Exploring Taxonomic and Genetic Relationships in the Pinus mugo Complex Using Genome Skimming Data
Abstract
:1. Introduction
2. Results
2.1. DNA Sequencing, Assembly, and Annotation
2.2. Genetic Characterization of Diversity in Data Sets
2.3. Delimitation of Taxa
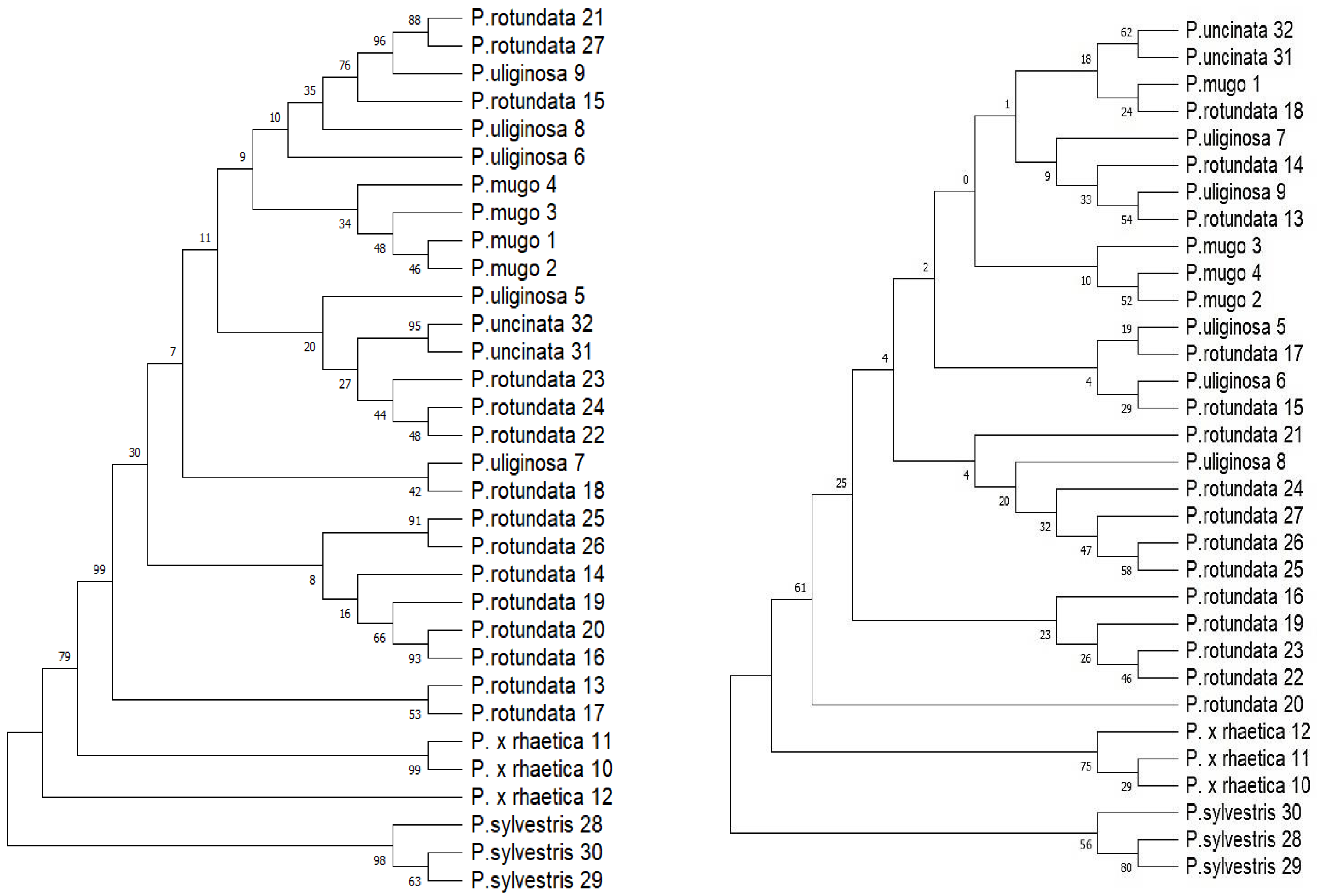
2.3.1. Tree-Based Method
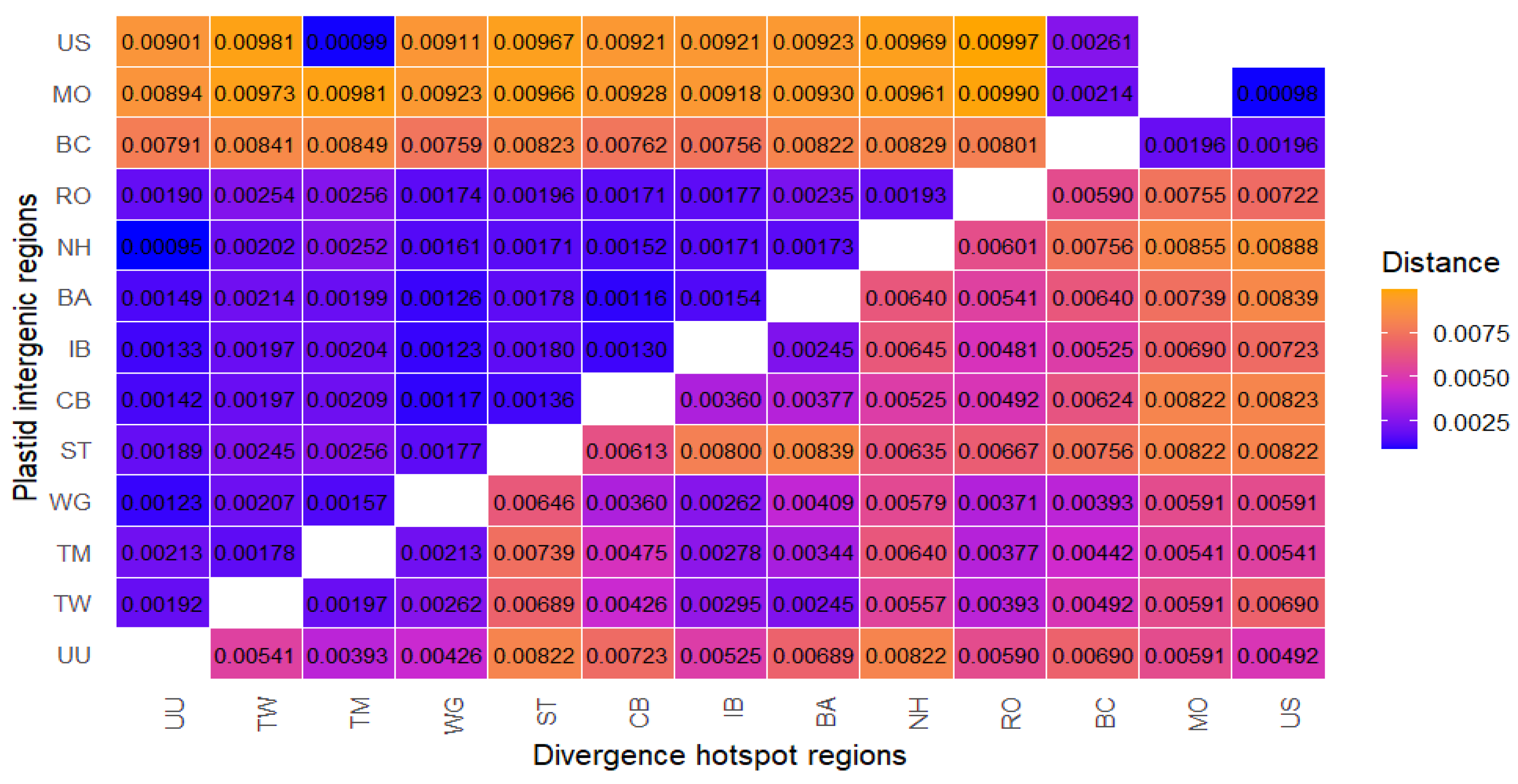
2.3.2. Distance-Based Methods
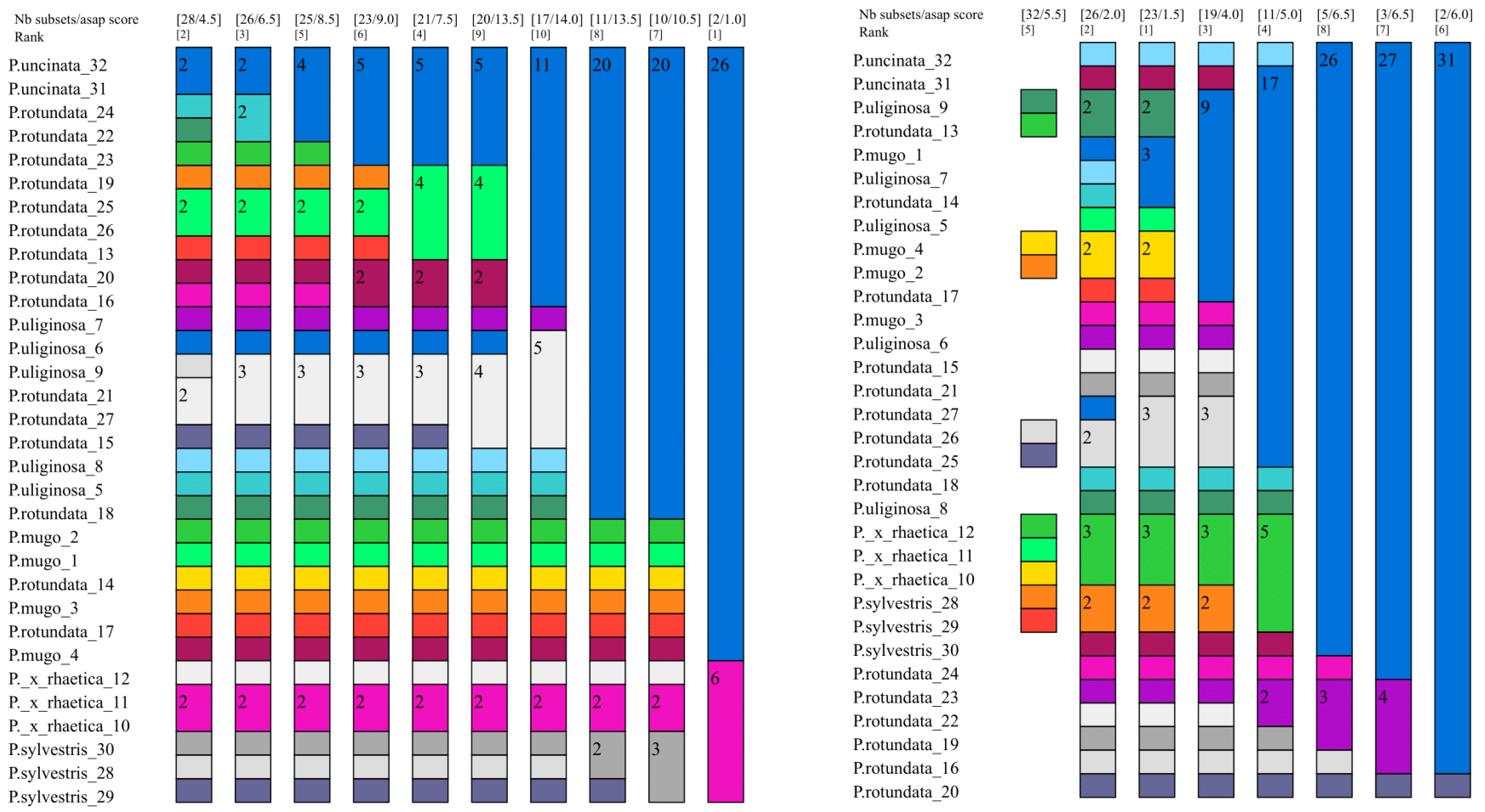
2.3.3. Assembling Species by the Automatic Partitioning Method (ASAP)
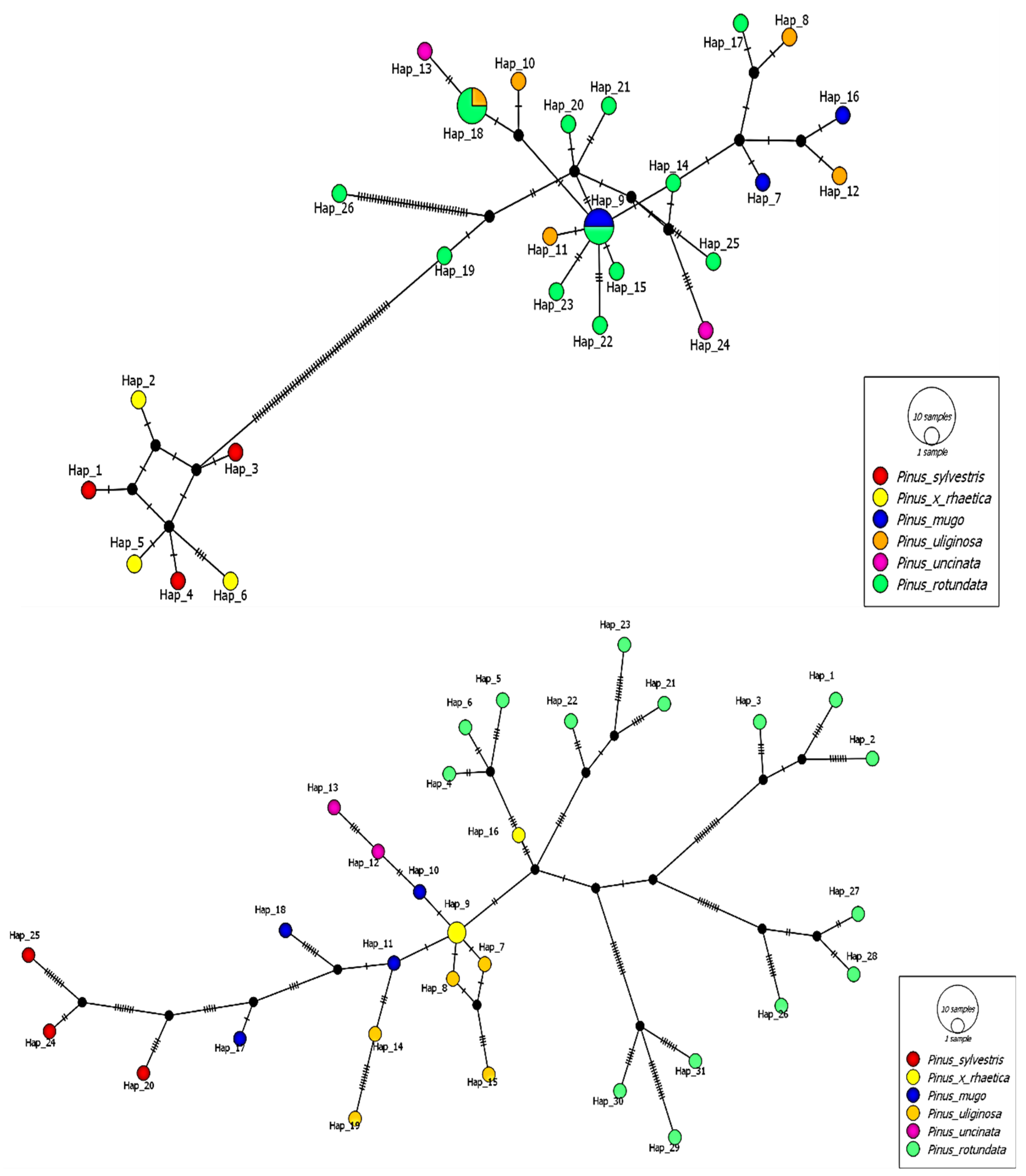
2.4. Genetic Relationships of Taxa and Populations
3. Discussion
4. Materials and Methods
4.1. Plant Material and DNA Extraction
4.2. DNA Sequencing, Assembly, and Annotation
4.3. Data Analysis
4.3.1. Genetic Characterization of Diversity in Data Sets
4.3.2. Delimitation of Taxa
4.3.3. Genetic Diversity of Taxa and Populations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, J.; McLenachan, P.A.; Biggs, P.J.; Winder, L.H.; Schoenfeld, B.I.K.; Narayan, V.V.; Phiri, B.J.; Lockhart, P.J. Environmental bio-monitoring with high-throughput sequencing. Briefings Bioinform. 2013, 14, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Shafer, A.B.A.; Northrup, J.M.; Wikelski, M.; Wittemyer, G.; Wolf, J.B.W. Forecasting Ecological Genomics: High-Tech Animal Instrumentation Meets High-Throughput Sequencing. PLoS Biol. 2016, 14, e1002350. [Google Scholar] [CrossRef] [PubMed]
- Titcomb, G.C.; Jerde, C.L.; Young, H.S. High-Throughput Sequencing for Understanding the Ecology of Emerging Infectious Diseases at the Wildlife-Human Interface. Front. Ecol. Evol. 2019, 7, 126. [Google Scholar] [CrossRef]
- Ríos-Castro, R.; Romero, A.; Aranguren, R.; Pallavicini, A.; Banchi, E.; Novoa, B.; Figueras, A. High-Throughput Sequencing of Environmental DNA as a Tool for Monitoring Eukaryotic Communities and Potential Pathogens in a Coastal Upwelling Ecosystem. Front. Veter-Sci. 2021, 8, 765606. [Google Scholar] [CrossRef]
- Wang, S.; Schneider, D.; Hartke, T.R.; Ballauff, J.; Moura, C.C.d.M.; Schulz, G.; Li, Z.; Polle, A.; Daniel, R.; Gailing, O.; et al. Optimising high-throughput sequencing data analysis, from gene database selection to the analysis of compositional data: A case study on tropical soil nematodes. Front. Ecol. Evol. 2024, 12, 1168288. [Google Scholar] [CrossRef]
- Turner, B.; Paun, O.; Munzinger, J.; Chase, M.W.; Samuel, R. Sequencing of whole plastid genomes and nuclear ribosomal DNA of Diospyros species (Ebenaceae) endemic to New Caledonia: Many species, little divergence. Ann. Bot. 2016, 117, 1175–1185. [Google Scholar] [CrossRef]
