HNF1B Transcription Factor: Key Regulator in Renal Physiology and Pathogenesis
Abstract
:1. Introduction
2. HNF1B-Associated Diseases and Clinical Significance
3. Hnf1b Protein Domains and Transcriptional Complexes
4. Hnf1b Renal Functions in Kidney Development and Physiology
4.1. Hnf1b in Nephrogenesis
- 1.
- Induction of nephron progenitor cells to form the RV: Hnf1b is implicated in several stages of renal development, regulating a significant proportion of the mechanisms underlying these processes (Figure 3c). Hnf1b conditional inactivation in murine nephron progenitors showed that Hnf1b acts upstream of Wnt9b in mice [3,48]. However, research by Niborski L et al. [21] showed minimal impact on Wnt9b expression in a mouse model with an identified HNF1B human mutation at the intron-2 splice donor site [21]. Wnt9b plays a crucial role in nephrogenesis, particularly in inducing the mesenchyme-to-epithelium transition (MET). Wnt9b is expressed uniformly in the UB epithelium, with increased expression in areas where RVs will form [47,51]. MET is responsible for forming the RV from nephron progenitor cells [2,9,47,51,52,53,54] and involves a stepwise assembly of intercellular junctions and de novo establishment of apical–basal polarity to form the RV, the first polarized epithelial precursor of the nephron [55]. This suggests that Hnf1b plays a crucial role in initiating nephrogenesis [3,48].Wnt9b signaling activates the expression of the differentiation markers Lef1, Fgf8, and Wnt4 in the surrounding MM [47]. Lef1 and Wnt4 display polarized expression patterns in the distal RV, regulating its early polarity [47]; Wnt4 also triggers the expression of Lhx1, the next transcriptional regulator [47,52,56,57]. Lhx1 drives RV progression to the CSB by activating Dll1 and Pou3f3 (also known as Brn1) expression in the RV. Lhx1 is also involved in the proximo-distal differentiation of the RV, CSB, and SSB [47,48,58]. Hnf1b also regulates PAX2, a transcription factor critical for the MET of nephron progenitors, maintaining nephric duct epithelial polarity and SSB differentiation [3,8,47,48,59,60,61,62]. PAX2 defects have been linked to focal segmental glomerulosclerosis (FSGS) [23].
- 2.
- Differentiation into CSB and its progression to SSB: in the CSB, Pou3f3 expression is regulated by Lhx1 in the distal RV and by Hnf1b in the distal CSB regions, as well as the proximal and bulge regions of the SSB [8,47,52]. Pou3f3 is involved in elongating and differentiating the loop of Henle and forming the distal convoluted tubule (DCT) [8,48,52]. Hnf1b also activates Notch pathway components such as Dll1, Jag1, Lfng [52], and Hes5 [8], which are crucial for inducing differentiation and polarization in nephrogenesis [8,47]. Dll1 and Jag1 are ligands for Notch receptors, Dll1 expression is regulated by both Lhx1 and Hnf1b [8,47,48,52], and Jag1 is regulated by Hnf1a and Hnf1b [8,52]. Lfng, regulated by Hnf1b, enhances its expression in the distal region of the CSB and in the proximal region of the SSB [47,48,52]. Defective expression in Notch components significantly reduces proximal tubule formation [8]. In addition, Hes5 expression was observed to change in murine Hnf1b mutants [8], and it is specially expressed in the CSB and epithelial cells that form the bulge between mid and lower limb of SSBs [8,48] (Figure 3c).
- 3.
- Progression from SSB to mature nephron: Hnf1b regulates the expression of IRX1, IRX2, and OSR2 genes in the SSB intermediate region, these are involved in tubule differentiation and expansion [8,47,48,52]. Hnf1b binds to the promoter regions of genes expressed in the proximal/intermediate tubule of SSB, including CDH6, PCSK9, and TFAP2B [48]. CDH6 is similarly expressed in RV and proximal tubule precursor cells [8]. Hnf1b activates HNF4A transcription in the distal region of the CSB and the proximal region of the SSB [2,8,48,52]. HNF4A encodes a nuclear transcription factor required for proximal tubule development [48,52]. In the Hnf1b-altered model, Hnf4a is downregulated during all nephrogenesis stages and in mature kidneys [21]. This gene has been associated with diabetes and renal cyst development [36,38,63,64].In the Heliot C. et al. [48] study, the conditional inactivation of Hnf1b in murine nephron progenitors led to rudimentary nephrons comprising a glomerulus connected to the collecting system by a short tubule with distal fates. This defect was preceded by strong downregulation of the Notch pathway components (Lfng, Dll1, and Jag1) and Irx1/2 factors, which are potential regulators of proximal and loop of Henle segment fates (Figure 3c).
4.2. Hnf1b Implication in Apical-Basolateral Polarity, Tight Junctions, Primary Cilia Development and Cyst Formation
- PKD2: encodes Polycystin 2, a Ca2+-permeable cation channel [1,2]. PKD2 mutations cause Autosomal Dominant Polycystic Kidney Disease (ADPKD) [3,23]. This is due to reduced Ca2+ entry and activation of the Ca2+-inhibitable adenylyl cyclases (AC5 and AC6), which elevates cAMP levels. This rise in cAMP stimulates cell proliferation and fluid secretion, promoting cyst growth [3,74].
- HNF4A: plays a role in various kidney processes, including PT development and cystogenesis, by modulating PKD1 expression [63].
4.3. Hnf1b Regulates Ion Transport in Kidney
- CASR (calcium-sensing receptor, CaSR), a negative regulator of UMOD [2,91]. Decreased CaSR expression is expected to elevate blood calcium levels [2,3,11,92]. In the parathyroid gland, PTH expression can be repressed by Hnf1b or CaSR [2,93]. PTH also inhibits uric acid secretion via ABCG2 downregulation [2].
