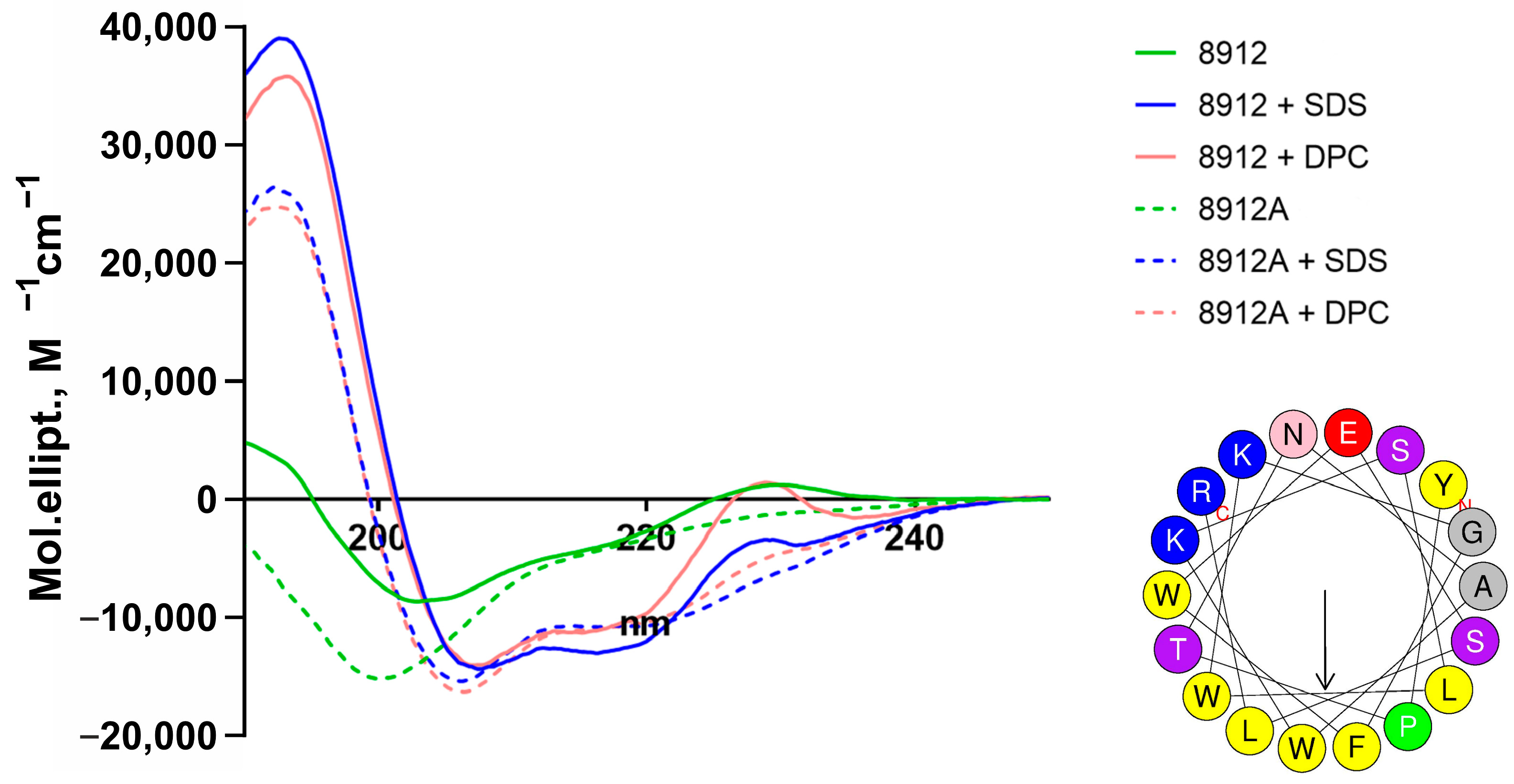


| Bacteriocin | Condition | α-Helix, % | β-Sheet, % | β-Turn, % | Random, % | NRMSD |
|---|---|---|---|---|---|---|
| Acidocin 8912 | Aqueous solution | 4.5 | 40.5 | 23.9 | 31.2 | 0.05 |
| DPC micelles | 41.4 | 15.9 | 22.4 | 20.3 | 0.03 | |
| SDS micelles | 49.6 | 4.5 | 17.0 | 28.9 | 0.08 | |
| Acidocin 8912A | Aqueous solution | 7.9 | 30.7 | 23.8 | 37.5 | 0.03 |
| DPC micelles | 44.9 | 7.3 | 25.2 | 22.6 | 0.08 | |
| SDS micelles | 44.0 | 10.1 | 25.4 | 20.4 | 0.04 | |
| Acidocin A | Aqueous solution | 5.7 | 33.7 | 22.6 | 37.9 | 0.02 |
| DPC micelles | 44.0 | 8.1 | 19.2 | 28.6 | 0.01 | |
| SDS micelles | 40.2 | 10.6 | 20.3 | 28.8 | 0.01 | |
| Avicin A | Aqueous solution | 5.8 | 32.8 | 22.1 | 39.3 | 0.02 |
| DPC micelles | 19.4 | 28.6 | 21.6 | 27.8 | 0.02 | |
| SDS micelles | 19.7 | 29.3 | 23.1 | 30.4 | 0.02 |
| Bacterial Strains | Minimum Inhibitory Concentration (µM) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AcdA | Acd 8912 | AcdA (1–31) | AcdA (10–33) | (10–33, C31S) | AcdA (10–39) | AcdA (10–48) | AcdA (19–58) | AcdA (32–58) | AcdA 8912A | AviA | |
| Listeria monocytogenes EGD | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | <0.125 |
| Lactococcus lactis ssp. lactis bv. diacetylactis MK66 | 0.5 | 4 | 4 | 4 | nd | 8 | 2 | 2 | 2 | nd | >32 |
| Lactococcus lactis ssp. lactis MK43 | 1 | 8 | 8 | 16 | 8 | 8 | 8 | 8 | nd | nd | >32 |
| Lactococcus cremoris B-1569 | 4 | 32 | >32 | nd | nd | nd | 8 | nd | >32 | nd | >32 |
| Enterococcus faecium E19 | 1 | 32 | nd | 16 | nd | nd | 8 | nd | nd | 32 | >32 * |
| Enterococcus faecium E62 | 2 | 16 | nd | >32 | nd | nd | nd | nd | nd | >32 | >32 * |
| Enterococcus faecium E63 | 2 | 32 | nd | nd | nd | nd | nd | nd | nd | >16 | nd * |
| Bacillus subtilis B-886 | 2 | 32 | 8 | 8 | 8 | 8 | 4 | 8 | >32 | 32 | >32 |