- Li, Y.; Sylvester, S.P.; Li, M.; Zhang, C.; Li, X.; Duan, Y.; Wang, X. The Complete Plastid Genome of Magnolia zenii and Genetic Comparison to Magnoliaceae species. Molecules 2019, 24, 261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, Y.; Huang, J.; Ya, J.; Zhe, M.; Zeng, C.; Zhang, Z.; Zhang, S.; Li, D.; Li, H.; et al. DNA barcoding of Cymbidium by genome skimming: Call for next-generation nuclear barcodes. Mol. Ecol. Resour. 2022, 23, 424–439. [Google Scholar] [CrossRef]
- Hamerník, J.; Musil, I. The Pinus mugo complex—Its structuring and general overview of the used nomenclature. J. For. Sci. 2007, 53, 253–266. [Google Scholar] [CrossRef]
- Christensen, K.L. Taxonomic revision of the Pinus mugo complex and P. rhaetica (P. mugo sylvestris) (Pinaceae). Nord. J. Bot. 1987, 7, 383–408. [Google Scholar] [CrossRef]
- Bastl, M.; Burian, M.; Kučera, J.; Prach, K.; Rektoris, L.; Štech, M. Central European Pine Bogs Change along an Altitudinal Gradient. Preslia 2008, 80, 349–363. [Google Scholar]
- Wachowiak, W.; Celiński, K.; Prus-Głowacki, W. Evidence of natural reciprocal hybridisation between Pinus uliginosa and P. sylvestris in the sympatric population of the species. Flora-Morphol. Distrib. Funct. Ecol. Plants 2005, 200, 563–568. [Google Scholar] [CrossRef]
- Wachowiak, W.; Prus-Głowacki, W. Hybridisation processes in sympatric populations of pines Pinus sylvestris L., P. mugo Turra and P. uliginosa Neumann. Plant Syst. Evol. 2007, 271, 29–40. [Google Scholar] [CrossRef]
- Lewandowski, A.; Wiśniewska, M. Short Note: Crossability Between Pinus uliginosa and its Putative Parental Species Pinus sylvestris and Pinus mugo. Silvae Genet. 2006, 55, 52–54. [Google Scholar] [CrossRef]
- Danusevičius, D.; Marozas, V.; Brazaitis, G.; Petrokas, R.; Christensen, K.I. Spontaneous Hybridization between Pinus mugo and Pinus sylvestris at the Lithuanian Seaside: A Morphological Survey. Sci. World J. 2012, 2012, 172407. [Google Scholar] [CrossRef]
- Kormutak, A.; Demankova, B.; Gömöry, D. Spontaneous Hybridization between Pinus sylvestris L. and P. mugo Turra in Slovakia. Silvae Genet. 2008, 57, 76–82. [Google Scholar] [CrossRef]
- Wachowiak, W.; Żukowska, W.B.; Wójkiewicz, B.; Cavers, S.; Litkowiec, M. Hybridization in contact zone between temperate European pine species. Tree Genet. Genomes 2016, 12, 48. [Google Scholar] [CrossRef]
- Sobierajska, K.; Wachowiak, W.; Zaborowska, J.; Łabiszak, B.; Wójkiewicz, B.; Sękiewicz, M.; Jasińska, A.K.; Sękiewicz, K.; Boratyńska, K.; Marcysiak, K.; et al. Genetic Consequences of Hybridization in Relict Isolated Trees Pinus sylvestris and the Pinus mugo Complex. Forests 2020, 11, 1086. [Google Scholar] [CrossRef]
- Łabiszak, B.; Wachowiak, W. Molecular Signatures of Reticulate Evolution within the Complex of European Pine Taxa. Forests 2021, 12, 489. [Google Scholar] [CrossRef]