4.4. Role of Hnf1b in Intrarenal Metabolism: Mitochondrial Respiration and Cholesterol
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- GeneCards—Human Genes|Gene Database|Gene Search. Available online: https://www.genecards.org/ (accessed on 23 May 2024).
- Tholen, L.E.; Hoenderop, J.G.J.; de Baaij, J.H.F. Mechanisms of ion transport regulation by HNF1β in the kidney: Beyond transcriptional regulation of channels and transporters. Pflugers Arch. 2022, 474, 901–916. [Google Scholar] [CrossRef] [PubMed]
- Ferrè, S.; Igarashi, P. New insights into the role of HNF-1β in kidney (patho)physiology. Pediatr. Nephrol. 2019, 34, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Rauluseviciute, I.; Riudavets-Puig, R.; Blanc-Mathieu, R.; Castro-Mondragon, J.A.; Ferenc, K.; Kumar, V.; Lemma, R.B.; Lucas, J.; Chèneby, J.; Baranasic, D.; et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2024, 52, D174–D182. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Resource. Available online: https://geneontology.org/ (accessed on 9 May 2024).
- Coffinier, C.; Barra, J.; Babinet, C.; Yaniv, M. Expression of the vHNF1/HNF1β homeoprotein gene during mouse organogenesis. Mech. Dev. 1999, 89, 211–213. [Google Scholar] [CrossRef]
- Coffinier, C.; Gresh, L.; Fiette, L.; Tronche, F.; Schütz, G.; Babinet, C.; Pontoglio, M.; Yaniv, M.; Barra, J. Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF1β. Development 2002, 129, 1829–1838. [Google Scholar] [CrossRef]
- Massa, F.; Garbay, S.; Bouvier, R.; Sugitani, Y.; Noda, T.; Gubler, M.-C.; Heidet, L.; Pontoglio, M.; Fischer, E. Hepatocyte nuclear factor 1β controls nephron tubular development. Development 2013, 140, 886–896. [Google Scholar] [CrossRef]
- Lokmane, L.; Heliot, C.; Garcia-Villalba, P.; Fabre, M.; Cereghini, S. vHNF1 functions in distinct regulatory circuits to control ureteric bud branching and early nephrogenesis. Development 2010, 137, 347–357. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (NCBI) Gene: HNF1B HNF1 Homeobox B [Homo sapiens (human)]. Available online: https://www.ncbi.nlm.nih.gov/gene/6928 (accessed on 20 May 2024).
- Viering, D.H.H.M.; de Baaij, J.H.F.; Walsh, S.B.; Kleta, R.; Bockenhauer, D. Genetic causes of hypomagnesemia, a clinical overview. Pediatr. Nephrol. 2017, 32, 1123–1135. [Google Scholar] [CrossRef]
- Mefford, H.C.; Clauin, S.; Sharp, A.J.; Moller, R.S.; Ullmann, R.; Kapur, R.; Pinkel, D.; Cooper, G.M.; Ventura, M.; Ropers, H.H.; et al. Recurrent Reciprocal Genomic Rearrangements of 17q12 Are Associated with Renal Disease, Diabetes, and Epilepsy. Am. J. Hum. Genet. 2007, 81, 1057–1069. [Google Scholar] [CrossRef]
- Bellanné-Chantelot, C.; Clauin, S.; Chauveau, D.; Collin, P.; Daumont, M.; Douillard, C.; Dubois-Laforgue, D.; Dusselier, L.; Gautier, J.-F.; Jadoul, M.; et al. Large Genomic Rearrangements in the Hepatocyte Nuclear Factor-1β (TCF2) Gene Are the Most Frequent Cause of Maturity-Onset Diabetes of the Young Type 5. Diabetes 2005, 54, 3126–3132. [Google Scholar] [CrossRef]
- Faguer, S.; Decramer, S.; Chassaing, N.; Bellanné-Chantelot, C.; Calvas, P.; Beaufils, S.; Bessenay, L.; Lengelé, J.P.; Dahan, K.; Ronco, P.; et al. Diagnosis, management, and prognosis of HNF1B nephropathy in adulthood. Kidney Int. 2011, 80, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Dubois-Laforgue, D.; Cornu, E.; Saint-Martin, C.; Coste, J.; Bellanné-Chantelot, C.; Timsit, J. Diabetes, Associated Clinical Spectrum, Long-term Prognosis, and Genotype/Phenotype Correlations in 201 Adult Patients With Hepatocyte Nuclear Factor 1B ( HNF1B ) Molecular Defects. Diabetes Care 2017, 40, 1436–1443. [Google Scholar] [CrossRef] [PubMed]
- Nassar, L.R.; Barber, G.P.; Benet-Pagès, A.; Casper, J.; Clawson, H.; Diekhans, M.; Fischer, C.; Gonzalez, J.N.; Hinrichs, A.S.; Lee, B.T.; et al. The UCSC Genome Browser database: 2023 update. Nucleic Acids Res. 2023, 51, D1188–D1195. [Google Scholar] [CrossRef] [PubMed]
- Bockenhauer, D.; Jaureguiberry, G. HNF1B-associated clinical phenotypes: The kidney and beyond. Pediatr. Nephrol. 2016, 31, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Hojny, J.; Bartu, M.; Krkavcova, E.; Nemejcova, K.; Sevcik, J.; Cibula, D.; Fryba, V.; Plincelnerova, L.; Dundr, P.; Struzinska, I. Identification of novel HNF1B mRNA splicing variants and their qualitative and semi-quantitative profile in selected healthy and tumour tissues. Sci. Rep. 2020, 10, 6958. [Google Scholar] [CrossRef]
- Buffin-Meyer, B.; Richard, J.; Guigonis, V.; Weber, S.; König, J.; Heidet, L.; Moussaoui, N.; Vu, J.-P.; Faguer, S.; Casemayou, A.; et al. Renal and extra-renal phenotypes in patients with HNF1B variants and chromosome 17q12 micro deletions. Kidney Int. Rep. 2024, 8, 2514–2526. [Google Scholar] [CrossRef]
- Vasileiou, G.; Hoyer, J.; Thiel, C.T.; Schaefer, J.; Zapke, M.; Krumbiegel, M.; Kraus, C.; Zweier, M.; Uebe, S.; Ekici, A.B.; et al. Prenatal diagnosis of HNF1B-associated renal cysts: Is there a need to differentiate intragenic variants from 17q12 microdeletion syndrome? Prenat. Diagn. 2019, 39, 1136–1147. [Google Scholar] [CrossRef]
- Niborski, L.L.; Paces-Fessy, M.; Ricci, P.; Bourgeois, A.; Magalhães, P.; Kuzma-Kuzniarska, M.; Lesaulnier, C.; Reczko, M.; Declercq, E.; Zürbig, P.; et al. Hnf1b haploinsufficiency differentially affects developmental target genes in a new renal cysts and diabetes mouse model. Dis. Model Mech. 2021, 14, dmm047498. [Google Scholar] [CrossRef]
- Clissold, R.L.; Hamilton, A.J.; Hattersley, A.T.; Ellard, S.; Bingham, C. HNF1B-associated renal and extra-renal disease—An expanding clinical spectrum. Nat. Rev. Nephrol. 2015, 11, 102–112. [Google Scholar] [CrossRef]
- Home—OMIM. Available online: https://www.omim.org/ (accessed on 19 April 2024).
