Vitamin B6 Pathway Maintains Glioblastoma Cell Survival in 3D Spheroid Cultures
Abstract
:1. Introduction
2. Results
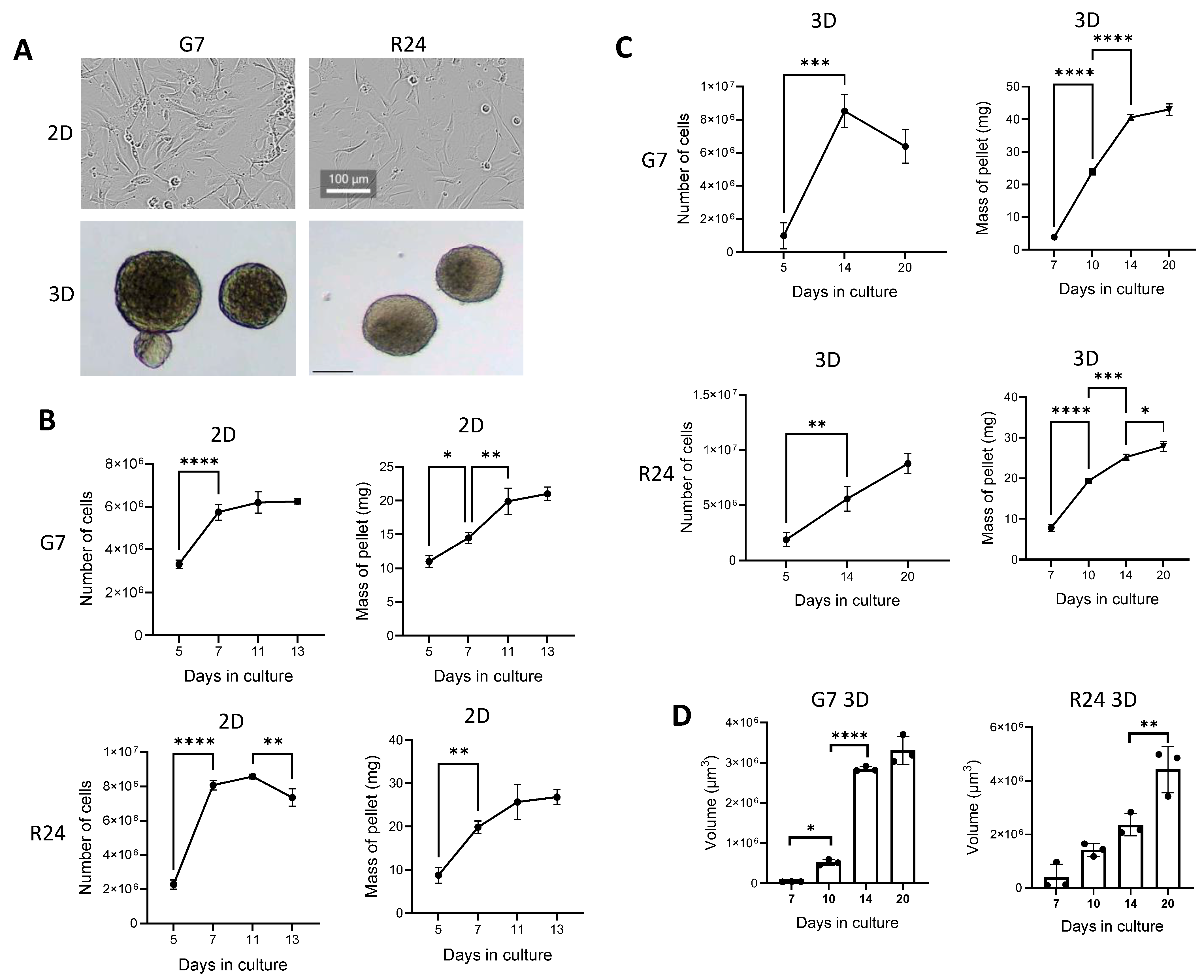
2.1. Characterisation of Cells under 2D and 3D Cell Culture Conditions
2.2. Validation and Molecular Analysis of 2D and 3D Spheroid Cultures
2.3. Metabolomic Analysis of 2D Monolayer and 3D Spheroid Cultures
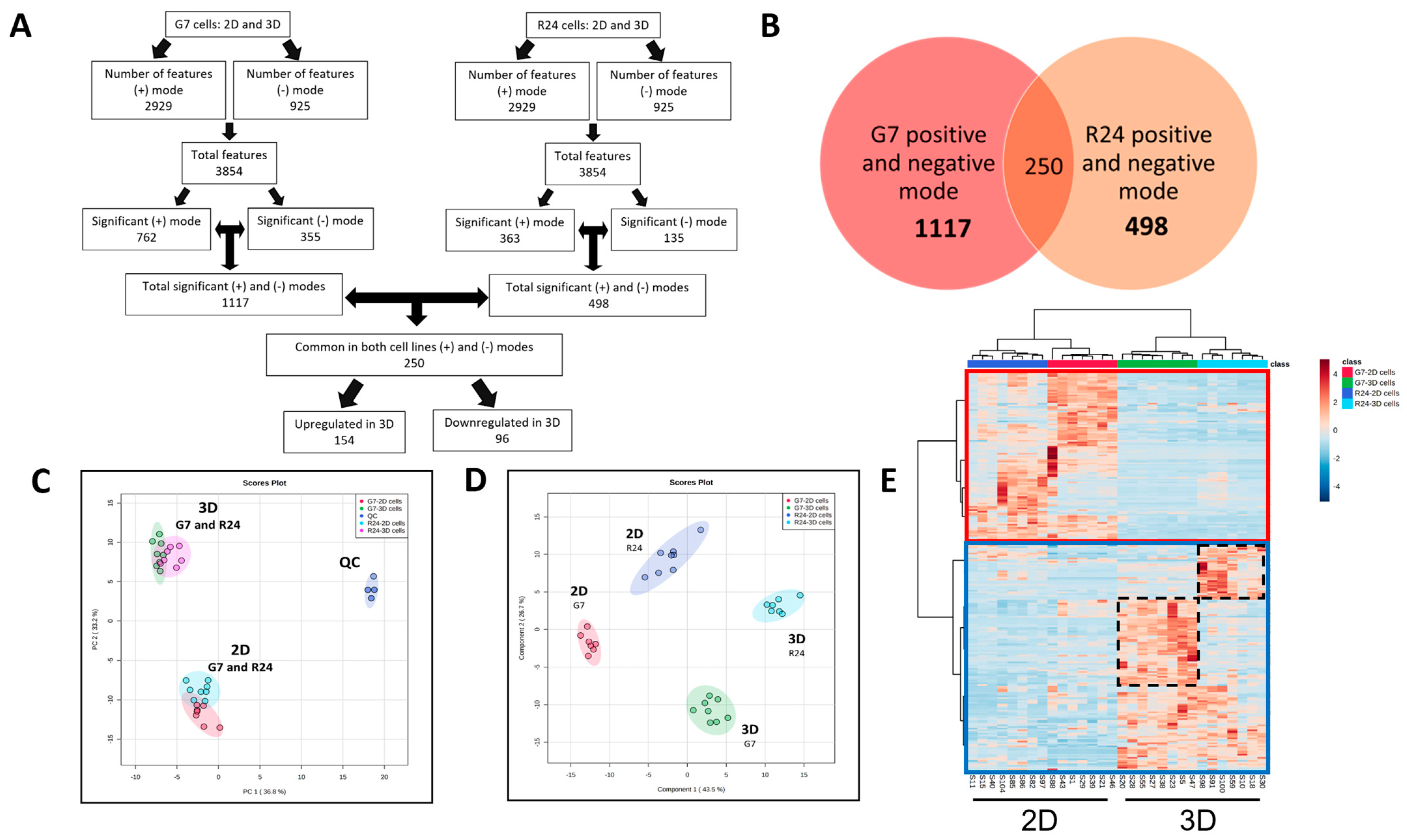
2.4. Multivariate Analysis of Significantly Altered Features in 2D vs. 3D GBM Cultures
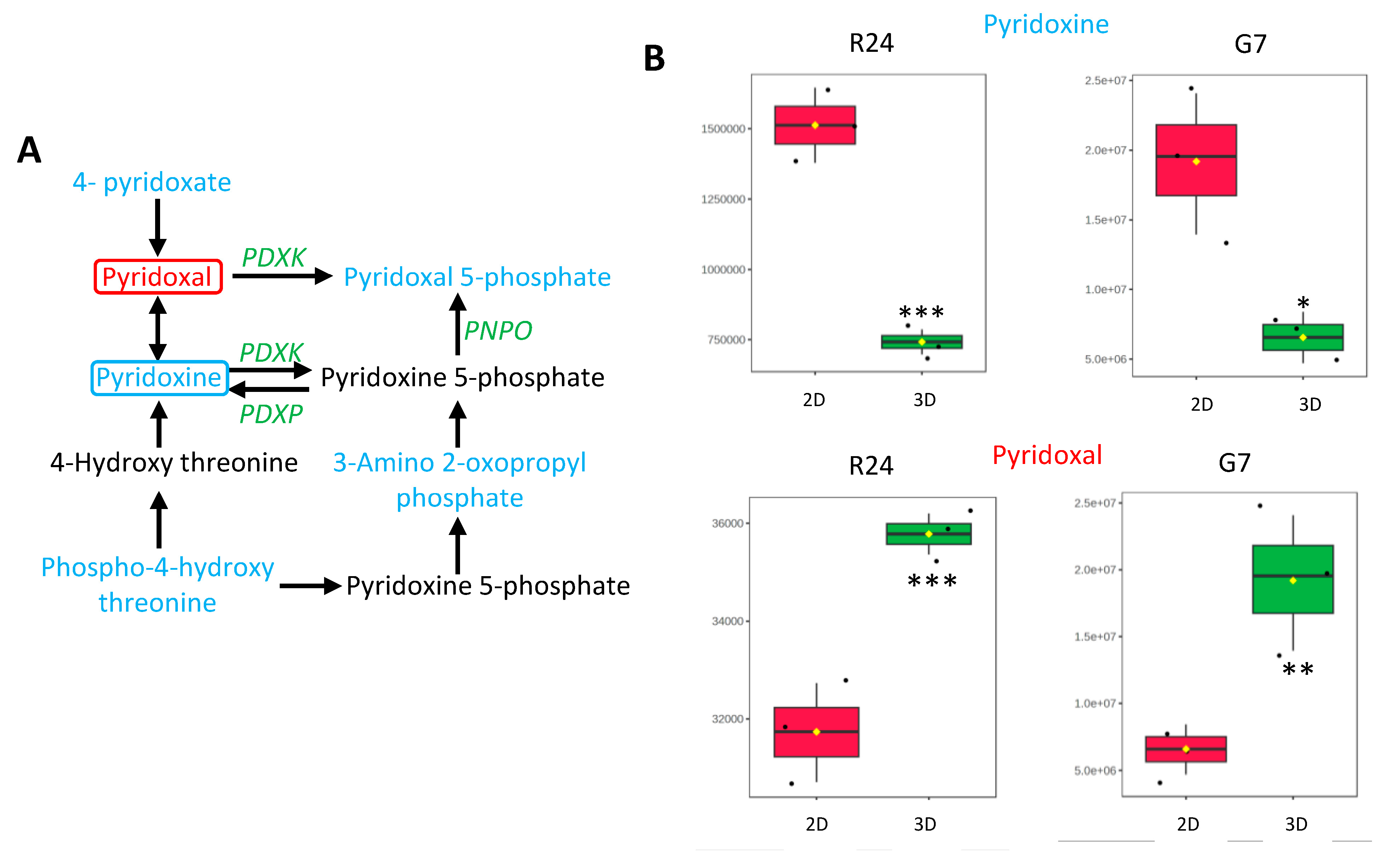
2.5. Identification and Validation of Significantly Altered Pathways in GBM 3D Spheroid Cultures
2.6. Targeting the Vitamin B6 Pathway in GBMs
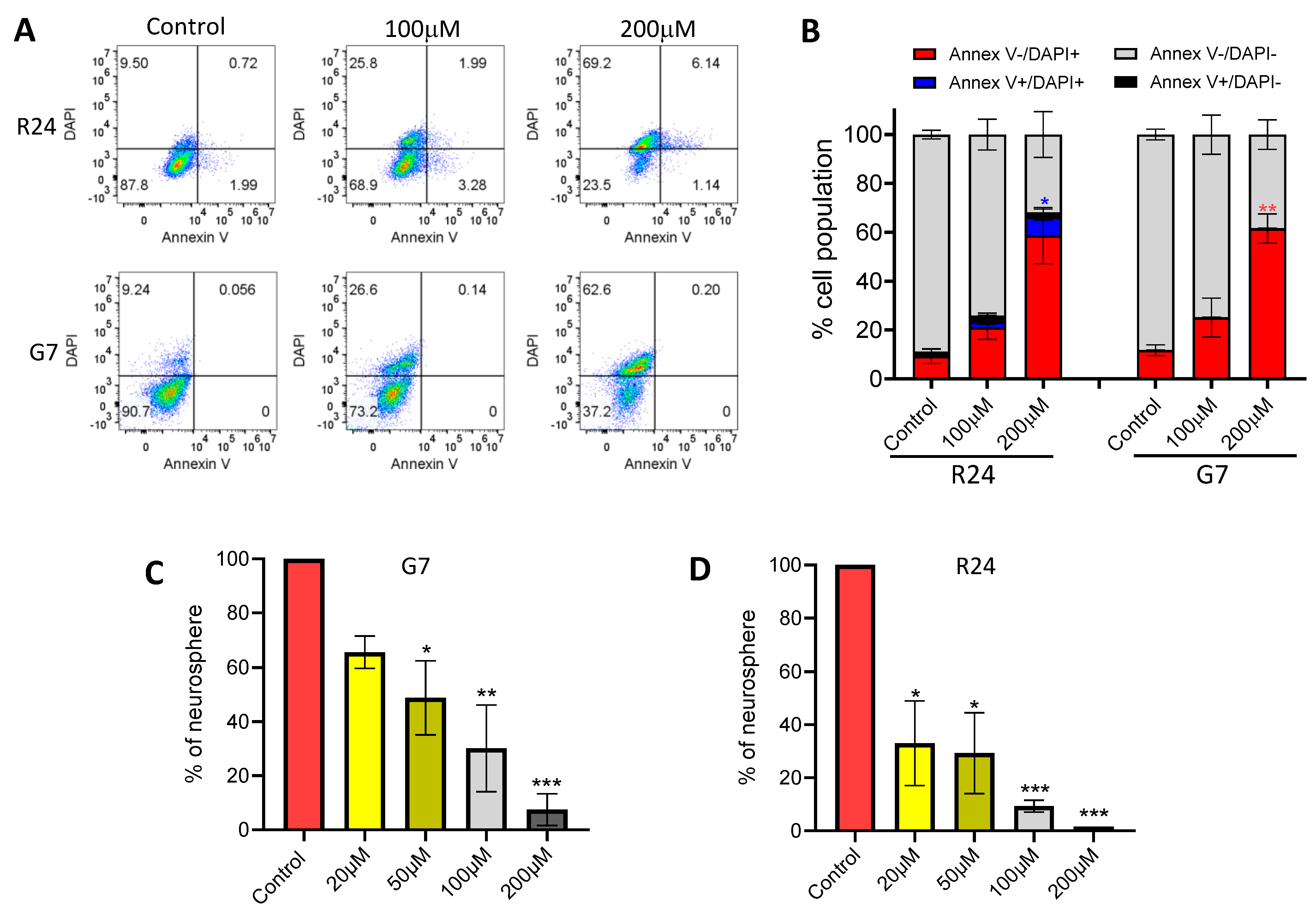
2.7. Mechanistic Studies Investigating the Effects of Hydralazine in 3D Spheroid Cultures
3. Discussion
4. Materials and Methods
4.1. Glioblastoma Cell Culture
4.2. 2D and 3D Cell Proliferation Studies
4.3. Analysis of Cell Death and Apoptosis
4.4. Analysis of Hypoxia in 2D and 3D Cell Cultures
4.5. Analysis of Reactive Oxygen Species
4.6. Analysis of Stem Cell Markers
4.7. Generation and Preparation of 2D and 3D Samples for Metabolomics Analysis
4.8. High Performance Liquid Chromatography (HPLC) Mass Spectrometry (HPLC-MS)-Based Metabolomics Analysis
4.9. Metabolomics Data Analysis
4.10. Statistical Analysis
4.11. LC-MS Validation of Metabolites
4.12. Effect of Hydralazine on 2D and 3D Cell Proliferation
4.13. Neurosphere Formation Assay
4.14. Western Blot Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.B.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of Radiotherapy with Concomitant and Adjuvant Temozolomide versus Radiotherapy Alone on Survival in Glioblastoma in a Randomised Phase III Study: 5-Year Analysis of the EORTC-NCIC Trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-Cell RNA-Seq Highlights Intratumoral Heterogeneity in Primary Glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef]
- Behnan, J.; Finocchiaro, G.; Hanna, G. The Landscape of the Mesenchymal Signature in Brain Tumours. Brain 2019, 142, 847–866. [Google Scholar] [CrossRef]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma Stem Cells Promote Radioresistance by Preferential Activation of the DNA Damage Response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.U.; Carruthers, R.; Gilmour, L.; Yildirim, S.; Watts, C.; Chalmers, A.J. Selective Inhibition of Parallel DNA Damage Response Pathways Optimizes Radiosensitization of Glioblastoma Stem-like Cells. Cancer Res. 2015, 75, 4416–4428. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Tu, R.; Liu, H.; Qing, G. Regulation of Cancer Cell Metabolism: Oncogenic MYC in the Driver’s Seat. Signal Transduct. Target. Ther. 2020, 5, 124. [Google Scholar] [CrossRef]
