Molecular and Systems Biology Approaches for Harnessing the Symbiotic Interaction in Mycorrhizal Symbiosis for Grain and Oil Crop Cultivation
Abstract
:1. Introduction
2. Molecular Basis of Cereal and Oilseed Crop Responsiveness in the Presence of AM Colonization
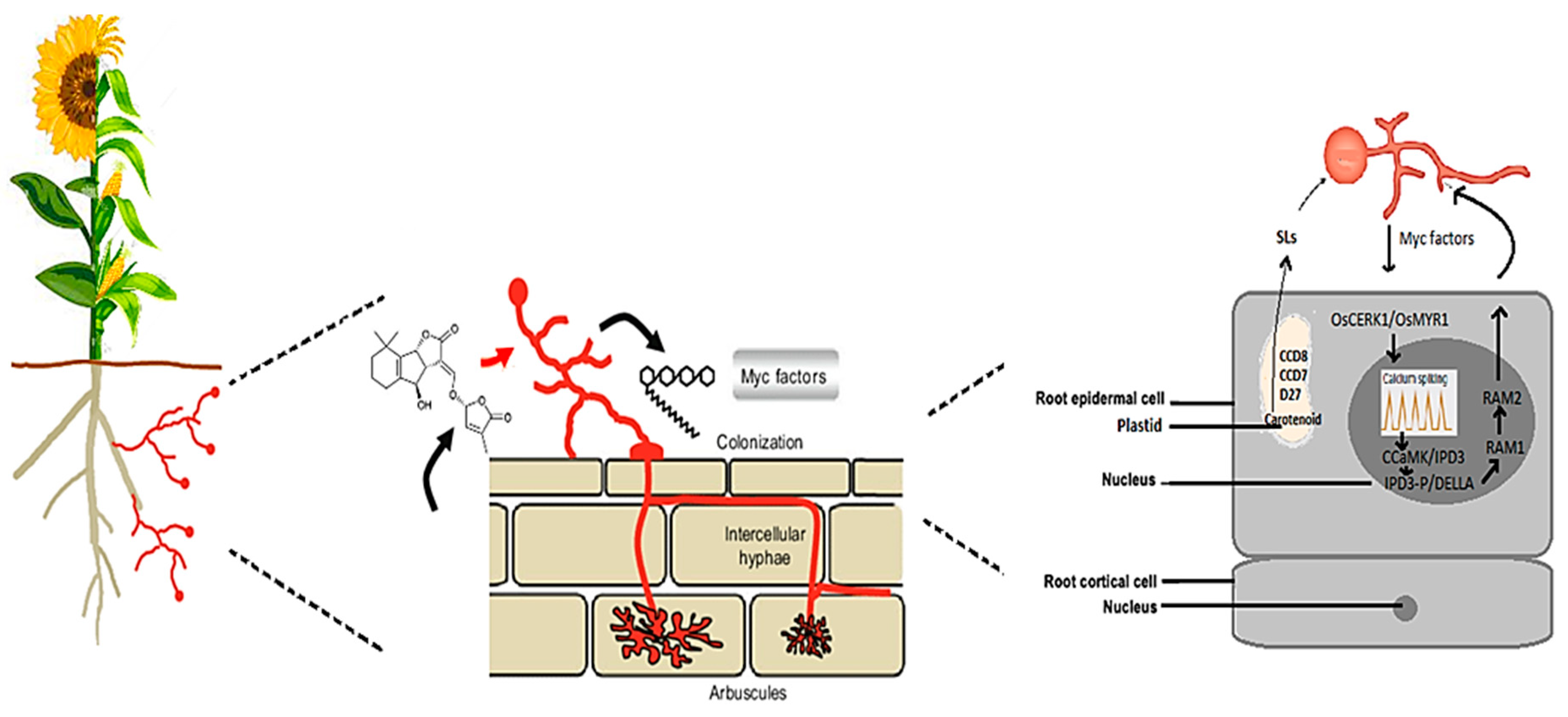
2.1. Recognition and Signaling between AMF and Cereal and Oilseed Crops
2.1.1. Receptor-Mediated Recognition of AMF Signals in Cereals and Oilseeds
2.1.2. Advances in Signal Transduction Pathways of AMF in Cereals and Oilseeds
2.2. Nutrient Exchange and Transport
2.2.1. Nutrient Uptake
2.2.2. Transporters Involved in Nutrient Exchange in the Symbiosis of AMF and Cereal and Oilseed Crops
| Nutrient/Metabolite | Plant Species | Mycorrhiza-Specific/Inducible Transporters | Ref. |
| Phosphorus | Oryza sativa | OsPT11 | [99] |
| OsPT13 | [81] | ||
| Hordeum vulgare | HvPt8 | [100] | |
| Zea mays | ZmPHT1;2, ZmPHT1;4, ZmPHT1;6, ZmPHT1;7, ZmPHT1;9, ZmPHT1;11 | [79] | |
| Glycine max | GmPHT1;6, GmPHT1;7, GmPHT1;10, GmPHT1;12, GmPHT1;13 | [101] | |
| Nitrogen | Glycine max | GmAMT4.1 | [85] |
| Sorghum bicolor | SbAMT3;1 | [87] | |
| SbNPF4.5 | [88] | ||
| Oryza sativa | OsNPF4.5 | [88] | |
| Zea mays | ZmNPF4.5 | [88] | |
| Zinc | H. vulgare | ZIP13 | [92] |
| Sugars | Glycine max | GmSWEET6, GmSWEET15 | [102] |
| Lipids | Oryza sativa | STR1, STR2 | [60] |
| Nutrient | Fungal transporters | Fungal species | Ref. |
| Phosphorus | GmosPT | Funneliformis mosseae | [80] |
| GiPT | Rhizophagus intraradices | [81] | |
| GigmPT | Gigaspora margarita | [82] | |
| Nitrogen | GmosAAP1 | Funneliformis mosseae | [89] |
| RiPTR2 | Rhizophagus irregularis | [90] | |
| Zinc | GintZnT1 | Rhizophagus irregularis | [91] |
| Sugars | RiMST2 | Rhizophagus irregularis | [103] |
| RiMST5, RiMST6 | Rhizophagus irregularis | [93] |
2.3. Genetic Regulation of Mycorrhizal Symbiosis in Cereal and Oilseed Crops
2.3.1. Transcription Factors (TFs) and microRNAs
2.3.2. Expression Profiling and Functional Genomics Studies
- Genomics-based approaches in mycorrhizal symbiosis in cereals/oilseeds: a brief insight
- AMF symbiosis establishment
- Pre-infection stage: many ‘molecules’ recognize each other
- Physical Contact, nutrient exchange, and associated events in AMF Symbiosis
- N and P acquisition in AMF symbiosis: an insight into cereal/oilseed transporter genes in AMF symbiosis
- AMF symbiosis regulation
3. Unlocking the Potential: Maximizing Nutrient Uptake and Growth through Mycorrhizal Symbiosis in Cereal and Oilseed Crops
3.1. Unleashing Nutrient Power: Enhancing Nutrient Acquisition and Growth in Cereal and Oilseed Crops
3.1.1. Manipulating Symbiotic Genes for Increased Nutrient Acquisition
3.1.2. Optimizing Plant-Mycorrhizal Associations for Improved Yield
| AMF Species | Host Plant | Main Affected Traits | Ref. |
|---|---|---|---|
| F. mosseae | Wheat | Regulation of genes involved in carbohydrate, lipid, N metabolism, cellulose synthase and chitinase activities, and membrane transport. | [221] |
| S. calospora, A. laevis, G. margarita, G. aggregatum, R. intraradices, F. mosseae, G. fasciculatum, G. etunicatum, and G. deserticola. | Durum wheat | Up-regulation of N (NRT1.1, NRT2, and NAR2.2) and Pi (Pht2) transporter genes. | [222] |
| R. irregularis | Rice | Induction of genes involved in N transport and metabolism (OsNPF4.5 and OsAMT3.1). | [88] |
| R. irregularis | Barley | Down-regulation of ZIP transporter genes (HvZIP3 and HvZIP8) and up-regulation of HvZIP13 | [92] |
| R. irregularis, F.mosseae, G. aggregatum, and G. etunicatum | Sunflower | Up-regulation of Fe and Zn transporter genes (HaIRT1, HaFRO1, and HaZIP1). | [223] |
| F. mosseae | Durum wheat | Enhancement of gene transcripts involved in the water stress response (TdSHN1 and TdDRF1). | [226] |
3.2. Moving toward Systems Biology for Mycorrhizal Management in Cereal and Oilseed Crops
4. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Khoury, C.K.; Bjorkman, A.D.; Dempewolf, H.; Ramirez-Villegas, J.; Guarino, L.; Jarvis, A.; Rieseberg, L.H.; Struik, P.C. Increasing Homogeneity in Global Food Supplies and the Implications for Food Security. Proc. Natl. Acad. Sci. USA 2014, 111, 4001–4006. [Google Scholar] [CrossRef]
- Renard, D.; Tilman, D. National Food Production Stabilized by Crop Diversity. Nature 2019, 571, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Fones, H.N.; Bebber, D.P.; Chaloner, T.M.; Kay, W.T.; Steinberg, G.; Gurr, S.J. Threats to Global Food Security from Emerging Fungal and Oomycete Crop Pathogens. Nat. Food 2020, 1, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Grassini, P.; Eskridge, K.M.; Cassman, K.G. Distinguishing between Yield Advances and Yield Plateaus in Historical Crop Production Trends. Nat. Commun. 2013, 4, 2918. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lehmann, A.; Zheng, W.; You, Z.; Rillig, M.C. Arbuscular Mycorrhizal Fungi Increase Grain Yields: A Meta-analysis. New Phytol. 2019, 222, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Kakouridis, A.; Hagen, J.A.; Kan, M.P.; Mambelli, S.; Feldman, L.J.; Herman, D.J.; Weber, P.K.; Pett-Ridge, J.; Firestone, M.K. Routes to Roots: Direct Evidence of Water Transport by Arbuscular Mycorrhizal Fungi to Host Plants. New Phytol. 2022, 236, 210–221. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, W.; Xie, Q.; Liu, N.; Liu, L.; Wang, D.; Zhang, X.; Yang, C.; Chen, X.; Tang, D.; et al. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 2017, 356, 1172–1175. [Google Scholar] [CrossRef] [PubMed]
- Luginbuehl, L.H.; Menard, G.N.; Kurup, S.; Van Erp, H.; Radhakrishnan, G.V.; Breakspear, A.; Oldroyd, G.E.D.; Eastmond, P.J. Fatty Acids in Arbuscular Mycorrhizal Fungi Are Synthesized by the Host Plant. Science 2017, 356, 1175–1178. [Google Scholar] [CrossRef]
- Luginbuehl, L.H.; Oldroyd, G.E. Understanding the arbuscule at the heart of endomycorrhizal symbioses in plants. Curr. Biol. 2017, 27, R952–R963. [Google Scholar] [CrossRef]
- Choi, J.; Summers, W.; Paszkowski, U. Mechanisms underlying establishment of arbuscular mycorrhizal symbioses. Annu. Rev. Phytopathol. 2018, 56, 135–160. [Google Scholar] [CrossRef]
- Maillet, F.; Poinsot, V.; André, O.; Puech-Pagès, V.; Haouy, A.; Gueunier, M.; Cromer, L.; Giraudet, D.; Formey, D.; Niebel, A. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 2011, 469, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Genre, A.; Chabaud, M.; Balzergue, C.; Puech-Pages, V.; Novero, M.; Rey, T.; Fournier, J.; Rochange, S.; Becard, G.; Bonfante, P.; et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013, 198, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Buendia, L.; Wang, T.; Girardin, A.; Lefebvre, B. The LysM receptor-like kinase SlLYK10 regulates the arbuscular mycorrhizal symbiosis in tomato. New Phytol. 2016, 210, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Girardin, A.; Wang, T.; Ding, Y.; Keller, J.; Buendia, L.; Gaston, M.; Ribeyre, C.; Gasciolli, V.; Auriac, M.C.; Vernie, T.; et al. LCO receptors involved in arbuscular mycorrhiza are functional for rhizobia perception in legumes. Curr. Biol. 2019, 29, 4249–4259. [Google Scholar] [CrossRef]
- Parniske, M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat. Rev. Genet. 2008, 6, 763–775. [Google Scholar] [CrossRef]
- Akiyama, K.; Matsuzaki, K.I.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef]
- Shi, J.; Wang, X.; Wang, E. Mycorrhizal symbiosis in Plant Growth and Stress Adaptation: From Genes to Ecosystems. Annu. Rev. Plant Biol. 2023, 74, 569–607. [Google Scholar] [CrossRef]