- Grand, K.; Stoltz, M.; Rizzo, L.; Röck, R.; Kaminski, M.M.; Salinas, G.; Getwan, M.; Naert, T.; Pichler, R.; Lienkamp, S.S. HNF1B Alters an Evolutionarily Conserved Nephrogenic Program of Target Genes. J. Am. Soc. Nephrol. 2023, 34, 412–432. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Verscaj, C.P.; Velez-Bartolomei, F.; Bodle, E.; Chan, K.; Lyons, M.J.; Thorson, W.; Tan, W.; Rodig, N.; Graham, J.M.; Peron, A.; et al. Characterization of the prenatal renal phenotype associated with 17q12, HNF1B, microdeletions. Prenat. Diagn. 2024, 44, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Cleper, R.; Reches, A.; Shapira, D.; Simchoni, S.; Reisman, L.; Ben-Sira, L.; Yaron, Y.; Wolman, I.; Malinger, G.; Brabbing-Goldstein, D.; et al. Improving renal phenotype and evolving extra-renal features of 17q12 deletion encompassing the HNF1B gene. Transl. Pediatr. 2021, 10, 3130–3139. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, K.U.; Alper, S.L.; Antignac, C.; Bleyer, A.J.; Chauveau, D.; Dahan, K.; Deltas, C.; Hosking, A.; Kmoch, S.; Rampoldi, L.; et al. Autosomal dominant tubulointerstitial kidney disease: Diagnosis, classification, and management—A KDIGO consensus report. Kidney Int. 2015, 88, 676–683. [Google Scholar] [CrossRef]
- Adalat, S.; Hayes, W.N.; Bryant, W.A.; Booth, J.; Woolf, A.S.; Kleta, R.; Subtil, S.; Clissold, R.; Colclough, K.; Ellard, S.; et al. HNF1B Mutations Are Associated with a Gitelman-like Tubulopathy That Develops During Childhood. Kidney Int. Rep. 2019, 4, 1304–1311. [Google Scholar] [CrossRef]
- Moreno-De-Luca, D.; Mulle, J.G.; Kaminsky, E.B.; Sanders, S.J.; Myers, S.M.; Adam, M.P.; Pakula, A.T.; Eisenhauer, N.J.; Uhas, K.; Weik, L.; et al. Deletion 17q12 Is a Recurrent Copy Number Variant that Confers High Risk of Autism and Schizophrenia. Am. J. Hum. Genet. 2010, 87, 618–630. [Google Scholar] [CrossRef]
- Lim, S.H.; Kim, J.H.; Han, K.H.; Ahn, Y.H.; Kang, H.G.; Ha, I.-S.; Cheong, H., II. Genotype and Phenotype Analyses in Pediatric Patients with HNF1B Mutations. J. Clin. Med. 2020, 9, 2320. [Google Scholar] [CrossRef]
- Kato, T.; Tanaka, D.; Muro, S.; Jambaljav, B.; Mori, E.; Yonemitsu, S.; Oki, S.; Inagaki, N. A Novel p.L145Q Mutation in the HNF1B Gene in a Case of Maturity-onset Diabetes of the Young Type 5 (MODY5). Intern. Med. 2018, 57, 2035–2039. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Rha, G.B.; Chi, Y.-I. Structural Basis of Disease-Causing Mutations in Hepatocyte Nuclear Factor 1β. Biochemistry 2007, 46, 12071–12080. [Google Scholar] [CrossRef]
- Tholen, L.E.; Latta, F.; Martens, J.H.A.; Hoenderop, J.G.J.; de Baaij, J.H.F. Transcription factor HNF1β controls a transcriptional network regulating kidney cell structure and tight junction integrity. Am. J. Physiol. Ren. Physiol. 2023, 324, F211–F224. [Google Scholar] [CrossRef]
- Wiedmann, M.M.; Aibara, S.; Spring, D.R.; Stewart, M.; Brenton, J.D. Structural and calorimetric studies demonstrate that the hepatocyte nuclear factor 1β (HNF1β) transcription factor is imported into the nucleus via a monopartite NLS sequence. J. Struct. Biol. 2016, 195, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Bouvet, D.; Blondel, A.; de Sainte Agathe, J.-M.; Leroy, G.; Saint-Martin, C.; Bellanné-Chantelot, C. Evaluation in Monogenic Diabetes of the Impact of GCK, HNF1A, and HNF4A Variants on Splicing through the Combined Use of In Silico Tools and Minigene Assays. Hum. Mutat. 2023, 2023, 6661013. [Google Scholar] [CrossRef]