- Zaborowska, J.; Łabiszak, B.; Wachowiak, W. Population history of European mountain pines Pinus mugo and Pinus uncinata revealed by mitochondrial DNA markers. J. Syst. Evol. 2019, 58, 474–486. [Google Scholar] [CrossRef]
- Staszkiewicz, J.; Tyszkiewicz, M. Variability of natural hybrids of Pinus sylvestris L. × P. mugo Turra (= P. × rotundata Link) in south-western Poland and in selected localities in Bohemia and Moravia. Fragm. Florist. Et Geobot. Pol. 1972, 18, 173–191. [Google Scholar]
- Boratyńska, K.; Bobowicz, M.A. Pinus uncinata Ramond Taxonomy Based on Needle Characters. Plant Syst. Evol. 2001, 227, 183–194. [Google Scholar] [CrossRef]
- Boratyńska, K.; Boratyński, A.; Lewandowski, A. Morphology of Pinus uliginosa (Pinaceae) needles from populations exposed to and isolated from the direct influence of Pinus sylvestris. Bot. J. Linn. Soc. 2003, 142, 83–91. [Google Scholar] [CrossRef]
- Monteleone, I.; Ferrazzini, D.; Belletti, P. Effectiveness of neutral RAPD markers to detect genetic divergence between the subspecies uncinata and mugo of Pinus mugo Turra. Silva Fenn. 2006, 40, 391. [Google Scholar] [CrossRef]
- Boratyńska, K.; Boratyński, A. Taxonomic differences among closely related pines Pinus sylvestris, P. mugo, P. uncinata, P. rotundata and P. uliginosa as revealed in needle sclerenchyma cells. Flora 2007, 202, 555–569. [Google Scholar] [CrossRef]
- Prus-Głowacki, W.; Bajus, E.; Ratyńska, H. Taxonomic position of Pinus uliginosa Neumann as related to other taxa of Pinus mugo complex. Acta Soc. Bot. Pol. 1998, 67, 269–274. [Google Scholar] [CrossRef]
- Lewandowski, A.; Boratyński, A.; Mejnartowicz, L. Allozyme investigations on the genetic differentiation between closely related pines —Pinus sylvestris, P. mugo, P. uncinata, andP. uliginosa (Pinaceae). Plant Syst. Evol. 2000, 221, 15–24. [Google Scholar] [CrossRef]
- Siedlewska, A.; Prus-Głowacki, W. Genetic structure and taxonomic position of Pinus uliginosa Neumann population from Wielkie Torfowisko Batorowskie in Stołowe Mts. (locus classicus). Acta Soc. Bot. Pol. 1995, 64, 51–58. [Google Scholar] [CrossRef]
- Celiński, K.; Sokołowska, J.; Zemleduch-Barylska, A.; Kuna, R.; Kijak, H.; Staszak, A.M.; Wojnicka-Półtorak, A.; Chudzińska, E. Seed Total Protein Profiling in Discrimination of Closely Related Pines: Evidence from the Pinus mugo Complex. Plants 2020, 9, 872. [Google Scholar] [CrossRef]
- Bonikowski, R.; Celiński, K.; Wojnicka-Półtorak, A.; Maliński, T. Composition of Essential Oils Isolated from the Needles of Pinus uncinata and P. uliginosa Grown in Poland. Nat. Prod. Commun. 2015, 10, 371–373. [Google Scholar] [CrossRef]
