Circular RNAs: Novel Players in Cancer Mechanisms and Therapeutic Strategies
Abstract
1. Overview of Circular RNAs (circRNAs)
2. Mechanisms of circRNA Formation
2.1. Biogenesis of circRNAs
2.2. Aberrant Biogenesis of circRNAs in Cancers
3. Functional Roles of circRNAs in Cancer
3.1. miRNA Sponging
3.2. Modulation of Protein Function
3.3. Production of circRNA-Derived Peptides or Proteins
3.4. Emerging Role of circRNA as a Chromosome Translocation Diver
4. CircRNAs as Biomarkers in Cancers
4.1. Diagnostic and Predictive Value of circRNAs in Cancers
4.2. Ongoing Clinical Trials
5. circRNAs as Therapeutic Platform in Cancer
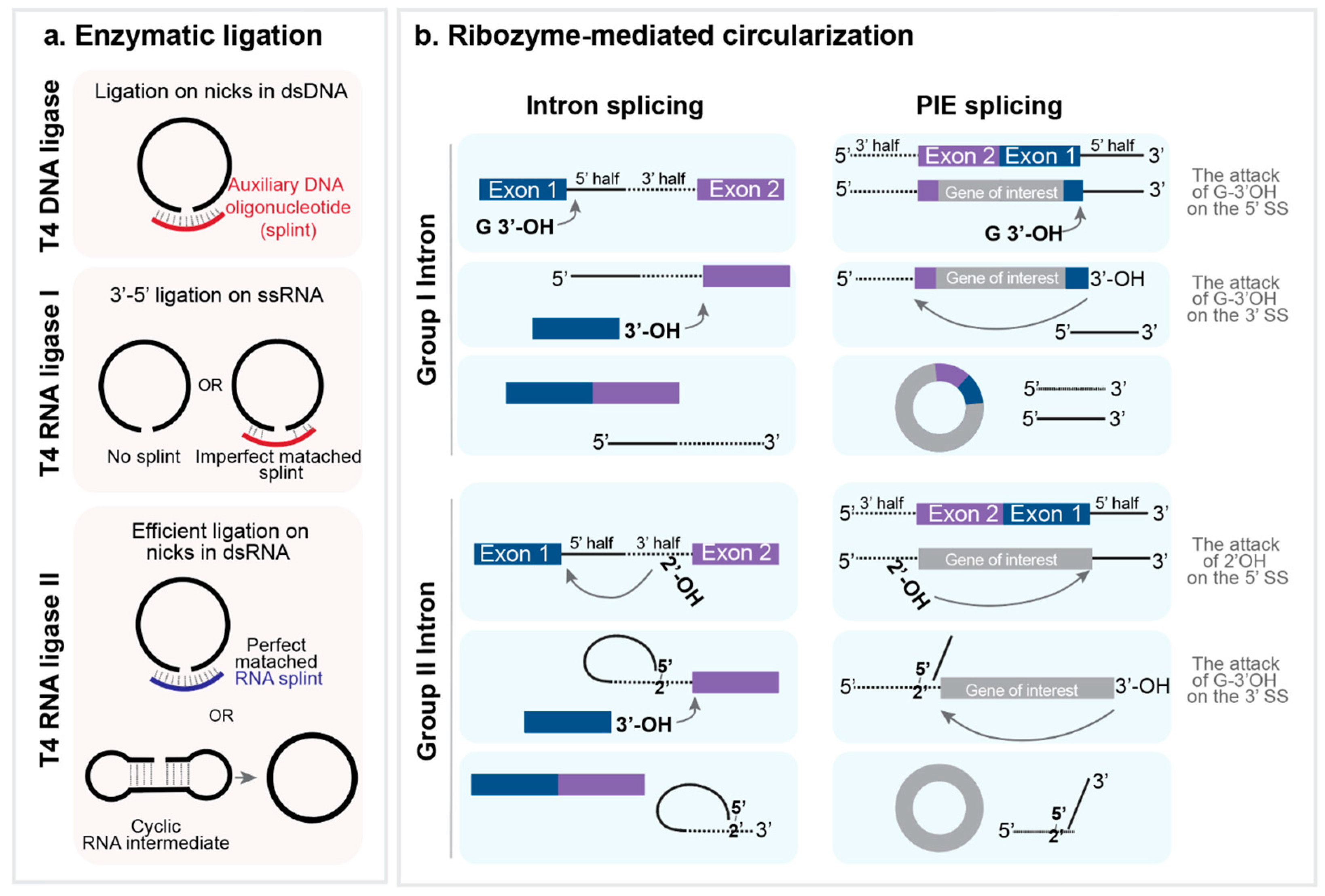
5.1. In Vitro Synthesis of circRNAs
5.1.1. In Vitro Synthesis of RNA Precursor
5.1.2. Ligase-Mediated Circularization
5.1.3. Ribozyme-Mediated Circularization
5.2. Engineering Circular RNA Translation
5.2.1. IRES-Mediated Translation
5.2.2. M6A-Mediated Translation
5.3. Applications in Cancer Therapy
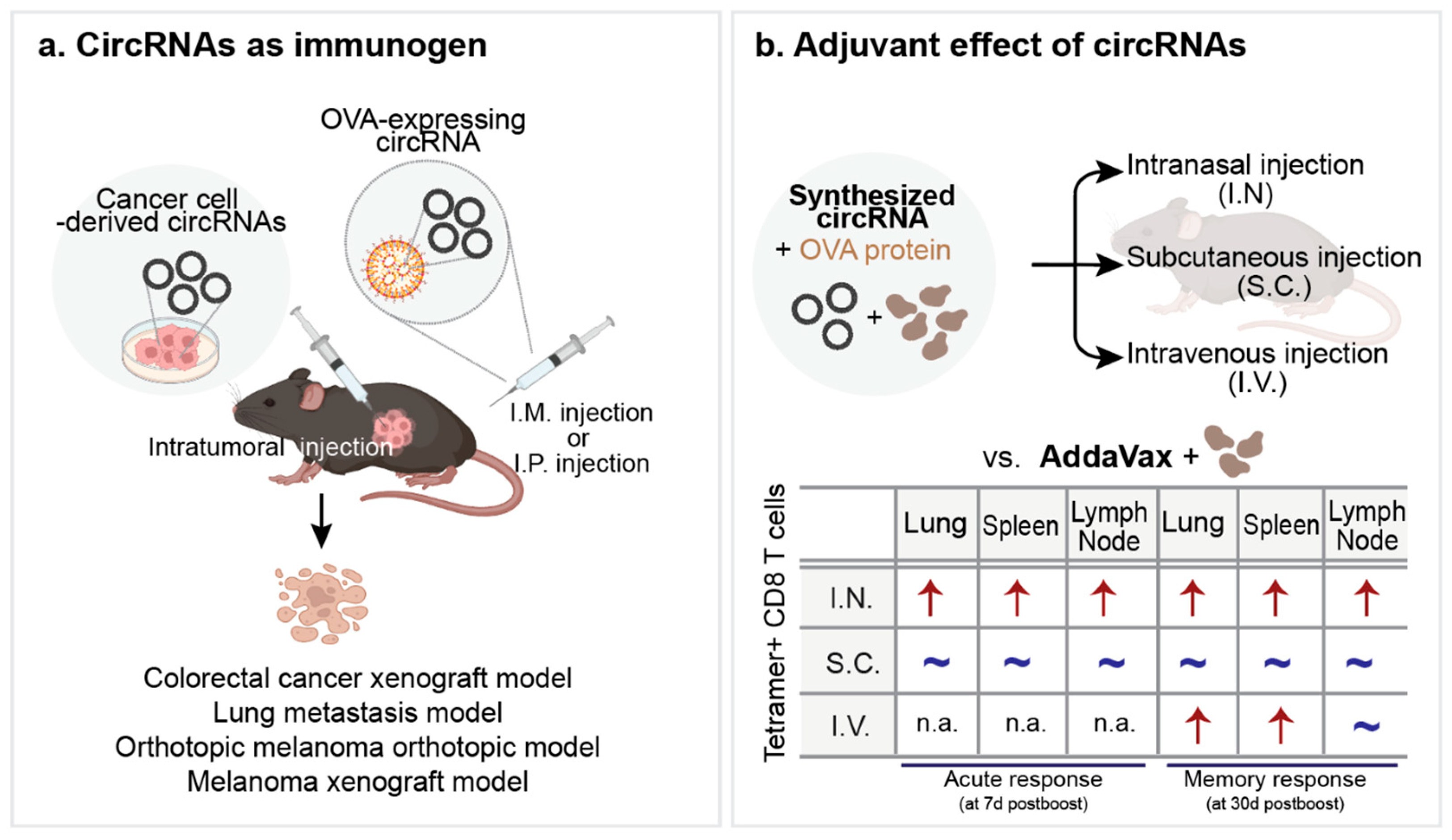
6. Conclusions and Future Perspective
Funding
Conflicts of Interest
References
- Cocquerelle, C.; Mascrez, B.; Hetuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993, 73, 1019–1030. [Google Scholar] [CrossRef]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Gruner, H.; Cortes-Lopez, M.; Cooper, D.A.; Bauer, M.; Miura, P. CircRNA accumulation in the aging mouse brain. Sci. Rep. 2016, 6, 38907. [Google Scholar] [CrossRef]
- Maass, P.G.; Glazar, P.; Memczak, S.; Dittmar, G.; Hollfinger, I.; Schreyer, L.; Sauer, A.V.; Toka, O.; Aiuti, A.; Luft, F.C.; et al. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. 2017, 95, 1179–1189. [Google Scholar] [CrossRef]
- Memczak, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Hansen, T.B.; Kjems, J.; Damgaard, C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013, 73, 5609–5612. [Google Scholar] [CrossRef]
- Medicine, H.M.L. Promising New ‘circRNA’ Vaccines Explored in CEPI-HMRI Collaboration. 2024. Available online: https://www.houstonmethodist.org/newsroom/promising-new-circrna-vaccines-explored-in-cepi-hmri-collaboration/ (accessed on 31 July 2024).