- Yamagata, K.; Oda, N.; Kaisaki, P.J.; Menzel, S.; Furuta, H.; Vaxillaire, M.; Southam, L.; Cox, R.D.; Lathrop, G.M.; Boriraj, V.V.; et al. Mutations in the hepatocyte nuclear factor-1α gene in maturity-onset diabetes of the young (MODY3). Nature 1996, 384, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Locke, J.M.; Ellard, S.; Norwood, V.F.; Harries, L.W. Variants in the isoform-specific coding regions of the HNF1A, HNF4A and HNF1B genes are not a common cause of familial, young-onset diabetes or renal cysts and diabetes (RCAD). Diabet. Med. 2009, 26, 569–570. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gonzalez, M.A.; Carette, C.; Bagattin, A.; Chiral, M.; Makinistoglu, M.P.; Garbay, S.; Prévost, G.; Madaras, C.; Hérault, Y.; Leibovici, M.; et al. A suppressor locus for MODY3-diabetes. Sci. Rep. 2016, 6, 33087. [Google Scholar] [CrossRef] [PubMed]
- Ferrè, S.; de Baaij, J.H.F.; Ferreira, P.; Germann, R.; de Klerk, J.B.C.; Lavrijsen, M.; van Zeeland, F.; Venselaar, H.; Kluijtmans, L.A.J.; Hoenderop, J.G.J.; et al. Mutations in PCBD1 Cause Hypomagnesemia and Renal Magnesium Wasting. J. Am. Soc. Nephrol. 2014, 25, 574–586. [Google Scholar] [CrossRef]
- Choi, Y.-H.; McNally, B.T.; Igarashi, P. Zyxin regulates migration of renal epithelial cells through activation of hepatocyte nuclear factor-1β. Am. J. Physiol. Ren. Physiol. 2013, 305, F100–F110. [Google Scholar] [CrossRef]
- Soutoglou, E. Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J. 2001, 20, 1984–1992. [Google Scholar] [CrossRef]
- Simaite, D.; Kofent, J.; Gong, M.; Rüschendorf, F.; Jia, S.; Arn, P.; Bentler, K.; Ellaway, C.; Kühnen, P.; Hoffmann, G.F.; et al. Recessive Mutations in PCBD1 Cause a New Type of Early-Onset Diabetes. Diabetes 2014, 63, 3557–3564. [Google Scholar] [CrossRef]
- Tholen, L.E.; Bos, C.; Jansen, P.W.T.C.; Venselaar, H.; Vermeulen, M.; Hoenderop, J.G.J.; de Baaij, J.H.F. Bifunctional protein PCBD2 operates as a co-factor for hepatocyte nuclear factor 1β and modulates gene transcription. FASEB J. 2021, 35, e21366. [Google Scholar] [CrossRef]
- Piedrafita, A.; Balayssac, S.; Casemayou, A.; Saulnier-Blache, J.; Lucas, A.; Iacovoni, J.S.; Breuil, B.; Chauveau, D.; Decramer, S.; Malet-Martino, M.; et al. Hepatocyte nuclear factor-1β shapes the energetic homeostasis of kidney tubule cells. FASEB J. 2021, 35, e21931. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, A.; Bluteau, O.; Garcia-Gonzalez, M.A.; Gresh, L.; Doyen, A.; Garbay, S.; Robine, S.; Pontoglio, M. Hepatocyte nuclear factor 1α and β control terminal differentiation and cell fate commitment in the gut epithelium. Development 2010, 137, 1573–1582. [Google Scholar] [CrossRef]
- O’Brien, L.L.; McMahon, A.P. Induction and patterning of the metanephric nephron. Semin. Cell Dev. Biol. 2014, 36, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Heliot, C.; Desgrange, A.; Buisson, I.; Prunskaite-Hyyryläinen, R.; Shan, J.; Vainio, S.; Umbhauer, M.; Cereghini, S. HNF1B controls proximal-intermediate nephron segment identity in vertebrates by regulating Notch signalling components and Irx1/2. Development 2013, 140, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Casemayou, A.; Fournel, A.; Bagattin, A.; Schanstra, J.; Belliere, J.; Decramer, S.; Marsal, D.; Gillet, M.; Chassaing, N.; Huart, A.; et al. Hepatocyte Nuclear Factor-1β Controls Mitochondrial Respiration in Renal Tubular Cells. J. Am. Soc. Nephrol. 2017, 28, 3205–3217. [Google Scholar] [CrossRef] [PubMed]
- Costantini, F. Genetic controls and cellular behaviors in branching morphogenesis of the renal collecting system. WIREs Dev. Biol. 2012, 1, 693–713. [Google Scholar] [CrossRef]
- Carroll, T.J.; Park, J.-S.; Hayashi, S.; Majumdar, A.; McMahon, A.P. Wnt9b Plays a Central Role in the Regulation of Mesenchymal to Epithelial Transitions Underlying Organogenesis of the Mammalian Urogenital System. Dev. Cell 2005, 9, 283–292. [Google Scholar] [CrossRef]
- Home—Reactome Pathway Database. Available online: https://reactome.org/ (accessed on 25 April 2024).
