The Use of Patient-Derived Organoids in the Study of Molecular Metabolic Adaptation in Breast Cancer
Abstract
1. Introduction
2. Metabolic Dysregulation in Cancer
2.1. Dysregulation in Glycolysis
2.2. Pentose Phosphate Pathway (PPP)
2.3. Mitochondrial Changes and TCA Dysregulation
2.4. Dysregulated Lipid Metabolism
2.5. Dysregulated Glutamine Metabolism
2.6. Cell Signaling Pathway Dysregulation Affecting Glucose Metabolism
2.7. Metabolic Alterations Due to Hypoxia-Inducible Factor (HIF) Activation
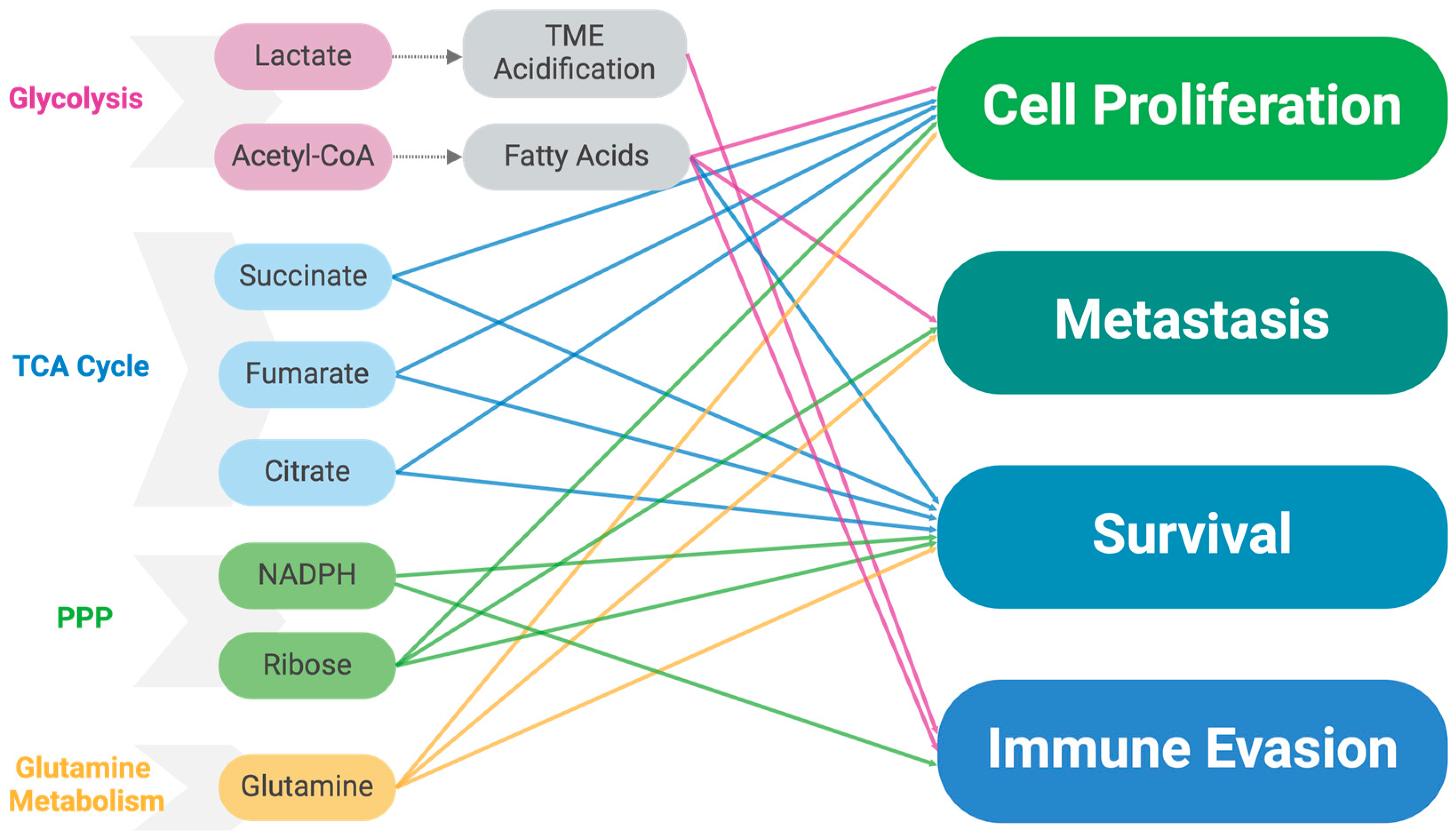
2.8. Alterations in Metabolite Levels
2.9. Effects of Altered Metabolism on the Tumor Microenvironment
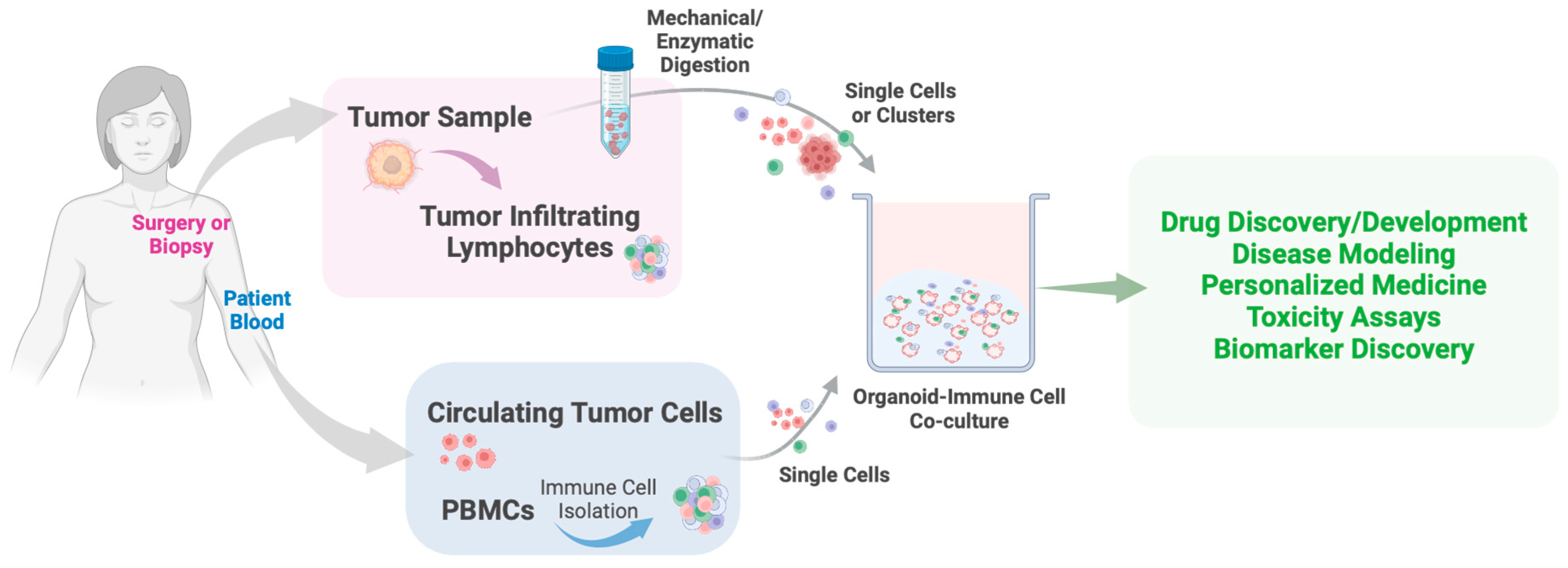
3. Organoid Models in Cancer Research
4. Organoid-Based Approaches in Studying Breast Cancer Metabolism
4.1. Optical Metabolic Imaging (OMI)
4.2. Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging (MALDI-MSI)
4.3. High-Throughput Analytic Chemistry-Based Metabolic and Lipidomic Profiling
4.4. Other Technologies
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- National Cancer Institute. Surveillance, Epidemiology, and End Results Program. Breast Cancer—Breast Cancer Risk from Birth over Time, by Sex, All Races/Ethnicities, Risk of Being Diagnosed with Cancer (2018–2019, 2021). Available online: https://seer.cancer.gov/statistics-network/explorer/ (accessed on 23 August 2024).
- Centers for Disease Control and Prevention; U.S. Department of Health and Human Services. U.S. Cancer Statistics Female Breast Cancer Stat Bite. 2024. Available online: https://www.cdc.gov/united-states-cancer-statistics/publications/breast-cancer-stat-bite.html (accessed on 23 August 2024).
