After the Hurricane: Anti-COVID-19 Drugs Development, Molecular Mechanisms of Action and Future Perspectives
Abstract
:1. Introduction
2. The Origin of COVID-19
3. Viral Structure
4. SARS-CoV-2 Replication
4.1. Attachment and Entry
4.2. Replication
5. Anti-COVID-19 Drug Development
5.1. Solidarity Trial
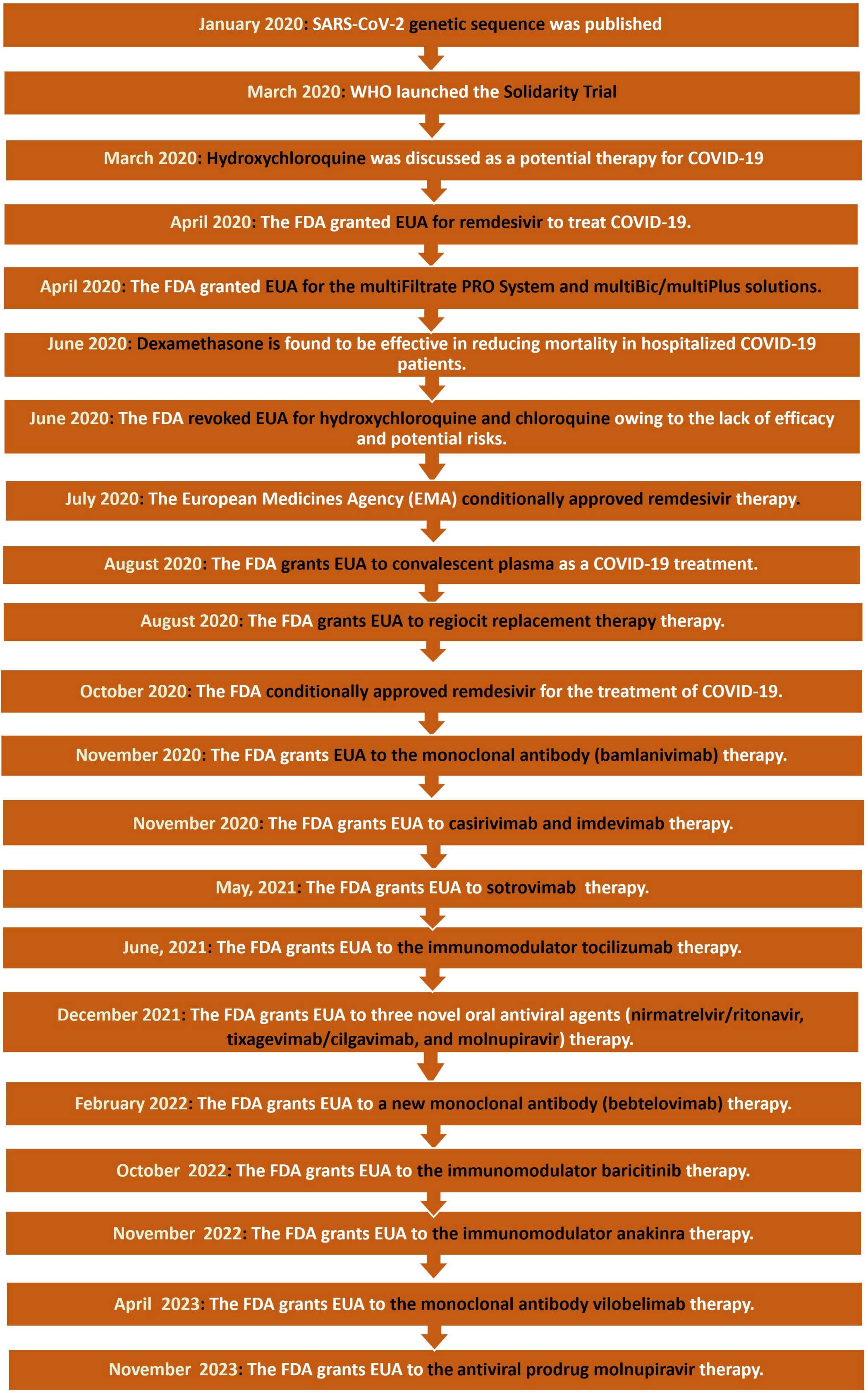
5.2. Timeline of Drug Development
6. Molecular Mechanisms of Action of Anti-COVID-19 Medications
6.1. Inhibitors of Viral Entry into the Human Cell
6.1.1. S Protein Inhibitors: Convalescent Plasma and Monoclonal Antibodies
6.1.2. Inhibitors of Fusional Entry: TMPRSS2 Inhibitors
6.2. Inhibitors of Endosomal Entry: Hydroxychloroquine
6.3. Inhibitors of Viral Proteases
6.3.1. Inhibitors of Viral Main Protease (Mpro): Rupintrivir, Lopinavir/Ritonavir and Nirmatrelvir
6.3.2. Inhibitors of Viral Papain-like Protease (PLpro)
6.4. Inhibitors of Viral RNA
6.4.1. Inhibitors of RNA Dependent RNA Polymerase (Rdrp): Remdesivir
6.4.2. Replication Inhibitor: Molnupiravir
| Drug | Application | Route of Administration | Target | MOA vs. SARS-CoV-2 | References |
|---|---|---|---|---|---|
| Convalescent plasma | - | Intravenous | S Protein | Binds to the S protein, which prevents viral attachment. | [47,53] |
| Monoclonal antibodies | - | Intravenous, subcutaneous, intramuscular | S protein | Binds to the S protein, which prevents viral attachment. | [46,48,49,50,51,52] |
| Camostat | Chronic pancreatitis | Oral | TMPRSS2 | A protease inhibitor that prevents SARS-CoV-2 lung cell infection by inhibiting the virus-activating host cell protease TMPRSS2. | [60] |
| Hydroxychloroquine | Malarial infections | Oral | Multiple | Different mechanisms of action have been proposed involving endocytic pathway interference, sialic acid receptor blockage, restriction of pH-mediated spike (S) protein cleavage at the angiotensin-converting enzyme 2 (ACE2) binding site, and cytokine storm prevention | [61,62,63,64,65] |
| Rupintrivir | Human rhinoviral (HRV) infections | Nasal | Mpro | Inhibitors of Viral Main protease (Mpro). | [67,68,69] |
| Lopinavir/ritonavir | HIV infections | Oral | Mpro | Inhibitors of Viral Main protease (Mpro). | [70,71] |
| Nirmatrelvir/ritonavir | - | Oral | Mpro | Inhibitors of Viral Main protease (Mpro). | [71,72] |
| 6-Thioguanine | Leukaemia | Oral | PLpro | Inhibitors of viral papain-like protease (PLpro). | [73,75] |
| Simeprevir, vaniprevir, paritaprevir, and grazoprevir | Chronic HCV infection | Oral | PLpro | Inhibitors of viral papain-like protease (PLpro). | [76] |
| Remdesivir | Ebola virus | Intravenous | Rdrp | Inhibitors of RNA Dependent RNA Polymerase (Rdrp) | [78,79,80] |
| Molnupiravir | Influenza | Oral | viral RNA | Disrupts the replication process because the viral RNA is copied incorrectly | [81,82,83,85,86,87] |
7. Future Directions and Areas of Research
7.1. Flavonoids
7.2. Inhaled Drugs
7.3. Aptamers