- Desgrange, A.; Cereghini, S. Nephron Patterning: Lessons from Xenopus, Zebrafish, and Mouse Studies. Cells 2015, 4, 483–499. [Google Scholar] [CrossRef]
- Park, J.-S.; Valerius, M.T.; McMahon, A.P. Wnt/β-catenin signaling regulates nephron induction during mouse kidney development. Development 2007, 134, 2533–2539. [Google Scholar] [CrossRef]
- Ng-Blichfeldt, J.-P.; Stewart, B.J.; Clatworthy, M.R.; Williams, J.M.; Röper, K. Identification of a core transcriptional program driving the human renal mesenchymal-to-epithelial transition. Dev. Cell 2024, 59, 595–612.e8. [Google Scholar] [CrossRef]
- Brown, A.C.; Muthukrishnan, S.D.; Guay, J.A.; Adams, D.C.; Schafer, D.A.; Fetting, J.L.; Oxburgh, L. Role for compartmentalization in nephron progenitor differentiation. Proc. Natl. Acad. Sci. USA 2013, 110, 4640–4645. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Kwan, K.-M.; Carroll, T.J.; McMahon, A.P.; Mendelsohn, C.L.; Behringer, R.R. Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development 2005, 132, 2809–2823. [Google Scholar] [CrossRef]
- Georgas, K.; Rumballe, B.; Valerius, M.T.; Chiu, H.S.; Thiagarajan, R.D.; Lesieur, E.; Aronow, B.J.; Brunskill, E.W.; Combes, A.N.; Tang, D.; et al. Analysis of early nephron patterning reveals a role for distal RV proliferation in fusion to the ureteric tip via a cap mesenchyme-derived connecting segment. Dev. Biol. 2009, 332, 273–286. [Google Scholar] [CrossRef]
- Naylor, R.W.; Davidson, A.J. Hnf1beta and nephron segmentation. Pediatr. Nephrol. 2014, 29, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Desgrange, A.; Heliot, C.; Skovorodkin, I.; Akram, S.U.; Heikkilä, J.; Ronkainen, V.-P.; Miinalainen, I.; Vainio, S.J.; Cereghini, S. HNF1B controls epithelial organization and cell polarity during ureteric bud branching and collecting duct morphogenesis. Development 2017, 144, 4704–4719. [Google Scholar] [CrossRef]
- Snoek, R.; Nguyen, T.Q.; Van Der Zwaag, B.; Van Zuilen, A.D.; Kruis, H.M.E.; Van Gils-Verrij, L.A.; Goldschmeding, R.; Knoers, N.V.A.M.; Rookmaaker, M.B.; Van Eerde, A.M. Importance of Genetic Diagnostics in Adult-Onset Focal Segmental Glomerulosclerosis. Nephron 2019, 142, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Paces-Fessy, M.; Fabre, M.; Lesaulnier, C.; Cereghini, S. Hnf1b and Pax2 cooperate to control different pathways in kidney and ureter morphogenesis. Hum. Mol. Genet. 2012, 21, 3143–3155. [Google Scholar] [CrossRef]
- Menezes, L.F.; Zhou, F.; Patterson, A.D.; Piontek, K.B.; Krausz, K.W.; Gonzalez, F.J.; Germino, G.G. Network Analysis of a Pkd1-Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier. PLoS Genet. 2012, 8, e1003053. [Google Scholar] [CrossRef]
- Yamagata, K.; Furuta, H.; Oda, N.; Kaisaki, P.J.; Menzel, S.; Cox, N.J.; Fajans, S.S.; Signorini, S.; Stoffel, M.; Bell, G.I. Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1). Nature 1996, 384, 458–460. [Google Scholar] [CrossRef]
- Mae, S.-I.; Ryosaka, M.; Sakamoto, S.; Matsuse, K.; Nozaki, A.; Igami, M.; Kabai, R.; Watanabe, A.; Osafune, K. Expansion of Human iPSC-Derived Ureteric Bud Organoids with Repeated Branching Potential. Cell Rep. 2020, 32, 107963. [Google Scholar] [CrossRef]
- Martin, E.; Girardello, R.; Dittmar, G.; Ludwig, A. New insights into the organization and regulation of the apical polarity network in mammalian epithelial cells. FEBS J. 2021, 288, 7073–7095. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Yatim, S.M.J.M.; Peng, S.; Gunaratne, J.; Hunziker, W.; Ludwig, A. The Mammalian Crumbs Complex Defines a Distinct Polarity Domain Apical of Epithelial Tight Junctions. Curr. Biol. 2020, 30, 2791–2804.e6. [Google Scholar] [CrossRef] [PubMed]
- Pieczynski, J.; Margolis, B. Protein complexes that control renal epithelial polarity. Am. J. Physiol. Ren. Physiol. 2011, 300, F589–F601. [Google Scholar] [CrossRef] [PubMed]