- Nadal, M.; Sawers, R.; Naseem, S.; Bassin, B.; Kulicke, C.; Sharman, A.; An, G.; An, K.; Ahern, K.R.; Romag, A.; et al. An N-Acetylglucosamine Transporter Required for Arbuscular Mycorrhizal Symbioses in Rice and Maize. Nat. Plants 2017, 3, 17073. [Google Scholar] [CrossRef]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The kinaze LYK5 is major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 2014, 3, e03766. [Google Scholar] [CrossRef]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Miyata, K.; Hayafune, M.; Kobae, Y.; Kaku, H.; Nishizawa, Y.; Masuda, Y.; Shibuya, N.; Nakagawa, T. Evaluation of the role of the LysM receptor-like kinase, OsNFR5/OsRLK2 for AM symbiosis in rice. Plant Cell Physiol. 2016, 57, 2283–2290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, W.; Sun, J.; Feng, F.; Deng, Y.; He, Z.; Oldroyd, G.; Wang, E. The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 2015, 81, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Carotenuto, G.; Chabaud, M.; Miyata, K.; Capozzi, M.; Takeda, N.; Kaku, H.; Shibuya, N.; Nakagawa, T.; Barker, D.G.; Genre, A. The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol. 2017, 214, 1440–1446. [Google Scholar] [CrossRef] [PubMed]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef]
- Miyata, K.; Kozaki, T.; Kouzai, Y.; Ozawa, K.; Ishii, K.; Asamizu, E.; Okabe, Y.; Umehara, Y.; Miyamoto, A.; Kobae, Y.; et al. The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol. 2014, 55, 1864–1872. [Google Scholar] [CrossRef]
- He, J.; Zhang, C.; Dai, H.; Liu, H.; Zhang, X.; Yang, J.; Chen, X.; Zhu, Y.; Wang, D.; Qi, X. A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol. Plant 2019, 12, 1561–1576. [Google Scholar] [CrossRef]
- Zhang, C.; He, J.; Dai, H.; Wang, G.; Zhang, X.; Wang, C.; Shi, J.; Chen, X.; Wang, D.; Wang, E. Discriminating symbiosis and immunity signals by receptor competition in rice. Proc. Natl. Acad. Sci. USA 2021, 118, e2023738118. [Google Scholar] [CrossRef]
- Zipfel, C.; Oldroyd, G.E. Plant signalling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef]
- Gutjahr, C.; Gobbato, E.; Choi, J.; Riemann, M.; Johnston, M.G.; Summers, W.; Carbonnel, S.; Mansfield, C.; Yang, S.Y.; Nadal, M.; et al. Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 2015, 350, 1521–1524. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, N.; Jiang, L.; Li, P.; Sharma, A.K.; Luo, X.; Wu, S.; Pandey, R.; Gao, Q.; Lou, B. Proteomic approach to understand the molecular physiology of symbiotic interaction between Piriformospora indica and Brassica napus. Sci. Rep. 2018, 8, 5773. [Google Scholar] [CrossRef] [PubMed]
- Antolín-Llovera, M.; Petutsching, E.K.; Ried, M.K.; Lipka, V.; Nürnberger, T.; Robatzek, S.; Parniske, M. Knowing your friends and foes—Plant receptor-like kinases as initiators of symbiosis or defence. New Phytol. 2014, 204, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.-Q.; Xue, J.; Zhang, N.; Xu, L.; Yao, X.; Yang, Q.-J.; Yu, Y.; Wang, H.-B.; Zhang, D.; Li, J.-F. Rice Chitin Receptor OsCEBiP Is Not a Transmembrane Protein but Targets the Plasma Membrane via a GPI Anchor. Mol. Plant 2017, 10, 767–770. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.C.; Scaffidi, A.; Dun, E.A.; Waters, M.T.; Flematti, G.R.; Dixon, K.W.; Beveridge, C.A.; Ghisalberti, E.L.; Smith, S.M. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 8897–8902. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Scaffidi, A.; Flematti, G.; Smith, S.M. Substrate-induced degradation of the α/β- fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Mol. Plant 2015, 8, 814–817. [Google Scholar] [CrossRef] [PubMed]
- Flematti, G.; Dixon, K.; Smith, S.M. What are karrikins and how were they ‘discovered’ by plants? BMC Biol. 2015, 13, 108. [Google Scholar] [CrossRef]
- Conn, C.E.; Nelson, D.C. Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front. Plant Sci. 2016, 6, 1219. [Google Scholar] [CrossRef]
- Yoshida, S.; Kameoka, H.; Tempo, M.; Akiyama, K.; Umehara, M.; Yamaguchi, S.; Hayashi, H.; Kyozuka, J.; Shirasu, K. The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytol. 2012, 196, 1208–1216. [Google Scholar] [CrossRef]
- Ho-Plágaro, T.; Morcillo, R.J.; Tamayo-Navarrete, M.I.; Huertas, R.; Molinero-Rosales, N.; López-Ráez, J.A.; Macho, A.P.; García-Garrido, J.M. DLK2 regulates arbuscule hyphal branching during arbuscular mycorrhizal symbiosis. New Phytol. 2021, 229, 548–562. [Google Scholar] [CrossRef]
- Erickson, J.; Weckwerth, P.; Romeis, T.; Lee, J. What’s new in protein kinase/phosphatase signalling in the control of plant immunity? Essays Biochem. 2022, 66, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Yamada, K.; Ishikawa, K.; Yoshimura, S.; Hayashi, N.; Uchihashi, K.; Ishihama, N.; Kishi-Kaboshi, M.; Takahashi, A.; Tsuge, S. A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 2013, 13, 347–357. [Google Scholar] [CrossRef]
- Ao, Y.; Li, Z.; Feng, D.; Xiong, F.; Liu, J.; Li, J.F.; Wang, M.; Wang, J.; Liu, B.; Wang, H.B. Os CERK 1 and Os RLCK 176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 2014, 80, 1072–1084. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Berkowitz, G.A. The grateful dead: Calcium and cell death in plant innate immunity. Cell Microbiol. 2007, 9, 2571–2585. [Google Scholar] [CrossRef]
- Kadota, Y.; Shirasu, K.; Zipfel, C. Regulation of the NADPH Oxidase RBOHD during Plant Immunity. Plant Cell Physiol. 2015, 56, 1472–1480. [Google Scholar] [CrossRef] [PubMed]
- Iriti, M.; Varoni, E.M. Chitosan-induced antiviral activity and innate immunity in plants. Environ. Sci. Pollut. Res. 2015, 22, 2935–2944. [Google Scholar] [CrossRef] [PubMed]
- Lizama-Uc, G.; Estrada-Mota, I.A.; Caamal-Chan, M.G.; Souza-Perera, R.; Oropeza-Salín, C.; Islas-Flores, I.; Zúñiga-Aguilar, J.J. Chitosan Activates a MAP-Kinase Pathway and Modifies Abundance of Defense-Related Transcripts in Calli of Cocos nucifera L. Physiol. Mol. Plant Pathol. 2007, 70, 130–141. [Google Scholar] [CrossRef]
- Zuppini, A.; Navazio, L.; Sella, L.; Castiglioni, C.; Favaron, F.; Mariani, P. An Endopolygalacturonase from Sclerotinia Sclerotiorum Induces Calcium-Mediated Signaling and Programmed Cell Death in Soybean Cells. MPMI 2005, 18, 849–855. [Google Scholar] [CrossRef]
- Hogekamp, C.; Küster, H. A Roadmap of Cell-Type Specific Gene Expression during Sequential Stages of the Arbuscular Mycorrhiza Symbiosis. Genomics 2013, 14, 306. [Google Scholar] [CrossRef]
- Park, H.J.; Floss, D.S.; Levesque-Tremblay, V.; Bravo, A.; Harrison, M.J. Hyphal Branching during Arbuscule Development Requires Reduced Arbuscular Mycorrhiza1. Plant Physiol. 2015, 169, 2774–2788. [Google Scholar] [CrossRef]
- Huisman, R.; Hontelez, J.; Mysore, K.S.; Wen, J.; Bisseling, T.; Limpens, E. A symbiosis- dedicated SYNTAXIN OF PLANTS 13II isoform controls the formation of a stable host-microbe interface in symbiosis. New Phytol. 2016, 211, 1338–1351. [Google Scholar] [CrossRef]
- Pan, H.; Oztas, O.; Zhang, X.; Wu, X.; Stonoha, C.; Wang, E.; Wang, B.; Wang, D. A symbiotic SNARE protein generated by alternative termination of transcription. Nat. Plants 2016, 2, 15197. [Google Scholar] [CrossRef] [PubMed]
- Feddermann, N.; Duvvuru Muni, R.R.; Zeier, T.; Stuurman, J.; Ercolin, F.; Schorderet, M.; Reinhardt, D. The PAM1 Gene of Petunia, Required for Intracellular Accommodation and Morphogenesis of Arbuscular Mycorrhizal Fungi, Encodes a Homologue of VAPYRIN: Intracellular Accommodation of AM Fungi. Plant J. 2010, 64, 470–481. [Google Scholar] [CrossRef]
- Pumplin, N.; Mondo, S.J.; Topp, S.; Starker, C.G.; Gantt, J.S.; Harrison, M.J. Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 2010, 61, 482–494. [Google Scholar] [CrossRef]
- Murray, J.D.; Muni, R.R.D.; Torres-Jerez, I.; Tang, Y.; Allen, S.; Andriankaja, M.; Li, G.; Laxmi, A.; Cheng, X.; Wen, J.; et al. Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 2011, 65, 244–252. [Google Scholar] [CrossRef]