- Celiński, K.; Bonikowski, R.; Wojnicka-Półtorak, A.; Chudzińska, E.; Maliński, T. Volatiles as Chemosystematic Markers for Distinguishing Closely Related Species within the Pinus mugo Complex. Chem. Biodivers. 2015, 12, 1208–1213. [Google Scholar] [CrossRef] [PubMed]
- Bogunić, F.; Siljak-Yakovlev, S.; Muratović, E.; Pustahija, F.; Medjedović, S. Molecular cytogenetics and flow cytometry reveal conserved genome organization in Pinus mugo and P. uncinata. Ann. For. Sci. 2011, 68, 179–187. [Google Scholar] [CrossRef]
- Celiński, K.; Chudzińska, E.; Gmur, A.; Piosik, Ł.; Wojnicka-Półtorak, A. Cytological characterization of three closely related pines—Pinus mugo, P. uliginosa and P. × rhaetica from the Pinus mugo complex (Pinaceae). Biologia 2019, 74, 751–756. [Google Scholar] [CrossRef]
- Celiński, K.; Zbránková, V.; Wojnicka-Półtorak, A.; Chudzińska, E. Biogeography and evolutionary factors determine genetic differentiation of Pinus mugo (Turra) in the Tatra Mountains (Central Europe). J. Mt. Sci. 2015, 12, 549–557. [Google Scholar] [CrossRef]
- Celiński, K.; Pawlaczyk, E.M.; Wojnicka-Półtorak, A.; Chudzińska, E.; Prus-Głowacki, W. Cross-species amplification and characterization of microsatellite loci in Pinus mugo Turra. Biologia 2013, 68, 621–626. [Google Scholar] [CrossRef]
- Celiński, K.; Kijak, H.; Wojnicka-Półtorak, A.; Buczkowska-Chmielewska, K.; Sokołowska, J.; Chudzińska, E. Effectiveness of the DNA barcoding approach for closely related conifers discrimination: A case study of the Pinus mugo complex. Comptes Rendus Biol. 2017, 340, 339–348. [Google Scholar] [CrossRef]
- Sokołowska, J.; Fuchs, H.; Celiński, K. Assessment of ITS2 Region Relevance for Taxa Discrimination and Phylogenetic Inference among Pinaceae. Plants 2022, 11, 1078. [Google Scholar] [CrossRef]
- Sokołowska, J.; Fuchs, H.; Celiński, K. New Insight into Taxonomy of European Mountain Pines, Pinus mugo Complex, Based on Complete Chloroplast Genomes Sequencing. Plants 2021, 10, 1331. [Google Scholar] [CrossRef]
- Straub, S.C.K.; Parks, M.; Weitemier, K.; Fishbein, M.; Cronn, R.C.; Liston, A. Navigating the tip of the genomic iceberg: Next-generation sequencing for plant systematics. Am. J. Bot. 2012, 99, 349–364. [Google Scholar] [CrossRef]
- Dodsworth, S. Genome skimming for next-generation biodiversity analysis. Trends Plant Sci. 2015, 20, 525–527. [Google Scholar] [CrossRef]
- Weitemier, K.; Straub, S.C.K.; Cronn, R.C.; Fishbein, M.; Schmickl, R.; McDonnell, A.; Liston, A. Hyb-Seq: Combining target enrichment and genome skimming for plant phylogenomics. Appl. Plant Sci. 2014, 2, 1400042. [Google Scholar] [CrossRef] [PubMed]