- Newswire, P. Flagship Pioneering Announces the Merger of Two Leading Programmable Medicine Platforms to Form Sail Biomedicines. 2023. Available online: https://www.prnewswire.com/news-releases/flagship-pioneering-announces-the-merger-of-two-leading-programmable-medicine-platforms-to-form-sail-biomedicines-301961581.html (accessed on 31 July 2024).
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Wilusz, J.E. Short intronic repeat sequences facilitate circular RNA production. Genes. Dev. 2014, 28, 2233–2247. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, C.X.; Xue, W.; Zhang, Y.; Jiang, S.; Yin, Q.F.; Wei, J.; Yao, R.W.; Yang, L.; Chen, L.L. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol. Cell 2017, 67, 214–227.e7. [Google Scholar] [CrossRef]
- Fei, T.; Chen, Y.; Xiao, T.; Li, W.; Cato, L.; Zhang, P.; Cotter, M.B.; Bowden, M.; Lis, R.T.; Zhao, S.G.; et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc. Natl. Acad. Sci. USA 2017, 114, E5207–E5215. [Google Scholar] [CrossRef]
- Kelly, S.; Greenman, C.; Cook, P.R.; Papantonis, A. Exon Skipping Is Correlated with Exon Circularization. J. Mol. Biol. 2015, 427, 2414–2417. [Google Scholar] [CrossRef]
- Errichelli, L.; Dini Modigliani, S.; Laneve, P.; Colantoni, A.; Legnini, I.; Capauto, D.; Rosa, A.; De Santis, R.; Scarfo, R.; Peruzzi, G.; et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017, 8, 14741. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Aktas, T.; Avsar Ilik, I.; Maticzka, D.; Bhardwaj, V.; Pessoa Rodrigues, C.; Mittler, G.; Manke, T.; Backofen, R.; Akhtar, A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 2017, 544, 115–119. [Google Scholar] [CrossRef]
- Shi, L.; Yan, P.; Liang, Y.; Sun, Y.; Shen, J.; Zhou, S.; Lin, H.; Liang, X.; Cai, X. Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 2017, 8, e3171. [Google Scholar] [CrossRef] [PubMed]
- Barrett, S.P.; Wang, P.L.; Salzman, J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 2015, 4, e07540. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Vidal, A.F. Read-through circular RNAs reveal the plasticity of RNA processing mechanisms in human cells. RNA Biol. 2020, 17, 1823–1826. [Google Scholar] [CrossRef]
- Liang, D.; Tatomer, D.C.; Luo, Z.; Wu, H.; Yang, L.; Chen, L.L.; Cherry, S.; Wilusz, J.E. The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Mol. Cell 2017, 68, 940–954.e3. [Google Scholar] [CrossRef] [PubMed]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e3. [Google Scholar] [CrossRef] [PubMed]
- Grosso, A.R.; Leite, A.P.; Carvalho, S.; Matos, M.R.; Martins, F.B.; Vitor, A.C.; Desterro, J.M.; Carmo-Fonseca, M.; de Almeida, S.F. Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. Elife 2015, 4, e09214. [Google Scholar] [CrossRef]
- Guarnerio, J.; Bezzi, M.; Jeong, J.C.; Paffenholz, S.V.; Berry, K.; Naldini, M.M.; Lo-Coco, F.; Tay, Y.; Beck, A.H.; Pandolfi, P.P. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016, 165, 289–302. [Google Scholar] [CrossRef]
- Tan, S.; Gou, Q.; Pu, W.; Guo, C.; Yang, Y.; Wu, K.; Liu, Y.; Liu, L.; Wei, Y.Q.; Peng, Y. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. 2018, 28, 693–695. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Liao, X.; Gong, Y.; He, J.; Zhou, J.K.; Tan, S.; Pu, W.; Huang, C.; Wei, Y.Q.; Peng, Y. Circular RNA F-circSR derived from SLC34A2-ROS1 fusion gene promotes cell migration in non-small cell lung cancer. Mol. Cancer 2019, 18, 98. [Google Scholar] [CrossRef]