- Ivanov, S.; Fedorova, E.E.; Limpens, E.; De Mita, S.; Genre, A.; Bonfante, P.; Bisseling, T. Rhizobium-Legume Symbiosis Shares an Exocytotic Pathway Required for Arbuscule Formation. Proc. Natl. Acad. Sci. USA 2012, 109, 8316–8321. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.J.; Dewbre, G.R.; Liu, J. A Phosphate Transporter from Medicago truncatula Involved in the Acquisition of Phosphate Released by Arbuscular Mycorrhizal Fungi. Plant Cell 2002, 14, 2413–2429. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Yu, N.; Bano, S.A.; Liu, C.; Miller, A.J.; Cousins, D.; Zhang, X.; Ratet, P.; Tadege, M.; Mysore, K.S.; et al. A H+-ATPase That Energizes Nutrient Uptake during Mycorrhizal Symbioses in Rice and Medicago truncatula. Plant Cell 2014, 26, 1818–1830. [Google Scholar] [CrossRef]
- Garcia, K.; Doidy, J.; Zimmermann, S.D.; Wipf, D.; Courty, P.E. Take a Trip Through the Plant and Fungal Transportome of Mycorrhiza. Trends Plant Sci. 2016, 21, 937–950. [Google Scholar] [CrossRef]
- Zhang, Q.; Blaylock, L.A.; Harrison, M.J. Two Medicago truncatula Half-ABC Transporters Are Essential for Arbuscule Development in Arbuscular Mycorrhizal Symbiosis. Plant Cell 2010, 22, 1483–1497. [Google Scholar] [CrossRef]
- Gutjahr, C.; Radovanovic, D.; Geoffroy, J.; Zhang, Q.; Siegler, H.; Chiapello, M.; Casieri, L.; An, K.; An, G.; Guiderdoni, E.; et al. The Half-Size ABC Transporters STR1 and STR2 Are Indispensable for Mycorrhizal Arbuscule Formation in Rice. Plant J. 2012, 69, 906–920. [Google Scholar] [CrossRef]
- Pumplin, N.; Zhang, X.; Noar, R.D.; Harrison, M.J. Polar Localization of a Symbiosis-Specific Phosphate Transporter Is Mediated by a Transient Reorientation of Secretion. Proc. Natl. Acad. Sci. USA 2012, 109, 665–672. [Google Scholar] [CrossRef]
- Floss, D.S.; Levy, J.G.; Lévesque-Tremblay, V.; Pumplin, N.; Harrison, M.J. DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2013, 110, E5025–E5034. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, K.; Xie, X.; Kusumoto, D.; Sekimoto, H.; Sugimoto, Y.; Takeuchi, Y.; Yoneyama, K. Nitrogen Deficiency as Well as Phosphorus Deficiency in Sorghum Promotes the Production and Exudation of 5-Deoxystrigol, the Host Recognition Signal for Arbuscular Mycorrhizal Fungi and Root Parasites. Planta 2007, 227, 125–132. [Google Scholar] [CrossRef]
- Tang, H.; Hassan, M.U.; Feng, L.; Nawaz, M.; Shah, A.N.; Qari, S.H.; Liu, Y.; Miao, J. The Critical Role of Arbuscular Mycorrhizal Fungi to Improve Drought Tolerance and Nitrogen Use Efficiency in Crops. Front. Plant Sci. 2022, 13, 919166. [Google Scholar] [CrossRef] [PubMed]
- Roth, R.; Paszkowski, U. Plant carbon nourishment of arbuscular mycorrhizal fungi. Curr. Opin. Plant Biol. 2017, 39, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Igiehon, N.O.; Babalola, O.O. Biofertilizers and Sustainable Agriculture: Exploring Arbuscular Mycorrhizal Fungi. Appl. Microbiol. Biotechnol. 2017, 101, 4871–4881. [Google Scholar] [CrossRef] [PubMed]
- Slimani, A.; Oufdou, K.; Meddich, A. Intercropping with Alfalfa and Co-Inoculation of AMF and PGPR Improve Growth, Yield, Grain Bioactive Quality, and Soil Fertility of Barley. Arch. Agron. Soil Sci. 2023, 69, 3469–3483. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D. Mineral Nutrition, Toxic Element Accumulation and Water Relations of Arbuscular Mycorrhizal Plants. In Mycorrhizal Symbiosis; Elsevier: Amsterdam, The Netherlands, 2008; pp. 145–187. ISBN 978-0-12-370526-6. [Google Scholar]
- Dai, M.; Hamel, C.; Bainard, L.D.; Arnaud, M.S.; Grant, C.A.; Lupwayi, N.Z.; Malhi, S.S.; Lemke, R. Negative and Positive Contributions of Arbuscular Mycorrhizal Fungal Taxa to Wheat Production and Nutrient Uptake Efficiency in Organic and Conventional Systems in the Canadian Prairie. Soil Biol. Biochem. 2014, 74, 156–166. [Google Scholar] [CrossRef]
- Merlos, M.A.; Zitka, O.; Vojtech, A.; Azcon-Aguilar, C.; Ferrol, N. The arbuscular mycorrhizal fungus Rhizophagus irregularis differentially regulates the copper response of two maize cultivars differing in copper tolerance. Plant Sci. 2016, 253, 68–76. [Google Scholar] [CrossRef]
- Watts-Williams, S.J.; Gill, A.R.; Jewell, N.; Brien, C.J.; Berger, B.; Tran, B.T.T.; Mace, E.; Cruickshank, A.W.; Jordan, D.R.; Garnett, T.; et al. Enhancement of Sorghum Grain Yield and Nutrition: A Role for Arbuscular Mycorrhizal Fungi Regardless of Soil Phosphorus Availability. Plants People Planet 2022, 4, 143–156. [Google Scholar] [CrossRef]
- Kuila, D.; Ghosh, S. Aspects, problems and utilization of Arbuscular Mycorrhizal (AM) application as bio-fertilizer in sustainable agriculture. Curr. Res. Microb. Sci. 2022, 3, 100107. [Google Scholar] [CrossRef] [PubMed]
- Bellido, E.; De la Haba, P.; Aguera, E. Physiological Alteration in sunflower plants (Helianthus annuus L.) exposed to high CO2 and arbuscular mycorrhizal fungi. Plants 2021, 10, 937. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Saini, I.; Kaushik, P.; Ansari, M.A.; Khan, M.R.; Haq, N. Effects of Arbuscular Mycorrhizal Fungi and P-Solubilizing Pseudomonas fluorescence (ATCC-17400) on Morphological Traits and Mineral Content of Sesame. Saudi J. Biol. Sci. 2021, 28, 2649–2654. [Google Scholar] [CrossRef]
- Dabre, E.E.; Hijri, M.; Favret, C. Influence on Soybean Aphid by the Tripartite Interaction between Soybean, a Rhizobium Bacterium, and an Arbuscular Mycorrhizal Fungus. Microorganisms 2022, 10, 1196. [Google Scholar] [CrossRef]
- Wang, W.; Shi, J.; Xie, Q.; Jiang, Y.; Yu, N.; Wang, E. Nutrient Exchange and Regulation in Arbuscular Mycorrhizal Symbiosis. Mol. Plant. 2017, 10, 1147–1158. [Google Scholar] [CrossRef]
- Banasiak, J.; Jamruszka, T.; Murray, J.D.; Jasiński, M. A roadmap of plant membrane transporters in arbuscular mycorrhizal and legume-rhizobium symbioses. Plant Physiol. 2021, 187, 2071–2091. [Google Scholar] [CrossRef]
- Liu, F.; Xu, Y.J.; Jiang, H.H.; Jiang, C.S.; Du, Y.B.; Gong, C.; Wang, W.; Zhu, S.W.; Han, G.M.; Cheng, B.J. Systematic identification, evolution and expression analysis of the Zea mays PHT1 gene family reveals several new members involved in root colonization by arbuscular mycorrhizal fungi. Int. J. Mol. Sci. 2016, 17, 930. [Google Scholar] [CrossRef]
- Benedetto, A.; Magurno, F.; Bonfante, P.; Lanfranco, L. Expression profiles of a phosphate transporter gene (GmosPT) from the endomycorrhizal fungus Glomus mosseae. Mycorrhiza 2005, 15, 620–627. [Google Scholar] [CrossRef]
- Yang, S.-Y.; Grønlund, M.; Jakobsen, I.; Grotemeyer, M.S.; Rentsch, D.; Miyao, A.; Hirochika, H.; Kumar, C.S.; Sundaresan, V.; Salamin, N.; et al. Nonredundant Regulation of Rice Arbuscular Mycorrhizal Symbiosis by Two Members of the PHOSPHATE TRANSPORTER1 Gene Family. Plant Cell 2012, 24, 4236–4251. [Google Scholar] [CrossRef]
- Salvioli, A.; Ghignone, S.; Novero, M.; Navazio, L.; Venice, F.; Bagnaresi, P.; Bonfante, P. Symbiosis with an endobacterium increases the fitness of a mycorrhizal fungus, raising its bioenergetic potential. ISME J. 2016, 10, 130–144. [Google Scholar] [CrossRef] [PubMed]
- Salvioli Di Fossalunga, A.; Novero, M. To Trade in the Field: The Molecular Determinants of Arbuscular Mycorrhiza Nutrient Exchange. Chem. Biol. Technol. Agric. 2019, 6, 12. [Google Scholar] [CrossRef]
- Xie, X.; Lin, H.; Peng, X.; Xu, C.; Sun, Z.; Jiang, K.; Huang, A.; Wu, X.; Tang, N.; Salvioli, A.; et al. Arbuscular mycorrhizal symbiosis requires a phosphate transceptor in the Gigaspora margarita fungal symbiont. Mol. Plant 2016, 9, 1583–1608. [Google Scholar] [CrossRef] [PubMed]
- Kobae, Y.; Tamura, Y.; Takai, S.; Banba, M.; Hata, S. Localized expression of arbuscular mycorrhiza-inducible ammonium transporters in soybean. Plant Cell Physiol. 2010, 51, 1411–1415. [Google Scholar] [CrossRef] [PubMed]
- Drechsler, N.; Courty, P.-E.; Brulé, D.; Kunze, R. Identification of arbuscular mycorrhiza- inducible Nitrate Transporter 1/Peptide Transporter Family (NPF) genes in rice. Mycorrhiza 2017, 28, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Koegel, S.; Lahmidi, N.A.; Arnould, C.; Chatagnier, O.; Walder, F.; Ineichen, K.; Boller, T.; Wipf, D.; Wiemken, A.; Courty, P. The family of ammonium transporters (AMT) in Sorghum bicolor: Two AMT members are induced locally, but not systemically in roots colonized by arbuscular mycorrhizal fungi. New Phytol. 2013, 198, 853–865. [Google Scholar] [CrossRef]