- De Vas, M.G.; Kopp, J.L.; Heliot, C.; Sander, M.; Cereghini, S.; Haumaitre, C. Hnf1b controls pancreas morphogenesis and the generation of Ngn3+ endocrine progenitors. Development 2015, 142, 871–882. [Google Scholar] [CrossRef]
- Veikkolainen, V.; Naillat, F.; Railo, A.; Chi, L.; Manninen, A.; Hohenstein, P.; Hastie, N.; Vainio, S.; Elenius, K. ErbB4 Modulates Tubular Cell Polarity and Lumen Diameter during Kidney Development. J. Am. Soc. Nephrol. 2012, 23, 112–122. [Google Scholar] [CrossRef]
- Wilson, P.D. Apico-basal polarity in polycystic kidney disease epithelia. Biochim. Et Biophys. Acta (BBA) Mol. Basis Dis. 2011, 1812, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- Shao, A.; Chan, S.C.; Igarashi, P. Role of transcription factor hepatocyte nuclear factor-1β in polycystic kidney disease. Cell Signal 2020, 71, 109568. [Google Scholar] [CrossRef]
- Chan, S.C.; Zhang, Y.; Pontoglio, M.; Igarashi, P. Hepatocyte nuclear factor-1β regulates Wnt signaling through genome-wide competition with β-catenin/lymphoid enhancer binding factor. Proc. Natl. Acad. Sci. USA 2019, 116, 24133–24142. [Google Scholar] [CrossRef]
- Choi, Y.-H.; Suzuki, A.; Hajarnis, S.; Ma, Z.; Chapin, H.C.; Caplan, M.J.; Pontoglio, M.; Somlo, S.; Igarashi, P. Polycystin-2 and phosphodiesterase 4C are components of a ciliary A-kinase anchoring protein complex that is disrupted in cystic kidney diseases. Proc. Natl. Acad. Sci. USA 2011, 108, 10679–10684. [Google Scholar] [CrossRef]
- Song, X.; Di Giovanni, V.; He, N.; Wang, K.; Ingram, A.; Rosenblum, N.D.; Pei, Y. Systems biology of autosomal dominant polycystic kidney disease (ADPKD): Computational identification of gene expression pathways and integrated regulatory networks. Hum. Mol. Genet. 2009, 18, 2328–2343. [Google Scholar] [CrossRef]
- Verdeguer, F.; Le Corre, S.; Fischer, E.; Callens, C.; Garbay, S.; Doyen, A.; Igarashi, P.; Terzi, F.; Pontoglio, M. A mitotic transcriptional switch in polycystic kidney disease. Nat. Med. 2010, 16, 106–110. [Google Scholar] [CrossRef]
- Kang, H.S.; Beak, J.Y.; Kim, Y.-S.; Herbert, R.; Jetten, A.M. Glis3 Is Associated with Primary Cilia and Wwtr1/TAZ and Implicated in Polycystic Kidney Disease. Mol. Cell Biol. 2009, 29, 2556–2569. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.S.; Kim, Y.-S.; ZeRuth, G.; Beak, J.Y.; Gerrish, K.; Kilic, G.; Sosa-Pineda, B.; Jensen, J.; Foley, J.; Jetten, A.M. Transcription Factor Glis3, a Novel Critical Player in the Regulation of Pancreatic β-Cell Development and Insulin Gene Expression. Mol. Cell Biol. 2009, 29, 6366–6379. [Google Scholar] [CrossRef] [PubMed]
- Schlingmann, K.P.; de Baaij, J.H.F. The genetic spectrum of Gitelman(-like) syndromes. Curr. Opin. Nephrol. Hypertens. 2022, 31, 508–515. [Google Scholar] [CrossRef]
- Saji, T.; Kikuchi, R.; Kusuhara, H.; Kim, I.; Gonzalez, F.J.; Sugiyama, Y. Transcriptional Regulation of Human and Mouse Organic Anion Transporter 1 by Hepatocyte Nuclear Factor 1 α/β. J. Pharmacol. Exp. Ther. 2008, 324, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Kikuchi, R.; Saji, T.; Kusuhara, H.; Sugiyama, Y. Regulation of Tissue-Specific Expression of Renal Organic Anion Transporters by Hepatocyte Nuclear Factor 1 α/β and DNA Methylation. J. Pharmacol. Exp. Ther. 2012, 340, 648–655. [Google Scholar] [CrossRef]
- Kikuchi, R.; Kusuhara, H.; Hattori, N.; Shiota, K.; Kim, I.; Gonzalez, F.J.; Sugiyama, Y. Regulation of the Expression of Human Organic Anion Transporter 3 by Hepatocyte Nuclear Factor 1α/β and DNA Methylation. Mol. Pharmacol. 2006, 70, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Cheret, C.; Doyen, A.; Yaniv, M.; Pontoglio, M. Hepatocyte Nuclear Factor 1 α Controls Renal Expression of the Npt1-Npt4 Anionic Transporter Locus. J. Mol. Biol. 2002, 322, 929–941. [Google Scholar] [CrossRef]
- Kikuchi, R.; Kusuhara, H.; Hattori, N.; Kim, I.; Shiota, K.; Gonzalez, F.J.; Sugiyama, Y. Regulation of Tissue-Specific Expression of the Human and Mouse Urate Transporter 1 Gene by Hepatocyte Nuclear Factor 1 α/β and DNA Methylation. Mol. Pharmacol. 2007, 72, 1619–1625. [Google Scholar] [CrossRef]