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2017, 23, 27. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Camp, J.G.; Badsha, F.; Florio, M.; Kanton, S.; Gerber, T.; Wilsch-Bräuninger, M.; Lewitus, E.; Sykes, A.; Hevers, W.; Lancaster, M.; et al. Human Cerebral Organoids Recapitulate Gene Expression Programs of Fetal Neocortex Development. Proc. Natl. Acad. Sci. USA 2015, 112, 15672–15677. [Google Scholar] [CrossRef]
- Kaluthantrige Don, F.; Huch, M. Organoids, where We Stand and where We Go. Trends Mol. Med. 2021, 27, 416–418. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral Organoids Model Human Brain Development and Microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Eichmüller, O.L.; Knoblich, J.A. Human Cerebral Organoids—A New Tool for Clinical Neurology Research. Nat. Rev. Neurol. 2022, 18, 661–680. [Google Scholar] [CrossRef]
- Garcez, G.; Loiola, L.; Madeiro da Costa Madeiro, D.C.; Higa, H.; Trindade, T.; Delvecchio, D.; Nascimento, N.; Brindeiro, B.; Tanuri, T.; Rehen, R. Zika Virus Impairs Growth in Human Neurospheres and Brain Organoids. Science 2016, 352, 816–818. [Google Scholar] [CrossRef]
- Mansour, A.A.; Gonçalves, J.T.; Bloyd, C.W.; Li, H.; Fernandes, S.; Quang, D.; Johnston, S.; Parylak, S.L.; Jin, X.; Gage, F.H. An In Vivo Model of Functional and Vascularized Human Brain Organoids. Nat. Biotechnol. 2018, 36, 432. [Google Scholar] [CrossRef]
- Tian, C.; Yang, M.; Xu, H.; Zhu, M.; Yue, N.; Zhang, Y.; Shi, R.; Yao, J.; Wang, L.; Liang, Y.; et al. Stem Cell-Derived Intestinal Organoids: A Novel Modality for IBD. Cell Death Discov. 2023, 9, 255. [Google Scholar] [CrossRef]
- Günther, C.; Winner, B.; Neurath, M.F.; Stappenbeck, T.S. Organoids in Gastrointestinal Diseases: From Experimental Models to Clinical Translation. Gut 2022, 71, 1892. [Google Scholar] [CrossRef]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed Differentiation of Human Pluripotent Stem Cells into Intestinal Tissue In Vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef]
- Zietek, T.; Rath, E.; Haller, D.; Daniel, H. Intestinal Organoids for Assessing Nutrient Transport, Sensing and Incretin Secretion. Sci. Rep. 2015, 5, 16831. [Google Scholar] [CrossRef]
- Harrison, S.P.; Siller, R.; Tanaka, Y.; Chollet, M.E.; De La Morena-Barrio, M.E.; Xiang, Y.; Patterson, B.; Andersen, E.; Bravo-Pérez, C.; Kempf, H.; et al. Scalable Production of Tissue-Like Vascularized Liver Organoids from Human PSCs. Exp. Mol. Med. 2023, 55, 2005. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhang, B.; He, Y.; Bao, J. Liver Organoids: Formation Strategies and Biomedical Applications. Tissue Eng. Regen. Med. 2021, 18, 573. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.P.; Baumgarten, S.F.; Verma, R.; Lunov, O.; Dejneka, A.; Sullivan, G.J. Liver Organoids: Recent Developments, Limitations and Potential. Front. Med. 2021, 8, 574047. [Google Scholar] [CrossRef] [PubMed]
- Khoshdel-Rad, N.; Ahmadi, A.; Moghadasali, R. Kidney Organoids: Current Knowledge and Future Directions. Cell Tissue Res. 2022, 387, 207. [Google Scholar] [CrossRef]
- Zeng, Z.; Huang, B.; Parvez, R.K.; Li, Y.; Chen, J.; Vonk, A.C.; Thornton, M.E.; Patel, T.; Rutledge, E.A.; Kim, A.D.; et al. Generation of Patterned Kidney Organoids that Recapitulate the Adult Kidney Collecting Duct System from Expandable Ureteric Bud Progenitors. Nat. Commun. 2021, 12, 3641. [Google Scholar] [CrossRef]
- Wang, Q.; Xiong, Y.; Zhang, S.; Sui, Y.; Yu, C.; Liu, P.; Li, H.; Guo, W.; Gao, Y.; Przepiorski, A.; et al. The Dynamics of Metabolic Characterization in iPSC-Derived Kidney Organoid Differentiation Via a Comparative Omics Approach. Front. Genet. 2021, 12, 632810. [Google Scholar] [CrossRef]
- Homan, K.A.; Gupta, N.; Kroll, K.T.; Kolesky, D.B.; Skylar-Scott, M.; Miyoshi, T.; Mau, D.; Valerius, M.T.; Ferrante, T.; Bonventre, J.V.; et al. Flow-Enhanced Vascularization and Maturation of Kidney Organoids in Vitro. Nat. Methods 2019, 16, 255. [Google Scholar] [CrossRef] [PubMed]
- Boj, S.F.; Hwang, C.; Baker, L.A.; Chio, I.I.C.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S.; et al. Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell 2016, 160, 324. [Google Scholar] [CrossRef] [PubMed]
- Casamitjana, J.; Espinet, E.; Rovira, M. Pancreatic Organoids for Regenerative Medicine and Cancer Research. Front. Cell Dev. Biol. 2022, 10, 886153. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, H.J.; Yacob, B.W.; Brown, M.E.; Goldstein, B.R.; Arcaroli, J.J.; Bagby, S.M.; Hartman, S.J.; Macbeth, M.; Goodspeed, A.; Danhorn, T.; et al. Examination of Wnt Signaling as a Therapeutic Target for Pancreatic Ductal Adenocarcinoma (PDAC) using a Pancreatic Tumor Organoid Library (PTOL). PLoS ONE 2024, 19, e0298808. [Google Scholar] [CrossRef] [PubMed]
- Cheaito, K.; Bahmad, H.F.; Hadadeh, O.; Msheik, H.; Monzer, A.; Ballout, F.; Dagher, C.; Telvizian, T.; Saheb, N.; Tawil, A.; et al. Establishment and Characterization of Prostate Organoids from Treatment-naïve Patients with Prostate Cancer. Oncol. Lett. 2021, 23, 6. [Google Scholar] [CrossRef]
- Beshiri, M.; Agarwal, S.; Yin, J.J.; Kelly, K. Prostate Organoids: Emerging Experimental Tools for Translational Research. J. Clin. Investig. 2023, 133, e169616. [Google Scholar] [CrossRef]
- Pasch, C.A.; Favreau, P.F.; Yueh, A.E.; Babiarz, C.P.; Gillette, A.A.; Sharick, J.T.; Karim, M.R.; Nickel, K.P.; DeZeeuw, A.K.; Sprackling, C.M.; et al. Patient-Derived Cancer Organoid Cultures to Predict Sensitivity to Chemotherapy and Radiation. Clin. Cancer Res. 2019, 25, 5376–5387. [Google Scholar] [CrossRef]
- Sharick, J.T.; Walsh, C.M.; Sprackling, C.M.; Pasch, C.A.; Pham, D.L.; Esbona, K.; Choudhary, A.; Garcia-Valera, R.; Burkard, M.E.; McGregor, S.M.; et al. Metabolic Heterogeneity in Patient Tumor-Derived Organoids by Primary Site and Drug Treatment. Front. Oncol. 2020, 10, 553. [Google Scholar] [CrossRef]
- Dominijanni, A.; Mazzocchi, A.; Shelkey, E.; Forsythe, S.; Devarsetty, M.; Soker, S. Bioengineered Tumor Organoids. Curr. Opin. Biomed. Eng. 2020, 13, 168–173. [Google Scholar] [CrossRef]
- Lesavage, B.L.; Suhar, R.A.; Broguiere, N.; Lutolf, M.P.; Heilshorn, S.C. Next-Generation Cancer Organoids. Nat. Mater. 2021, 21, 143. [Google Scholar] [CrossRef]
- Qu, J.; Kalyani, F.S.; Liu, L.; Cheng, T.; Chen, L. Tumor Organoids: Synergistic Applications, Current Challenges, and Future Prospects in Cancer Therapy. Cancer Commun. 2021, 41, 1331. [Google Scholar] [CrossRef] [PubMed]
- Deberardinis, R.J. Is Cancer a Disease of Abnormal Cellular Metabolism? New Angles on an Old Idea. Genet. Med. 2008, 10, 767. [Google Scholar] [CrossRef] [PubMed]
- Deberardinis, R.J.; Chandel, N.S. Fundamentals of Cancer Metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2010, 324, 1029. [Google Scholar] [CrossRef]
- Arora, R.; Schmitt, D.; Karanam, B.; Tan, M.; Yates, C.; Dean-Colomb, W. Inhibition of the Warburg Effect with a Natural Compound Reveals a Novel Measurement for Determining the Metastatic Potential of Breast Cancers. Oncotarget 2015, 6, 662–678. [Google Scholar] [CrossRef]
- Altenberg, B.; Greulich, K.O. Genes of Glycolysis are Ubiquitously Overexpressed in 24 Cancer Classes. Genomics 2004, 84, 1014–1020. [Google Scholar] [CrossRef]
- Brahimi-Horn, M.C.; Pouyssegur, J. Oxygen, a Source of Life and Stress. FEBS Lett. 2007, 581, 3582–3591. [Google Scholar] [CrossRef]
- Fajas, L. Metabolic Control in Cancer Cells. Ann. Endocrinol. (Paris) 2013, 74, 71–73. [Google Scholar] [CrossRef]
- Pouysségur, J.; Dayan, F.; Mazure, N.M. Hypoxia Signalling in Cancer and Approaches to Enforce Tumour Regression. Nature 2006, 441, 437–443. [Google Scholar] [CrossRef]
- Wu, Z.; Wu, J.; Zhao, Q.; Fu, S.; Jin, J. Emerging Roles of Aerobic Glycolysis in Breast Cancer. Clin. Transl. Oncol. 2019, 22, 631–646. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Gillies, R.J. Why do Cancers have High Aerobic Glycolysis? Nat. Rev. Cancer 2014, 4, 891. [Google Scholar] [CrossRef] [PubMed]
- Lunt, S.Y.; Heiden, M.G.V. Aerobic Glycolysis: Meeting the Metabolic Requirements of Cell Proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy-Kanniappan, S. Molecular Intricacies of Aerobic Glycolysis in Cancer: Current Insights into the Classic Metabolic Phenotype. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 667–682. [Google Scholar] [CrossRef]
- Li, W.; Tanikawa, T.; Kryczek, I.; Xia, H.; Li, G.; Wu, K.; Wei, S.; Zhao, L.; Vatan, L.; Wen, B.; et al. Aerobic Glycolysis Controls Myeloid-Derived Suppressor Cells and Tumor Immunity Via a Specific CEBPB Isoform in Triple-Negative Breast Cancer. Cell Metab. 2018, 28, 87–1036. [Google Scholar] [CrossRef]
- Tan, V.P.; Miyamoto, S. HK2/Hexokinase-II Integrates Glycolysis and Autophagy to Confer Cellular Protection. Autophagy 2015, 11, 963–964. [Google Scholar] [CrossRef]
- Hennipman, A.; Smits, J.; van Oirschot, B.; van Houwelingen, J.C.; Rijksen, G.; Neyt, J.P.; Van Unnik, J.A.; Staal, G.E. Glycolytic Enzymes in Breast Cancer, Benign Breast Disease and Normal Breast Tissue. Tumour Biol. 1987, 8, 251–263. [Google Scholar] [CrossRef]