- Wang, S.; Chen, A.; Xie, K.; Yang, X.; Luo, Z.; Chen, J.; Zeng, D.; Ren, Y.; Yang, C.; Wang, L.; et al. Functional analysis of the OsNPF4.5 nitrate transporter reveals a conserved mycorrhizal pathway of nitrogen acquisition in plants. Proc. Natl. Acad. Sci. USA 2020, 117, 16649–16659. [Google Scholar] [CrossRef]
- Cappellazzo, G.; Lanfranco, L.; Fitz, M.; Wipf, D.; Bonfante, P. Characterization of an amino acid permease from the endomycorrhizal fungus Glomus mosseae. Plant Physiol. 2008, 147, 429–437. [Google Scholar] [CrossRef]
- Belmondo, S.; Fiorilli, V.; Perez-Tienda, J.; Ferrol, N.; Marmeisse, R.; Lanfranco, L. A dipeptide transporter from the arbuscular mycorrhizal fungus Rhizophagus irregularis is upregulated in the intraradical phase. Front. Plant Sci. 2014, 5, 436. [Google Scholar] [CrossRef]
- González-Guerrero, M.; Azcón-Aguilar, C.; Mooney, M.; Valderas, A.; MacDiarmid, C.W.; Eide, D.J.; Ferrol, N. Characterization of a Glomus intraradices gene encoding a putative Zn transporter of the cation diffusion facilitator family. Fungal Genet. Biol. 2005, 42, 130–140. [Google Scholar] [CrossRef]
- Watts-Williams, S.J.; Cavagnaro, T.R. Arbuscular mycorrhizal fungi increase grain zinc concentration and modify the expression of root ZIP transporter genes in a modern barley (Hordeum vulgare) cultivar. Plant Sci. 2018, 274, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Ait Lahmidi, N.; Courty, P.E.; Brulé, D.; Chatagnier, O.; Arnould, C.; Doidy, J.; Berta, G.; Lingua, G.; Wipf, D.; Bonneau, L. Sugar exchanges in arbuscular mycorrhiza: RiMST5 and RiMST6, two novel Rhizophagus irregularis monosaccharide transporters, are involved in both sugar uptake from the soil and from the plant partner. Plant Physiol. Biochem. 2016, 107, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Bago, B.; Zipfel, W.; Williams, R.M.; Jun, J.; Arreola, R.; Lammers, P.J.; Pfeffer, P.E.; Shachar-Hill, Y. Translocation and Utilization of Fungal Storage Lipid in the Arbuscular Mycorrhizal Symbiosis. Plant Physiol. 2002, 128, 108–124. [Google Scholar] [CrossRef] [PubMed]
- Wewer, V.; Brands, M.; Dörmann, P. Fatty acid synthesis and lipid metabolism in the obligate biotrophic fungus Rhizophagus irregularis during mycorrhization of Lotus japonicus. Plant J. 2014, 79, 398–412. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; San Clemente, H.; Roy, S.; Bécard, G.; Zhao, B.; Roux, C. A survey of the gene repertoire of Gigaspora rosea unravels conserved features among Glomeromycota for obligate biotrophy. Front. Microbiol. 2016, 7, 233. [Google Scholar] [CrossRef]
- Bravo, A.; Brands, M.; Wewer, V.; Dormann, P.; Harrison, M.J. Arbuscular mycorrhiza- specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol. 2017, 214, 1631–1645. [Google Scholar] [CrossRef]
- Radhakrishnan, G.V.; Keller, J.; Rich, M.K.; Vernié, T.; Mbadinga Mbadinga, D.L.; Vigneron, N.; Cottret, L.; Clemente, H.S.; Libourel, C.; Cheema, J.; et al. An Ancestral Signalling Pathway Is Conserved in Intracellular Symbioses-Forming Plant Lineages. Nat. Plants 2020, 6, 280–289. [Google Scholar] [CrossRef]
- Paszkowski, U.; Kroken, U.; Roux, C.; Briggs, S.P. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2002, 99, 13324–13329. [Google Scholar] [CrossRef]
- Glassop, D.; Smith, S.E.; Smith, F.W. Cereal phosphate transporters associated with the mycorrhizal pathway of phosphate uptake into roots. Planta 2005, 222, 688–698. [Google Scholar] [CrossRef]
- Bulgarelli, R.G.; De Oliveira, V.H.; De Andrade, S.A.L. Arbuscular Mycorrhizal Symbiosis Alters the Expression of PHT1 Phosphate Transporters in Roots and Nodules of P-Starved Soybean Plants. Theor. Exp. Plant Physiol. 2020, 32, 243–253. [Google Scholar] [CrossRef]
- Zhao, S.; Chen, A.; Chen, C.; Li, C.; Xia, R.; Wang, X. Transcriptomic analysis reveals the possible roles of sugar metabolism and export for positive mycorrhizal growth responses in soybean. Physiol. Plant. 2019, 166, 712–728. [Google Scholar] [CrossRef] [PubMed]
- Helber, N.; Wippel, K.; Sauer, N.; Schaarschmidt, S.; Hause, B.; Requena, N. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell 2011, 23, 3812–3823. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Cui, H.; Buer, B.; Vijayakumar, V.; Delaux, P.M.; Junkermann, S.; Bucher, M. Network of GRAS Transcription Factors Involved in the Control of Arbuscule Development in Lotus japonicus. Plant Physiol. 2015, 167, 854–871. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, R.M.; Schaepe, S.; Nübel, D.; Petersen, A.C.; Bertolini, M.; Vasilev, J.; Küster, H.; Hohnjec, N. Insights into the Complex Role of GRAS Transcription Factors in the Arbuscular Mycorrhiza Symbiosis. Sci. Rep. 2019, 9, 3360. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.B.; Pratap, A.; Miyahara, A.; Zhou, L.; Bornemann, S.; Morris, R.J.; Oldroyd, G.E.D. Calcium/Calmodulin-Dependent Protein Kinase Is Negatively and Positively Regulated by Calcium, Providing a Mechanism for Decoding Calcium Responses during Symbiosis Signaling. Plant Cell 2013, 25, 5053–5066. [Google Scholar] [CrossRef]
- Gong, X.; Jensen, E.; Bucerius, S.; Parniske, M. A CCaMK/Cyclops response element in the promoter of Lotus japonicus calcium-binding protein 1 (CBP1) mediates transcriptional activation in root symbioses. New Phytol. 2022, 235, 1196–1211. [Google Scholar] [CrossRef]
- Camps, C.; Jardinaud, M.F.; Rengel, D.; Carrère, S.; Hervé, C.; Debellé, F.; Gamas, P.; Bensmihen, S.; Gough, C. Combined Genetic and Transcriptomic Analysis Reveals Three Major Signalling Pathways Activated by Myc-LCOs in Medicago truncatula. New Phytol. 2015, 208, 224–240. [Google Scholar] [CrossRef]
- Delaux, P.M.; Bécard, G.; Combier, J.P. NSP1 Is a Component of the Myc Signaling Pathway. New Phytol. 2013, 199, 59–65. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, H.; Luo, D.; Yu, N.; Dong, W.; Wang, C.; Zhang, X.; Dai, H.; Yang, J.; Wang, E. DELLA Proteins Are Common Components of Symbiotic Rhizobial and Mycorrhizal Signalling Pathways. Nat. Commun. 2016, 7, 12433. [Google Scholar] [CrossRef]
- Shi, Z.; Zhang, J.; Lu, S.; Li, Y.; Wang, F. Arbuscular Mycorrhizal Fungi Improve the Performance of Sweet Sorghum Grown in a Mo-Contaminated Soil. J. Fungi 2020, 6, 44. [Google Scholar] [CrossRef]
- Das, D.; Gutjahr, C. Old Dog, New Trick: The PHR-SPX System Regulates Arbuscular Mycorrhizal Symbiosis. Mol. Plant 2022, 15, 225–227. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Wu, Y.N.; Liu, C.C.; Liu, Y.N.; Tian, L.; Cheng, J.F.; Pan, Z.; Wang, D.; Wang, B. OsADK1, a Novel Kinase Regulating Arbuscular Mycorrhizal Symbiosis in Rice. New Phytol. 2022, 234, 256–268. [Google Scholar] [CrossRef]
- Gu, L.; Zhao, M.; Ge, M.; Zhu, S.; Cheng, B.; Li, X. Transcriptome Analysis Reveals Comprehensive Responses to Cadmium Stress in Maize Inoculated with Arbuscular Mycorrhizal Fungi. Ecotoxicol. Environ. Saf. 2019, 186, 109744. [Google Scholar] [CrossRef]
- Lauressergues, D.; Delaux, P.M.; Formey, D.; Lelandais-Brière, C.; Fort, S.; Cottaz, S.; Bécard, G.; Niebel, A.; Roux, C.; Combier, J.P. The microRNA miR171h Modulates Arbuscular Mycorrhizal Colonization of Medicago truncatula by Targeting NSP2. Plant J. 2012, 72, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Couzigou, J.M.; Lauressergues, D.; André, O.; Gutjahr, C.; Guillotin, B.; Bécard, G.; Combier, J.P. Positive Gene Regulation by a Natural Protective miRNA Enables Arbuscular Mycorrhizal Symbiosis. Cell Host Microbe 2017, 21, 106–112. [Google Scholar] [CrossRef]