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Majumder, J.; Minko, T. Recent developments on therapeutic and diagnostic approaches for COVID-19. AAPS J. 2021, 23, 14. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-C.; Chen, C.-S.; Chan, Y.-J. The outbreak of COVID-19: An overview. J. Chin. Med. Assoc. 2020, 83, 217–220. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Cucinotta, D.; Vanelli, M. WHO declares COVID-19 a pandemic. Acta Bio Medica Atenei Parm. 2020, 91, 157. [Google Scholar]
- Lu, L.; Zhong, W.; Bian, Z.; Li, Z.; Zhang, K.; Liang, B.; Zhong, Y.; Hu, M.; Lin, L.; Liu, J.; et al. A comparison of mortality-related risk factors of COVID-19, SARS, and MERS: A systematic review and meta-analysis. J. Infect. 2020, 81, e18–e25. [Google Scholar] [CrossRef] [PubMed]
- Richards, F.; Kodjamanova, P.; Chen, X.; Li, N.; Atanasov, P.; Bennetts, L.; Patterson, B.J.; Yektashenas, B.; Mesa-Frias, M.; Tronczynski, K.; et al. Economic burden of COVID-19: A systematic review. Clin. Outcomes Res. 2022, 14, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Mulabbi, E.N.; Tweyongyere, R.; Byarugaba, D.K. The history of the emergence and transmission of human coronaviruses. Onderstepoort J. Vet. Res. 2021, 88, 1872. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, N.; Saxena, S.K. Prevention and Control Strategies for SARS-CoV-2 Infection. In Coronavirus Disease 2019 (COVID-19): Epidemiology, Pathogenesis, Diagnosis, and Therapeutics; Saxena, S.K., Ed.; Springer: Singapore, 2020; pp. 127–140. [Google Scholar]
- Holmes, E.C.; Goldstein, S.A.; Rasmussen, A.L.; Robertson, D.L.; Crits-Christoph, A.; Wertheim, J.O.; Anthony, S.J.; Barclay, W.S.; Boni, M.F.; Doherty, P.C.; et al. The origins of SARS-CoV-2: A critical review. Cell 2021, 184, 4848–4856. [Google Scholar] [CrossRef]
- Umakanthan, S.; Sahu, P.; Ranade, A.V.; Bukelo, M.M.; Rao, J.S.; Abrahao-Machado, L.F.; Dahal, S.; Kumar, H.; Kv, D. Origin, transmission, diagnosis and management of coronavirus disease 2019 (COVID-19). Postgrad. Med. J. 2020, 96, 753–758. [Google Scholar]
- World Health Organization. WHO-Convened Global Study of Origins of SARS-CoV-2: China Part; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Wang, M.Y.; Zhao, R.; Gao, L.J.; Gao, X.F.; Wang, D.P.; Cao, J.M. SARS-CoV-2: Structure, biology, and structure-based therapeutics development. Front. Cell. Infect. Microbiol. 2020, 10, 587269. [Google Scholar] [CrossRef]
- Singh, S.P.; Pritam, M.; Pandey, B.; Yadav, T.P. Microstructure, pathophysiology, and potential therapeutics of COVID-19: A comprehensive review. J. Med. Virol. 2021, 93, 275–299. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S. The structure of the membrane protein of SARS-CoV-2 resembles the sugar transporter semisweet. Pathog. Immun. 2020, 5, 342. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Moustaqil, M.; Ollivier, E.; Chiu, H.P.; Van Tol, S.; Rudolffi-Soto, P.; Stevens, C.; Bhumkar, A.; Hunter, D.J.; Freiberg, A.N.; Jacques, D.; et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): Implications for disease presentation across species. Emerg. Microbes Infect. 2021, 10, 178–195. [Google Scholar] [CrossRef] [PubMed]