- Yang, T.; Ren, C.; Qiao, P.; Han, X.; Wang, L.; Lv, S.; Sun, Y.; Liu, Z.; Du, Y.; Yu, Z. PIM2-Mediated Phosphorylation of Hexokinase 2 is Critical for Tumor Growth and Paclitaxel Resistance in Breast Cancer. Oncogene 2018, 37, 5997–6009. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, L.; Zhang, H.; Hu, S.; Lu, M.; Liang, S.; Li, B.; Li, Y.; Li, D.; Wang, E.; et al. A Novel miR-155/miR-143 Cascade Controls Glycolysis by Regulating Hexokinase 2 in Breast Cancer Cells. EMBO J. 2012, 31, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yu, Y.; Li, H.; Hu, Q.; Chen, X.; He, Y.; Xue, C.; Ren, F.; Ren, Z.; Li, J.; et al. Long Non-Coding RNA PVT1 Promotes Tumor Progression by Regulating the miR-143/HK2 Axis in Gallbladder Cancer. Mol. Cancer 2019, 18, 33. [Google Scholar] [CrossRef]
- Sang, R.; Fan, R.; Deng, A.; Gou, J.; Lin, R.; Zhao, T.; Hai, Y.; Song, J.; Liu, Y.; Qi, B.; et al. Degradation of Hexokinase 2 Blocks Glycolysis and Induces GSDME-Dependent Pyroptosis to Amplify Immunogenic Cell Death for Breast Cancer Therapy. J. Med. Chem. 2023, 66, 8464–8483. [Google Scholar] [CrossRef]
- Kim, S.; Manes, N.P.; El-Maghrabi, M.R.; Lee, Y. Crystal Structure of the Hypoxia-Inducible Form of 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase (PFKFB3): A Possible New Target for Cancer Therapy. J. Biol. Chem. 2006, 281, 2939–2944. [Google Scholar] [CrossRef] [PubMed]
- O’Neal, J.; Clem, A.; Reynolds, L.; Dougherty, S.; Imbert-Fernandez, Y.; Telang, S.; Chesney, J.; Clem, B.F. Inhibition of 6-Phosphofructo-2-Kinase (PFKFB3) Suppresses Glucose Metabolism and the Growth of HER2+ Breast Cancer. Breast Cancer Res. Treat. 2016, 160, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.A.; Gupta, V.; Gopinath, P.; Mazurek, S.; Bamezai, R.N.K. Pyruvate Kinase M2 and Cancer: An Updated Assessment. FEBS Lett. 2014, 588, 2685–2692. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, S. Pyruvate Kinase Type M2: A Key Regulator of the Metabolic Budget System in Tumor Cells. Int. J. Biochem. Cell Biol. 2011, 43, 969–980. [Google Scholar] [CrossRef]
- Tamada, M.; Suematsu, M.; Saya, H. Pyruvate Kinase M2: Multiple Faces for Conferring Benefits on Cancer Cells. Clin. Cancer Res. 2012, 18, 5554–5561. [Google Scholar] [CrossRef]
- Tamada, M.; Nagano, O.; Tateyama, S.; Ohmura, M.; Yae, T.; Ishimoto, T.; Sugihara, E.; Onishi, N.; Yamamoto, T.; Yanagawa, H.; et al. Modulation of Glucose Metabolism by CD44 Contributes to Antioxidant Status and Drug Resistance in Cancer Cells. Cancer Res. 2012, 72, 1438–1448. [Google Scholar] [CrossRef]
- Luo, W.; Hu, H.; Chang, R.; Zhong, J.; Knabel, M.; O’Meally, R.; Cole, R.N.; Pandey, A.; Semenza, G.L. Pyruvate Kinase M2 is a PHD3-Stimulated Coactivator for Hypoxia-Inducible Factor 1. Cell 2011, 145, 732–744. [Google Scholar] [CrossRef]
- Anwar, S.; Shamsi, A.; Mohammad, T.; Islam, A.; Hassan, M.I. Targeting Pyruvate Dehydrogenase Kinase Signaling in the Development of Effective Cancer Therapy. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188568. [Google Scholar] [CrossRef]
- Hitosugi, T.; Fan, J.; Chung, T.; Lythgoe, K.; Wang, X.; Xie, J.; Ge, Q.; Gu, T.; Polakiewicz, R.D.; Roesel, J.L.; et al. Tyrosine Phosphorylation of Mitochondrial Pyruvate Dehydrogenase Kinase 1 is Important for Cancer Metabolism. Mol. Cell 2011, 44, 864–877. [Google Scholar] [CrossRef]
- Saunier, E.; Benelli, C.; Bortoli, S. The Pyruvate Dehydrogenase Complex in Cancer: An Old Metabolic Gatekeeper Regulated by New Pathways and Pharmacological Agents. Int. J. Cancer 2016, 138, 809–817. [Google Scholar] [CrossRef]
- Jiang, P.; Du, W.; Wu, M. Regulation of the Pentose Phosphate Pathway in Cancer. Protein Cell 2014, 5, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Patra, K.C.; Hay, N. The Pentose Phosphate Pathway and Cancer. Trends Biochem. Sci. 2014, 39, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Benito, A.; Polat, I.H.; Noé, V.; Ciudad, C.J.; Marin, S.; Cascante, M. Glucose-6-Phosphate Dehydrogenase and Transketolase Modulate Breast Cancer Cell Metabolic Reprogramming and Correlate with Poor Patient Outcome. Oncotarget 2017, 8, 106693–106706. [Google Scholar] [CrossRef] [PubMed]
- Polat, I.H.; Tarrado-Castellarnau, M.; Bharat, R.; Perarnau, J.; Benito, A.; Cortés, R.; Sabatier, P.; Cascante, M. Oxidative Pentose Phosphate Pathway Enzyme 6-Phosphogluconate Dehydrogenase Plays a Key Role in Breast Cancer Metabolism. Biology 2021, 10, 85. [Google Scholar] [CrossRef]
- Li, Y.; Yao, C.; Xu, F.; Qu, Y.; Li, J.; Lin, Y.; Cao, Z.; Lin, P.; Xu, W.; Zhao, S.; et al. APC/C(CDH1) Synchronizes Ribose-5-Phosphate Levels and DNA Synthesis to Cell Cycle Progression. Nat. Commun. 2019, 10, 2502. [Google Scholar] [CrossRef]
- Cho, E.S.; Cha, Y.H.; Kim, H.S.; Kim, N.H.; Yook, J.I. The Pentose Phosphate Pathway as a Potential Target for Cancer Therapy. Biomol. Ther. 2018, 26, 29–38. [Google Scholar] [CrossRef]
- Wu, S.; Wang, H.; Li, Y.; Xie, Y.; Huang, C.; Zhao, H.; Miyagishi, M.; Kasim, V. Transcription Factor YY1 Promotes Cell Proliferation by Directly Activating the Pentose Phosphate Pathway. Cancer Res. 2018, 78, 4549–4562. [Google Scholar] [CrossRef]
- Yadav, N.; Chandra, D. Mitochondrial DNA Mutations and Breast Tumorigenesis. Biochim. Biophys. Acta 2013, 1836, 336–344. [Google Scholar] [CrossRef]
- Singh, K.K.; Ayyasamy, V.; Owens, K.M.; Koul, M.S.; Vujcic, M. Mutations in Mitochondrial DNA Polymerase-Gamma Promote Breast Tumorigenesis. J. Hum. Genet. 2009, 54, 516–524. [Google Scholar] [CrossRef]
- Brandon, M.; Baldi, P.; Wallace, D.C. Mitochondrial Mutations in Cancer. Oncogene 2006, 25, 4647–4662. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, X. Significance of Mitochondria DNA Mutations in Diseases. Adv. Exp. Med. Biol. 2017, 1038, 219–230. [Google Scholar] [PubMed]
- Yu, M.; Shi, Y.; Wei, X.; Yang, Y.; Zang, F.; Niu, R. Mitochondrial DNA Depletion Promotes Impaired Oxidative Status and Adaptive Resistance to Apoptosis in T47D Breast Cancer Cells. Eur. J. Cancer Prev. 2009, 18, 445–457. [Google Scholar] [CrossRef]
- Ciccarone, F.; Vegliante, R.; Di Leo, L.; Ciriolo, M.R. The TCA Cycle as a Bridge between Oncometabolism and DNA Transactions in Cancer. Semin. Cancer Biol. 2017, 47, 50–56. [Google Scholar] [CrossRef]
- Desideri, E.; Vegliante, R.; Ciriolo, M.R. Mitochondrial Dysfunctions in Cancer: Genetic Defects and Oncogenic Signaling Impinging on TCA Cycle Activity. Cancer Lett. 2015, 356, 217–223. [Google Scholar] [CrossRef]
- Eniafe, J.; Jiang, S. The Functional Roles of TCA Cycle Metabolites in Cancer. Oncogene 2021, 40, 3351–3363. [Google Scholar] [CrossRef] [PubMed]
- Laurenti, G.; Tennant, D.A. Isocitrate Dehydrogenase (IDH), Succinate Dehydrogenase (SDH), Fumarate Hydratase (FH): Three Players for One Phenotype in Cancer? Biochem. Soc. Trans. 2016, 44, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Simões, R.V.; Serganova, I.S.; Kruchevsky, N.; Leftin, A.; Shestov, A.A.; Thaler, H.T.; Sukenick, G.; Locasale, J.W.; Blasberg, R.G.; Koutcher, J.A.; et al. Metabolic Plasticity of Metastatic Breast Cancer Cells: Adaptation to Changes in the Microenvironment. Neoplasia 2015, 17, 671–684. [Google Scholar] [CrossRef]
- Sengupta, S.; de Oliviera, K.A.; Jin, L.; Clarke, R. Abstract 2327: Dysregulated TCA Cycle is Associated with Endocrine Therapy Resistance in ER+ Breast Cancer Cells. Cancer Res. 2022, 82, 2327. [Google Scholar] [CrossRef]
- Privat, M.; Radosevic-Robin, N.; Aubel, C.; Cayre, A.; Penault-Llorca, F.; Marceau, G.; Sapin, V.; Bignon, Y.; Morvan, D. BRCA1 Induces Major Energetic Metabolism Reprogramming in Breast Cancer Cells. PLoS ONE 2014, 9, e102438. [Google Scholar] [CrossRef]
- Swinnen, J.V.; Brusselmans, K.; Verhoeven, G. Increased Lipogenesis in Cancer Cells: New Players, Novel Targets. Curr. Opin. Clin. Nutr. Metab. Care 2006, 9, 358–365. [Google Scholar] [CrossRef]
- Hilvo, M.; Denkert, C.; Lehtinen, L.; Müller, B.; Brockmöller, S.; Seppänen-Laakso, T.; Budczies, J.; Bucher, E.; Yetukuri, L.; Castillo, S.; et al. Novel Theranostic Opportunities Offered by Characterization of Altered Membrane Lipid Metabolism in Breast Cancer Progression. Cancer Res. 2011, 71, 3236–3245. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Chen, C.; Li, X.; Zhang, X.; Su, C.; Liu, Y.; Cao, T.; Hao, L.; Wang, M.; Kang, J.X. Increased Lipogenesis is Critical for Self-Renewal and Growth of Breast Cancer Stem Cells: Impact of Omega-3 Fatty Acids. Stem Cells 2021, 39, 1660–1670. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, H.; Zhang, Y.; Li, L.; Fang, R.; Li, Y.; Liu, Q.; Zhang, W.; Qiu, L.; Liu, F.; et al. Oncoprotein HBXIP Modulates Abnormal Lipid Metabolism and Growth of Breast Cancer Cells by Activating the LXRs/SREBP-1c/FAS Signaling Cascade. Cancer Res. 2016, 76, 4696–4707. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, N.; Lupien, L.; Kuemmerle, N.B.; Kinlaw, W.B.; Swinnen, J.V.; Smans, K. Lipogenesis and Lipolysis: The Pathways Exploited by the Cancer Cells to Acquire Fatty Acids. Prog. Lipid Res. 2013, 52, 585–589. [Google Scholar] [CrossRef]