- Ferreira, H.J.; Davalos, V.; de Moura, M.C.; Soler, M.; Perez-Salvia, M.; Bueno-Costa, A.; Setien, F.; Moran, S.; Villanueva, A.; Esteller, M. Circular RNA CpG island hypermethylation-associated silencing in human cancer. Oncotarget 2018, 9, 29208–29219. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, M.; Xue, C.; Chen, S.; Zheng, L.; Deng, H.; Tang, F.; Li, G.; Xiong, W.; Zeng, Z.; et al. Understanding the roles and regulation patterns of circRNA on its host gene in tumorigenesis and tumor progression. J. Exp. Clin. Cancer Res. 2023, 42, 86. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Jakobsen, T.; Hager, H.; Kjems, J. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 2022, 19, 188–206. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Tao, Y.; Zhou, Y.; Qin, N.; Chen, C.; Tian, D.; Xu, L. MicroRNA-7: A promising new target in cancer therapy. Cancer Cell Int. 2015, 15, 103. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Han, Y.; Zhang, H.; Li, Y.; Yi, L.; Wang, X.; Zhou, S.; Yu, D.; Song, X.; Xiao, N.; et al. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-kappaB signaling. J. Cell. Mol. Med. 2018, 22, 3097–3107. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, M.; Zheng, X.; Yi, P.; Lan, C.; Xu, M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2017, 143, 17–27. [Google Scholar] [CrossRef]
- Rahmati, Y.; Asemani, Y.; Aghamiri, S.; Ezzatifar, F.; Najafi, S. CiRS-7/CDR1as; An oncogenic circular RNA as a potential cancer biomarker. Pathol. Res. Pract. 2021, 227, 153639. [Google Scholar] [CrossRef]
- Liu, L.; Liu, F.B.; Huang, M.; Xie, K.; Xie, Q.S.; Liu, C.H.; Shen, M.J.; Huang, Q. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat. Dis. Int. 2019, 18, 580–586. [Google Scholar] [CrossRef]
- Huang, H.; Wei, L.; Qin, T.; Yang, N.; Li, Z.; Xu, Z. Circular RNA ciRS-7 triggers the migration and invasion of esophageal squamous cell carcinoma via miR-7/KLF4 and NF-kappaB signals. Cancer Biol. Ther. 2019, 20, 73–80. [Google Scholar] [CrossRef]
- Weng, W.; Wei, Q.; Toden, S.; Yoshida, K.; Nagasaka, T.; Fujiwara, T.; Cai, S.; Qin, H.; Ma, Y.; Goel, A. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin. Cancer Res. 2017, 23, 3918–3928. [Google Scholar] [CrossRef]
- Zheng, Q.P.; Bao, C.Y.; Guo, W.J.; Li, S.Y.; Chen, J.; Chen, B.; Luo, Y.T.; Lyu, D.B.; Li, Y.; Shi, G.H.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Shi, Y.; Liu, M.; Sun, J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 175. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Khoshbakht, T.; Taheri, M.; Jamali, E. CircITCH: A Circular RNA With Eminent Roles in the Carcinogenesis. Front. Oncol. 2021, 11, 774979. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.D.; Yuan, W.B.; Yang, X.; Li, P.; Wang, J.Z.; Han, J.; Tao, J.; Li, P.C.; Yang, H.W.; Lv, Q.; et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol. Cancer 2018, 17, 19. [Google Scholar] [CrossRef]
- Drula, R.; Pirlog, R.; Trif, M.; Slaby, O.; Braicu, C.; Berindan-Neagoe, I. circFOXO3: Going around the mechanistic networks in cancer by interfering with miRNAs regulatory networks. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166045. [Google Scholar] [CrossRef]
- Rao, D.; Yu, C.; Sheng, J.; Lv, E.; Huang, W. The Emerging Roles of circFOXO3 in Cancer. Front. Cell Dev. Biol. 2021, 9, 659417. [Google Scholar] [CrossRef]
- Du, W.W.; Fang, L.; Yang, W.N.; Wu, N.; Awan, F.M.; Yang, Z.G.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef]
- Yang, Q.; Du, W.W.; Wu, N.; Yang, W.; Awan, F.M.; Fang, L.; Ma, J.; Li, X.; Zeng, Y.; Yang, Z.; et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017, 24, 1609–1620. [Google Scholar] [CrossRef]
- Wang, X.; Li, J.; Bian, X.; Wu, C.; Hua, J.; Chang, S.; Yu, T.; Li, H.; Li, Y.; Hu, S.; et al. CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc. Natl. Acad. Sci. USA 2021, 118, e2012881118. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, D.; Caponnetto, A.; Brex, D.; Mirabella, F.; Barbagallo, C.; Lauretta, G.; Morrone, A.; Certo, F.; Broggi, G.; Caltabiano, R.; et al. CircSMARCA5 Regulates VEGFA mRNA Splicing and Angiogenesis in Glioblastoma Multiforme Through the Binding of SRSF1. Cancers 2019, 11, 194. [Google Scholar] [CrossRef]