- Hillen, H.S. Structure and function of SARS-CoV-2 polymerase. Curr. Opin. Virol. 2021, 48, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Chams, N.; Chams, S.; Badran, R.; Shams, A.; Araji, A.; Raad, M.; Mukhopadhyay, S.; Stroberg, E.; Duval, E.J.; Barton, L.M.; et al. COVID-19: A multidisciplinary review. Front. Public Health 2020, 8, 383. [Google Scholar] [CrossRef] [PubMed]
- Saraste, J.; Prydz, K. Assembly and cellular exit of coronaviruses: Hijacking an unconventional secretory pathway from the pre-Golgi intermediate compartment via the Golgi ribbon to the extracellular space. Cells 2021, 10, 503. [Google Scholar] [CrossRef] [PubMed]
- Güner, H.R.; Hasanoğlu, İ.; Aktaş, F. COVID-19: Prevention and control measures in community. Turk. J. Med. Sci. 2020, 50, 571–577. [Google Scholar] [CrossRef]
- WHO Solidarity Trial Consortium Repurposed antiviral drugs for COVID-19—Interim WHO solidarity trial results. N. Engl. J. Med. 2021, 384, 497–511. [CrossRef]
- Safiabadi Tali, S.H.; LeBlanc, J.J.; Sadiq, Z.; Oyewunmi, O.D.; Camargo, C.; Nikpour, B.; Armanfard, N.; Sagan, S.M.; Jahanshahi-Anbuhi, S. Tools and techniques for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)/COVID-19 detection. Clin. Microbiol. Rev. 2021, 34, 1110–1128. [Google Scholar] [CrossRef]
- Harilal, D.; Ramaswamy, S.; Loney, T.; Suwaidi, H.A.; Khansaheb, H.; Alkhaja, A.; Varghese, R.; Deesi, Z.; Nowotny, N.; Alsheikh-Ali, A.; et al. SARS-CoV-2 whole genome amplification and sequencing for effective population-based surveillance and control of viral transmission. Clin. Chem. 2020, 66, 1450–1458. [Google Scholar] [CrossRef]
- Dima, A.; Jurcut, C.; Chasset, F.; Felten, R.; Arnaud, L. Hydroxychloroquine in systemic lupus erythematosus: Overview of current knowledge. Ther. Adv. Musculoskelet. Dis. 2022, 14, 1759720X211073001. [Google Scholar] [CrossRef] [PubMed]
- Gautret, P.; Lagier, J.C.; Parola, P.; Meddeb, L.; Sevestre, J.; Mailhe, M.; Doudier, B.; Aubry, C.; Amrane, S.; Seng, P.; et al. Clinical and microbiological effect of a combination of hydroxychloroquine and azithromycin in 80 COVID-19 patients with at least a six-day follow up: A pilot observational study. Travel Med. Infect. Dis. 2020, 34, 101663. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, S.; Martínez, O.; Valenzuela, F.; Silva, F.; Valenzuela, O. Hydroxychloroquine and chloroquine in COVID-19: Should they be used as standard therapy? Clin. Rheumatol. 2020, 39, 2461–2465. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration. Coronavirus (COVID-19) Update: FDA Revokes Emergency Use Authorization for Chloroquine and Hydroxychloroquine. FDA News Release. Available online: www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-chloroquine (accessed on 28 December 2020).