- Simeone, P.; Tacconi, S.; Longo, S.; Lanuti, P.; Bravaccini, S.; Pirini, F.; Ravaioli, S.; Dini, L.; Giudetti, A.M. Expanding Roles of De Novo Lipogenesis in Breast Cancer. Int. J. Environ. Res. Public Health 2021, 18, 3575. [Google Scholar] [CrossRef] [PubMed]
- Camarda, R.; Zhou, A.Y.; Kohnz, R.A.; Balakrishnan, S.; Mahieu, C.; Anderton, B.; Eyob, H.; Kajimura, S.; Tward, A.; Krings, G.; et al. Inhibition of Fatty Acid Oxidation as a Therapy for MYC-Overexpressing Triple-Negative Breast Cancer. Nat. Med. 2016, 22, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Fahrmann, J.F.; Lee, H.; Li, Y.; Tripathi, S.C.; Yue, C.; Zhang, C.; Lifshitz, V.; Song, J.; Yuan, Y.; et al. JAK/STAT3-Regulated Fatty Acid Β-Oxidation is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance. Cell Metab. 2018, 27, 136–150.e5. [Google Scholar] [CrossRef]
- Carracedo, A.; Cantley, L.C.; Pandolfi, P.P. Cancer Metabolism: Fatty Acid Oxidation in the Limelight. Nat. Rev. Cancer 2013, 13, 227–232. [Google Scholar] [CrossRef]
- Tang, Y.; Tian, W.; Xie, J.; Zou, Y.; Wang, Z.; Li, N.; Zeng, Y.; Wu, L.; Zhang, Y.; Wu, S.; et al. Prognosis and Dissection of Immunosuppressive Microenvironment in Breast Cancer Based on Fatty Acid Metabolism-Related Signature. Front. Immunol. 2022, 13, 843515. [Google Scholar] [CrossRef]
- Hensley, C.T.; Wasti, A.T.; DeBerardinis, R.J. Glutamine and Cancer: Cell Biology, Physiology, and Clinical Opportunities. J. Clin. Investig. 2013, 123, 3678–3684. [Google Scholar] [CrossRef]
- Masisi, B.K.; El Ansari, R.; Alfarsi, L.; Rakha, E.A.; Green, A.R.; Craze, M.L. The Role of Glutaminase in Cancer. Histopathology 2020, 76, 498–508. [Google Scholar] [CrossRef] [PubMed]
- Lampa, M.; Arlt, H.; He, T.; Ospina, B.; Reeves, J.; Zhang, B.; Murtie, J.; Deng, G.; Barberis, C.; Hoffmann, D.; et al. Glutaminase is Essential for the Growth of Triple-Negative Breast Cancer Cells with a Deregulated Glutamine Metabolism Pathway and its Suppression Synergizes with mTOR Inhibition. PLoS ONE 2017, 12, e0185092. [Google Scholar] [CrossRef]
- Yu, W.; Yang, X.; Zhang, Q.; Sun, L.; Yuan, S.; Xin, Y. Targeting GLS1 to Cancer Therapy through Glutamine Metabolism. Clin. Transl. Oncol. 2021, 23, 2253–2268. [Google Scholar] [CrossRef] [PubMed]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to Clinic: Glutamine Metabolism to Cancer Therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef]
- Matés, J.M.; Di Paola, F.J.; Campos-Sandoval, J.A.; Mazurek, S.; Márquez, J. Therapeutic Targeting of Glutaminolysis as an Essential Strategy to Combat Cancer. Semin. Cell Dev. Biol. 2020, 98, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Alesi, G.N.; Kang, S. Glutaminolysis as a Target for Cancer Therapy. Oncogene 2016, 35, 3619–3625. [Google Scholar] [CrossRef]
- Dornier, E.; Rabas, N.; Mitchell, L.; Novo, D.; Dhayade, S.; Marco, S.; Mackay, G.; Sumpton, D.; Pallares, M.; Nixon, C.; et al. Glutaminolysis Drives Membrane Trafficking to Promote Invasiveness of Breast Cancer Cells. Nat. Commun. 2017, 8, 2255. [Google Scholar] [CrossRef]
- Fu, A.; Yu, Z.; Song, Y.; Zhang, E. Silencing of Glutaminase 1 Resensitizes Taxol-Resistant Breast Cancer Cells to Taxol. Mol. Med. Rep. 2015, 11, 4727–4733. [Google Scholar] [CrossRef]
- Shajahan-Haq, A.; Cook, K.; Schwartz-Roberts, J.; Eltayeb, A.; Demas, D.; Warri, A.; Hilakivi-Clarke, L.; Clarke, R. Glutamine Metabolism and the Unfolded Protein Response in MYC-Driven Breast Cancer. Cancer Metab. 2014, 2, P66. [Google Scholar] [CrossRef][Green Version]
- Kung, H.; Marks, J.R.; Chi, J. Glutamine Synthetase is a Genetic Determinant of Cell Type-Specific Glutamine Independence in Breast Epithelia. PLoS Genet. 2011, 7, e1002229. [Google Scholar] [CrossRef]
- Ramirez-Peña, E.; Arnold, J.; Shivakumar, V.; Joseph, R.; Vidhya Vijay, G.; den Hollander, P.; Bhangre, N.; Allegakoen, P.; Prasad, R.; Conley, Z.; et al. The Epithelial to Mesenchymal Transition Promotes Glutamine Independence by Suppressing GLS2 Expression. Cancers 2019, 11, 1610. [Google Scholar] [CrossRef] [PubMed]
- Beaudin, S.; Welsh, J. 1,25-Dihydroxyvitamin D Regulation of Glutamine Synthetase and Glutamine Metabolism in Human Mammary Epithelial Cells. Endocrinology 2017, 158, 4174–4188. [Google Scholar] [CrossRef]
- Schömel, N.; Hancock, S.E.; Gruber, L.; Olzomer, E.M.; Byrne, F.L.; Shah, D.; Hoehn, K.L.; Turner, N.; Grösch, S.; Geisslinger, G.; et al. UGCG Influences Glutamine Metabolism of Breast Cancer Cells. Sci. Rep. 2019, 9, 15665–15667. [Google Scholar] [CrossRef]
- Youngblood, V.M.; Kim, L.C.; Edwards, D.N.; Hwang, Y.; Santapuram, P.R.; Stirdivant, S.M.; Lu, P.; Ye, F.; Brantley-Sieders, D.M.; Chen, J. The Ephrin-A1/EPHA2 Signaling Axis Regulates Glutamine Metabolism in HER2-Positive Breast Cancer. Cancer Res. 2016, 76, 1825–1836. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Kim, H. Oncogenes and Tumor Suppressors Regulate Glutamine Metabolism in Cancer Cells. J. Cancer Prev. 2013, 18, 221–226. [Google Scholar] [CrossRef]
- Edwards, D.N.; Ngwa, V.M.; Wang, S.; Shiuan, E.; Brantley-Sieders, D.M.; Kim, L.C.; Reynolds, A.B.; Chen, J. The Receptor Tyrosine Kinase EphA2 Promotes Glutamine Metabolism in Tumors by Activating the Transcriptional Coactivators YAP and TAZ. Sci. Signal. 2017, 10, eaan4667. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, R.; Talana, C.A.; Song, C.; Dixon, A.; Uehara, K.; Weichhaus, M. Β-hydroxybutyrate does Not Alter the Effects of Glucose Deprivation on Breast Cancer Cells. Oncol. Lett. 2020, 21, 65. [Google Scholar] [CrossRef]
- Leto, D.; Saltiel, A.R. Regulation of Glucose Transport by Insulin: Traffic Control of GLUT4. Nat. Rev. Mol. Cell Biol. 2012, 13, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Pascoe, W.S.; Inukai, K.; Oka, Y.; Slot, J.W.; James, D.E. Differential Targeting of Facilitative Glucose Transporters in Polarized Epithelial Cells. Am. J. Physiol. 1996, 271, 547. [Google Scholar] [CrossRef]
- Tsuchiya, A.; Kanno, T.; Nishizaki, T. PI3 Kinase Directly Phosphorylates Akt1/2 at Ser473/474 in the Insulin Signal Transduction Pathway. J. Endocrinol. 2013, 220, 49–59. [Google Scholar] [CrossRef]
- Miricescu, D.; Totan, A.; Stanescu-Spinu, I.; Badoiu, S.C.; Stefani, C.; Greabu, M. PI3K/AKT/mTOR Signaling Pathway in Breast Cancer: From Molecular Landscape to Clinical Aspects. Int. J. Mol. Sci. 2020, 22, 173. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.A.; Fraile-Martínez, O.; Asúnsolo, Á.; Buján, J.; García-Honduvilla, N.; Coca, S. Signal Transduction Pathways in Breast Cancer: The Important Role of PI3K/Akt/mTOR. J. Oncol. 2020, 2020, 9258396. [Google Scholar] [CrossRef] [PubMed]
- McAuliffe, P.F.; Meric-Bernstam, F.; Mills, G.B.; Gonzalez-Angulo, A.M. Deciphering the Role of PI3K/Akt/mTOR Pathway in Breast Cancer Biology and Pathogenesis. Clin. Breast Cancer 2010, 10 (Suppl. S3), 59. [Google Scholar] [CrossRef]
- Lauring, J.; Park, B.H.; Wolff, A.C. The Phosphoinositide-3-Kinase-Akt-mTOR Pathway as a Therapeutic Target in Breast Cancer. J. Natl. Compr. Cancer Netw. 2013, 11, 670–678. [Google Scholar] [CrossRef]
- Cidado, J.; Park, B.H. Targeting the PI3K/Akt/mTOR Pathway for Breast Cancer Therapy. J. Mammary Gland Biol. Neoplasia 2012, 17, 205–216. [Google Scholar] [CrossRef]
- Paplomata, E.; O’Regan, R. The PI3K/AKT/mTOR Pathway in Breast Cancer: Targets, Trials and Biomarkers. Ther. Adv. Med. Oncol. 2014, 6, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Prever, L.; Hirsch, E.; Gulluni, F. Targeting PI3K/AKT/mTOR Signaling Pathway in Breast Cancer. Cancers 2021, 13, 3517. [Google Scholar] [CrossRef]
- Ghayad, S.E.; Cohen, P.A. Inhibitors of the PI3K/Akt/mTOR Pathway: New Hope for Breast Cancer Patients. Recent. Pat. Anticancer Drug Discov. 2010, 5, 29–57. [Google Scholar] [CrossRef]
- Ellis, H.; Ma, C.X. PI3K Inhibitors in Breast Cancer Therapy. Curr. Oncol. Rep. 2019, 21, 110–117. [Google Scholar] [CrossRef]
- Massacesi, C.; Di Tomaso, E.; Urban, P.; Germa, C.; Quadt, C.; Trandafir, L.; Aimone, P.; Fretault, N.; Dharan, B.; Tavorath, R.; et al. PI3K Inhibitors as New Cancer Therapeutics: Implications for Clinical Trial Design. OncoTargets Ther. 2016, 9, 203–210. [Google Scholar] [CrossRef]
- Osthus, R.C.; Shim, H.; Kim, S.; Li, Q.; Reddy, R.; Mukherjee, M.; Xu, Y.; Wonsey, D.; Lee, L.A.; Dang, C.V. Deregulation of Glucose Transporter 1 and Glycolytic Gene Expression by C-Myc. J. Biol. Chem. 2000, 275, 21797–21800. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Metukuri, M.R.; Bindom, S.M.; Prochownik, E.V.; O’Doherty, R.M.; Scott, D.K. C-Myc is Required for the CHREBP-Dependent Activation of Glucose-Responsive Genes. Mol. Endocrinol. 2010, 24, 1274–1286. [Google Scholar] [CrossRef]
- Briata, P.; Laurino, C.; Gherzi, R. C-Myc Gene Expression in Human Cells is Controlled by Glucose. Biochem. Biophys. Res. Commun. 1989, 165, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Morrish, F.; Isern, N.; Sadilek, M.; Jeffrey, M.; Hockenbery, D.M. C-Myc Activates Multiple Metabolic Networks to Generate Substrates for Cell-Cycle Entry. Oncogene 2009, 28, 2485–2491. [Google Scholar] [CrossRef] [PubMed]