- Kumar, R.; Mishra, A.; Gautam, P.; Feroz, Z.; Vijayaraghavalu, S.; Likos, E.M.; Shukla, G.C.; Kumar, M. Metabolic Pathways, Enzymes, and Metabolites: Opportunities in Cancer Therapy. Cancers 2022, 14, 5268. [Google Scholar] [CrossRef]
- Stine, Z.E.; Schug, Z.T.; Salvino, J.M.; Dang, C.V. Targeting Cancer Metabolism in the Era of Precision Oncology. Nat. Rev. Drug Discov. 2022, 21, 141–162. [Google Scholar] [CrossRef]
- Han, S.; Liu, Y.; Cai, S.J.; Qian, M.; Ding, J.; Larion, M.; Gilbert, M.R.; Yang, C. IDH Mutation in Glioma: Molecular Mechanisms and Potential Therapeutic Targets. Br. J. Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef]
- Murugan, A.K.; Alzahrani, A.S. Isocitrate Dehydrogenase IDH1 and IDH2 Mutations in Human Cancer: Prognostic Implications for Gliomas. Br. J. Biomed. Sci. 2022, 79, 10208. [Google Scholar] [CrossRef] [PubMed]
- Reitman, Z.J.; Jin, G.; Karoly, E.D.; Spasojevic, I.; Yang, J.; Kinzler, K.W.; He, Y.; Bigner, D.D.; Vogelstein, B.; Yan, H. Profiling the Effects of Isocitrate Dehydrogenase 1 and 2 Mutations on the Cellular Metabolome. Proc. Natl. Acad. Sci. USA 2011, 108, 3270–3275. [Google Scholar] [CrossRef]
- Intlekofer, A.M.; Dematteo, R.G.; Venneti, S.; Finley, L.W.S.; Lu, C.; Judkins, A.R.; Rustenburg, A.S.; Grinaway, P.B.; Chodera, J.D.; Cross, J.R.; et al. Hypoxia Induces Production of L-2-Hydroxyglutarate. Cell Metab. 2015, 22, 304–311. [Google Scholar] [CrossRef]
- Kickingereder, P.; Sahm, F.; Radbruch, A.; Wick, W.; Heiland, S.; von Deimling, A.; Bendszus, M.; Wiestler, B. IDH Mutation Status Is Associated with a Distinct Hypoxia/Angiogenesis Transcriptome Signature Which Is Non-Invasively Predictable with rCBV Imaging in Human Glioma. Sci. Rep. 2015, 5, 16238. [Google Scholar] [CrossRef] [PubMed]
- Givechian, K.B.; Garner, C.; Benz, S.; Rabizadeh, S.; Soon-Shiong, P. Glycolytic Expression in Lower-Grade Glioma Reveals an Epigenetic Association between IDH Mutation Status and PDL1/2 Expression. Neuro-Oncol. Adv. 2021, 3, vdaa162. [Google Scholar] [CrossRef]
- Pathmanapan, S.; Poon, R.; De Renshaw, T.B.; Nadesan, P.; Nakagawa, M.; Seesankar, G.A.; Loe, A.K.H.; Zhang, H.H.; Guinovart, J.J.; Duran, J.; et al. Mutant IDH Regulates Glycogen Metabolism from Early Cartilage Development to Malignant Chondrosarcoma Formation. Cell Rep. 2023, 42, 112578. [Google Scholar] [CrossRef] [PubMed]