- Fukui, K.; Yang, Q.; Cao, Y.; Takahashi, N.; Hatakeyama, H.; Wang, H.; Wada, J.; Zhang, Y.; Marselli, L.; Nammo, T.; et al. The HNF-1 target Collectrin controls insulin exocytosis by SNARE complex formation. Cell Metab. 2005, 2, 373–384. [Google Scholar] [CrossRef]
- Terryn, S.; Tanaka, K.; Lengelé, J.-P.; Olinger, E.; Dubois-Laforgue, D.; Garbay, S.; Kozyraki, R.; Van Der Smissen, P.; Christensen, E.I.; Courtoy, P.J.; et al. Tubular proteinuria in patients with HNF1α mutations: HNF1α drives endocytosis in the proximal tubule. Kidney Int. 2016, 89, 1075–1089. [Google Scholar] [CrossRef] [PubMed]
- Tokonami, N.; Takata, T.; Beyeler, J.; Ehrbar, I.; Yoshifuji, A.; Christensen, E.I.; Loffing, J.; Devuyst, O.; Olinger, E.G. Uromodulin is expressed in the distal convoluted tubule where it is critical for regulation of the sodium chloride cotransporter, N.C.C. Kidney Int. 2018, 94, 701–715. [Google Scholar] [CrossRef]
- Nie, M.; Bal, M.S.; Liu, J.; Yang, Z.; Rivera, C.; Wu, X.-R.; Hoenderop, J.G.J.; Bindels, R.J.M.; Marciano, D.K.; Wolf, M.T.F. Uromodulin regulates renal magnesium homeostasis through the ion channel transient receptor potential melastatin 6 (TRPM6). J. Biol. Chem. 2018, 293, 16488–16502. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.T.F.; Wu, X.-R.; Huang, C.-L. Uromodulin upregulates TRPV5 by impairing caveolin-mediated endocytosis. Kidney Int. 2013, 84, 130–137. [Google Scholar] [CrossRef]
- Trudu, M.; Janas, S.; Lanzani, C.; Debaix, H.; Schaeffer, C.; Ikehata, M.; Citterio, L.; Demaretz, S.; Trevisani, F.; Ristagno, G.; et al. Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat. Med. 2013, 19, 1655–1660. [Google Scholar] [CrossRef]
- Tokonami, N.; Olinger, E.; Debaix, H.; Houillier, P.; Devuyst, O. The excretion of uromodulin is modulated by the calcium-sensing receptor. Kidney Int. 2018, 94, 882–886. [Google Scholar] [CrossRef]
- Kompatscher, A.; de Baaij, J.H.F.; Aboudehen, K.; Farahani, S.; van Son, L.H.J.; Milatz, S.; Himmerkus, N.; Veenstra, G.C.; Bindels, R.J.M.; Hoenderop, J.G.J. Transcription factor HNF1β regulates expression of the calcium-sensing receptor in the thick ascending limb of the kidney. Am. J. Physiol. Ren. Physiol. 2018, 315, F27–F35. [Google Scholar] [CrossRef] [PubMed]
- Ferrè, S.; Bongers, E.M.H.F.; Sonneveld, R.; Cornelissen, E.A.M.; van der Vlag, J.; van Boekel, G.A.J.; Wetzels, J.F.M.; Hoenderop, J.G.J.; Bindels, R.J.M.; Nijenhuis, T. Early Development of Hyperparathyroidism Due to Loss of PTH Transcriptional Repression in Patients With HNF1β Mutations? J. Clin. Endocrinol. Metab. 2013, 98, 4089–4096. [Google Scholar] [CrossRef]
- Ferrè, S.; Veenstra, G.J.C.; Bouwmeester, R.; Hoenderop, J.G.J.; Bindels, R.J.M. HNF-1B specifically regulates the transcription of the γa-subunit of the Na+/K+-ATPase. Biochem. Biophys. Res. Commun. 2011, 404, 284–290. [Google Scholar] [CrossRef]
- de Baaij, J.H.F.; Dorresteijn, E.M.; Hennekam, E.A.M.; Kamsteeg, E.-J.; Meijer, R.; Dahan, K.; Muller, M.; van den Dorpel, M.A.; Bindels, R.J.M.; Hoenderop, J.G.J.; et al. Recurrent FXYD2 p.Gly41Arg mutation in patients with isolated dominant hypomagnesaemia. Nephrol. Dial. Transplant. 2015, 30, 952–957. [Google Scholar] [CrossRef]
- Kompatscher, A.; de Baaij, J.H.F.; Aboudehen, K.; Hoefnagels, A.P.W.M.; Igarashi, P.; Bindels, R.J.M.; Veenstra, G.J.C.; Hoenderop, J.G.J. Loss of transcriptional activation of the potassium channel Kir5.1 by HNF1β drives autosomal dominant tubulointerstitial kidney disease. Kidney Int. 2017, 92, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Schlingmann, K.P.; Renigunta, A.; Hoorn, E.J.; Forst, A.-L.; Renigunta, V.; Atanasov, V.; Mahendran, S.; Barakat, T.S.; Gillion, V.; Godefroid, N.; et al. Defects in KCNJ16 Cause a Novel Tubulopathy with Hypokalemia, Salt Wasting, Disturbed Acid-Base Homeostasis, and Sensorineural Deafness. J. Am. Soc. Nephrol. 2021, 32, 1498–1512. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, S.; Gao, M.; Liu, J.; Jia, X.; Han, Q.; Zheng, S.; Miao, Y.; Li, S.; Weng, H.; et al. Farnesoid X receptor (FXR) gene deficiency impairs urine concentration in mice. Proc. Natl. Acad. Sci. USA 2014, 111, 2277–2282. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Lee, J.M. Transcriptional Control of Trpm6 by the Nuclear Receptor, F.X.R. Int. J. Mol. Sci. 2022, 23, 1980. [Google Scholar] [CrossRef]