- Bott, A.J.; Peng, I.; Fan, Y.; Faubert, B.; Zhao, L.; Li, J.; Neidler, S.; Sun, Y.; Jaber, N.; Krokowski, D.; et al. Oncogenic Myc Induces Expression of Glutamine Synthetase through Promoter Demethylation. Cell Metab. 2015, 22, 1068–1077. [Google Scholar] [CrossRef]
- Gao, P.; Tchernyshyov, I.; Chang, T.; Lee, Y.; Kita, K.; Ochi, T.; Zeller, K.I.; De Marzo, A.M.; Van Eyk, J.E.; Mendell, J.T.; et al. C-Myc Suppression of miR-23a/B Enhances Mitochondrial Glutaminase Expression and Glutamine Metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef]
- Wise, D.R.; DeBerardinis, R.J.; Mancuso, A.; Sayed, N.; Zhang, X.; Pfeiffer, H.K.; Nissim, I.; Daikhin, E.; Yudkoff, M.; McMahon, S.B.; et al. Myc Regulates a Transcriptional Program that Stimulates Mitochondrial Glutaminolysis and Leads to Glutamine Addiction. Proc. Natl. Acad. Sci. USA 2008, 105, 18782–18787. [Google Scholar] [CrossRef]
- Thai, M.; Thaker, S.K.; Feng, J.; Du, Y.; Hu, H.; Ting Wu, T.; Graeber, T.G.; Braas, D.; Christofk, H.R. MYC-Induced Reprogramming of Glutamine Catabolism Supports Optimal Virus Replication. Nat. Commun. 2015, 6, 8873. [Google Scholar] [CrossRef]
- Yoshida, G.J. Beyond the Warburg Effect: N-Myc Contributes to Metabolic Reprogramming in Cancer Cells. Front. Oncol. 2020, 10, 791. [Google Scholar] [CrossRef]
- Dang, C.V.; Le, A.; Gao, P. MYC-Induced Cancer Cell Energy Metabolism and Therapeutic Opportunities. Clin. Cancer Res. 2009, 15, 6479–6483. [Google Scholar] [CrossRef]
- Wahlström, T.; Henriksson, M.A. Impact of MYC in Regulation of Tumor Cell Metabolism. Biochim. Biophys. Acta 2015, 1849, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Korangath, P.; Teo, W.W.; Sadik, H.; Han, L.; Mori, N.; Huijts, C.M.; Wildes, F.; Bharti, S.; Zhang, Z.; Santa-Maria, C.A.; et al. Targeting Glutamine Metabolism in Breast Cancer with Aminooxyacetate. Clin. Cancer Res. 2015, 21, 3263–3273. [Google Scholar] [CrossRef] [PubMed]
- Laughner, E.; Taghavi, P.; Chiles, K.; Mahon, P.C.; Semenza, G.L. HER2 (Neu) Signaling Increases the Rate of Hypoxia-Inducible Factor 1alpha (HIF-1alpha) Synthesis: Novel Mechanism for HIF-1-Mediated Vascular Endothelial Growth Factor Expression. Mol. Cell. Biol. 2001, 21, 3995–4004. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhu, Q.; Hu, L.; Chen, H.; Wu, Z.; Li, D. Anterior Gradient 2 is a Binding Stabilizer of Hypoxia Inducible Factor-1α that Enhances CoCl2 -Induced Doxorubicin Resistance in Breast Cancer Cells. Cancer Sci. 2015, 106, 1041–1049. [Google Scholar] [CrossRef]
- Semenza, G.L. Regulation of Cancer Cell Metabolism by Hypoxia-Inducible Factor 1. Semin. Cancer Biol. 2009, 19, 12–16. [Google Scholar] [CrossRef]
- Zeng, W.; Liu, P.; Pan, W.; Singh, S.R.; Wei, Y. Hypoxia and Hypoxia Inducible Factors in Tumor Metabolism. Cancer Lett. 2015, 356, 263–267. [Google Scholar] [CrossRef]
- Lv, X.; Li, J.; Zhang, C.; Hu, T.; Li, S.; He, S.; Yan, H.; Tan, Y.; Lei, M.; Wen, M.; et al. The Role of Hypoxia-Inducible Factors in Tumor Angiogenesis and Cell Metabolism. Genes Dis. 2016, 4, 19–24. [Google Scholar] [CrossRef]
- Gort, E.H.; Groot, A.J.; van der Wall, E.; van Diest, P.J.; Vooijs, M.A. Hypoxic Regulation of Metastasis Via Hypoxia-Inducible Factors. Curr. Mol. Med. 2008, 8, 60–67. [Google Scholar]
- Dong, T.; Yan, Y.; Chai, H.; Chen, S.; Xiong, X.; Sun, D.; Yu, Y.; Deng, L.; Cheng, F. Pyruvate Kinase M2 Affects Liver Cancer Cell Behavior through Up-Regulation of HIF-1α and Bcl-xL in Culture. Biomed. Pharmacother. 2015, 69, 277–284. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, P.; Fukuda, R.; Kumar, G.; Krishnamachary, B.; Zeller, K.I.; Dang, C.V.; Semenza, G.L. HIF-1 Inhibits Mitochondrial Biogenesis and Cellular Respiration in VHL-Deficient Renal Cell Carcinoma by Repression of C-MYC Activity. Cancer Cell 2007, 11, 407–420. [Google Scholar] [CrossRef]
- Kuo, C.; Cheng, C.; Hou, P.; Lin, Y.; Ma, H.; Chung, Y.; Chi, K.; Chen, Y.; Li, W.; Kung, H.; et al. HIF-1-Alpha Links Mitochondrial Perturbation to the Dynamic Acquisition of Breast Cancer Tumorigenicity. Oncotarget 2016, 7, 34052–34069. [Google Scholar] [CrossRef] [PubMed]
- Taub, M.; Mahmoudzadeh, N.H.; Tennessen, J.M.; Sudarshan, S. Renal Oncometabolite L-2-Hydroxyglutarate Imposes a Block in Kidney Tubulogenesis: Evidence for an Epigenetic Basis for the L-2HG-Induced Impairment of Differentiation. Front. Endocrinol. 2022, 13, 932286. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yang, H.; Liu, Y.; Yang, Y.; Wang, P.; Kim, S.; Ito, S.; Yang, C.; Wang, P.; Xiao, M.; et al. Oncometabolite 2-Hydroxyglutarate is a Competitive Inhibitor of A-Ketoglutarate-Dependent Dioxygenases. Cancer Cell 2011, 19, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Janke, R.; Iavarone, A.T.; Rine, J. Oncometabolite D-2-Hydroxyglutarate Enhances Gene Silencing through Inhibition of Specific H3K36 Histone Demethylases. eLife 2017, 6, e22451. [Google Scholar] [CrossRef]
- Xiao, M.; Yang, H.; Xu, W.; Ma, S.; Lin, H.; Zhu, H.; Liu, L.; Liu, Y.; Yang, C.; Xu, Y.; et al. Inhibition of A-KG-Dependent Histone and DNA Demethylases by Fumarate and Succinate that are Accumulated in Mutations of FH and SDH Tumor Suppressors. Genes Dev. 2012, 26, 1326–1338. [Google Scholar] [CrossRef]
- Terunuma, A.; Putluri, N.; Mishra, P.; Mathé, E.A.; Dorsey, T.H.; Yi, M.; Wallace, T.A.; Issaq, H.J.; Zhou, M.; Killian, J.K.; et al. MYC-Driven Accumulation of 2-Hydroxyglutarate is Associated with Breast Cancer Prognosis. J. Clin. Investig. 2014, 124, 398–412. [Google Scholar] [CrossRef]
- Mishra, P.; Tang, W.; Ambs, S. ADHFE1 is a MYC-Linked Oncogene that Induces Metabolic Reprogramming and Cellular De-Differentiation in Breast Cancer. Mol. Cell. Oncol. 2018, 5, e1432260. [Google Scholar] [CrossRef]
- Mishra, P.; Tang, W.; Putluri, V.; Dorsey, T.H.; Jin, F.; Wang, F.; Zhu, D.; Amable, L.; Deng, T.; Zhang, S.; et al. ADHFE1 is a Breast Cancer Oncogene and Induces Metabolic Reprogramming. J. Clin. Investig. 2018, 128, 323–340. [Google Scholar] [CrossRef] [PubMed]
- Grassian, A.R.; Lin, F.; Barrett, R.; Liu, Y.; Jiang, W.; Korpal, M.; Astley, H.; Gitterman, D.; Henley, T.; Howes, R.; et al. Isocitrate Dehydrogenase (IDH) Mutations Promote a Reversible ZEB1/microRNA (miR)-200-Dependent Epithelial-Mesenchymal Transition (EMT). J. Biol. Chem. 2012, 287, 42180–42194. [Google Scholar] [CrossRef]
- May, C.D.; Sphyris, N.; Evans, K.W.; Werden, S.J.; Guo, W.; Mani, S.A. Epithelial-Mesenchymal Transition and Cancer Stem Cells: A Dangerously Dynamic Duo in Breast Cancer Progression. Breast Cancer Res. 2011, 13, 202. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Mboge, M.Y.; Chen, Z.; Khokhar, D.; Wolff, A.; Ai, L.; Heldermon, C.D.; Bozdag, M.; Carta, F.; Supuran, C.T.; Brown, K.D.; et al. A Non-Catalytic Function of Carbonic Anhydrase IX Contributes to the Glycolytic Phenotype and pH Regulation in Human Breast Cancer Cells. Biochem. J. 2019, 476, 1497–1513. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, C.; Longatto-Filho, A.; Azevedo-Silva, J.; Casal, M.; Schmitt, F.C.; Baltazar, F. Role of Monocarboxylate Transporters in Human Cancers: State of the Art. J. Bioenerg. Biomembr. 2012, 44, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Ames, S.; Andring, J.T.; McKenna, R.; Becker, H.M. CAIX Forms a Transport Metabolon with Monocarboxylate Transporters in Human Breast Cancer Cells. Oncogene 2020, 39, 1710–1723. [Google Scholar] [CrossRef] [PubMed]
- Granja, S.; Tavares-Valente, D.; Queirós, O.; Baltazar, F. Value of pH Regulators in the Diagnosis, Prognosis and Treatment of Cancer. Semin. Cancer Biol. 2017, 43, 17–34. [Google Scholar] [CrossRef]
- Liao, S.; Wu, G.; Xie, Z.; Lei, X.; Yang, X.; Huang, S.; Deng, X.; Wang, Z.; Tang, G. pH Regulators and their Inhibitors in Tumor Microenvironment. Eur. J. Med. Chem. 2024, 267, 116170. [Google Scholar] [CrossRef]
- Wang, J.X.; Choi, S.Y.C.; Niu, X.; Kang, N.; Xue, H.; Killam, J.; Wang, Y. Lactic Acid and an Acidic Tumor Microenvironment Suppress Anticancer Immunity. Int. J. Mol. Sci. 2020, 21, 8363. [Google Scholar] [CrossRef]
- Santos, N.; Pereira-Nunes, A.; Baltazar, F.; Granja, S. Lactate as a Regulator of Cancer Inflammation and Immunity. Immunometabolism 2019, 1, e190015. [Google Scholar] [CrossRef]
- Hayes, C.; Donohoe, C.L.; Davern, M.; Donlon, N.E. The Oncogenic and Clinical Implications of Lactate Induced Immunosuppression in the Tumour Microenvironment. Cancer Lett. 2021, 500, 75–86. [Google Scholar] [CrossRef]
- Wang, Z.; Peng, W.; Zhang, P.; Yang, X.; Zhou, Q. Lactate in the Tumour Microenvironment: From Immune Modulation to Therapy. eBioMedicine 2021, 73, 103627. [Google Scholar] [CrossRef]
- Calcinotto, A.; Filipazzi, P.; Grioni, M.; Iero, M.; De Milito, A.; Ricupito, A.; Cova, A.; Canese, R.; Jachetti, E.; Rossetti, M.; et al. Modulation of Microenvironment Acidity Reverses Anergy in Human and Murine Tumor-Infiltrating T Lymphocytes. Cancer Res. 2012, 72, 2746–2756. [Google Scholar] [CrossRef] [PubMed]