- Quartuccio, N.; Laudicella, R.; Vento, A.; Pignata, S.; Mattoli, M.V.; Filice, R.; Comis, A.D.; Arnone, A.; Baldari, S.; Cabria, M.; et al. The Additional Value of 18F-FDG PET and MRI in Patients with Glioma: A Review of the Literature from 2015 to 2020. Diagnostics 2020, 10, 357. [Google Scholar] [CrossRef]
- Son, B.; Lee, S.; Kim, H.; Kang, H.; Jeon, J.; Jo, S.; Seong, K.M.; Lee, S.-J.; Youn, H.; Youn, B. Decreased FBP1 Expression Rewires Metabolic Processes Affecting Aggressiveness of Glioblastoma. Oncogene 2020, 39, 36–49. [Google Scholar] [CrossRef]
- Sohrabi, A.; Lefebvre, A.E.Y.T.; Harrison, M.J.; Condro, M.C.; Sanazzaro, T.M.; Safarians, G.; Solomon, I.; Bastola, S.; Kordbacheh, S.; Toh, N.; et al. Microenvironmental Stiffness Induces Metabolic Reprogramming in Glioblastoma. Cell Rep. 2023, 42, 113175. [Google Scholar] [CrossRef]
- Patil, M.D.; Bhaumik, J.; Babykutty, S.; Banerjee, U.C.; Fukumura, D. Arginine Dependence of Tumor Cells: Targeting a Chink in Cancer’s Armor. Oncogene 2016, 35, 4957–4972. [Google Scholar] [CrossRef]
- Sawicka, M.M.; Sawicki, K.; Łysoń, T.; Polityńska, B.; Miltyk, W. Proline Metabolism in Malignant Gliomas: A Systematic Literature Review. Cancers 2022, 14, 2030. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cui, L.; Lu, S.; Xu, S. Amino Acid Metabolism in Tumor Biology and Therapy. Cell Death Dis. 2024, 15, 42. [Google Scholar] [CrossRef] [PubMed]
- Mörén, L.; Bergenheim, A.T.; Ghasimi, S.; Brännström, T.; Johansson, M.; Antti, H. Metabolomic Screening of Tumor Tissue and Serum in Glioma Patients Reveals Diagnostic and Prognostic Information. Metabolites 2015, 5, 502–520. [Google Scholar] [CrossRef]
- Tardito, S.; Oudin, A.; Ahmed, S.U.; Fack, F.; Keunen, O.; Zheng, L.; Miletic, H.; Sakariassen, P.Ø.; Weinstock, A.; Wagner, A.; et al. Glutamine Synthetase Activity Fuels Nucleotide Biosynthesis and Supports Growth of Glutamine-Restricted Glioblastoma. Nat. Cell Biol. 2015, 17, 1556–1568. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Weinstein, S.J.; Kitahara, C.M.; Karoly, E.D.; Sampson, J.N.; Albanes, D. A Prospective Study of Serum Metabolites and Glioma Risk. Oncotarget 2017, 8, 70366–70377. [Google Scholar] [CrossRef]
- Mörén, L.; Perryman, R.; Crook, T.; Langer, J.K.; Oneill, K.; Syed, N.; Antti, H. Metabolomic Profiling Identifies Distinct Phenotypes for ASS1 Positive and Negative GBM. BMC Cancer 2018, 18, 167. [Google Scholar] [CrossRef]
- Masters, J.R. Human Cancer Cell Lines: Fact and Fantasy. Nat. Rev. Mol. Cell Biol. 2000, 1, 233–236. [Google Scholar] [CrossRef]