- Bhargava, P.; Schnellmann, R.G. Mitochondrial energetics in the kidney. Nat. Rev. Nephrol. 2017, 13, 629–646. [Google Scholar] [CrossRef]
- Thomas, J.L.; Pham, H.; Li, Y.; Hall, E.; Perkins, G.A.; Ali, S.S.; Patel, H.H.; Singh, P. Hypoxia-inducible factor-1α activation improves renal oxygenation and mitochondrial function in early chronic kidney disease. Am. J. Physiol. Ren. Physiol. 2017, 313, F282–F290. [Google Scholar] [CrossRef]
- Viering, D.; Schlingmann, K.P.; Hureaux, M.; Nijenhuis, T.; Mallett, A.; Chan, M.M.Y.; van Beek, A.; van Eerde, A.M.; Coulibaly, J.-M.; Vallet, M.; et al. Gitelman-Like Syndrome Caused by Pathogenic Variants in mtDNA. J. Am. Soc. Nephrol. 2022, 33, 305–325. [Google Scholar] [CrossRef]
- Wang, Z.; Ying, Z.; Bosy-Westphal, A.; Zhang, J.; Schautz, B.; Later, W.; Heymsfield, S.B.; Müller, M.J. Specific metabolic rates of major organs and tissues across adulthood: Evaluation by mechanistic model of resting energy expenditure. Am. J. Clin. Nutr. 2010, 92, 1369–1377. [Google Scholar] [CrossRef]
- Kang, H.M.; Ahn, S.H.; Choi, P.; Ko, Y.-A.; Han, S.H.; Chinga, F.; Park, A.S.D.; Tao, J.; Sharma, K.; Pullman, J.; et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 2015, 21, 37–46. [Google Scholar] [CrossRef]
- Aboudehen, K.; Kim, M.S.; Mitsche, M.; Garland, K.; Anderson, N.; Noureddine, L.; Pontoglio, M.; Patel, V.; Xie, Y.; DeBose-Boyd, R.; et al. Transcription Factor Hepatocyte Nuclear Factor–1β Regulates Renal Cholesterol Metabolism. J. Am. Soc. Nephrol. 2016, 27, 2408–2421. [Google Scholar] [CrossRef]
- Horton, J.D.; Cohen, J.C.; Hobbs, H.H. PCSK9: A convertase that coordinates LDL catabolism. J. Lipid Res. 2009, 50, S172–S177. [Google Scholar] [CrossRef] [PubMed]

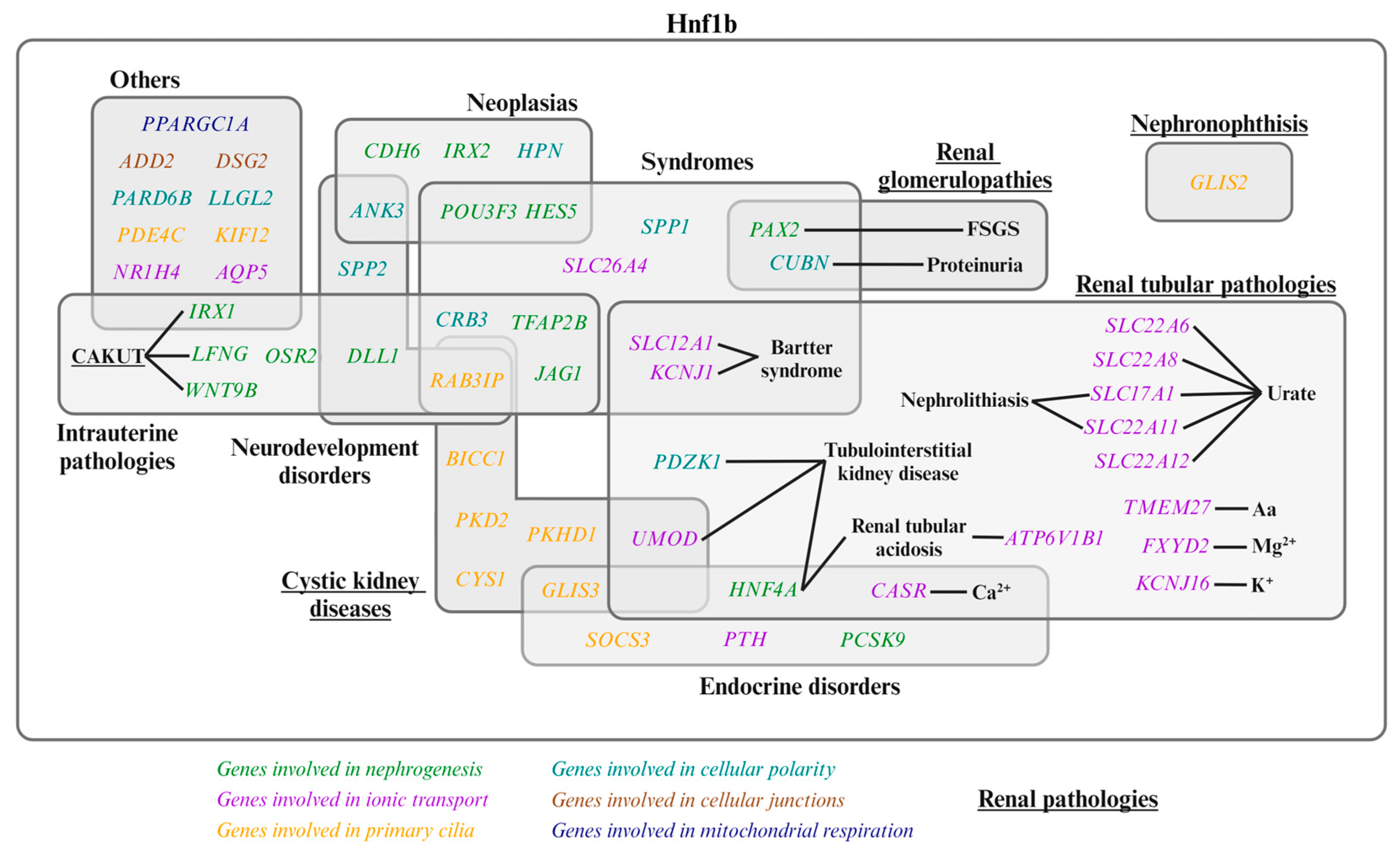


| Subsection | Mechanistic Action |
|---|---|
| 4.1. | Nephrogenesis. Identification of the genes regulated by Hnf1b at each stage of nephron formation and their contributions to kidney development. |
| 4.2. | Apical-basolateral polarity, tight junctions, primary cilia development, and cyst formation. Role of Hnf1b in these processes. |
| 4.3. | Ion transport. Hnf1b-regulated genes involved in tubular ion transport, a process crucial for maintaining electrolyte balance and renal function. |
| 4.4. | Intrarenal metabolism. Hnf1b involvement in mitochondrial respiration and cholesterol metabolism. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Cazorla, E.; Carrera, N.; García-González, M.Á. HNF1B Transcription Factor: Key Regulator in Renal Physiology and Pathogenesis. Int. J. Mol. Sci. 2024, 25, 10609. https://doi.org/10.3390/ijms251910609
Sánchez-Cazorla E, Carrera N, García-González MÁ. HNF1B Transcription Factor: Key Regulator in Renal Physiology and Pathogenesis. International Journal of Molecular Sciences. 2024; 25(19):10609. https://doi.org/10.3390/ijms251910609
Chicago/Turabian StyleSánchez-Cazorla, Eloísa, Noa Carrera, and Miguel Ángel García-González. 2024. "HNF1B Transcription Factor: Key Regulator in Renal Physiology and Pathogenesis" International Journal of Molecular Sciences 25, no. 19: 10609. https://doi.org/10.3390/ijms251910609
APA StyleSánchez-Cazorla, E., Carrera, N., & García-González, M. Á. (2024). HNF1B Transcription Factor: Key Regulator in Renal Physiology and Pathogenesis. International Journal of Molecular Sciences, 25(19), 10609. https://doi.org/10.3390/ijms251910609