- Lin, H.X.J.; Cho, S.; Meyyur Aravamudan, V.; Sanda, H.Y.; Palraj, R.; Molton, J.S.; Venkatachalam, I. Remdesivir in Coronavirus Disease 2019 (COVID-19) treatment: A review of evidence. Infection 2021, 49, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, A.; Ali Asgari, A.; Norouzi, A.; Kheiri, Z.; Anushirvani, A.; Montazeri, M.; Hosamirudsai, H.; Afhami, S.; Akbarpour, E.; Aliannejad, R.; et al. Sofosbuvir and daclatasvir compared with standard of care in the treatment of patients admitted to hospital with moderate or severe coronavirus infection (COVID-19): A randomized controlled trial. J. Antimicrob. Chemother. 2020, 75, 3379–3385. [Google Scholar] [CrossRef] [PubMed]
- Joseph, B.A.; Dibas, M.; Evanson, K.W.; Paranjape, G.; Vegivinti, C.T.R.; Selvan, P.T.; Saravu, K.; Gupta, N.; Pulakurthi, Y.S.; Keesari, P.R.; et al. Efficacy and safety of lopinavir/ritonavir in the treatment of COVID-19: A systematic review. Expert. Rev. Anti Infect. Ther. 2021, 19, 679–687. [Google Scholar] [CrossRef] [PubMed]
- RECOVERY Collaborative Group. Lopinavir-ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial. Lancet 2020, 396, 1345–1352. [Google Scholar] [CrossRef] [PubMed]
- Ader, F.; Peiffer-Smadja, N.; Poissy, J.; Bouscambert-Duchamp, M.; Belhadi, D.; Diallo, A.; Delmas, C.; Saillard, J.; Dechanet, A.; Mercier, N.; et al. An open-label randomized controlled trial of the effect of lopinavir/ritonavir, lopinavir/ritonavir plus IFN-β-1a and hydroxychloroquine in hospitalized patients with COVID-19. Clin. Microbiol. Infect. 2021, 27, 1826–1837. [Google Scholar] [CrossRef]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef]
- Bosaeed, M.; Mahmoud, E.; Alharbi, A.; Altayib, H.; Albayat, H.; Alharbi, F.; Ghalilah, K.; Al Arfaj, A.; AlJishi, J.; Alarfaj, A.; et al. Favipiravir and hydroxychloroquine combination therapy in patients with moderate to severe COVID-19 (FACCT Trial): An open-label, multicenter, randomized. Control. Trial Infect. Dis. Ther. 2021, 10, 2291–2307. [Google Scholar] [CrossRef] [PubMed]
- RECOVERY Collaborative Group. Dexamethasone in hospitalized patients with COVID-19. N. Engl. J. Med. 2021, 384, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Tomazini, B.M.; Maia, I.S.; Cavalcanti, A.B.; Berwanger, O.; Rosa, R.G.; Veiga, V.C.; Avezum, A.; Lopes, R.D.; Bueno, F.R.; Silva, M.V.A.; et al. Effect of dexamethasone on days alive and ventilator-free in patients with moderate or severe acute respiratory distress syndrome and COVID-19: The CoDEX randomized clinical trial. JAMA 2020, 324, 1307–1316. [Google Scholar] [CrossRef] [PubMed]
- FDA US. FDA Issues Emergency Use Authorization for Convalescent Plasma as Potential Promising COVID–19 Treatment, Another Achievement in Administration’s Fight Against Pandemic. 2020. Available online: https://www.fda.gov/news-events/press-announcements/fda-issues-emergency-use-authorization-convalescent-plasma-potential-promising-covid-19-treatment (accessed on 22 December 2023).
- Brown, B.L.; McCullough, J. Treatment for emerging viruses: Convalescent plasma and COVID-19. Transfus. Apher. Sci. 2020, 59, 102790. [Google Scholar] [CrossRef] [PubMed]
- Corti, D.; Purcell, L.A.; Snell, G.; Veesler, D. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell 2021, 184, 3086–3108. [Google Scholar] [CrossRef]
- Simşek Yavuz, S.; Komşuoğlu Celikyurt, I. An update of anti-viral treatment of COVID-19. Turk. J. Med. Sci. 2021, 51, 3372–3390. [Google Scholar] [CrossRef] [PubMed]
- Butt, A.A.; Yan, P.; Shaikh, O.S.; Omer, S.B.; Mayr, F.B.; Talisa, V.B. Molnupiravir use and 30-day hospitalizations or death in a previously uninfected nonhospitalized high-risk population with COVID-19. J. Infect. Dis. 2023, 228, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- FDA. Coronavirus (COVID-19). Drugs. Available online: https://www.fda.gov/drugs/emergency-preparedness-drugs/coronavirus-covid-19-drugs (accessed on 22 December 2023).
- FDA. Emergency Use Authorization (EUA) for Bebtelovimab (LY-CoV1404) Center for Drug Evaluation and Research (CDER) Review. Available online: https://www.fda.gov/media/156396/download (accessed on 22 December 2023).
- FDA. FDA Authorizes Gohibic (Vilobelimab) Injection for the Treatment of COVID-19. Available online: https://www.fda.gov/media/166823/download?attachment (accessed on 22 December 2023).