- Mirabelli, P.; Coppola, L.; Salvatore, M. Cancer Cell Lines Are Useful Model Systems for Medical Research. Cancers 2019, 11, 1098. [Google Scholar] [CrossRef]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef]
- Witusik-Perkowska, M.; Rieske, P.; Hułas-Bigoszewska, K.; Zakrzewska, M.; Stawski, R.; Kulczycka-Wojdala, D.; Bieńkowski, M.; Stoczyńska-Fidelus, E.; Grešner, S.M.; Piaskowski, S.; et al. Glioblastoma-Derived Spheroid Cultures as an Experimental Model for Analysis of EGFR Anomalies. J. Neurooncol. 2011, 102, 395–407. [Google Scholar] [CrossRef]
- Akay, M.; Hite, J.; Avci, N.G.; Fan, Y.; Akay, Y.; Lu, G.; Zhu, J.-J. Drug Screening of Human GBM Spheroids in Brain Cancer Chip. Sci. Rep. 2018, 8, 15423. [Google Scholar] [CrossRef] [PubMed]
- Paolillo, M.; Comincini, S.; Schinelli, S. In Vitro Glioblastoma Models: A Journey into the Third Dimension. Cancers 2021, 13, 2449. [Google Scholar] [CrossRef] [PubMed]
- Rybin, M.J.; Ivan, M.E.; Ayad, N.G.; Zeier, Z. Organoid Models of Glioblastoma and Their Role in Drug Discovery. Front. Cell. Neurosci. 2021, 15, 605255. [Google Scholar] [CrossRef]
- Manikandan, C.; Jaiswal, A.K. Scaffold-Based Spheroid Models of Glioblastoma Multiforme and Its Use in Drug Screening. Biotechnol. Bioeng. 2023, 120, 2117–2132. [Google Scholar] [CrossRef] [PubMed]
- Riffle, S.; Pandey, R.N.; Albert, M.; Hegde, R.S. Linking Hypoxia, DNA Damage and Proliferation in Multicellular Tumor Spheroids. BMC Cancer 2017, 17, 338. [Google Scholar] [CrossRef]
- Raskin, N.H.; Fishman, R.A. Pyridoxine-Deficiency Neuropathy Due to Hydralazine. N. Engl. J. Med. 1965, 273, 1182–1185. [Google Scholar] [CrossRef]
- Vidrio, H. Interaction with Pyridoxal as a Possible Mechanism of Hydralazine Hypotension. J. Cardiovasc. Pharmacol. 1990, 15, 150–156. [Google Scholar] [CrossRef]
- Jung, J.; Zhang, Y.; Celiku, O.; Zhang, W.; Song, H.; Williams, B.J.; Giles, A.J.; Rich, J.N.; Abounader, R.; Gilbert, M.R.; et al. Mitochondrial NIX Promotes Tumor Survival in the Hypoxic Niche of Glioblastoma. Cancer Res. 2019, 79, 5218–5232. [Google Scholar] [CrossRef]
- Krawczynski, K.; Godlewski, J.; Bronisz, A. Oxidative Stress—Part of the Solution or Part of the Problem in the Hypoxic Environment of a Brain Tumor. Antioxidants 2020, 9, 747. [Google Scholar] [CrossRef]
- Peixoto, J.; Janaki-Raman, S.; Schlicker, L.; Schmitz, W.; Walz, S.; Winkelkotte, A.M.; Herold-Mende, C.; Soares, P.; Schulze, A.; Lima, J. Integrated Metabolomics and Transcriptomics Analysis of Monolayer and Neurospheres from Established Glioblastoma Cell Lines. Cancers 2021, 13, 1327. [Google Scholar] [CrossRef]