- Barbagallo, D.; Caponnetto, A.; Cirnigliaro, M.; Brex, D.; Barbagallo, C.; D’Angeli, F.; Morrone, A.; Caltabiano, R.; Barbagallo, G.M.; Ragusa, M.; et al. CircSMARCA5 Inhibits Migration of Glioblastoma Multiforme Cells by Regulating a Molecular Axis Involving Splicing Factors SRSF1/SRSF3/PTB. Int. J. Mol. Sci. 2018, 19, 480. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Xu, C.; Jun, E.; Okugawa, Y.; Toiyama, Y.; Borazanci, E.; Bolton, J.; Taketomi, A.; Kim, S.C.; Shang, D.; Von Hoff, D.; et al. A Circulating Panel of circRNA Biomarkers for the Noninvasive and Early Detection of Pancreatic Ductal Adenocarcinoma. Gastroenterology 2024, 166, 178–190.e6. [Google Scholar] [CrossRef]
- AbouHaidar, M.G.; Venkataraman, S.; Golshani, A.; Liu, B.; Ahmad, T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc. Natl. Acad. Sci. USA 2014, 111, 14542–14547. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef]
- Yang, Y.; Gao, X.; Zhang, M.; Yan, S.; Sun, C.; Xiao, F.; Huang, N.; Yang, X.; Zhao, K.; Zhou, H.; et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J. Natl. Cancer Inst. 2018, 110, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Ma, S.; Xu, J.; Ren, X.; Guo, P.; Liu, H.; Li, P.; Yin, F.; Liu, M.; Wang, Q.; et al. A novel polypeptide encoded by the circular RNA ZKSCAN1 suppresses HCC via degradation of mTOR. Mol. Cancer 2023, 22, 16. [Google Scholar] [CrossRef]
- Liang, W.C.; Wong, C.W.; Liang, P.P.; Shi, M.; Cao, Y.; Rao, S.T.; Tsui, S.K.; Waye, M.M.; Zhang, Q.; Fu, W.M.; et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019, 20, 84. [Google Scholar] [CrossRef]
- Jiang, T.L.; Xia, Y.W.; Lv, J.L.; Li, B.W.; Li, Y.; Wang, S.; Xuan, Z.; Xie, L.; Qiu, S.K.; He, Z.Y.; et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol. Cancer 2021, 20, 66. [Google Scholar] [CrossRef]
- Huang, D.; Zhu, X.; Ye, S.; Zhang, J.; Liao, J.; Zhang, N.; Zeng, X.; Wang, J.; Yang, B.; Zhang, Y.; et al. Tumour circular RNAs elicit anti-tumour immunity by encoding cryptic peptides. Nature 2024, 625, 593–602. [Google Scholar] [CrossRef]
- Garcia-Muse, T.; Aguilera, A. R Loops: From Physiological to Pathological Roles. Cell 2019, 179, 604–618. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Gabryelska, M.; Toubia, J.; Kirk, K.; Gantley, L.; Powell, J.A.; Cildir, G.; Marri, S.; Liu, R.; Stringer, B.W.; et al. Circular RNAs drive oncogenic chromosomal translocations within the MLL recombinome in leukemia. Cancer Cell 2023, 41, 1309–1326.e10. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Bao, Y.; Deng, L.; Su, D.; Zheng, H.; Zhang, W. Circular RNA hsa_circ_0014130 Inhibits Apoptosis in Non-Small Cell Lung Cancer by Sponging miR-136-5p and Upregulating BCL2. Mol. Cancer Res. 2020, 18, 1110. [Google Scholar] [CrossRef]
- Verduci, L.; Ferraiuolo, M.; Sacconi, A.; Ganci, F.; Vitale, J.; Colombo, T.; Paci, P.; Strano, S.; Macino, G.; Rajewsky, N.; et al. The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription-competent complex. Genome Biol. 2017, 18, 237. [Google Scholar] [CrossRef]
- Wang, S.; Tong, H.; Su, T.; Zhou, D.; Shi, W.; Tang, Z.; Quan, Z. CircTP63 promotes cell proliferation and invasion by regulating EZH2 via sponging miR-217 in gallbladder cancer. Cancer Cell Int. 2021, 21, 608. [Google Scholar] [CrossRef]
- Wang, J.; Che, J. CircTP63 promotes hepatocellular carcinoma progression by sponging miR-155-5p and upregulating ZBTB18. Cancer Cell Int. 2021, 21, 156. [Google Scholar] [CrossRef]
- Liu, S.; Yang, N.; Jiang, X.; Wang, J.; Dong, J.; Gao, Y. FUS-induced circular RNA ZNF609 promotes tumorigenesis and progression via sponging miR-142-3p in lung cancer. J. Cell Physiol. 2021, 236, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Danac, J.M.C.; Garcia, R.L. CircPVT1 attenuates negative regulation of NRAS by let-7 and drives cancer cells towards oncogenicity. Sci. Rep. 2021, 11, 9021. [Google Scholar] [CrossRef]