- FDA. Lagevrio Letter of Authorization. Available online: https://www.fda.gov/media/155053/download?attachment (accessed on 22 December 2023).
- Taylor, P.C.; Adams, A.C.; Hufford, M.M.; De La Torre, I.; Winthrop, K.; Gottlieb, R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021, 21, 382–393. [Google Scholar] [CrossRef]
- Casadevall, A.; Pirofski, L.A. The convalescent sera option for containing COVID-19. J. Clin. Investig. 2020, 130, 1545–1548. [Google Scholar] [CrossRef]
- Casadevall, A.; Dadachova, E.; Pirofski, L.A. Passive antibody therapy for infectious diseases. Nat. Rev. Microbiol. 2004, 2, 695–703. [Google Scholar] [CrossRef]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.-J.; Jiang, S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009, 7, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef] [PubMed]
- Chaigne, B.; Mouthon, L. Mechanisms of action of intravenous immunoglobulin. Transfus. Apher. Sci. 2017, 56, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Rokni, M.; Ghasemi, V.; Tavakoli, Z. Immune responses and pathogenesis of SARS-CoV-2 during an outbreak in Iran: Comparison with SARS and MERS. Rev. Med. Virol. 2020, 30, e2107. [Google Scholar] [CrossRef] [PubMed]
- Bloch, E.M.; Shoham, S.; Casadevall, A.; Sachais, B.S.; Shaz, B.; Winters, J.L. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Investig. 2020, 130, 2757–2765. [Google Scholar] [CrossRef] [PubMed]
- Hempel, T.; Raich, L.; Olsson, S.; Azouz, N.P.; Klingler, A.M.; Hoffmann, M.; Pöhlmann, S.; Rothenberg, M.E.; Noé, F. Molecular mechanism of inhibiting the SARS-CoV-2 cell entry facilitator TMPRSS2 with camostat and nafamostat. Chem. Sci. 2021, 12, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Stopsack, K.H.; Mucci, L.A.; Mph, S.; Antonarakis, E.S.; Nelson, P.S.; Kantoff, P.W. TMPRSS2 and COVID-19: Serendipity or opportunity for intervention? Cancer Discov. 2020, 10, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.W.; Mao, H.J.; Wu, Y.L.; Tanaka, Y.; Zhang, W. TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections. Biochimie 2017, 142, 1–10. [Google Scholar] [CrossRef]
- Hussain, M.; Jabeen, N.; Amanullah, A.; Baig, A.A.; Aziz, B.; Shabbir, S.; Raza, F.; Uddin, N. Molecular docking between human tmprss2 and SARS-CoV-2 spike protein: Conformation and intermolecular interactions. AIMS Microbiol. 2020, 6, 350–360. [Google Scholar] [CrossRef]
- Touret, F.; Gilles, M.; Barral, K.; Nougairède, A.; Van Helden, J.; Decroly, E.; De Lamballerie, X.; Coutard, B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 2020, 10, 13093. [Google Scholar] [CrossRef]
- Alzain, A.A.; Elbadwi, F.A.; Alsamani, F.O. Discovery of novel TMPRSS2 inhibitors for COVID-19 using in silico fragment-based drug design, molecular docking, molecular dynamics, and quantum mechanics studies. Inform. Med. Unlocked 2022, 29, 100870. [Google Scholar] [CrossRef]
- Mahoney, M.; Damalanka, V.C.; Tartell, M.A.; Chung, D.H.; Lourenço, A.L.; Pwee, D.; Mayer Bridwell, A.E.; Hoffmann, M.; Voss, J.; Karmakar, P.; et al. A novel class of TMPRSS2 inhibitors potently block SARS-CoV-2 and MERS-CoV viral entry and protect human epithelial lung cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2108728118. [Google Scholar] [CrossRef]
- Satarker, S.; Ahuja, T.; Banerjee, M.; E, V.B.; Dogra, S.; Agarwal, T.; Nampoothiri, M. Hydroxychloroquine in COVID-19: Potential mechanism of action against SARS-CoV-2. Curr. Pharmacol. Rep. 2020, 6, 203–211. [Google Scholar] [CrossRef]
- Yang, N.; Shen, H.M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020, 16, 1724–1731. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, R.; Xu, M.; Wang, X.; Zhang, H.; Hu, H.; Li, Y.; Hu, Z.; Zhong, W.; Wang, M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020, 6, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Di Scala, C.; Chahinian, H.; Yahi, N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J Antimicrob. Agents 2020, 55, 105960. [Google Scholar] [CrossRef] [PubMed]