- Phang, J.M. Proline Metabolism in Cell Regulation and Cancer Biology: Recent Advances and Hypotheses. Antioxid. Redox Signal. 2019, 30, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Elia, I.; Broekaert, D.; Christen, S.; Boon, R.; Radaelli, E.; Orth, M.F.; Verfaillie, C.; Grünewald, T.G.P.; Fendt, S.-M. Proline Metabolism Supports Metastasis Formation and Could Be Inhibited to Selectively Target Metastasizing Cancer Cells. Nat. Commun. 2017, 8, 15267. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Glunde, K.; Bhujwalla, Z.M.; Raman, V.; Sharma, A.; Phang, J.M. Proline Oxidase Promotes Tumor Cell Survival in Hypoxic Tumor Microenvironments. Cancer Res. 2012, 72, 3677–3686. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-L.; Hsu, S.-C.; Ann, D.K.; Yen, Y.; Kung, H.-J. Arginine Signaling and Cancer Metabolism. Cancers 2021, 13, 3541. [Google Scholar] [CrossRef]
- Syed, N.; Langer, J.; Janczar, K.; Singh, P.; Lo Nigro, C.; Lattanzio, L.; Coley, H.M.; Hatzimichael, E.; Bomalaski, J.; Szlosarek, P.; et al. Epigenetic Status of Argininosuccinate Synthetase and Argininosuccinate Lyase Modulates Autophagy and Cell Death in Glioblastoma. Cell Death Dis. 2013, 4, e458. [Google Scholar] [CrossRef]
- Wu, G.; Bazer, F.W.; Davis, T.A.; Kim, S.W.; Li, P.; Rhoads, J.M.; Satterfield, M.C.; Smith, S.B.; Spencer, T.E.; Yin, Y. Arginine Metabolism and Nutrition in Growth, Health and Disease. Amino Acids 2009, 37, 153–168. [Google Scholar] [CrossRef]
- Geeraerts, S.L.; Heylen, E.; De Keersmaecker, K.; Kampen, K.R. The Ins and Outs of Serine and Glycine Metabolism in Cancer. Nat. Metab. 2021, 3, 131–141. [Google Scholar] [CrossRef]
- Ganini, C.; Amelio, I.; Bertolo, R.; Candi, E.; Cappello, A.; Cipriani, C.; Mauriello, A.; Marani, C.; Melino, G.; Montanaro, M.; et al. Serine and One-Carbon Metabolisms Bring New Therapeutic Venues in Prostate Cancer. Discov. Oncol. 2021, 12, 45. [Google Scholar] [CrossRef]
- Shunxi, W.; Xiaoxue, Y.; Guanbin, S.; Li, Y.; Junyu, J.; Wanqian, L. Serine Metabolic Reprogramming in Tumorigenesis, Tumor Immunity, and Clinical Treatment. Adv. Nutr. 2023, 14, 1050–1066. [Google Scholar] [CrossRef]
- Singh, C.; Sharma, A.; Hoppe, G.; Song, W.; Bolok, Y.; Sears, J.E. 3-Hydroxypyruvate Destabilizes Hypoxia Inducible Factor and Induces Angiostasis. Investig. Ophthalmol. Vis. Sci. 2018, 59, 3440–3448. [Google Scholar] [CrossRef]