- Shtark, O.Y.; Sulima, A.S.; Zhernakov, A.I.; Kliukova, M.S.; Fedorina, J.V.; Pinaev, A.G.; Kryukov, A.A.; Akhtemova, G.A.; Tikhonovich, I.A.; Zhukov, V.A. Arbuscular mycorrhiza development in pea (Pisum sativum L.) mutants impaired in five early nodulation genes including putative orthologs of NSP1 and NSP2. Symbiosis 2016, 68, 129–144. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, S.; Liu, F.; Wang, W.; Wang, X.; Han, G.; Cheng, B. Identification of Arbuscular Mycorrhiza Fungi Responsive microRNAs and Their Regulatory Network in Maize. Int. J. Mol. Sci. 2018, 19, 3201. [Google Scholar] [CrossRef]
- López-Raez, J.A.; Fernández, I.; García, J.M.; Berrio, E.; Bonfante, P.; Walter, M.H.; Pozo, M.J. Differential spatio-temporal expression of carotenoid cleavage dioxygenases regulates apocarotenoid fluxes during AM symbiosis. Plant Sci. 2015, 230, 59–69. [Google Scholar] [CrossRef]
- Balestrini, R.; Lanfranco, L. Fungal and plant gene expression in arbuscular mycorrhizal symbiosis. Mycorrhiza 2006, 16, 509–524. [Google Scholar] [CrossRef]
- Marquez, N.; Giachero, M.L.; Gallou, A.; Debat, H.J.; Declerck, S.; Ducasse, D.A. Transcriptome analysis of mycorrhizal and nonmycorrhizal soybean plantlets upon infection with Fusarium virguliforme, one causal agent of sudden death syndrome. Plant Pathol. 2019, 68, 470–480. [Google Scholar] [CrossRef]
- Bonfante, P.; Requena, N. Dating in the dark: How roots respond to fungal signals to establish arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 2011, 14, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.J. Signaling in the arbuscular mycorrhizal symbiosis. Annu. Rev. Microbiol. 2005, 59, 19–42. [Google Scholar] [CrossRef] [PubMed]
- Güimil, S.; Chang, H.S.; Zhu, T.; Sesma, A.; Osbourn, A.; Roux, C.; Ioannidis, V.; Oakeley, E.J.; Docquier, M.; Descombes, P.; et al. Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc. Natl. Acad. Sci. USA 2005, 102, 8066–8070. [Google Scholar] [CrossRef] [PubMed]
- Roth, R.; Chiapello, M.; Montero, H.; Gehrig, P.; Grossmann, J.; O’Holleran, K.; Hartken, D.; Walters, F.; Yang, S.Y.; Hillmer, S.; et al. A rice Serine/Threonine receptor-like kinase regulates arbuscular mycorrhizal symbiosis at the peri-arbuscular membrane. Nat. Commun. 2018, 9, 4677. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.; Liu, G.; Zhang, Z.; Lu, X.; Liang, C.; Tian, J. Mechanisms underlying soybean response to phosphorus deficiency through integration of omics analysis. Int. J. Mol. Sci. 2022, 23, 4592. [Google Scholar] [CrossRef]
- Vangelisti, A.; Natali, L.; Bernardi, R.; Sbrana, C.; Turrini, A.; Hassani-Pak, K.; Hughes, D.; Cavallini, A.; Giovannetti, M.; Giordani, T. Transcriptome changes induced by arbuscular mycorrhizal fungi in sunflower (Helianthus annuus L.) roots. Sci. Rep. 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Jain, M. Next-generation sequencing technologies for gene expression profiling in plants. Brief. Funct. Genom. 2012, 11, 63–70. [Google Scholar] [CrossRef]
- Kogenaru, S.; Qing, Y.; Guo, Y.; Wang, N. RNA-seq and microarray complement each other in transcriptome profiling. BMC Genom. 2012, 13, 629. [Google Scholar] [CrossRef]
- Fiorilli, V.; Vallino, M.; Biselli, C.; Faccio, A.; Bagnaresi, P.; Bonfante, P. Host and non-host roots in rice: Cellular and molecular approaches reveal differential responses to arbuscular mycorrhizal fungi. Front. Plant Sci. 2015, 6, 636. [Google Scholar] [CrossRef]
- Schultz, C.J.; Wu, Y.; Baumann, U. A Targeted Bioinformatics Approach Identifies Highly Variable Cell Surface Proteins That Are Unique to Glomeromycotina. Mycorrhiza 2022, 32, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Brechenmacher, L.; Weidmann, S.; Van Tuinen, D.; Chatagnier, O.; Gianinazzi, S.; Franken, P.; Gianinazzi-Pearson, V. Expression Profiling of Up-Regulated Plant and Fungal Genes in Early and Late Stages of Medicago truncatula-Glomus mosseae Interactions. Mycorrhiza 2004, 14, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Salvioli, A.; Bonfante, P. Systems biology and “omics” tools: A cooperation for next- generation mycorrhizal studies. Plant Sci. 2013, 203, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Ran, Z.; Yang, X.; Zhang, Y.; Zhou, J.; Guo, L. Transcriptional Responses for Biosynthesis of Ginsenoside in Arbuscular Mycorrhizal Fungi-Treated Panax quinquefolius L. Seedlings Using RNA-Seq. Plant Growth Regul 2021, 95, 83–96. [Google Scholar] [CrossRef]
- Schliesky, S.; Gowik, U.; Weber, A.P.; Brautigam, A. RNA-seq assembly—Are we there yet? Front. Plant Sci 2012, 3, 220. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Sun, M.; van Velzen, R.; Ji, C.; Zheng, Z.; Limpens, E.; Bisseling, T.; Deng, X.; Xiao, S.; Pan, Z. Comparative transcriptome analysis of Poncirus trifoliata identifies a core set of genes involved in arbuscular mycorrhizal symbiosis. J. Exp. Bot. 2018, 69, 5255–5264. [Google Scholar] [CrossRef]
- Bravo, A.; York, T.; Pumplin, N.; Mueller, L.A.; Harrison, M.J. Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nat. Plants 2016, 2, 15208. [Google Scholar] [CrossRef] [PubMed]
- Koegel, S.; Mieulet, D.; Baday, S.; Chatagnier, O.; Lehmann, M.F.; Wiemken, A.; Courty, P.E. Phylogenetic, structural, and functional characterization of AMT3;1, an ammonium transporter induced by mycorrhization among model grasses. Mycorrhiza 2017, 27, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Yang, Y.; Futrell, S.; Kelly, E.A.; Lorts, C.M.; Nebie, B.; Runo, S.; Yang, J.; Alvarez, S.; Lasky, J.R.; et al. CRISPR/Cas9-Mediated Mutagenesis of Carotenoid Cleavage Dioxygenase (CCD) Genes in Sorghum Alters Strigolactone Biosynthesis and Plant Biotic Interactions. Phytobiomes J. 2023, 7, 339–351. [Google Scholar] [CrossRef]
- Liu, Y.-N.; Liu, C.-C.; Zhu, A.-Q.; Niu, K.-X.; Guo, R.; Tian, L.; Wu, Y.-N.; Sun, B.; Wang, B. OsRAM2 function in lipid biosynthesis is required for arbuscular mycorrhizal symbiosis in rice. Mol. Plant-Microbe Interact. 2022, 35, 187–199. [Google Scholar] [CrossRef]
- Bucher, M.; Hause, B.; Krajinski, F.; Küster, H. Through the doors of perception to function in arbuscular mycorrhizal symbioses. New Phytol. 2014, 204, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Rabara, R.C.; Negi, S. AMF: The future prospect for sustainable agriculture. Physiol. Mol. Plant Pathol. 2018, 102, 36–45. [Google Scholar] [CrossRef]
- Paszkowski, U. A journey through signaling in arbuscular mycorrhizal symbioses. New Phytol. 2006, 172, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Lanfranco, L.; Fiorilli, V.; Gutjahr, C. Partner communication and role of nutrients in the arbuscular mycorrhizal symbiosis. New Phytol. 2018, 220, 1031–1046. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, K.; Mori, N.; Sato, T.; Yoda, A.; Xie, X.; Okamoto, M.; Iwanaga, M.; Ohnishi, T.; Nishiwaki, H.; Asami, T.; et al. Conversion of carlactone to carlactonoic acid is a conserved function of MAX1 homologs in strigolactone biosynthesis. New Phytol. 2018, 218, 1522–1533. [Google Scholar] [CrossRef] [PubMed]
- Arite, T.; Kameoka, H.; Kyozuka, J. Strigolactone Positively Controls Crown Root Elongation in Rice. J. Plant Growth Regul. 2012, 31, 165–172. [Google Scholar] [CrossRef]
- Dutta, S.; Muthusamy, V.; Zunjare, R.U. Analysis of Paralogous Genes of Carotenoid Dioxygenase Affecting Carotenoid Biosynthesis Pathway in Maize (Zea mays L.). J. Pharmacogn. Phytochem. 2019, 8, 524–530. [Google Scholar]
- Kyozuka, J.; Nomura, T. Origins and evolution of the dual functions of strigolactones as rhizosphere signaling molecules and plant hormones. Curr. Opin. Plant Biol. 2022, 65, 102154. [Google Scholar] [CrossRef]
- Arite, T.; Umehara, M.; Ishikawa, S.; Hanada, A.; Maekawa, M.; Yamaguchi, S.; Kyozuka, J. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009, 50, 1416–1424. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.D.; Cousins, D.R.; Jackson, K.J.; Liu, C. Signaling at the root surface: The role of cutin monomers in mycorrhization. Mol. Plant 2013, 6, 1381–1383. [Google Scholar] [CrossRef]