- Pahan, P.; Pahan, K. Smooth or risky revisit of an old malaria drug for COVID-19? J. Neuroimmune Pharmacol. 2020, 19, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Xu, W.; Liu, Y.; Li, H.; Chen, L. The research progress of SARS-CoV-2 main protease inhibitors from 2020 to 2022. Eur. J. Med. Chem. 2023, 257, 115491. [Google Scholar] [CrossRef] [PubMed]
- Vatansever, E.C.; Yang, K.; Kratch, K.C.; Drelich, A.; Cho, C.-C.; Mellott, D.M.; Xu, S.; Tseng, C.-T.K.; Liu, W.R. Bepridil is potent against SARS-CoV-2 in vitro. Proc. Natl. Acad. Sci. USA 2021, 118, e2012201118. [Google Scholar] [CrossRef]
- Lockbaum, G.J.; Henes, M.; Lee, J.M.; Timm, J.; Nalivaika, E.A.; Thompson, P.R.; Yilmaz, N.K.; Schiffer, C.A. Pan-3C protease inhibitor rupintrivir binds SARS-CoV-2 main protease in a unique binding mode. Biochemistry 2021, 60, 2925–2931. [Google Scholar] [CrossRef]
- Ma, C.; Sacco, M.D.; Hurst, B.; Townsend, J.A.; Hu, Y.; Szeto, T.; Zhang, X.; Tarbet, B.; Marty, M.T.; Chen, Y.; et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020, 30, 678–692. [Google Scholar] [CrossRef] [PubMed]
- Nutho, B.; Mahalapbutr, P.; Hengphasatporn, K.; Pattaranggoon, N.C.; Simanon, N.; Shigeta, Y.; Hannongbua, S.; Rungrotmongkol, T. Why are lopinavir and ritonavir effective against the newly emerged coronavirus 2019? Atomistic insights into the inhibitory mechanisms. Biochemistry 2020, 59, 1769–1779. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.W.; Yap, Y.L. Old drugs as lead compounds for a new disease? Binding analysis of SARS coronavirus main proteinase with HIV, psychotic and parasite drugs. Bioorg. Med. Chem. 2004, 12, 2517–2521. [Google Scholar] [CrossRef] [PubMed]
- Lam, C.; Patel, P. Nirmatrelvir-Ritonavir; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Swaim, C.D.; Dwivedi, V.; Perng, Y.-C.; Zhao, X.; Canadeo, L.A.; Harastani, H.H.; Darling, T.L.; Boon, A.C.M.; Lenschow, D.J.; Kulkarni, V.; et al. 6-Thioguanine blocks SARS-CoV-2 replication by inhibition of PLpro. iScience 2021, 24, 103213. [Google Scholar] [CrossRef] [PubMed]
- Bafna, K.; White, K.; Harish, B.; Rosales, R.; Ramelot, T.A.; Acton, T.B.; Moreno, E.; Kehrer, T.; Miorin, L.; Royer, C.A.; et al. Hepatitis C virus drugs that inhibit SARS-CoV-2 papain-like protease synergize with remdesivir to suppress viral replication in cell culture. Cell Rep. 2021, 35, 109133. [Google Scholar] [CrossRef] [PubMed]
- Swaim, C.D.; Perng, Y.C.; Zhao, X.; Canadeo, L.A.; Harastani, H.H.; Darling, T.L.; Boon, A.C.; Lenschow, D.J.; Huibregtse, J.M. 6-Thioguanine blocks SARS-CoV-2 replication by inhibition of PLpro protease activities. BioRxiv 2020. [Google Scholar] [CrossRef]
- Gammeltoft, K.A.; Zhou, Y.; Duarte Hernandez, C.R.; Galli, A.; Offersgaard, A.; Costa, R.; Pham, L.V.; Fahnøe, U.; Feng, S.; Scheel, T.K.; et al. Hepatitis C virus protease inhibitors show differential efficacy and interactions with remdesivir for treatment of SARS-CoV-2 in vitro. Antimicrob. Agents Chemother. 2021, 65, e0268020. [Google Scholar] [CrossRef]
- Klemm, T.; Ebert, G.; Calleja, D.J.; Allison, C.C.; Richardson, L.W.; Bernardini, J.P.; Lu, B.G.C.; Kuchel, N.W.; Grohmann, C.; Shibata, Y.; et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020, 39, e106275. [Google Scholar] [CrossRef]
- Brown, A.J.; Won, J.J.; Graham, R.L.; Dinnon, K.H., III; Sims, A.C.; Feng, J.Y.; Cihlar, T.; Denison, M.R.; Baric, R.S.; Sheahan, T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir. Res. 2019, 169, 104541. [Google Scholar] [CrossRef]
- Elfiky, A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020, 248, 117477. [Google Scholar] [CrossRef]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef] [PubMed]
- Teli, D.; Balar, P.; Patel, K.; Sharma, A.; Chavda, V.; Vora, L. Molnupiravir: A versatile prodrug against SARS-CoV-2 variants. Metabolites 2023, 13, 309. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, L. Broad-spectrum prodrugs with anti-SARS-CoV-2 activities: Strategies, benefits, and challenges. J. Med. Virol. 2022, 94, 1373–1390. [Google Scholar] [CrossRef] [PubMed]