- Labuschagne, C.F.; van den Broek, N.J.F.; Mackay, G.M.; Vousden, K.H.; Maddocks, O.D.K. Serine, but Not Glycine, Supports One-Carbon Metabolism and Proliferation of Cancer Cells. Cell Rep. 2014, 7, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Mentch, S.J.; Locasale, J.W. One Carbon Metabolism and Epigenetics: Understanding the Specificity. Ann. N. Y. Acad. Sci. 2016, 1363, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.; Kim, J.H.; Lee, E.; Jang, Y.J.; Son, J.E.; Kwon, J.Y.; Lim, T.-G.; Kim, S.; Park, J.H.Y.; Kim, J.-E.; et al. Methionine Deprivation Suppresses Triple-Negative Breast Cancer Metastasis in Vitro and in Vivo. Oncotarget 2016, 7, 67223–67234. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, W. Correlation between Amino Acid Metabolism and Self-Renewal of Cancer Stem Cells: Perspectives in Cancer Therapy. World J. Stem Cells 2022, 14, 267–286. [Google Scholar] [CrossRef]
- Palanichamy, K.; Thirumoorthy, K.; Kanji, S.; Gordon, N.; Singh, R.; Jacob, J.R.; Sebastian, N.; Litzenberg, K.T.; Patel, D.; Bassett, E.; et al. Methionine and Kynurenine Activate Oncogenic Kinases in Glioblastoma, and Methionine Deprivation Compromises Proliferation. Clin. Cancer Res. 2016, 22, 3513–3523. [Google Scholar] [CrossRef]
- Pascale, R.M.; Peitta, G.; Simile, M.M.; Feo, F. Alterations of Methionine Metabolism as Potential Targets for the Prevention and Therapy of Hepatocellular Carcinoma. Medicina 2019, 55, 296. [Google Scholar] [CrossRef]
- Zgheib, R.; Battaglia-Hsu, S.-F.; Hergalant, S.; Quéré, M.; Alberto, J.-M.; Chéry, C.; Rouyer, P.; Gauchotte, G.; Guéant, J.-L.; Namour, F. Folate Can Promote the Methionine-Dependent Reprogramming of Glioblastoma Cells towards Pluripotency. Cell Death Dis. 2019, 10, 596. [Google Scholar] [CrossRef]
- Jin, J.; Byun, J.-K.; Choi, Y.-K.; Park, K.-G. Targeting Glutamine Metabolism as a Therapeutic Strategy for Cancer. Exp. Mol. Med. 2023, 55, 706–715. [Google Scholar] [CrossRef]
- Martins, F.; Gonçalves, L.G.; Pojo, M.; Serpa, J. Take Advantage of Glutamine Anaplerosis, the Kernel of the Metabolic Rewiring in Malignant Gliomas. Biomolecules 2020, 10, 1370. [Google Scholar] [CrossRef]
- Jin, L.; Alesi, G.; Kang, S. Glutaminolysis as a Target for Cancer Therapy. Oncogene 2016, 35, 3619–3625. [Google Scholar] [CrossRef]
- Mukherjee, P.; Augur, Z.M.; Li, M.; Hill, C.; Greenwood, B.; Domin, M.A.; Kondakci, G.; Narain, N.R.; Kiebish, M.A.; Bronson, R.T.; et al. Therapeutic Benefit of Combining Calorie-Restricted Ketogenic Diet and Glutamine Targeting in Late-Stage Experimental Glioblastoma. Commun. Biol. 2019, 2, 200. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.; Hartmann, R.; Tsiampali, J.; Uhlmann, C.; Nickel, A.