- Vuillefroy de Silly, R.; Pericou, L.; Seijo, B.; Crespo, I.; Coukos, G.; Irving, M. Acidity-Induced Dysfunction of CD8+ T Cells is Characterized by Impaired IL-2 Responsiveness and Perturbations to mTORC1 Signaling and C-Myc Levels. bioRxiv 2023. [Google Scholar] [CrossRef]
- Bellone, M.; Calcinotto, A.; Filipazzi, P.; De Milito, A.; Fais, S.; Rivoltini, L. The Acidity of the Tumor Microenvironment is a Mechanism of Immune Escape that can be Overcome by Proton Pump Inhibitors. Oncoimmunology 2013, 2, e22058. [Google Scholar] [CrossRef] [PubMed]
- Erra Díaz, F.; Dantas, E.; Geffner, J. Unravelling the Interplay between Extracellular Acidosis and Immune Cells. Mediat. Inflamm. 2018, 2018, 1218297. [Google Scholar] [CrossRef]
- Netea-Maier, R.T.; Smit, J.W.A.; Netea, M.G. Metabolic Changes in Tumor Cells and Tumor-Associated Macrophages: A Mutual Relationship. Cancer Lett. 2018, 413, 102–109. [Google Scholar] [CrossRef]
- Kes, M.M.G.; Van den Bossche, J.; Griffioen, A.W.; Huijbers, E.J.M. Oncometabolites Lactate and Succinate Drive Pro-Angiogenic Macrophage Response in Tumors. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188427. [Google Scholar] [CrossRef]
- Tao, H.; Zhong, X.; Zeng, A.; Song, L. Unveiling the Veil of Lactate in Tumor-Associated Macrophages: A Successful Strategy for Immunometabolic Therapy. Front. Immunol. 2023, 14, 1208870. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.N.; Capuk, O.; Patel, S.M.; Sun, D. The Role of Metabolic Plasticity of Tumor-Associated Macrophages in Shaping the Tumor Microenvironment Immunity. Cancers 2022, 14, 3331. [Google Scholar] [CrossRef]
- Tuomela, K.; Levings, M.K. Acidity Promotes the Differentiation of Immunosuppressive Regulatory T Cells. Eur. J. Immunol. 2023, 53, e2350511. [Google Scholar] [CrossRef]
- Rao, D.; Stunnenberg, J.A.; Lacroix, R.; Dimitriadis, P.; Kaplon, J.; Verburg, F.; van van Royen, P.T.; Hoefsmit, E.P.; Renner, K.; Blank, C.U.; et al. Acidity-Mediated Induction of FoxP3(+) Regulatory T Cells. Eur. J. Immunol. 2023, 53, e2250258. [Google Scholar] [CrossRef]
- Watson, M.J.; Vignali, P.D.A.; Mullett, S.J.; Overacre-Delgoffe, A.E.; Peralta, R.M.; Grebinoski, S.; Menk, A.V.; Rittenhouse, N.L.; DePeaux, K.; Whetstone, R.D.; et al. Metabolic Support of Tumour-Infiltrating Regulatory T Cells by Lactic Acid. Nature 2021, 591, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, M.; Korechika, A.; Yamakawa, H.; Kawabe, N.; Nakai, K.; Muragaki, Y. Acidic Microenvironment Induction of Interleukin-8 Expression and Matrix Metalloproteinase-2/-9 Activation Via Acid-Sensing Ion Channel 1 Promotes Breast Cancer Cell Progression. Oncol. Rep. 2021, 45, 1284–1294. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.C.; Singh, R.; Pochampally, R.; Watabe, K.; Mo, Y. Acidosis Promotes Invasiveness of Breast Cancer Cells through ROS-AKT-NF-κB Pathway. Oncotarget 2014, 5, 12070–12082. [Google Scholar] [CrossRef] [PubMed]
- Espinoza-Sánchez, N.A.; Chimal-Ramírez, G.K.; Mantilla, A.; Fuentes-Pananá, E.M. IL-1β, IL-8, and Matrix Metalloproteinases-1, -2, and -10 are Enriched upon Monocyte-Breast Cancer Cell Cocultivation in a Matrigel-Based Three-Dimensional System. Front. Immunol. 2017, 8, 205. [Google Scholar]
- Todorović-Raković, N.; Milovanović, J. Interleukin-8 in Breast Cancer Progression. J. Interferon Cytokine Res. 2013, 33, 563–570. [Google Scholar] [CrossRef]
- Xie, Q.; Zhu, Z.; He, Y.; Zhang, Z.; Zhang, Y.; Wang, Y.; Luo, J.; Peng, T.; Cheng, F.; Gao, J.; et al. A Lactate-Induced Snail/STAT3 Pathway Drives GPR81 Expression in Lung Cancer Cells. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165576. [Google Scholar] [CrossRef]
- Su, J.; Mao, X.; Wang, L.; Chen, Z.; Wang, W.; Zhao, C.; Li, G.; Guo, W.; Hu, Y. Lactate/GPR81 Recruits Regulatory T Cells by Modulating CX3CL1 to Promote Immune Resistance in a Highly Glycolytic Gastric Cancer. Oncoimmunology 2024, 13, 2320951. [Google Scholar] [CrossRef]
- Lee, Y.J.; Shin, K.J.; Park, S.; Park, K.S.; Park, S.; Heo, K.; Seo, Y.; Noh, D.; Ryu, S.H.; Suh, P. G-Protein-Coupled Receptor 81 Promotes a Malignant Phenotype in Breast Cancer through Angiogenic Factor Secretion. Oncotarget 2016, 7, 70898–70911. [Google Scholar] [CrossRef]
- Brown, T.P.; Ganapathy, V. Lactate/GPR81 Signaling and Proton Motive Force in Cancer: Role in Angiogenesis, Immune Escape, Nutrition, and Warburg Phenomenon. Pharmacol. Ther. 2020, 206, 107451. [Google Scholar] [CrossRef]
- Yin, X.; Mead, B.E.; Safaee, H.; Langer, R.; Karp, J.M.; Levy, O. Engineering Stem Cell Organoids. Cell Stem Cell 2016, 18, 25–38. [Google Scholar] [CrossRef]
- Rossi, G.; Manfrin, A.; Lutolf, M.P. Progress and Potential in Organoid Research. Nat. Rev. Genet. 2018, 19, 671–687. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Oyang, L.; Peng, Q.; Liu, Q.; Xu, X.; Wu, N.; Tan, S.; Yang, W.; Han, Y.; Lin, J.; et al. Organoids: Opportunities and Challenges of Cancer Therapy. Front. Cell Dev. Biol. 2023, 11, 1232528. [Google Scholar] [CrossRef] [PubMed]
- Bartfeld, S.; Bayram, T.; van de Wetering, M.; Huch, M.; Begthel, H.; Kujala, P.; Vries, R.; Peters, P.J.; Clevers, H. In Vitro Expansion of Human Gastric Epithelial Stem Cells and their Responses to Bacterial Infection. Gastroenterology 2015, 148, 126–136.e6. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, R.; Lee, C.M.; Shugart, E.C.; Benedetti, M.; Charo, R.A.; Gartner, Z.; Hogan, B.; Knoblich, J.; Nelson, C.M.; Wilson, K.M. Human Organoids: A New Dimension in Cell Biology. Mol. Biol. Cell 2019, 30, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid Cultures Derived from Patients with Advanced Prostate Cancer. Cell 2014, 159, 176. [Google Scholar] [CrossRef]
- Kiwaki, T.; Kataoka, H. Patient-Derived Organoids of Colorectal Cancer: A Useful Tool for Personalized Medicine. J. Pers. Med. 2022, 12, 695. [Google Scholar] [CrossRef]
- Martini, G.; Belli, V.; Napolitano, S.; Ciaramella, V.; Ciardiello, D.; Belli, A.; Izzo, F.; Avallone, A.; Selvaggi, F.; Menegon Tasselli, F.; et al. Establishment of Patient-Derived Tumor Organoids to Functionally Inform Treatment Decisions in Metastatic Colorectal Cancer. ESMO Open 2023, 8, 101198. [Google Scholar] [CrossRef]
- Nayak, B.; Balachander, G.M.; Manjunath, S.; Rangarajan, A.; Chatterjee, K. Tissue Mimetic 3D Scaffold for Breast Tumor-Derived Organoid Culture Toward Personalized Chemotherapy. Colloids Surf. B Biointerfaces 2019, 180, 334–343. [Google Scholar] [CrossRef]
- Prince, E.; Cruickshank, J.; Ba-Alawi, W.; Hodgson, K.; Haight, J.; Tobin, C.; Wakeman, A.; Avoulov, A.; Topolskaia, V.; Elliott, M.J.; et al. Biomimetic Hydrogel Supports Initiation and Growth of Patient-Derived Breast Tumor Organoids. Nat. Commun. 2022, 13, 1466. [Google Scholar] [CrossRef]
- Guillen, K.P.; Fujita, M.; Butterfield, A.J.; Scherer, S.D.; Bailey, M.H.; Chu, Z.; Derose, Y.S.; Zhao, L.; Cortes-Sanchez, E.; Yang, C.; et al. A Human Breast Cancer-Derived Xenograft and Organoid Platform for Drug Discovery and Precision Oncology. Nat. Cancer 2022, 3, 232. [Google Scholar] [CrossRef]
- Aggarwal, D.; Russo, S.; Naik, P.; Bhatia, S.; Spector, D.L. Establishment and Culture of Patient-Derived Breast Organoids. JoVE 2023, e64889. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Dai, W.; Xia, X.; Wang, R.; Zhao, J.; Han, L.; Mo, S.; Xiang, W.; Du, L.; Zhu, G.; et al. Modeling Tumor Development and Metastasis using Paired Organoids Derived from Patients with Colorectal Cancer Liver Metastases. J. Hematol. Oncol. 2020, 13, 119. [Google Scholar] [CrossRef] [PubMed]
- Maru, Y.; Tanaka, N.; Tatsumi, Y.; Nakamura, Y.; Itami, M.; Hippo, Y. Kras Activation in Endometrial Organoids Drives Cellular Transformation and Epithelial-Mesenchymal Transition. Oncogenesis 2021, 10, 46. [Google Scholar] [CrossRef]
- Stokes, K.; Nunes, M.; Trombley, C.; Flôres, D.E.F.L.; Wu, G.; Taleb, Z.; Alkhateeb, A.; Banskota, S.; Harris, C.; Love, O.P.; et al. The Circadian Clock Gene, Bmal1, Regulates Intestinal Stem Cell Signaling and Represses Tumor Initiation. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 1847–1872.e0. [Google Scholar] [CrossRef]
- Xiao, Y.; Ma, D.; Yang, Y.; Yang, F.; Ding, J.; Gong, Y.; Jiang, L.; Ge, L.; Wu, S.; Yu, Q.; et al. Comprehensive Metabolomics Expands Precision Medicine for Triple-Negative Breast Cancer. Cell Res. 2022, 32, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Sharick, J.T.; Jeffery, J.J.; Karim, M.R.; Walsh, C.M.; Esbona, K.; Cook, R.S.; Skala, M.C. Cellular Metabolic Heterogeneity In Vivo is Recapitulated in Tumor Organoids. Neoplasia 2019, 21, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Sachs, N.; De Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H.; et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 2018, 172, 373. [Google Scholar] [CrossRef]
- Guan, D.; Liu, X.; Shi, Q.; He, B.; Zheng, C.; Meng, X. Breast Cancer Organoids and their Applications for Precision Cancer Immunotherapy. World J. Surg. Oncol. 2023, 21, 343. [Google Scholar] [CrossRef]