- Zarenezhad, E.; Marzi, M. Review on molnupiravir as a promising oral drug for the treatment of COVID-19. Med. Chem. Res. 2022, 31, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Parra-Lucares, A.; Segura, P.; Rojas, V.; Pumarino, C.; Saint-Pierre, G.; Toro, L. Emergence of SARS-CoV-2 variants in the world: How could this happen? Life 2022, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Menéndez-Arias, L. Decoding molnupiravir-induced mutagenesis in SARS-CoV-2. J. Biol. Chem. 2021, 297, 100867. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.J.; Tchesnokov, E.P.; Schinazi, R.F.; Götte, M. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J. Biol. Chem. 2021, 297, 100770. [Google Scholar] [CrossRef]
- Kabinger, F.; Stiller, C.; Schmitzová, J.; Dienemann, C.; Kokic, G.; Hillen, H.S.; Höbartner, C.; Cramer, P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021, 28, 740–746. [Google Scholar] [CrossRef]
- Khalifa, H.O.; Kamimoto, M.; Shimamoto, T.; Shimamoto, T. Antimicrobial effects of blueberry, raspberry, and strawberry aqueous extracts and their effects on virulence gene expression in Vibrio cholerae. Phytother. Res. 2015, 29, 1791–1797. [Google Scholar] [CrossRef]
- Sorour, S.S.; Abou Asa, S.; Elhawary, N.M.; Ghazy, E.W.; Abd El Latif, A.; El-Abasy, M.A.; Khalifa, H.O. Anticoccidial and hepatoprotective effects of artemisinin liquid extract, cinnamon essential oil and clove essential oil against Eimeria stiedae infection in rabbits. Trop. Biomed. 2018, 35, 926–943. [Google Scholar]
- Abd El-Hafeez, A.A.; Khalifa, H.O.; Elgawish, R.A.; Shouman, S.A.; Abd El-Twab, M.H.; Kawamoto, S. Melilotus indicus extract induces apoptosis in hepatocellular carcinoma cells via a mechanism involving mitochondria-mediated pathways. Cytotechnology 2018, 70, 831–842. [Google Scholar] [CrossRef]
- Abd El-Hafeez, A.A.; Marzouk, H.M.M.; Abdelhamid, M.A.; Khalifa, H.O.; Hasanin, T.H.; Habib, A.G.; Abdelwahed, F.M.; Barakat, F.M.; Bastawy, E.M.; Abdelghani, E.M.; et al. Anti-cancer effect of Hyoscyamus muticus extract via its activation of Fas/FasL-ASK1-p38 pathway. Biotechnol. Bioprocess Eng. 2022, 27, 833–845. [Google Scholar] [CrossRef]
- Khalifa, H.O.A. Molecular Pharmacological Studies on Multidrug-Resistant Bacteria: Analysis of Antimicrobial Resistance Mechanisms and Evaluation of Antimicrobial and Antivirulence Activities of Novel Plant Extracts. Ph.D. Thesis, Hiroshima University, Hiroshima, Japan, 2016. [Google Scholar]
- Mbikay, M.; Chrétien, M. Isoquercetin as an anti-COVID-19 medication: A potential to realize. Front. Pharmacol. 2022, 13, 830205. [Google Scholar] [CrossRef] [PubMed]
- Aghababaei, F.; Hadidi, M. Recent advances in potential health benefits of quercetin. Pharmaceuticals 2023, 16, 1020. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Yuk, H.J.; Ryu, H.W.; Lim, S.H.; Kim, K.S.; Park, K.H.; Ryu, Y.B.; Lee, W.S. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzyme Inhib. Med. Chem. 2017, 32, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Hiremath, S.; Kumar, H.V.; Nandan, N.; Mantesh, M. In silico docking analysis revealed the potential of phytochemicals present in Phyllanthus amarus and Andrographis paniculata, used in Ayurveda medicine in inhibiting SARS-CoV-2. 3 Biotech 2021, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Ta, W.; Tang, W.; Hua, R.; Wang, J.; Wang, C.; Lu, W. Potential antiviral activity of isorhamnetin against SARS-CoV-2 spike pseudotyped virus in vitro. Drug Dev. Res. 2021, 82, 1124–1130. [Google Scholar] [CrossRef] [PubMed]
- Ramalho, S.D.; de Sousa, L.R.; Burger, M.C.; Lima, M.I.S.; da Silva, M.F.D.G.; Fernandes, J.B.; Vieira, P.C. Evaluation of flavonols and derivatives as human cathepsin B inhibitor. Nat. Prod. Res. 2015, 29, 2212–2214. [Google Scholar] [CrossRef]
- Pasqua, E.; Hamblin, N.; Edwards, C.; Baker-Glenn, C.; Hurley, C. Developing inhaled drugs for respiratory diseases: A medicinal chemistry perspective. Drug Discov. Today 2022, 27, 134–150. [Google Scholar] [CrossRef]