- Gopikrishnan, M.; Ashour, H.M.; Pintus, G.; Hammad, M.; Kashyap, M.K.; Zayed, H. Therapeutic and diagnostic applications of exosomal circRNAs in breast cancer. Funct. Integr. Genom. 2023, 23, 184. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, J.; Yang, W.; Ye, W.C. CircRNAs in colorectal cancer: Potential biomarkers and therapeutic targets. Cell Death Dis. 2023, 14, 353. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, K.; Tan, S.; Xin, J.; Yuan, Q.; Xu, H.; Xu, X.; Liang, Q.; Christiani, D.C.; Wang, M.; et al. Circular RNAs in body fluids as cancer biomarkers: The new frontier of liquid biopsies. Mol. Cancer 2021, 20, 13. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Yang, Q.; He, A.T.; Yang, B.B. Circular RNAs in cancer: Limitations in functional studies and diagnostic potential. Semin. Cancer Biol. 2021, 75, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Chen, M.; Zhang, L.; Huang, S.; Xiao, F.; Zou, L. Circular RNAs: Biomarkers of cancer. Cancer Innov. 2022, 1, 197–206. [Google Scholar] [CrossRef] [PubMed]
- He, Y.D.; Tao, W.; He, T.; Wang, B.Y.; Tang, X.M.; Zhang, L.M.; Wu, Z.Q.; Deng, W.M.; Zhang, L.X.; Shao, C.K.; et al. A urine extracellular vesicle circRNA classifier for detection of high-grade prostate cancer in patients with prostate-specific antigen 2–10 ng/mL at initial biopsy. Mol. Cancer 2021, 20, 96. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, Q.; Qin, S.; Ju, S. CircBRIP1: A plasma diagnostic marker for non-small-cell lung cancer. J. Cancer Res. Clin. Oncol. 2024, 150, 83. [Google Scholar] [CrossRef]
- Gupta, S.K.; Garg, A.; Bar, C.; Chatterjee, S.; Foinquinos, A.; Milting, H.; Streckfuss-Bomeke, K.; Fiedler, J.; Thum, T. Quaking Inhibits Doxorubicin-Mediated Cardiotoxicity Through Regulation of Cardiac Circular RNA Expression. Circ. Res. 2018, 122, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Li, Y.; Ming, Z.; Wang, H.; Dong, Z.; Qiu, L.; Wang, T. Comprehensive circular RNA expression profile in radiation-treated HeLa cells and analysis of radioresistance-related circRNAs. PeerJ 2018, 6, e5011. [Google Scholar] [CrossRef]
- Wu, G.; Sun, Y.; Xiang, Z.; Wang, K.; Liu, B.; Xiao, G.; Niu, Y.; Wu, D.; Chang, C. Preclinical study using circular RNA 17 and micro RNA 181c-5p to suppress the enzalutamide-resistant prostate cancer progression. Cell Death Dis. 2019, 10, 37. [Google Scholar] [CrossRef]
- Greene, J.; Baird, A.M.; Casey, O.; Brady, L.; Blackshields, G.; Lim, M.; O’Brien, O.; Gray, S.G.; McDermott, R.; Finn, S.P. Circular RNAs are differentially expressed in prostate cancer and are potentially associated with resistance to enzalutamide. Sci. Rep. 2019, 9, 10739. [Google Scholar] [CrossRef]
- Sang, Y.; Chen, B.; Song, X.; Li, Y.; Liang, Y.; Han, D.; Zhang, N.; Zhang, H.; Liu, Y.; Chen, T.; et al. circRNA_0025202 Regulates Tamoxifen Sensitivity and Tumor Progression via Regulating the miR-182-5p/FOXO3a Axis in Breast Cancer. Mol. Ther. 2021, 29, 3525–3527. [Google Scholar] [CrossRef]
- Li, H.; Li, Q.; He, S. Hsa_circ_0025202 suppresses cell tumorigenesis and tamoxifen resistance via miR-197-3p/HIPK3 axis in breast cancer. World J. Surg. Oncol. 2021, 19, 39. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Chinnaiyan, A.M. The Potential of Circular RNAs as Cancer Biomarkers. Cancer Epidem. Biomar. 2020, 29, 2541–2555. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Gao, Q.; Chen, Y.; Zhang, Z.; Du, Y.H.; Liu, Y.; Zhang, G.X.; Li, S.L.; Wang, G.Y.; Chen, X.; et al. Identification of CircRNA signature associated with tumor immune infiltration to predict therapeutic efficacy of immunotherapy. Nat. Commun. 2023, 14, 2540. [Google Scholar] [CrossRef]
- Qu, L.; Yi, Z.; Shen, Y.; Lin, L.; Chen, F.; Xu, Y.; Wu, Z.; Tang, H.; Zhang, X.; Tian, F.; et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 2022, 185, 1728–1744.e16. [Google Scholar] [CrossRef]
- Medince, H.M.L. CircRNA Breakthrough: Houston Methodist and CEPI Forge Alliance to Revolutionize Vaccine Technology. Available online: https://www.houstonmethodist.org/leading-medicine-blog/articles/2024/feb/circrna-breakthrough-houston-methodist-and-cepi-forge-alliance-to-revolutionize-vaccine-technology/ (accessed on 31 July 2024).