-C.; He, X.; Kamp, M.A.; Sabel, M.; Barker, R.A.; Steiger, H.-J.; et al. A Comparative Pharmaco-Metabolomic Study of Glutaminase Inhibitors in Glioma Stem-like Cells Confirms Biological Effectiveness but Reveals Differences in Target-Specificity. Cell Death Discov. 2020, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- De Los Santos-Jiménez, J.; Rosales, T.; Ko, B.; Campos-Sandoval, J.A.; Alonso, F.J.; Márquez, J.; DeBerardinis, R.J.; Matés, J.M. Metabolic Adjustments Following Glutaminase Inhibition by CB-839 in Glioblastoma Cell Lines. Cancers 2023, 15, 531. [Google Scholar] [CrossRef] [PubMed]
- Rabinovich, S.; Adler, L.; Yizhak, K.; Sarver, A.; Silberman, A.; Agron, S.; Stettner, N.; Sun, Q.; Brandis, A.; Helbling, D.; et al. Diversion of Aspartate in ASS1-Deficient Tumours Fosters de Novo Pyrimidine Synthesis. Nature 2015, 527, 379–383. [Google Scholar] [CrossRef]
- Mullen, N.J.; Singh, P.K. Nucleotide Metabolism: A Pan-Cancer Metabolic Dependency. Nat. Rev. Cancer 2023, 23, 275–294. [Google Scholar] [CrossRef]
- Jung, J.; Kim, L.J.; Wang, X.; Wu, Q.; Sanvoranart, T.; Hubert, C.G.; Prager, B.C.; Wallace, L.C.; Jin, X.; Mack, S.C.; et al. Nicotinamide Metabolism Regulates Glioblastoma Stem Cell Maintenance. JCI Insight 2017, 2, e90019. [Google Scholar] [CrossRef]
- Wang, X.; Yang, K.; Xie, Q.; Wu, Q.; Mack, S.C.; Shi, Y.; Kim, L.J.Y.; Prager, B.C.; Flavahan, W.A.; Liu, X.; et al. Purine Synthesis Promotes Maintenance of Brain Tumor Initiating Cells in Glioma. Nat. Neurosci. 2017, 20, 661–673. [Google Scholar] [CrossRef]
- Talib, W.H.; Ahmed Jum’AH, D.A.; Attallah, Z.S.; Jallad, M.S.; Al Kury, L.T.; Hadi, R.W.; Mahmod, A.I. Role of Vitamins A, C, D, E in Cancer Prevention and Therapy: Therapeutic Potentials and Mechanisms of Action. Front. Nutr. 2024, 10, 1281879. [Google Scholar] [CrossRef]
- Jabbari, P.; Yazdanpanah, O.; Benjamin, D.J.; Kalebasty, A.R. Supplement Use and Increased Risks of Cancer: Unveiling the Other Side of the Coin. Cancers 2024, 16, 880. [Google Scholar] [CrossRef]
- Stach, K.; Stach, W.; Augoff, K. Vitamin B6 in Health and Disease. Nutrients 2021, 13, 3229. [Google Scholar] [CrossRef]
- Ellershaw, D.C.; Gurney, A.M. Mechanisms of Hydralazine Induced Vasodilation in Rabbit Aorta and Pulmonary Artery. Br. J. Pharmacol. 2001, 134, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Lee, H.; Kim, D.; Kim, B.; Kang, J.; Kim, H.Y.; Youn, H.; Youn, B. Targeting Glioblastoma Stem Cells to Overcome Chemoresistance: An Overview of Current Therapeutic Strategies. Biomedicines 2022, 10, 1308. [Google Scholar] [CrossRef] [PubMed]