- Nevill, P.G.; Zhong, X.; Tonti-Filippini, J.; Byrne, M.; Hislop, M.; Thiele, K.; van Leeuwen, S.; Boykin, L.M.; Small, I. Large scale genome skimming from herbarium material for accurate plant identification and phylogenomics. Plant Methods 2020, 16, 1. [Google Scholar] [CrossRef]
- Meena, R.K.; Negi, N.; Uniyal, N.; Bhandari, M.S.; Sharma, R.; Ginwal, H.S. Genome skimming-based STMS marker discovery and its validation in temperate hill bamboo Drepanostachyum falcatum. J. Genet. 2021, 100, 28. [Google Scholar] [CrossRef] [PubMed]
- Kane, N.; Sveinsson, S.; Dempewolf, H.; Yang, J.Y.; Zhang, D.; Engels, J.M.M.; Cronk, Q. Ultra-barcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. Am. J. Bot. 2012, 99, 320–329. [Google Scholar] [CrossRef]
- Ruhsam, M.; Rai, H.S.; Mathews, S.; Ross, T.G.; Graham, S.W.; Raubeson, L.A.; Mei, W.; Thomas, P.I.; Gardner, M.F.; Ennos, R.A.; et al. Does complete plastid genome sequencing improve species discrimination and phylogenetic resolution in Araucaria? Mol. Ecol. Resour. 2015, 15, 1067–1078. [Google Scholar] [CrossRef]
- Ji, Y.; Liu, C.; Yang, Z.; Yang, L.; He, Z.; Wang, H.; Yang, J.; Yi, T. Testing and using complete plastomes and ribosomal DNA sequences as the next generation DNA barcodes in Panax (Araliaceae). Mol. Ecol. Resour. 2019, 19, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. Popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Celiński, K.; Kijak, H.; Barylski, J.; Grabsztunowicz, M.; Wojnicka-Półtorak, A.; Chudzińska, E. Characterization of the complete chloroplast genome of Pinus uliginosa (Neumann) from the Pinus mugo complex. Conserv. Genet. Resour. 2016, 9, 209–212. [Google Scholar] [CrossRef]
- Heuertz, M.; Teufel, J.; González-Martínez, S.C.; Soto, A.; Fady, B.; Alía, R.; Vendramin, G.G. Geography determines genetic relationships between species of mountain pine (Pinus mugo complex) in western Europe. J. Biogeogr. 2010, 37, 541–556. [Google Scholar] [CrossRef]
- Lv, S.-Y.; Ye, X.-Y.; Li, Z.-H.; Ma, P.-F.; Li, D.-Z. Testing complete plastomes and nuclear ribosomal DNA sequences for species identification in a taxonomically difficult bamboo genus Fargesia. Plant Divers. 2023, 45, 147–155. [Google Scholar] [CrossRef]
- Ji, Y.; Liu, C.; Yang, J.; Jin, L.; Yang, Z.; Yang, J.-B. Ultra-Barcoding Discovers a Cryptic Species in Paris yunnanensis (Melanthiaceae), a Medicinally Important Plant. Front. Plant Sci. 2020, 11, 411. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Mo, Z.; Yang, J.; Cai, J.; Ye, L.; Zou, J.; Qin, H.; Zheng, W.; Hollingsworth, P.M.; Li, D.; et al. Testing genome skimming for species discrimination in the large and taxonomically difficult genus Rhododendron. Mol. Ecol. Resour. 2021, 22, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Raimondeau, P.; Manzi, S.; Brucato, N.; Kinipi, C.; Leavesley, M.; Ricaut, F.-X.; Besnard, G. Genome skims analysis of betel palms (Areca spp., Arecaceae) and development of a profiling method to assess their plastome diversity. Gene 2021, 800, 145845. [Google Scholar] [CrossRef]