- Niu, D.; Wu, Y.; Lian, J. Circular RNA vaccine in disease prevention and treatment. Signal Transduct. Target. Ther. 2023, 8, 341. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Liu, D.; He, Q.; Liu, J.; Mao, Q.; Liang, Z. Research progress on circular RNA vaccines. Front. Immunol. 2022, 13, 1091797. [Google Scholar] [CrossRef]
- Petkovic, S.; Muller, S. Synthesis and Engineering of Circular RNAs. Methods Mol. Biol. 2018, 1724, 167–180. [Google Scholar] [CrossRef]
- Muller, S.; Appel, B. In vitro circularization of RNA. RNA Biol. 2017, 14, 1018–1027. [Google Scholar] [CrossRef]
- Obi, P.; Chen, Y.G. The design and synthesis of circular RNAs. Methods 2021, 196, 85–103. [Google Scholar] [CrossRef]
- Gholamalipour, Y.; Karunanayake Mudiyanselage, A.; Martin, C.T. 3’ end additions by T7 RNA polymerase are RNA self-templated, distributive and diverse in character-RNA-Seq analyses. Nucleic Acids Res. 2018, 46, 9253–9263. [Google Scholar] [CrossRef]
- Western, L.M.; Rose, S.J. A Novel DNA Joining Activity Catalyzed by T4 DNA-Ligase. Nucleic Acids Res. 1991, 19, 809–813. [Google Scholar] [CrossRef][Green Version]
- Moore, M.J.; Query, C.C. Joining of RNAs by splinted ligation. Methods Enzymol. 2000, 317, 109–123. [Google Scholar]
- Wang, L.; Ruffner, D.E. Oligoribonucleotide circularization by ‘template-mediated’ ligation with T4 RNA ligase: Synthesis of circular hammerhead ribozymes. Nucleic Acids Res. 1998, 26, 2502–2504. [Google Scholar] [CrossRef]
- Petkovic, S.; Muller, S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015, 43, 2454–2465. [Google Scholar] [CrossRef]
- Chen, H.; Cheng, K.; Liu, X.L.; An, R.; Komiyama, M.; Liang, X.G. Preferential production of RNA rings by T4 RNA ligase 2 without any splint through rational design of precursor strand. Nucleic Acids Res. 2020, 48, e54. [Google Scholar] [CrossRef]
- Gomes, R.M.O.D.; da Silva, K.J.G.; Theodoro, R.C. Group I introns: Structure, splicing and their applications in medical mycology. Genet. Mol. Biol. 2024, 47, e20230228. [Google Scholar] [CrossRef]
- Saldanha, R.; Mohr, G.; Belfort, M.; Lambowitz, A.M. Group-I and Group-Ii Introns. FASEB J. 1993, 7, 15–24. [Google Scholar] [CrossRef]
- Puttaraju, M.; Been, M.D. Group-I Permuted Intron Exon (Pie) Sequences Self-Splice to Produce Circular Exons. Nucleic Acids Res. 1992, 20, 5357–5364. [Google Scholar] [CrossRef]
- Wesselhoeft, R.A.; Kowalski, P.S.; Anderson, D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018, 9, 2629. [Google Scholar] [CrossRef] [PubMed]
- Jacquier, A.; Rosbash, M. Efficient trans-splicing of a yeast mitochondrial RNA group II intron implicates a strong 5’ exon-intron interaction. Science 1986, 234, 1099–1104. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mikheeva, S.; Hakim-Zargar, M.; Carlson, D.; Jarrell, K. Use of an engineered ribozyme to produce a circular human exon. Nucleic Acids Res. 1997, 25, 5085–5094. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wei, H.; Zhang, K.; Li, Z.; Wei, T.; Tang, C.; Yang, Y.; Wang, Z. A flexible, efficeint, and scalable platform to produce circular RNAs as new therapeutics. bioRxiv 2022. [Google Scholar]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e7. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.Y.; Qadir, J.; Yang, B.B. Circular RNA translation: Novel protein isoforms and clinical significance. Trends Mol. Med. 2022, 28, 405–420. [Google Scholar] [CrossRef]
- Chen, C.K.; Cheng, R.; Demeter, J.; Chen, J.; Weingarten-Gabbay, S.; Jiang, L.; Snyder, M.P.; Weissman, J.S.; Segal, E.; Jackson, P.K.; et al. Structured elements drive extensive circular RNA translation. Mol. Cell 2021, 81, 4300–4318.e13. [Google Scholar] [CrossRef]
- Chen, R.; Wang, S.K.; Belk, J.A.; Amaya, L.; Li, Z.; Cardenas, A.; Abe, B.T.; Chen, C.K.; Wender, P.A.; Chang, H.Y. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 2023, 41, 262–272. [Google Scholar] [CrossRef]