- Tzeng, Y.T.; Hsiao, J.; Tseng, L.; Hou, M.; Li, C. Breast Cancer Organoids Derived from Patients: A Platform for Tailored Drug Screening. Biochem. Pharmacol. 2023, 217, 115803. [Google Scholar] [CrossRef]
- Dekkers, J.F.; van Vliet, E.J.; Sachs, N.; Rosenbluth, J.M.; Kopper, O.; Rebel, H.G.; Wehrens, E.J.; Piani, C.; Visvader, J.E.; Verissimo, C.S.; et al. Long-Term Culture, Genetic Manipulation and Xenotransplantation of Human Normal and Breast Cancer Organoids. Nat. Protoc. 2021, 16, 1936–1965. [Google Scholar] [CrossRef]
- Han, Z.; Yao, L.; Fang, Y.; Chen, S.; Lian, R.; Yao, Y.; Chen, H.; Ji, X.; Yu, W.; Wang, Z.; et al. Patient-Derived Organoid Elucidates the Identical Clonal Origin of Bilateral Breast Cancer with Diverse Molecular Subtypes. Front. Oncol. 2024, 14, 1361603. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Ji, P.; Yang, Y.; Xie, S.; Yu, T.; Xiao, Y.; Jin, M.; Ma, D.; Guo, L.; Pei, Y.; et al. Metabolic-Pathway-Based Subtyping of Triple-Negative Breast Cancer Reveals Potential Therapeutic Targets. Cell Metab. 2021, 33, 51. [Google Scholar] [CrossRef] [PubMed]
- Magré, L.; Verstegen, M.M.A.; Buschow, S.; Van Der Laan, L.J.W.; Peppelenbosch, M.; Desai, J. Emerging Organoid-Immune Co-Culture Models for Cancer Research: From Oncoimmunology to Personalized Immunotherapies. J. Immunother. Cancer 2023, 11, e006290. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Kang, M. Exploring Tumor–Immune Interactions in Co-Culture Models of T Cells and Tumor Organoids Derived from Patients. Int. J. Mol. Sci. 2023, 24, 14609. [Google Scholar] [CrossRef] [PubMed]
- Subtil, B.; Iyer, K.K.; Poel, D.; Bakkerus, L.; Gorris, M.A.J.; Escalona, J.C.; Dries, K.V.D.; Cambi, A.; Verheul, H.M.W.; De Vries, I.J.M.; et al. Dendritic Cell Phenotype and Function in a 3D Co-Culture Model of Patient-Derived Metastatic Colorectal Cancer Organoids. Front. Immunol. 2023, 14, 1105244. [Google Scholar] [CrossRef]
- Grassi, L.; Alfonsi, R.; Francescangeli, F.; Signore, M.; De Angelis, M.L.; Addario, A.; Costantini, M.; Flex, E.; Ciolfi, A.; Pizzi, S.; et al. Organoids as a New Model for Improving Regenerative Medicine and Cancer Personalized Therapy in Renal Diseases. Cell Death Dis. 2019, 10, 201. [Google Scholar] [CrossRef]
- Kim, M.; Mun, H.; Sung, C.O.; Cho, E.J.; Jeon, H.; Chun, S.; Jung, D.J.; Shin, T.H.; Jeong, G.S.; Kim, D.K.; et al. Patient-Derived Lung Cancer Organoids as In Vitro Cancer Models for Therapeutic Screening. Nat. Commun. 2019, 10, 3991. [Google Scholar] [CrossRef]
- Balážová, K.; Clevers, H.; Dost, A.F. The Role of Macrophages in Non-Small Cell Lung Cancer and Advancements in 3D Co-Cultures. eLife 2023, 12, e82998. [Google Scholar] [CrossRef]
- Lee, S.H.; Hu, W.; Matulay, J.T.; Silva, M.V.; Owczarek, T.B.; Kim, K.; Chua, C.W.; Barlow, L.J.; Kandoth, C.; Williams, A.B.; et al. Tumor Evolution and Drug Response in Patient-Derived Organoid Models of Bladder Cancer. Cell 2019, 173, 515. [Google Scholar] [CrossRef]
- Aung, A.; Kumar, V.; Theprungsirikul, J.; Davey, S.K.; Varghese, S. An Engineered Tumor-on-a-Chip Device with Breast Cancer–Immune Cell Interactions for Assessing T-Cell Recruitment. Cancer Res. 2021, 80, 263. [Google Scholar] [CrossRef]
- Raffo-Romero, A.; Ziane-Chaouche, L.; Salomé-Desnoulez, S.; Hajjaji, N.; Fournier, I.; Salzet, M.; Duhamel, M. A Co-Culture System of Macrophages with Breast Cancer Tumoroids to Study Cell Interactions and Therapeutic Responses. Cell Rep. Methods 2024, 4, 100792. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.Y.; Li, J.; Wang, M.L.; Chen, X.Y.; Tang, R.; Liu, X.Q. Fabrication of a Coculture Organoid Model in the Biomimetic Matrix of Alginate to Investigate Breast Cancer Progression in a TAMs-Leading Immune Microenvironment. ACS Appl. Mater. Interfaces 2024, 16, 11275–11288. [Google Scholar] [CrossRef] [PubMed]
- Walsh, A.J.; Cook, R.S.; Manning, H.C.; Hicks, D.J.; Lafontant, A.; Arteaga, C.L.; Skala, M.C. Optical Metabolic Imaging Identifies Glycolytic Levels, Sub-Types and Early Treatment Response in Breast Cancer. Cancer Res. 2013, 73, 6164–6174. [Google Scholar] [CrossRef]
- Gillette, A.A.; Babiarz, C.P.; VanDommelen, A.R.; Pasch, C.A.; Clipson, L.; Matkowskyj, K.A.; Deming, D.A.; Skala, M.C. Autofluorescence Imaging of Treatment Response in Neuroendocrine Tumor Organoids. Cancers 2021, 13, 1873. [Google Scholar] [CrossRef]
- Walsh, A.J.; Castellanos, J.A.; Nagathihalli, N.S.; Merchant, N.; Skala, M.C. Optical Imaging of Drug-Induced Metabolism Changes in Murine and Human Pancreatic Cancer Organoids Reveals Heterogeneous Drug Response. Pancreas 2016, 45, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.T.; Heaster, T.M.; Skala, M.C. Metabolic Imaging of Head and Neck Cancer Organoids. PLoS ONE 2017, 12, e0170415. [Google Scholar] [CrossRef] [PubMed]
- Walsh, A.J.; Cook, R.S.; Sanders, M.E.; Aurisicchio, L.; Ciliberto, G.; Arteaga, C.L.; Skala, M.C. Quantitative Optical Imaging of Primary Tumor Organoid Metabolism Predicts Drug Response in Breast Cancer. Cancer Res. 2014, 74, 5184. [Google Scholar] [CrossRef]
- Walsh, A.J.; Skala, M.C. Optical Metabolic Imaging Quantifies Heterogeneous Cell Populations. Biomed. Opt. Express 2015, 6, 559–573. [Google Scholar] [CrossRef]
- Heaster, T.M.; Humayun, M.; Yu, J.; Beebe, D.J.; Skala, M.C. Autofluorescence Imaging of 3D Tumor–Macrophage Microscale Cultures Resolves Spatial and Temporal Dynamics of Macrophage Metabolism. Cancer Res. 2024, 80, 5408. [Google Scholar] [CrossRef]
- He, M.J.; Pu, W.; Wang, X.; Zhang, W.; Tang, D.; Dai, Y. Comparing DESI-MSI and MALDI-MSI Mediated Spatial Metabolomics and their Applications in Cancer Studies. Front. Oncol. 2022, 12, 891018. [Google Scholar] [CrossRef]
- Li, H.; Hummon, A.B. Imaging Mass Spectrometry of Three-Dimensional Cell Culture. Anal. Chem. 2011, 83, 8794–8801. [Google Scholar] [CrossRef]
- Hiraide, T.; Ikegami, K.; Sakaguchi, T.; Morita, Y.; Hayasaka, T.; Masaki, N.; Waki, M.; Sugiyama, E.; Shinriki, S.; Takeda, M.; et al. Accumulation of Arachidonic Acid-Containing Phosphatidylinositol at the Outer Edge of Colorectal Cancer. Sci. Rep. 2016, 6, 29935. [Google Scholar] [CrossRef]
- Liu, X.; Hummon, A.B. Chemical Imaging of Platinum-Based Drugs and their Metabolites. Sci. Rep. 2016, 6, 38507. [Google Scholar] [CrossRef]
- Liu, X.; Weaver, E.M.; Hummon, A.B. Evaluation of Therapeutics in Three-Dimensional Cell Culture Systems by MALDI Imaging Mass Spectrometry. Anal. Chem. 2014, 85, 6295. [Google Scholar] [CrossRef]
- Feist, P.E.; Sidoli, S.; Liu, X.; Schroll, M.M.; Rahmy, S.; Fujiwara, R.; Garcia, B.A.; Hummon, A.B. Multicellular Tumor Spheroids Combined with Mass Spectrometric Histone Analysis to Evaluate Epigenetic Drugs. Anal. Chem. 2018, 89, 2773. [Google Scholar] [CrossRef]
- Lukowski, J.K.; Weaver, E.M.; Hummon, A.B. Analyzing Liposomal Drug Delivery Systems in Three-Dimensional Cell Culture Models using MALDI Imaging Mass Spectrometry. Anal. Chem. 2017, 89, 8453. [Google Scholar] [CrossRef]
- Ahlf Wheatcraft, D.R.; Liu, X.; Hummon, A.B. Sample Preparation Strategies for Mass Spectrometry Imaging of 3D Cell Culture Models. JoVE 2014, e52313. [Google Scholar] [CrossRef]
- Spencer, C.E.; Flint, L.E.; Duckett, C.J.; Cole, L.M.; Cross, N.; Smith, D.P.; Clench, M.R. Role of MALDI-MSI in Combination with 3D Tissue Models for Early Stage Efficacy and Safety Testing of Drugs and Toxicants. Expert Rev. Proteom. 2020, 17, 827. [Google Scholar] [CrossRef]
- Francese, S.; Bradshaw, R.; Flinders, B.; Mitchell, C.; Bleay, S.; Cicero, L.; Clench, M.R. Curcumin: A Multipurpose Matrix for MALDI Mass Spectrometry Imaging Applications. Anal. Chem. 2013, 85, 5240–5248. [Google Scholar] [CrossRef]
- Avery, J.L.; McEwen, A.; Flinders, B.; Francese, S.; Clench, M.R. Matrix-Assisted Laser Desorption Mass Spectrometry Imaging for the Examination of Imipramine Absorption by Straticell-RHE-EPI/001 an Artificial Model of the Human Epidermis. Xenobiotica 2011, 41, 735–742. [Google Scholar] [CrossRef]
- Bergmann, S.; Lawler, S.E.; Qu, Y.; Fadzen, C.M.; Wolfe, J.M.; Regan, M.S.; Pentelute, B.L.; Agar, N.Y.R.; Cho, C. Blood–brain-Barrier Organoids for Investigating the Permeability of CNS Therapeutics. Nat. Protoc. 2019, 13, 2827. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.; Sharick, J.T.; Skala, M.C.; Li, L. Sample Preparation Strategies for High-throughput Mass Spectrometry Imaging of Primary Tumor Organoids. J. Mass Spectrom. 2020, 55, e4452. [Google Scholar] [CrossRef] [PubMed]
- David, B.P.; Dubrovskyi, O.; Speltz, T.E.; Wolff, J.J.; Frasor, J.; Sanchez, L.M.; Moore, T.W. Using Tumor Explants for Imaging Mass Spectrometry Visualization of Unlabeled Peptides and Small Molecules. ACS Med. Chem. Lett. 2018, 9, 768. [Google Scholar] [CrossRef] [PubMed]
- Gross, M.I.; Demo, S.D.; Dennison, J.B.; Chen, L.; Chernov-Rogan, T.; Goyal, B.; Janes, J.R.; Laidig, G.J.; Lewis, E.R.; Li, J.; et al. Antitumor Activity of the Glutaminase Inhibitor CB-839 in Triple-Negative Breast Cancer. Mol. Cancer Ther. 2014, 13, 890. [Google Scholar] [CrossRef] [PubMed]