- de Reus, Y.A.; Hagedoorn, P.; Sturkenboom, M.G.G.; Grasmeijer, F.; Bolhuis, M.S.; Sibum, I.; Kerstjens, H.A.M.; Frijlink, H.W.; Akkerman, O.W. Tolerability and pharmacokinetic evaluation of inhaled dry powder hydroxychloroquine in healthy volunteers. PLoS ONE 2022, 17, e0272034. [Google Scholar] [CrossRef]
- Monk, P.D.; Marsden, R.J.; Tear, V.J.; Brookes, J.; Batten, T.N.; Mankowski, M.; Gabbay, F.J.; Davies, D.E.; Gabbay, F.J.; Davies, D.E.; et al. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: A randomised, double-blind, placebo- controlled, phase 2 trial. Lancet Respir. Med. 2021, 9, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Al Sulaiman, K.; Aljuhani, O.; Al Aamer, K.; Al Shaya, O.; Al Shaya, A.; Alsaeedi, A.S.; Alhubaishi, A.; Altebainawi, A.F.; Al Harthi, A.; Albelwi, S.; et al. The role of inhaled corticosteroids (ICS) in critically ill patients with COVID-19: A multicentre, cohort study. J. Intensive Care Med. 2022, 37, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.-M.; Bafadhel, M.; Dorward, J.; Hayward, G.; Saville, B.R.; Gbinigie, O.; Van Hecke, O.; Ogburn, E.; Evans, P.H.; Thomas, N.P.B.; et al. Inhaled budesonide for COVID-19 in people at high risk of complications in the community in the UK (PRINCIPLE): A randomised, controlled, open- label, adaptive platform trial. Lancet 2021, 398, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Song, J.Y.; Yoon, J.G.; Seo, Y.B.; Lee, J.; Eom, J.S.; Lee, J.S.; Choi, W.S.; Lee, E.Y.; Choi, Y.A.; Hyun, H.J.; et al. Ciclesonide inhaler treatment for mild-to-moderate COVID-19: A randomized, open-label, Phase 2 trial. J. Clin. Med. 2021, 10, 3545. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.C.; Liew, D.F.; Tanner, H.L.; Grainger, J.R.; Dwek, R.A.; Reisler, R.B.; Steinman, L.; Feldmann, M.; Ho, L.P.; Hussell, T.; et al. COVID-19 therapeutics: Challenges and directions for the future. Proc. Natl. Acad. Sci. USA 2022, 119, e2119893119. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, A.; Alavizadeh, S.H.; Hosseini, S.A.; Meidany, P.; Doagooyan, M.; Abolhasani, Y.; Saadat, Z.; Amani, F.; Kesharwani, P.; Gheybi, F.; et al. Harnessing aptamers against COVID-19: A therapeutic strategy. Drug Discov. Today 2023, 28, 103663. [Google Scholar] [CrossRef] [PubMed]
- Becker, N.-P.; Haberland, A.; Wenzel, K.; Göttel, P.; Wallukat, G.; Davideit, H.; Schulze-Rothe, S.; Hönicke, A.-S.; Schimke, I.; Bartel, S.; et al. Three-Part, randomised study to investigate the safety, Tolerability, Pharmacokinetics and Mode of action of BC 007, neutraliser of pathogenic auto-antibodies against G-Protein coupled receptors in healthy, young and elderly subjects. Clin. Drug Investig. 2020, 40, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.M.; Drabkin, H.A.; Lara, P.N., Jr.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Investig. New Drugs 2014, 32, 178–187. [Google Scholar] [CrossRef]
- Haberland, A.; Müller, J. Aptamers against COVID-19: An untested opportunity. Mini Rev. Med. Chem. 2022, 22, 1708–1715. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalifa, H.O.; Al Ramahi, Y.M. After the Hurricane: Anti-COVID-19 Drugs Development, Molecular Mechanisms of Action and Future Perspectives. Int. J. Mol. Sci. 2024, 25, 739. https://doi.org/10.3390/ijms25020739
Khalifa HO, Al Ramahi YM. After the Hurricane: Anti-COVID-19 Drugs Development, Molecular Mechanisms of Action and Future Perspectives. International Journal of Molecular Sciences. 2024; 25(2):739. https://doi.org/10.3390/ijms25020739
Chicago/Turabian StyleKhalifa, Hazim O., and Yousef M. Al Ramahi. 2024. "After the Hurricane: Anti-COVID-19 Drugs Development, Molecular Mechanisms of Action and Future Perspectives" International Journal of Molecular Sciences 25, no. 2: 739. https://doi.org/10.3390/ijms25020739
APA StyleKhalifa, H. O., & Al Ramahi, Y. M. (2024). After the Hurricane: Anti-COVID-19 Drugs Development, Molecular Mechanisms of Action and Future Perspectives. International Journal of Molecular Sciences, 25(2), 739. https://doi.org/10.3390/ijms25020739