| Bacillus subtilis B-2895 | 4 | nd | 16 | 8 | 8 | 16 | 4 | 8 | 32 | nd | >32 |
| Bacillus licheniformis B-511 | 2 | 32 | 8 | 8 | 8 | 8 | 4 | 8 | >32 | 8 | >32 |
| Mycobacterium phlei Ac-1221 | 2 | >32 | 16 | 8 | 4 | 16 | 4 | 16 | 32 | nd | >32 |
| Mycobacterium smegmatis MC2 155 | 1 | >32 | nd | nd | nd | nd | nd | nd | nd | >32 | nd |
| Micrococcus luteus Ac-2229 | 8 | 16 | >32 | >32 | 32 | >32 | nd | 32 | >32 | nd | >32 |
| Staphylococcus aureus 209P | 8 | nd | >32 | >32 | >32 | >32 | nd | nd | nd | nd | >32 |
| Bacterial Strains | Minimum Inhibitory Concentration (µM) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AcdA | Acd 8912 | AcdA (1–31) | AcdA (10–33) | (10–33, C31S) | AcdA (10–39) | AcdA (10–48) | AcdA (19–58) | AcdA (32–58) | AcdA 8912A | AviA | |
| E. coli SQ110 | 2 | 4 | >32 | >32 | 16 | >32 | 4 | 4 | >32 | 8 | >32 |
| E. coli ML-35p | 2 | 16 | >32 | >32 | 16 | >32 | >32 | 8 | >32 | 16 | >32 |
| E. coli ATCC 25922 | 4 | 32 | >32 | >32 | 32 | >32 | 32 | 16 | >32 | nd | >32 |
| E. coli XDR CI 1057 | 8 | 32 | >32 | >32 | >32 | >32 | 16 | >32 | >32 | nd | >32 |
| E. coli CI 214 | 8 | >32 | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| E. coli SBS 1936 | 8 | >32 | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| Pseudomonas aeruginosa PAO1 | 16 | >32 | nd | nd | nd | nd | nd | nd | nd | nd | >32 |
| Acinetobacter baumannii XDR CI 2675 | 4 | nd | >32 | >32 | 32 | >32 | nd | 8 | nd | nd | >32 |
| Fungal Strains | Acidocin A | Acidocin 8912 | Acidocin 8912A | Avicin A | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | MFC | FI | MIC | MFC | FI | MIC | MFC | FI | MIC | MFC | FI | |
| C. albicans ATCC 18804 * | 4 | 8 | 2 | 32 | >32 | nd | 32 | >32 | nd | >32 | nd | nd |
| C. albicans ATCC 10231 * | 8 | 16 | 2 | 16 | 32 | 2 | 32 | >32 | nd | 32 | >32 | nd |
| C. albicans v47a | 4 | 4 | 1 | 16 | >32 | nd | 32 | >32 | nd | 32 | >32 | nd |