- Heredia-Guerrero, J.A.; Benítez, J.J.; Heredia, A. Self-assembled polyhydroxy fatty acids vesicles: A mechanism for plant cutin synthesis. BioEssays 2008, 30, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Bonfante, P.; Genre, A. Arbuscular mycorrhizal dialogues: Do you speak ‘plantish’ or ‘fungish’? Trends Plant Sci. 2015, 20, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Rush, T.A.; Puech-Pagès, V.; Bascaules, A.; Jargeat, P.; Maillet, F.; Haouy, A.; Maës, A.Q.; Carriel, C.C.; Khokhani, D.; Keller-Pearson, M.; et al. Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat. Commun. 2020, 11, 3897. [Google Scholar] [CrossRef]
- Sun, J.; Miller, J.B.; Granqvist, E.; Wiley-Kalil, A.; Gobbato, E.; Maillet, F.; Cottaz, S.; Samain, E.; Venkateshwaran, M.; Fort, S.; et al. Activation of Symbiosis Signaling by Arbuscular Mycorrhizal Fungi in Legumes and Rice. Plant Cell 2015, 27, 823–838. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zang, X.; Zhou, J. Synthetic Biology: A Powerful Booster for Future Agriculture. Adv. Agrochem 2022, 1, 7–11. [Google Scholar] [CrossRef]
- Chiu, C.H.; Choi, J.; Paszkowski, U. Independent signalling cues underpin arbuscular mycorrhizal symbiosis and large lateral root induction in rice. New Phytol. 2018, 217, 552–557. [Google Scholar] [CrossRef]
- Jung, S.C.; Martinez-Medina, A.; Lopez-Raez, J.A.; Pozo, M.J. Mycorrhiza-Induced Resistance and Priming of Plant Defenses. J. Chem. Ecol. 2012, 38, 651–664. [Google Scholar] [CrossRef]
- Oldroyd, G.E. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252. [Google Scholar] [CrossRef] [PubMed]
- Besserer, A.; Puech-Pagès, V.; Kiefer, P.; Gomez-Roldan, V.; Jauneau, A.; Roy, S.; Portais, J.-C.; Roux, C.; Bécard, G.; Séjalon-Delmas, N. Strigolactones Stimulate Arbuscular Mycorrhizal Fungi by Activating Mitochondria. PLOS Biol. 2006, 4, e226. [Google Scholar] [CrossRef]
- Besserer, A.; Becard, G.; Roux, C.; Séjalon-Delmas, N. Role of Mitochondria in the Response of Arbuscular Mycorrhizal Fungi to Strigolactones. Plant Signal. Behav. 2009, 4, 75–77. [Google Scholar] [CrossRef]
- Tsuzuki, S.; Handa, Y.; Takeda, N.; Kawaguchi, M. Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mol. Plant-Microbe Interact. 2016, 29, 277–286. [Google Scholar] [CrossRef]
- Chen, C.; Fan, C.; Gao, M.; Zhu, H. Antiquity and Function of CASTOR and POLLUX, the Twin Ion Channel-Encoding Genes Key to the Evolution of Root Symbioses in Plants. Plant Physiol. 2008, 149, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Gutjahr, C.; Novero, M.; Guether, M.; Montanari, O.; Udvardi, M.; Bonfante, P. Presymbiotic factors released by the arbuscular mycorrhizal fungus Gigaspora margarita induce starch accumulation in Lotus japonicus roots. New Phytol. 2009, 183, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Luo, D.; Zhang, X.; Liu, J.; Wang, W.; Jin, Y.; Dong, W.; Liu, J.; Liu, H.; Yang, W. A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res. 2014, 24, 130. [Google Scholar] [CrossRef]
- Pimprikar, P.; Carbonnel, S.; Paries, M.; Katzer, K.; Klingl, V.; Bohmer, M.J.; Karl, L.; Floss, D.S.; Harrison, M.J.; Parniske, M. A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr. Biol. 2016, 26, 987–998. [Google Scholar] [CrossRef]
- Gobbato, E.; Marsh, J.F.; Vernié, T.; Wang, E.; Maillet, F.; Kim, J.; Miller, J.B.; Sun, J.; Bano, S.A.; Ratet, P. A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 2012, 22, 2236–2241. [Google Scholar] [CrossRef]
- Genre, A.; Chabaud, M.; Timmers, T.; Bonfante, P.; Barker, D.G. Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 2005, 17, 3489–3499. [Google Scholar] [CrossRef]
- Siciliano, V.; Genre, A.; Balestrini, R.; deWit, P.J.G.M.; Bonfante, P. Pre-Penetration Apparatus Formation During AM Infection Is Associated With a Specific Transcriptome Response in Epidermal Cells. Plant Signal. Behav. 2007, 2, 533–535. [Google Scholar] [CrossRef]
- Chen, C.; Gao, M.; Liu, J.; Zhu, H. Fungal symbiosis in rice requires an ortholog of a legume common symbiosis gene encoding a Ca2+/calmodulin-dependent protein kinase. Plant Physiol. 2007, 145, 1619–1628. [Google Scholar] [CrossRef]
- Genre, A.; Ortu, G.; Bertoldo, C.; Martino, E.; Bonfante, P. Biotic and Abiotic Stimulation of Root Epidermal Cells Reveals Common and Specific Responses to Arbuscular Mycorrhizal Fungi. Plant Physiol. 2009, 149, 1424–1434. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, B.; Jin, R.; Hou, L.; Zhang, X.; Dai, H.; Yu, N.; Wang, E. A phosphate starvation response-regulated receptor-like kinase, OsADK1, is required for mycorrhizal symbiosis and phosphate starvation responses. New Phytol. 2022, 236, 2282–2293. [Google Scholar] [CrossRef] [PubMed]
- Salmeron-Santiago, I.A.S.; Martinez-Trujillo, M.; Valdez-Alarcon, J.J.; Pedraza-Santos, M.E.; Santoyo, G.; Pozo, M.J.; Chavez-Barcenas, A.T. An updated review on the modulation of carbon partitioning and allocation in arbuscular mycorrhizal plants. Microorganisms 2022, 10, 75. [Google Scholar] [CrossRef] [PubMed]
- Gaude, N.; Bortfeld, S.; Duensing, N.; Lohse, M.; Krajinski, F. Arbuscule-containing and non-colonized cortical cells of mycorrhizal roots undergo extensive and specific reprogramming during arbuscular mycorrhizal development. Plant J. 2012, 69, 510–528. [Google Scholar] [CrossRef] [PubMed]
- Manck-Gotzenberger, J.; Requena, N. Arbuscular mycorrhiza Symbiosis Induces a Major Transcriptional Reprogramming of the Potato SWEET Sugar Transporter Family. Front. Plant Sci. 2016, 7, 14. [Google Scholar] [CrossRef]
- Bécard, G.; Kosuta, S.; Tamasloukht, M.; Séjalon-Delmas, N.; Roux, C. Partner Communication in the Arbuscular Mycorrhizal Interaction. Can. J. Bot. 2004, 82, 1186–1197. [Google Scholar] [CrossRef]
- Kameoka, H.; Gutjahr, C. Functions of Lipids in Development and Reproduction of Arbuscular Mycorrhizal Fungi. Plant Cell Physiol. 2022, 63, 1356–1365. [Google Scholar] [CrossRef]
- Keymer, A.; Pimprikar, P.; Wewer, V.; Huber, C.; Brands, M.; Bucerius, S.L.; Delaux, P.-M.; Klingl, V.; Röpenack-Lahaye, E.V.; Wang, T.L.; et al. Lipid Transfer from Plants to Arbuscular Mycorrhiza Fungi. eLife 2017, 6, e29107. [Google Scholar] [CrossRef]
- Jiang, Y.; Xie, Q.; Wang, W.; Yang, J.; Zhang, X.; Yu, N.; Zhou, Y.; Wang, E. Medicago AP2-domain transcription factor WRI5a is a master regulator of lipid biosynthesis and transfer during mycorrhizal symbiosis. Mol. Plant 2018, 11, 1344–1359. [Google Scholar] [CrossRef]
- Smith, F.A.; Smith, F.A. Roles of Arbuscular Mycorrhizas in Plant Nutrition and Growth: New Paradigms from Cellular to Ecosystem Scales. Annu. Rev. Plant. Biol. 2011, 62, 227–250. [Google Scholar] [CrossRef]
- Rausch, C.; Bucher, M. Molecular mechanisms of phosphate transport in plants. Planta 2002, 216, 23–37. [Google Scholar] [CrossRef]
- Wang, D.; Lv, S.; Jiang, P.; Li, Y. Roles, regulation, and agricultural application of plant phosphate transporters. Front. Plant Sci. 2017, 8, 817. [Google Scholar] [CrossRef]
- Nagy, R.; Vasconcelos, M.J.; Zhao, S.; McElver, J.; Bruce, W.; Amrhein, N.; Raghothama, K.G.; Bucher, M. Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant Biol. 2006, 8, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Ceasar, S.A.; Hodge, A.; Baker, A.; Baldwin, S.A. Phosphate Concentration and Arbuscular Mycorrhizal Colonisation Influence the Growth, Yield and Expression of Twelve PHT1 Family Phosphate Transporters in Foxtail Millet (Setaria italica). PLoS ONE 2014, 9, e108459. [Google Scholar] [CrossRef]
- Walder, F.; Brule, D.; Koegel, S.; Wiemken, A.; Boller, T.; Pierre-Emmanuel, C. Plant phosphorus acquisition in a common mycorrhizal network: Regulation of phosphate transporter genes of the Pht1 family in sorghum and flax. New Phytol. 2015, 205, 1632–1645. [Google Scholar] [CrossRef] [PubMed]