- Chavez-Blanco, A.; Perez-Plasencia, C.; Perez-Cardenas, E.; Carrasco-Legleu, C.; Rangel-Lopez, E.; Segura-Pacheco, B.; Taja-Chayeb, L.; Trejo-Becerril, C.; Gonzalez-Fierro, A.; Candelaria, M.; et al. Antineoplastic Effects of the DNA Methylation Inhibitor Hydralazine and the Histone Deacetylase Inhibitor Valproic Acid in Cancer Cell Lines. Cancer Cell Int. 2006, 6, 2. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, C. Hydralazine Inhibits Human Cervical Cancer Cell Growth in Vitro in Association with APC Demethylation and Re-Expression. Cancer Chemother. Pharmacol. 2009, 63, 605–613. [Google Scholar] [CrossRef]
- Knowles, H.J.; Tian, Y.-M.; Mole, D.R.; Harris, A.L. Novel Mechanism of Action for Hydralazine. Circ. Res. 2004, 95, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Graça, I.; Sousa, E.J.; Costa-Pinheiro, P.; Vieira, F.Q.; Torres-Ferreira, J.; Martins, M.G.; Henrique, R.; Jerónimo, C. Anti-Neoplastic Properties of Hydralazine in Prostate Cancer. Oncotarget 2014, 5, 5950–5964. [Google Scholar] [CrossRef]
- Lopes, N.; Pacheco, M.B.; Soares-Fernandes, D.; Correia, M.P.; Camilo, V.; Henrique, R.; Jerónimo, C. Hydralazine and Enzalutamide: Synergistic Partners against Prostate Cancer. Biomedicines 2021, 9, 976. [Google Scholar] [CrossRef]
- Pacheco, M.B.; Camilo, V.; Lopes, N.; Moreira-Silva, F.; Correia, M.P.; Henrique, R.; Jerónimo, C. Hydralazine and Panobinostat Attenuate Malignant Properties of Prostate Cancer Cell Lines. Pharmaceuticals 2021, 14, 670. [Google Scholar] [CrossRef]
- Chen, C.-C.; Li, B.; Millman, S.E.; Chen, C.; Li, X.; Morris, J.P.; Mayle, A.; Ho, Y.-J.; Loizou, E.; Liu, H.; et al. Vitamin B6 Addiction in Acute Myeloid Leukemia. Cancer Cell 2020, 37, 71–84.e7. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moosa, N.Y.; Azeem, S.A.; Lodge, J.K.; Cheung, W.; Ahmed, S.U. Vitamin B6 Pathway Maintains Glioblastoma Cell Survival in 3D Spheroid Cultures. Int. J. Mol. Sci. 2024, 25, 10428. https://doi.org/10.3390/ijms251910428
Moosa NY, Azeem SA, Lodge JK, Cheung W, Ahmed SU. Vitamin B6 Pathway Maintains Glioblastoma Cell Survival in 3D Spheroid Cultures. International Journal of Molecular Sciences. 2024; 25(19):10428. https://doi.org/10.3390/ijms251910428
Chicago/Turabian StyleMoosa, Najla Yussuf, Sara Abdullah Azeem, John K. Lodge, William Cheung, and Shafiq Uddin Ahmed. 2024. "Vitamin B6 Pathway Maintains Glioblastoma Cell Survival in 3D Spheroid Cultures" International Journal of Molecular Sciences 25, no. 19: 10428. https://doi.org/10.3390/ijms251910428