- Simmonds, S.E.; Smith, J.F.; Davidson, C.; Buerki, S. Phylogenetics and comparative plastome genomics of two of the largest genera of angiosperms, Piper and Peperomia (Piperaceae). Mol. Phylogenetics Evol. 2021, 163, 107229. [Google Scholar] [CrossRef]
- Wagner, N.D.; Volf, M.; Hörandl, E. Highly Diverse Shrub Willows (Salix L.) Share Highly Similar Plastomes. Front. Plant Sci. 2021, 12, 662715. [Google Scholar] [CrossRef]
- Bakker, F.T. Herbarium genomics: Skimming and plastomics from archival specimens. Webbia 2017, 72, 35–45. [Google Scholar] [CrossRef]
- Zeng, C.-X.; Hollingsworth, P.M.; Yang, J.; He, Z.-S.; Zhang, Z.-R.; Li, D.-Z.; Yang, J.-B. Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods 2018, 14, 43. [Google Scholar] [CrossRef]
- Morse, A.M.; Peterson, D.G.; Islam-Faridi, M.N.; Smith, K.E.; Magbanua, Z.; Garcia, S.A.; Kubisiak, T.L.; Amerson, H.V.; Carlson, J.E.; Nelson, C.D.; et al. Evolution of Genome Size and Complexity in Pinus. PLoS ONE 2009, 4, e4332. [Google Scholar] [CrossRef] [PubMed]
- Zonneveld, B.J.M. Conifer genome sizes of 172 species, covering 64 of 67 genera, range from 8 to 72 picogram. Nord. J. Bot. 2012, 30, 490–502. [Google Scholar] [CrossRef]
- De La Torre, A.R.; Birol, I.; Bousquet, J.; Ingvarsson, P.K.; Jansson, S.; Jones, S.J.M.; Keeling, C.I.; MacKay, J.; Nilsson, O.; Ritland, K.; et al. Insights into Conifer Giga-Genomes. Plant Physiol. 2014, 166, 1724–1732. [Google Scholar] [CrossRef]
- Álvarez, I.; Wendel, J. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenetics Evol. 2003, 29, 417–434. [Google Scholar] [CrossRef]
- Liston, A.; Robinson, W.A.; Oliphant, J.M.; Alvarez-Buylla, E.R. Length Variation in the Nuclear Ribosomal DNA Internal Transcribed Spacer Region of Non-Flowering Seed Plants. Syst. Bot. 1996, 21, 109–120. [Google Scholar] [CrossRef]
- Marrocco, R.; Gelati, M.T.; Maggini, F. Nucleotide sequence of the internal transcribed spacers and 5.8s region of ribosomal DNA in Pinus pinea L. DNA Seq. 1996, 6, 175–177. [Google Scholar] [CrossRef]
- Gernandt, D.S.; Liston, A. Internal transcribed spacer region evolution in Larix and Pseudotsuga (Pinaceae). Am. J. Bot. 1999, 86, 711–723. [Google Scholar] [CrossRef]
- Maggini, F.; Frediani, M.; Gelati, M.T. Nucleotide Sequence of the Internal Transcribed Spacers of Ribosomal DNA in Picea Abies Karst. DNA Seq. 2000, 11, 87–89. [Google Scholar] [CrossRef]