- Tamura, Y.; Kobae, Y.; Mizuno, T.; Hata, S. Identification and Expression Analysis of Arbuscular Mycorrhiza-Inducible Phosphate Transporter Genes of Soybean. Biosci. Biotechnol. Biochem. 2012, 76, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, J.; Zhao, J.; Tian, J.; Liao, H. Control of Phosphate Homeostasis through Gene Regulation in Crops. Curr. Opin. Plant Biol. 2014, 21, 59–66. [Google Scholar] [CrossRef]
- Ham, B.-K.; Chen, J.; Yan, Y.; Lucas, W.J. Insights into Plant Phosphate Sensing and Signaling. Curr. Opin. Biotechnol. 2018, 49, 1–9. [Google Scholar] [CrossRef]
- Nagy, R.; Drissner, D.; Amrhein, N.; Jakobsen, I.; Bucher, M. Mycorrhizal Phosphate Uptake Pathway in Tomato Is Phosphorus-repressible and Transcriptionally Regulated. New Phytol. 2009, 181, 950–959. [Google Scholar] [CrossRef]
- Tobar, R.; Azcón, R.; Barea, J.M. Improved Nitrogen Uptake and Transport from 15 N-labelled Nitrate by External Hyphae of Arbuscular Mycorrhiza under Water-stressed Conditions. New Phytol. 1994, 126, 119–122. [Google Scholar] [CrossRef]
- Govindarajulu, M.; Pfeffer, P.E.; Jin, H.; Abubaker, J.; Douds, D.D.; Allen, J.W.; Bücking, H.; Lammers, P.J.; Shachar-Hill, Y. Nitrogen Transfer in the Arbuscular Mycorrhizal Symbiosis. Nature 2005, 435, 819–823. [Google Scholar] [CrossRef]
- Tian, C.; Kasiborski, B.; Koul, R.; Lammers, P.J.; Bücking, H.; Shachar-Hill, Y. Regulation of the Nitrogen Transfer Pathway in the Arbuscular Mycorrhizal Symbiosis: Gene Characterization and the Coordination of Expression with Nitrogen Flux. Plant Physiol. 2010, 153, 1175–1187. [Google Scholar] [CrossRef]
- Hui, J.; An, X.; Li, Z.; Neuhäuser, B.; Ludewig, U.; Wu, X.; Schulze, W.X.; Chen, F.; Feng, G.; Lambers, H.; et al. The Mycorrhiza-Specific Ammonium Transporter ZmAMT3;1 Mediates Mycorrhiza-Dependent Nitrogen Uptake in Maize Roots. Plant Cell 2022, 34, 4066–4087. [Google Scholar] [CrossRef] [PubMed]
- Guttenberger, M. Arbuscules of Vesicular-Arbuscular Mycorrhizal Fungi Inhabit an Acidic Compartment within Plant Roots. Planta 2000, 211, 299–304. [Google Scholar] [CrossRef]
- Ariz, I.; Boeckstaens, M.; Gouveia, C.; Martins, A.P.; Sanz-Luque, E.; Fernández, E.; Soveral, G.; Von Wirén, N.; Marini, A.M.; Aparicio-Tejo, P.M.; et al. Nitrogen Isotope Signature Evidences Ammonium Deprotonation as a Common Transport Mechanism for the AMT-Mep-Rh Protein Superfamily. Sci. Adv. 2018, 4, eaar3599. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Yano, K. Nitrogen Delivery to Maize via Mycorrhizal Hyphae Depends on the Form of N Supplied. Plant Cell Environ. 2005, 28, 1247–1254. [Google Scholar] [CrossRef]
- Pérez-Tienda, J.; Corrêa, A.; Azcón-Aguilar, C.; Ferrol, N. Transcriptional Regulation of Host Transporters and GS/GOGAT Pathway in Arbuscular Mycorrhizal Rice Roots. Plant Physiol. Biochem. 2014, 75, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.; Wang, S.; Cui, M.; Liu, J.; Chen, A.; Xu, G. Phytohormones Regulate the Development of Arbuscular Mycorrhizal Symbiosis. IJMS 2018, 19, 3146. [Google Scholar] [CrossRef] [PubMed]
- Lanfranco, L.; Fiorilli, V.; Venice, F.; Bonfante, P. Strigolactones Cross the Kingdoms: Plants, Fungi, and Bacteria in the Arbuscular Mycorrhizal Symbiosis. J. Exp. Bot. 2018, 69, 2175–2188. [Google Scholar] [CrossRef]
- Foo, E.; Ferguson, B.J.; Reid, J.B. Common and Divergent Roles of Plant Hormones in Nodulation and Arbuscular Mycorrhizal Symbioses. Plant Signal. Behav. 2014, 9, e29593. [Google Scholar] [CrossRef]
- Müller, L.M.; Harrison, M.J. Phytohormones, miRNAs, and Peptide Signals Integrate Plant Phosphorus Status with Arbuscular Mycorrhizal Symbiosis. Curr. Opin. Plant Biol. 2019, 50, 132–139. [Google Scholar] [CrossRef]
- Sigalas, P.P.; Buchner, P.; Thomas, S.G.; Jamois, F.; Arkoun, M.; Yvin, J.C.; Bennett, M.J.; Hawkesford, M.J. Nutritional and tissue-specific regulation of cytochrome P450 CYP711A MAX1 homologues and strigolactone biosynthesis in wheat. J. Exp. Bot. 2023, 74, 1890–1910. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, K.; Xie, X.; Kim, H.I.; Kisugi, T.; Nomura, T.; Sekimoto, H.; Yokota, T.; Yoneyama, K. How Do Nitrogen and Phosphorus Deficiencies Affect Strigolactone Production and Exudation? Planta 2012, 235, 1197–1207. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Muszynska, A.; Gruszka, D. The Role of Strigolactones in Nutrient-Stress Responses in Plants. IJMS 2013, 14, 9286–9304. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Li, W.; Burritt, D.J.; Tian, H.; Zhang, H.; Liang, X.; Miao, Y.; Mostofa, M.G.; Tran, L.-S.P. Strigolactones Interact with Other Phytohormones to Modulate Plant Root Growth and Development. Crop J. 2022, 10, 1517–1527. [Google Scholar] [CrossRef]
- Tariq, A.; Ullah, I.; Sardans, J.; Graciano, C.; Mussarat, S.; Ullah, A.; Zeng, F.; Wang, W.; Al-Bakre, D.A.; Ahmed, Z.; et al. Strigolactones Can Be a Potential Tool to Fight Environmental Stresses in Arid Lands. Environ. Res. 2023, 229, 115966. [Google Scholar] [CrossRef]
- Kobae, Y.; Kameoka, H.; Sugimura, Y.; Saito, K.; Ohtomo, R.; Fujiwara, T.; Kyozuka, J. Strigolactone Biosynthesis Genes of Rice Are Required for the Punctual Entry of Arbuscular Mycorrhizal Fungi into the Roots. Plant Cell Physiol. 2018, 59, 544–553. [Google Scholar] [CrossRef]
- Al-Babili, S.; Bouwmeester, H.J. Strigolactones, a Novel Carotenoid-Derived Plant Hormone. Annu. Rev. Plant Biol. 2015, 66, 161–186. [Google Scholar] [CrossRef]
- Fiorilli, V.; Wang, J.Y.; Bonfante, P.; Lanfranco, L.; Al-Babili, S. Apocarotenoids: Old and New Mediators of the Arbuscular Mycorrhizal Symbiosis. Front. Plant Sci. 2019, 10, 1186. [Google Scholar] [CrossRef]
- Gamir, J.; Torres-Vera, R.; Rial, C.; Berrio, E.; De Souza Campos, P.M.; Varela, R.M.; Macías, F.A.; Pozo, M.J.; Flors, V.; López-Ráez, J.A. Exogenous Strigolactones Impact Metabolic Profiles and Phosphate Starvation Signalling in Roots. Plant Cell Environ. 2020, 43, 1655–1668. [Google Scholar] [CrossRef]
- Liu, W.; Kohlen, W.; Lillo, A.; Op Den Camp, R.; Ivanov, S.; Hartog, M.; Limpens, E.; Jamil, M.; Smaczniak, C.; Kaufmann, K.; et al. Strigolactone Biosynthesis in Medicago truncatula and Rice Requires the Symbiotic GRAS-Type Transcription Factors NSP1 and NSP2. Plant Cell 2011, 23, 3853–3865. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Madhuvanthi, R.; Karthikeyan, A.S.; Raghothama, K.G. Phosphate Starvation Responses and Gibberellic Acid Biosynthesis Are Regulated by the MYB62 Transcription Factor in Arabidopsis. Mol. Plant 2009, 2, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Guo, F.; Zhang, J.; Yang, S.; Meng, J.; Geng, Y.; Li, X.; Wan, S. Synergy of Arbuscular Mycorrhizal Symbiosis and Exogenous Ca 2+ Benefits Peanut (Arachis hypogaea L.) Growth through the Shared Hormone and Flavonoid Pathway. Sci Rep. 2019, 9, 16281. [Google Scholar] [CrossRef]
- Floss, D.S.; Gomez, S.K.; Park, H.-J.; MacLean, A.M.; Müller, L.M.; Bhattarai, K.K.; Lévesque-Tremblay, V.; Maldonado-Mendoza, I.E.; Harrison, M.J. A Transcriptional Program for Arbuscule Degeneration during AM Symbiosis Is Regulated by MYB1. Curr. Biol. 2017, 27, 1206–1212. [Google Scholar] [CrossRef]
- Nakamura, H.; Xue, Y.-L.; Miyakawa, T.; Hou, F.; Qin, H.-M.; Fukui, K.; Shi, X.; Ito, E.; Ito, S.; Park, S.-H.; et al. Molecular Mechanism of Strigolactone Perception by DWARF14. Nat. Commun. 2013, 4, 2613. [Google Scholar] [CrossRef] [PubMed]
- French, K.E. Engineering Mycorrhizal Symbioses to Alter Plant Metabolism and Improve Crop Health. Front. Microbiol. 2017, 8, 1403. [Google Scholar] [CrossRef] [PubMed]
- Campos, C.; Carvalho, M.; Brígido, C.; Goss, M.J.; Nobre, T. Symbiosis Specificity of the Preceding Host Plant Can Dominate but Not Obliterate the Association Between Wheat and Its Arbuscular Mycorrhizal Fungal Partners. Front. Microbiol. 2018, 9, 2920. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, L.; Yu, D.; Xu, K.; Zhang, J.; Li, X.; Wang, P.; Chen, G.; Liu, Z.; Peng, C.; et al. Integrative Analysis of the Wheat PHT1 Gene Family Reveals A Novel Member Involved in Arbuscular Mycorrhizal Phosphate Transport and Immunity. Cells 2019, 8, 490. [Google Scholar] [CrossRef]