- Denti, V.; Andersen, M.K.; Smith, A.; Bofin, A.M.; Nordborg, A.; Magni, F.; Moestue, S.A.; Giampà, M. Reproducible Lipid Alterations in Patient-Derived Breast Cancer Xenograft FFPE Tissue Identified with MALDI MSI for Pre-Clinical and Clinical Application. Metabolites 2021, 11, 577. [Google Scholar] [CrossRef]
- Torata, N.; Kubo, M.; Miura, D.; Ohuchida, K.; Mizuuchi, Y.; Fujimura, Y.; Hayakawa, E.; Kai, M.; Oda, Y.; Mizumoto, K.; et al. Visualizing Energy Charge in Breast Carcinoma Tissues by MALDI Mass-Spectrometry Imaging Profiles of Low-Molecular-Weight Metabolites. Anticancer Res. 2018, 38, 4267. [Google Scholar] [CrossRef]
- Tucker, L.H.; Hamm, G.R.; Sargeant, R.J.E.; Goodwin, R.J.A.; Mackay, C.L.; Campbell, C.J.; Clarke, D.J. Untargeted Metabolite Mapping in 3D Cell Culture Models using High Spectral Resolution FT-ICR Mass Spectrometry Imaging. Anal. Chem. 2019, 91, 9522–9529. [Google Scholar] [CrossRef]
- Yoshizaki, H.; Ogiso, H.; Okazaki, T.; Kiyokawa, E. Comparative Lipid Analysis in the Normal and Cancerous Organoids of MDCK Cells. J. Biochem. 2016, 159, 573. [Google Scholar] [CrossRef]
- Weygand, J.; Carter, S.E.; Salzillo, T.C.; Moussalli, M.; Dai, B.; Dutta, P.; Zuo, X.; Fleming, J.B.; Shureiqi, I.; Bhattacharya, P. Can an Organoid Recapitulate the Metabolome of its Parent Tissue? A Pilot NMR Spectroscopy Study. J. Cancer Prev. Curr. Res. 2017, 8, 00307. [Google Scholar]
- Lackner, M.; Neef, S.K.; Winter, S.; Beer-Hammer, S.; Nürnberg, B.; Schwab, M.; Hofmann, U.; Haag, M. Untargeted Stable Isotope-Resolved Metabolomics to Assess the Effect of PI3Kβ Inhibition on Metabolic Pathway Activities in a PTEN Null Breast Cancer Cell Line. Front. Mol. Biosci. 2022, 9, 1004602. [Google Scholar] [CrossRef]
- Neef, S.K.; Janssen, N.; Winter, S.; Wallisch, S.K.; Hofmann, U.; Dahlke, M.H.; Schwab, M.; Mürdter, T.E.; Haag, M. Metabolic Drug Response Phenotyping in Colorectal Cancer Organoids by LC-QTOF-MS. Metabolites 2020, 10, 494. [Google Scholar] [CrossRef]
- Reustle, A.; Menig, L.; Leuthold, P.; Hofmann, U.; Stühler, V.; Schmees, C.; Becker, M.; Haag, M.; Klumpp, V.; Winter, S.; et al. Nicotinamide-N-Methyltransferase is a Promising Metabolic Drug Target for Primary and Metastatic Clear Cell Renal Cell Carcinoma. Clin. Transl. Med. 2022, 12, e883. [Google Scholar] [CrossRef]
- Zhou, Q.; Alvarez, M.R.S.; Solakyildirim, K.; Tena, J.; Serrano, L.M.N.; Lam, M.; Nguyen, C.; Tobias, F.; Hummon, A.B.; Nacario, R.C.; et al. Multi-Glycomic Analysis of Spheroid Glycocalyx Differentiates 2- and 3-Dimensional Cell Models. Glycobiology 2022, 33, 2. [Google Scholar] [CrossRef] [PubMed]
- Mönch, D.; Koch, J.; Maaß, A.; Janssen, N.; Mürdter, T.; Renner, P.; Fallier-Becker, P.; Solaß, W.; Schwab, M.; Dahlke, N.H.; et al. A Human Ex Vivo Coculture Model to Investigate Peritoneal Metastasis and Innovative Treatment Options. Pleura Peritoneum 2021, 6, 121. [Google Scholar] [CrossRef] [PubMed]
- Campioni, G.; Pasquale, V.; Busti, S.; Ducci, G.; Sacco, E.; Vanoni, M. An Optimized Workflow for the Analysis of Metabolic Fluxes in Cancer Spheroids using Seahorse Technology. Cells 2022, 11, 866. [Google Scholar] [CrossRef]
- Grün, C.; Pfeifer, J.; Liebsch, G.; Gottwald, E. O2-Sensitive Microcavity Arrays: A New Platform for Oxygen Measurements in 3D Cell Cultures. Front. Bioeng. Biotechnol. 2023, 11, 1111316. [Google Scholar] [CrossRef]
- Nashimoto, Y.; Shishido, S.; Onuma, K.; Ino, K.; Inoue, M.; Shiku, H. Oxygen Metabolism Analysis of a Single Organoid for Non-Invasive Discrimination of Cancer Subpopulations with Different Growth Capabilities. Front. Bioeng. Biotechnol. 2023, 11, 1184325. [Google Scholar] [CrossRef]
- Ino, K.; Wachi, M.; Utagawa, Y.; Konno, A.; Takinoue, M.; Abe, H.; Shiku, H. Scanning Electrochemical Microscopy for Determining Oxygen Consumption Rates of Cells in Hydrogel Fibers Fabricated using an Extrusion 3D Bioprinter. Anal. Chim. Acta 2024, 1304, 342539. [Google Scholar] [CrossRef]
- Dornhof, J.; Kieninger, J.; Muralidharan, H.; Maurer, J.; Urban, G.A.; Weltin, A. Oxygen and Lactate Monitoring in 3D Breast Cancer Organoid Culture with Sensor-Integrated Microfluidic Platform. In Proceedings of the 2021 21st International Conference on Solid-State Sensors, Actuators and Microsystems (Transducers), Orlando, FL, USA, 20–24 June 2021; pp. 703–706. [Google Scholar]
- Shi, W.; Mirza, S.; Kuss, M.; Liu, B.; Hartin, A.; Wan, S.; Kong, Y.; Mohapatra, B.; Krishnan, M.; Band, H.; et al. Embedded Bioprinting of Breast Tumor Cells and Organoids using Low-Concentration Collagen-Based Bioinks. Adv. Healthc. Mater. 2023, 12, 2300905. [Google Scholar] [CrossRef]


| Methodology | Organoid Model | Reference | Summary |
|---|---|---|---|
| Agilent Seahorse Analyzer | MCF7 and MDA-MB-231 breast tumor organoids | Campioni et al. (2022) [247] | Optimized Seahorse metabolic analysis for high-resolution metabolic characterization of breast cancer spheroids. |
| LC-qTOF-MS | Metastatic clear cell renal cell carcinoma (ccRCC) PDOs | Reustle et al. (2022) [244] | Studied metabolite changes in metastatic renal carcinoma organoids using LC-qTOF-MS. |
| LC-qTOF-MS | HCT116 and HT29 CRC organoids | Zhou et al. (2022) [245] | Investigated metabolite changes in CRC organoids, revealing insights into metabolic reprogramming. |
| LC-qTOF-MS | CRC PDOs—Ex vivo peritoneum co-cultures | Mönch et al. (2021) [246] | Analyzed metabolic profiles in CRC PDO-peritoneum co-cultures |
| LC-qTOF-MS | CRC PDOs | Neef et al. (2020) [243] | Developed a novel protocol for metabolomic and lipidomic profiling, identifying dose-dependent changes in metabolic profiles of CRC PDOs. |
| MALDI-MSI | Patient-derived breast cancer xenograft FFPE tissue | Denti et al. (2021) [237] | Investigated lipid alterations and treatment responses in breast cancer xenografts, identifying specific metabolic lipid changes and reduced heterogeneity with treatment. |
| MALDI-MSI | MCF7 breast tumor organoids | Tucker et al. (2019) [239] | Used MALDI-MSI to image endogenous metabolite distribution, identifying markers of hypoxic and oxidative stress in breast cancer spheroids. |
| MALDI-MSI | Breast carcinoma tissues embedded in frozen tissue microarrays | Torata et al. (2018) [238] | Analyzed energy charge and adenosine phosphate compound values in breast carcinoma tissues, finding higher values compared to normal tissue. |
| Microcavity arrays for oxygen concentration measurements | HCC spheroids | Grün et al. (2023) [248] | Developed microcavity arrays for determining oxygen in the organoid microenvironment. |
| Microfluidic platform with electrochemical sensors | TNBC PDTOs | Dornhof et al. (2021) [251] | Created a microfluidic platform for real-time measurement of metabolic parameters in breast cancer spheroids. |
| OMI | Primary invasive ductal carcinoma breast PDTO-macrophage co-cultures | Heaster et al. (2020) [221] | Captured spatiotemporal changes in macrophage metabolism, polarization, and migration in breast cancer organoid models, revealing significant metabolic differences. |
| OMI | Breast cancer PDxOs | Sharick et al. (2019) [198] | Demonstrated distinct metabolic profiles within breast cancer organoids, correlating with drug responses and identifying potentially treatment-resistant cell populations. |
| OMI | Breast cancer PDX and PDxOs | Walsh et al. (2014) [219] | Used OMI to detect metabolic changes in breast cancer organoids upon anticancer drug treatment. |
| Scanning electrochemical microscopy | 3D breast cancer cell culture in hydrogel fibers | Kosuke et al. (2024) [250] | Determined oxygen consumption rates in breast cancer cells using scanning electrochemical microscopy in 3D bioprinted hydrogel fibers. |
| Scanning electrochemical microscopy | CRC PDOs | Nashimoto et al. (2023) [249] | Identified subpopulations with different growth capabilities based on oxygen metabolism in colorectal cancer organoids. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glibetic, N.; Bowman, S.; Skaggs, T.; Weichhaus, M. The Use of Patient-Derived Organoids in the Study of Molecular Metabolic Adaptation in Breast Cancer. Int. J. Mol. Sci. 2024, 25, 10503. https://doi.org/10.3390/ijms251910503
Glibetic N, Bowman S, Skaggs T, Weichhaus M. The Use of Patient-Derived Organoids in the Study of Molecular Metabolic Adaptation in Breast Cancer. International Journal of Molecular Sciences. 2024; 25(19):10503. https://doi.org/10.3390/ijms251910503
Chicago/Turabian StyleGlibetic, Natalija, Scott Bowman, Tia Skaggs, and Michael Weichhaus. 2024. "The Use of Patient-Derived Organoids in the Study of Molecular Metabolic Adaptation in Breast Cancer" International Journal of Molecular Sciences 25, no. 19: 10503. https://doi.org/10.3390/ijms251910503
APA StyleGlibetic, N., Bowman, S., Skaggs, T., & Weichhaus, M. (2024). The Use of Patient-Derived Organoids in the Study of Molecular Metabolic Adaptation in Breast Cancer. International Journal of Molecular Sciences, 25(19), 10503. https://doi.org/10.3390/ijms251910503