| C. albicans 9.1 * | 4 | 16 | 4 | 16 | 32 | 2 | 32 | 32 | 1 | >32 | nd | nd |
| C. tropicalis v13a4/2 | 4 | 4 | 1 | 16 | 16 | 1 | 4 | 8 | 2 | >32 | nd | nd |
| C. krusei 225/2 | 8 | 8 | 1 | 32 | 32 | 1 | >32 | nd | nd | >32 | nd | nd |
| C. glabrata 252/2 | 4 | 4 | 1 | 16 | 16 | 1 | 16 | 16 | 1 | >32 | nd | nd |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antoshina, D.V.; Balandin, S.V.; Finkina, E.I.; Bogdanov, I.V.; Eremchuk, S.I.; Kononova, D.V.; Kovrizhnykh, A.A.; Ovchinnikova, T.V. Acidocin A and Acidocin 8912 Belong to a Distinct Subfamily of Class II Bacteriocins with a Broad Spectrum of Antimicrobial Activity. Int. J. Mol. Sci. 2024, 25, 10059. https://doi.org/10.3390/ijms251810059
Antoshina DV, Balandin SV, Finkina EI, Bogdanov IV, Eremchuk SI, Kononova DV, Kovrizhnykh AA, Ovchinnikova TV. Acidocin A and Acidocin 8912 Belong to a Distinct Subfamily of Class II Bacteriocins with a Broad Spectrum of Antimicrobial Activity. International Journal of Molecular Sciences. 2024; 25(18):10059. https://doi.org/10.3390/ijms251810059
Chicago/Turabian StyleAntoshina, Daria V., Sergey V. Balandin, Ekaterina I. Finkina, Ivan V. Bogdanov, Sofia I. Eremchuk, Daria V. Kononova, Alena A. Kovrizhnykh, and Tatiana V. Ovchinnikova. 2024. "Acidocin A and Acidocin 8912 Belong to a Distinct Subfamily of Class II Bacteriocins with a Broad Spectrum of Antimicrobial Activity" International Journal of Molecular Sciences 25, no. 18: 10059. https://doi.org/10.3390/ijms251810059
APA StyleAntoshina, D. V., Balandin, S. V., Finkina, E. I., Bogdanov, I. V., Eremchuk, S. I., Kononova, D. V., Kovrizhnykh, A. A., & Ovchinnikova, T. V. (2024). Acidocin A and Acidocin 8912 Belong to a Distinct Subfamily of Class II Bacteriocins with a Broad Spectrum of Antimicrobial Activity. International Journal of Molecular Sciences, 25(18), 10059. https://doi.org/10.3390/ijms251810059