- Liu, Z.; Zeng, X.; Yang, D.; Chu, G.; Yuan, Z.; Chen, S. Applying DNA barcodes for identification of plant species in the family Araliaceae. Gene 2012, 499, 76–80. [Google Scholar] [CrossRef]
- Gao, T.; Sun, Z.; Yao, H.; Song, J.; Zhu, Y.; Ma, X.; Chen, S. Identification of Fabaceae plants using the DNA barcode matK. Planta Medica 2011, 77, 92–94. [Google Scholar] [CrossRef]
- Liu, B.; Ma, Z.; Ren, C.; Hodel, R.G.J.; Sun, M.; Liu, X.; Liu, G.; Hong, D.; Zimmer, E.A.; Wen, J. Capturing single-copy nuclear genes, organellar genomes, and nuclear ribosomal DNA from deep genome skimming data for plant phylogenetics: A case study in Vitaceae. J. Syst. Evol. 2021, 59, 1124–1138. [Google Scholar] [CrossRef]
- Su, N.; Liu, B.-B.; Wang, J.-R.; Tong, R.-C.; Ren, C.; Chang, Z.-Y.; Zhao, L.; Potter, D.; Wen, J. On the Species Delimitation of the Maddenia Group of Prunus (Rosaceae): Evidence From Plastome and Nuclear Sequences and Morphology. Front. Plant Sci. 2021, 12, 743643. [Google Scholar] [CrossRef]
- Kormutak, A.; Galgoci, M.; Sukenikova, D.; Bolecek, P.; Libantova, J.; Gőmőry, D. Maternal inheritance of chloroplast DNA in Pinus mugo Turra: A case study of Pinus mugo × Pinus sylvestris crossing. Plant Syst. Evol. 2017, 304, 71–76. [Google Scholar] [CrossRef]
- Wang, B.; Wang, X.-R. Mitochondrial DNA capture and divergence in Pinus provide new insights into the evolution of the genus. Mol. Phylogenetics Evol. 2014, 80, 20–30. [Google Scholar] [CrossRef]
- Kane, N.C.; Cronk, Q. Botany without borders: Barcoding in focus. Mol. Ecol. 2008, 17, 5175–5176. [Google Scholar] [CrossRef]
- Bucci, G.; Anzidei, M.; Madaghiele, A.; Vendramin, G.G. Detection of haplotypic variation and natural hybridization in halepensis-complex pine species using chloroplast simple sequence repeat (SSR) markers. Mol. Ecol. 1998, 7, 1633–1643. [Google Scholar] [CrossRef]
- Businský, R.; Frantík, T.; Vít, P. Morphological evaluation of the Pinus kesiya complex (Pinaceae). Plant Syst. Evol. 2013, 300, 273–285. [Google Scholar] [CrossRef]
- Neumann, C. Über Eine Auf Den Seefeldern Bei Reinerz u. Einigen Ähnlichen Gebirgsmooren Der Königl. Oberförsterei Karlsberg in Der Graftschaft Glatz Vorkommende Noch Unbeschrieben Form Der Gattung Pinus. Jahresber Schlesische Ges. Für Vaterländische Kult. 1837, 11, 52–57. [Google Scholar]
- Rozporządzenie Ministra Środowiska Z Dnia 9 Października 2014 R. W Sprawie Ochrony Gatunkowej Roślin. W: Dz.U. 2014 Poz. 1409.; Poland. Internetowy System Aktów Prawnych. Available online: https://isap.sejm.gov.pl/isap.nsf (accessed on 20 July 2024).