- Martinez-Salas, E.; Francisco-Velilla, R.; Fernandez-Chamorro, J.; Embarek, A.M. Insights into Structural and Mechanistic Features of Viral IRES Elements. Front. Microbiol. 2018, 8, 2629. [Google Scholar] [CrossRef]
- Hinnebusch, A.G.; Ivanov, I.P.; Sonenberg, N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 2016, 352, 1413–1416. [Google Scholar] [CrossRef]
- Seo, J.J.; Jung, S.J.; Yang, J.; Choi, D.E.; Kim, V.N. Functional viromic screens uncover regulatory RNA elements. Cell 2023, 186, 3291–3306.e21. [Google Scholar] [CrossRef]
- Leppek, K.; Byeon, G.W.; Kladwang, W.; Wayment-Steele, H.K.; Kerr, C.H.; Xu, A.F.; Kim, D.S.; Topkar, V.V.; Choe, C.; Rothschild, D.; et al. Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics. Nat. Commun. 2022, 13, 1536. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.K.; Han, D.L.; Ma, H.H.; Weng, X.C.; Chen, K.; Shi, H.L.; He, C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Peng, K.; Yang, K.; Ma, W.; Qi, S.; Yu, X.; He, J.; Lin, X.; Yu, G. Circular RNA cancer vaccines drive immunity in hard-to-treat malignancies. Theranostics 2022, 12, 6422–6436. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Fang, M.; Li, C.; Zuo, B.; Ren, J.; Zhang, Y. BORIS-mediated generation of circular RNAs induces inflammation. Transl. Oncol. 2022, 18, 101363. [Google Scholar] [CrossRef]
- Chen, Y.G.; Kim, M.V.; Chen, X.; Batista, P.J.; Aoyama, S.; Wilusz, J.E.; Iwasaki, A.; Chang, H.Y. Sensing Self and Foreign Circular RNAs by Intron Identity. Mol. Cell 2017, 67, 228–238.e5. [Google Scholar] [CrossRef]
- Chen, Y.G.; Chen, R.; Ahmad, S.; Verma, R.; Kasturi, S.P.; Amaya, L.; Broughton, J.P.; Kim, J.; Cadena, C.; Pulendran, B.; et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol. Cell 2019, 76, 96–109.e9. [Google Scholar] [CrossRef]
- Liu, C.X.; Guo, S.K.; Nan, F.; Xu, Y.F.; Yang, L.; Chen, L.L. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell 2022, 82, 420–434.e6. [Google Scholar] [CrossRef]
- Amaya, L.; Grigoryan, L.; Li, Z.J.; Lee, A.; Wender, P.A.; Pulendran, B.; Chang, H.Y. Circular RNA vaccine induces potent T cell responses. Proc. Natl. Acad. Sci. USA 2023, 120, e2302191120. [Google Scholar] [CrossRef]

| Study | Cancer | circRNAs | Objective | Sponsor |
|---|---|---|---|---|
| NCT 05934045 | ALK positive anaplastic large-cell lymphoma | circRNAs in patient blood | Prognostic and/or predictive markers/therapeutic targets | University Hospital, Toulouse |
| NCT 04464122 | Neuroendocrine Neoplasm | circRNAs in patient platelets | Diagnostic and predictive markers | University of Roma La Sapienza |
| NCT 06042842 | Hepatocellular carcinoma | has_circ_0004001 in patient blood | Diagnostic marker for early stages of disease | Assiut University |
| NCT 05771337 | Breast cancer | has_circ_0001785(circELP3) has_circ_100219(circFAF1) in patient blood | Diagnostic and prognostic biomarker | Assiut University |
| NCT 03334708 | Pancreatic adenocarcinoma | circRNAs in patient blood | Diagnostic marker for early stages of disease and predictive marker | Memorial Sloan Kettering Cancer Center |
| NCT 04584996 | Pancreaticobiliary cancer | circRNAs in patient blood | Diagnostic marker | Royal Surrey County Hospital NHS Foundation Trust |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J. Circular RNAs: Novel Players in Cancer Mechanisms and Therapeutic Strategies. Int. J. Mol. Sci. 2024, 25, 10121. https://doi.org/10.3390/ijms251810121
Kim J. Circular RNAs: Novel Players in Cancer Mechanisms and Therapeutic Strategies. International Journal of Molecular Sciences. 2024; 25(18):10121. https://doi.org/10.3390/ijms251810121
Chicago/Turabian StyleKim, Jimi. 2024. "Circular RNAs: Novel Players in Cancer Mechanisms and Therapeutic Strategies" International Journal of Molecular Sciences 25, no. 18: 10121. https://doi.org/10.3390/ijms251810121
APA StyleKim, J. (2024). Circular RNAs: Novel Players in Cancer Mechanisms and Therapeutic Strategies. International Journal of Molecular Sciences, 25(18), 10121. https://doi.org/10.3390/ijms251810121