- Bucher, M. Functional Biology of Plant Phosphate Uptake at Root and Mycorrhiza Interfaces. New Phytol. 2007, 173, 11–26. [Google Scholar] [CrossRef]
- Gutjahr, C.; Parniske, M. Cell and Developmental Biology of Arbuscular Mycorrhiza Symbiosis. Annu. Rev. Cell Dev. Biol. 2013, 29, 593–617. [Google Scholar] [CrossRef]
- Moradi Tarnabi, Z.; Iranbakhsh, A.; Mehregan, I.; Ahmadvand, R. Impact of Arbuscular Mycorrhizal Fungi (AMF) on Gene Expression of Some Cell Wall and Membrane Elements of Wheat (Triticum aestivum L.) under Water Deficit Using Transcriptome Analysis. Physiol. Mol. Biol. Plants 2020, 26, 143–162. [Google Scholar] [CrossRef]
- Saia, S.; Rappa, V.; Ruisi, P.; Abenavoli, M.R.; Sunseri, F.; Giambalvo, D.; Frenda, A.S.; Martinelli, F. Soil Inoculation with Symbiotic Microorganisms Promotes Plant Growth and Nutrient Transporter Genes Expression in Durum Wheat. Front. Plant Sci. 2015, 6, 815. [Google Scholar] [CrossRef]
- Kabir, A.H.; Debnath, T.; Das, U.; Prity, S.A.; Haque, A.; Rahman, M.M.; Parvez, M.S. Arbuscular Mycorrhizal Fungi Alleviate Fe-Deficiency Symptoms in Sunflower by Increasing Iron Uptake and Its Availability along with Antioxidant Defense. Plant Physiol. Biochem. 2020, 150, 254–262. [Google Scholar] [CrossRef]
- Sheteiwy, M.S.; Ali, D.F.I.; Xiong, Y.-C.; Brestic, M.; Skalicky, M.; Hamoud, Y.A.; Ulhassan, Z.; Shaghaleh, H.; AbdElgawad, H.; Farooq, M.; et al. Physiological and Biochemical Responses of Soybean Plants Inoculated with Arbuscular Mycorrhizal Fungi and Bradyrhizobium under Drought Stress. BMC Plant Biol. 2021, 21, 195. [Google Scholar] [CrossRef]
- Liao, Q.; Zhou, T.; Yao, J.; Han, Q.; Song, H.; Guan, C.; Hua, Y.; Zhang, Z. Genome-Scale Characterization of the Vacuole Nitrate Transporter Chloride Channel (CLC) Genes and Their Transcriptional Responses to Diverse Nutrient Stresses in Allotetraploid Rapeseed. PLoS ONE 2018, 13, e0208648. [Google Scholar] [CrossRef]
- Fiorilli, V.; Maghrebi, M.; Novero, M.; Votta, C.; Mazzarella, T.; Buffoni, B.; Astolfi, S.; Vigani, G. Arbuscular Mycorrhizal Symbiosis Differentially Affects the Nutritional Status of Two Durum Wheat Genotypes under Drought Conditions. Plants 2022, 11, 804. [Google Scholar] [CrossRef]
- Kitano, H. Systems Biology: A Brief Overview. Science 2002, 295, 1662–1664. [Google Scholar] [CrossRef]
- Selinger, D.W.; Wright, M.A.; Church, G.M. On the Complete Determination of Biological Systems. TRENDS Biotechnol. 2003, 21, 251–254. [Google Scholar] [CrossRef]
- Kitano, H. Systems Biology: Toward System-Level Understanding of Biological Systems. In Foundations of Systems Biology; Kitano, H., Ed.; The MIT Press: Cambridge, MA, USA, 2001; pp. 1–36. [Google Scholar]
- Damiani, I.; Baldacci-cresp, F.; Hopkins, J.; Andrio, E.; Balzergue, S.; Lecomte, P.; Puppo, A.; Abad, P.; Favery, B.; Herouart, D. Plant Genes Involved in Harbouring Symbiotic Rhizobia or Pathogenic Nematodes. New Phytol. 2012, 194, 511–522. [Google Scholar] [CrossRef]
- Aravind, L.; Anantharaman, V.; Zhang, D.; Souza, R.F.d.; Iyer, L.M. Gene Flow and Biological Conflict Systems in the Origin and Evolution of Eukaryotes. Front. Cell. Infect. Microbiol. 2012, 2, 89. [Google Scholar] [CrossRef]
- Franz Lang, B.; Hijri, M. The Complete Glomus intraradices Mitochondrial Genome Sequence—A Milestone in Mycorrhizal Research: Commentary. New Phytol. 2009, 183, 3–6. [Google Scholar] [CrossRef]
- Qin, M.; Zhang, Q.; Pan, J.; Jiang, S.; Liu, Y.; Bahadur, A.; Peng, Z.; Yang, Y.; Feng, H. Effect of Arbuscular Mycorrhizal Fungi on Soil Enzyme Activity Is Coupled with Increased Plant Biomass. Eur. J. Soil Sci. 2020, 71, 84–92. [Google Scholar] [CrossRef]
- Saxena, B.; Shukla, K.; Giri, B. Arbuscular Mycorrhizal Fungi and Tolerance of Salt Stress in Plants. In Arbuscular Mycorrhizas and Stress Tolerance of Plants; Springer: Singapore, 2017; pp. 67–97. ISBN 9789811041150. [Google Scholar]
- Diagne, N.; Ngom, M.; Djighaly, P.I.; Fall, D.; Hocher, V.; Svistoonoff, S. Roles of Arbuscular Mycorrhizal Fungi on Plant Growth and Performance: Importance in Biotic and Abiotic Stressed Regulation. Diversity 2020, 12, 370. [Google Scholar] [CrossRef]
- Fiorilli, V.; Vannini, C.; Ortolani, F.; Garcia-Seco, D.; Chiapello, M.; Novero, M.; Domingo, G.; Terzi, V.; Morcia, C.; Bagnaresi, P.; et al. Omics Approaches Revealed How Arbuscular Mycorrhizal Symbiosis Enhances Yield and Resistance to Leaf Pathogen in Wheat. Sci. Rep. 2018, 8, 9625. [Google Scholar] [CrossRef]
- George, N.P.; Ray, J.G. The inevitability of arbuscular mycorrhiza for sustainability in organic agriculture—A critical review. Front. Sustain. Food Syst. 2023, 7, 1124688. [Google Scholar] [CrossRef]
- Zhang, Y.; Malzahn, A.A.; Sretenovic, S.; Qi, Y. The Emerging and Uncultivated Potential of CRISPR Technology in Plant Science. Nat. Plants 2019, 5, 778–794. [Google Scholar] [CrossRef]

| AMF Strains | Host Plant | Genetic Factor | Main Function | Ref. |
|---|---|---|---|---|
| R. irregularis | Barrel medic | CCaMK/DMI3 | Calcium signaling | [106] |
| R. irregularis | Rice | OsADK1 | Arbuscule development | [113] |
| R. irregularis | Maize | CERK1 | Pre-symbiotic fungal perception | [157] |
| R. irregularis | Rice | CASTOR and POLLUX | AMF roots penetration/symbiosis | [163] |
| R. irregularis | Rice | OsDMI3 | Pre-penetration apparatus induction | [170] |
| R. irregularis + G. aggregatum | Soybean | GmSWEET6 and GmSWEET15 | Sugar metabolism and transport | [102] |
| R. irregularis | Rice | RAM2 | Lipid transfer | [141] |
| R. irregularis + S. calospora | Barley | HORvu; Pht1;8 | Pi transport | [100] |
| F. mosseae | Millet | SiPHT1;8 and SiPHT1;9 | Pi transport | [184] |
| R. irregularis | Soybean | GmPT10 and GmPT11 | Pi transport | [185] |
| R. irregularis | Rice | OsPT11 | Pi transport | [99] |
| F. mosseae | Soybean | GmAMT4.1 | NH4+ transport | [85] |
| R. irregularis + F. mosseae | Sorghum | SbAMT3;1 and SbAMT4 | NH4+ transport and N transfer | [87] |
| R. irregularis | Maize | ZmAMT3;1 | NH4+ transport | [193] |
| R. irregularis | Rice | OsNPF4,5 | NO3− transport | [88] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Slimani, A.; Ait-El-Mokhtar, M.; Ben-Laouane, R.; Boutasknit, A.; Anli, M.; Abouraicha, E.F.; Oufdou, K.; Meddich, A.; Baslam, M. Molecular and Systems Biology Approaches for Harnessing the Symbiotic Interaction in Mycorrhizal Symbiosis for Grain and Oil Crop Cultivation. Int. J. Mol. Sci. 2024, 25, 912. https://doi.org/10.3390/ijms25020912
Slimani A, Ait-El-Mokhtar M, Ben-Laouane R, Boutasknit A, Anli M, Abouraicha EF, Oufdou K, Meddich A, Baslam M. Molecular and Systems Biology Approaches for Harnessing the Symbiotic Interaction in Mycorrhizal Symbiosis for Grain and Oil Crop Cultivation. International Journal of Molecular Sciences. 2024; 25(2):912. https://doi.org/10.3390/ijms25020912
Chicago/Turabian StyleSlimani, Aiman, Mohamed Ait-El-Mokhtar, Raja Ben-Laouane, Abderrahim Boutasknit, Mohamed Anli, El Faiza Abouraicha, Khalid Oufdou, Abdelilah Meddich, and Marouane Baslam. 2024. "Molecular and Systems Biology Approaches for Harnessing the Symbiotic Interaction in Mycorrhizal Symbiosis for Grain and Oil Crop Cultivation" International Journal of Molecular Sciences 25, no. 2: 912. https://doi.org/10.3390/ijms25020912
APA StyleSlimani, A., Ait-El-Mokhtar, M., Ben-Laouane, R., Boutasknit, A., Anli, M., Abouraicha, E. F., Oufdou, K., Meddich, A., & Baslam, M. (2024). Molecular and Systems Biology Approaches for Harnessing the Symbiotic Interaction in Mycorrhizal Symbiosis for Grain and Oil Crop Cultivation. International Journal of Molecular Sciences, 25(2), 912. https://doi.org/10.3390/ijms25020912