- Bastl, M.; Štechová, T.; Prach, K. Effect of Disturbance on the Vegetation of Peat Bogs with Pinus Rotundata in the Třeboň Basin, Czech Republic. Preslia 2009, 81, 105–117. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kazutaka, K.; Misakwa, K.; Kei-ichi, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Zhang, M.-F.; Xue, J.; Dong, R.; Du, Y.-P.; Zhang, X.-H. Chloroplast genomic resources for phylogeny and DNA barcoding: A case study on Fritillaria. Sci. Rep. 2018, 8, 1184. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press (OUP): Oxford, UK, 2000; pp. 3–295. [Google Scholar]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 2020, 21, 609–620. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]


| Alignment | Data Set Code | Length (bp) | Variable Sites (% divergence) | Parsimony Informative Sites (%) |
|---|---|---|---|---|
| Plastid coding genes | A | 45,467 | 75 (0.165%) | 17 (0.037%) |
| Mitochondrial coding genes | B | 8378 | 128 (1.528%) | 68 (0.812%) |
| Plastid intergenic regions | C | 7092 | 59 (0.832%) | 33 (0.465%) |
| nrDNA cistron | D | 7849 | 10 (0.127%) | 2 (0.025%) |
| ITS | E | 2881 | 3 (0.104%) | 0 (0.000%) |
| matK + rbcL | F | 2976 | 8 (0.269%) | 1 (0.034%) |
| Divergence hotspot regions | G | 1021 | 20 (1.959%) | 15 (1.469%) |
| Alignment | Data Set Code | Tree-Based Method | Distance-Based Method | ASAP * |
|---|---|---|---|---|
| Plastid coding genes | A | 0/6 (0.00%) | 0/6 (0.0%) | 0/6 (0.00%) |
| Mitochondrial coding genes | B | 2/6 (33.33%) | 2/6 (33.33%) | 0/6 (0.00%) |
| Plastid intergenic regions | C | 2/6 (33.33%) | 2/6 (33.33%) | 0/6 (0.00%) |
| nr DNA cistron | D | 1/6 (16.67%) | 1/6 (16.67%) | 0/6 (0.00%) |
| ITS | E | 0/6 (0.00%) | 0/6 (0.00%) | 0/6 (0.00%) |
| matK + rbcL | F | 0/6 (0.00%) | 0/6 (0.00%) | 0/6 (0.00%) |
| Divergence hotspot regions | G | 3/6 (50.00%) | 3/6 (50.0%) | 1/6 (16.67%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sikora, J.; Celiński, K. Exploring Taxonomic and Genetic Relationships in the Pinus mugo Complex Using Genome Skimming Data. Int. J. Mol. Sci. 2024, 25, 10178. https://doi.org/10.3390/ijms251810178
Sikora J, Celiński K. Exploring Taxonomic and Genetic Relationships in the Pinus mugo Complex Using Genome Skimming Data. International Journal of Molecular Sciences. 2024; 25(18):10178. https://doi.org/10.3390/ijms251810178
Chicago/Turabian StyleSikora, Joanna, and Konrad Celiński. 2024. "Exploring Taxonomic and Genetic Relationships in the Pinus mugo Complex Using Genome Skimming Data" International Journal of Molecular Sciences 25, no. 18: 10178. https://doi.org/10.3390/ijms251810178
APA StyleSikora, J., & Celiński, K. (2024). Exploring Taxonomic and Genetic Relationships in the Pinus mugo Complex Using Genome Skimming Data. International Journal of Molecular Sciences, 25(18), 10178. https://doi.org/10.3390/ijms251810178





