Analyzing Amylin Aggregation Inhibition Through Quantum Dot Fluorescence Imaging
Abstract
1. Introduction
2. Results
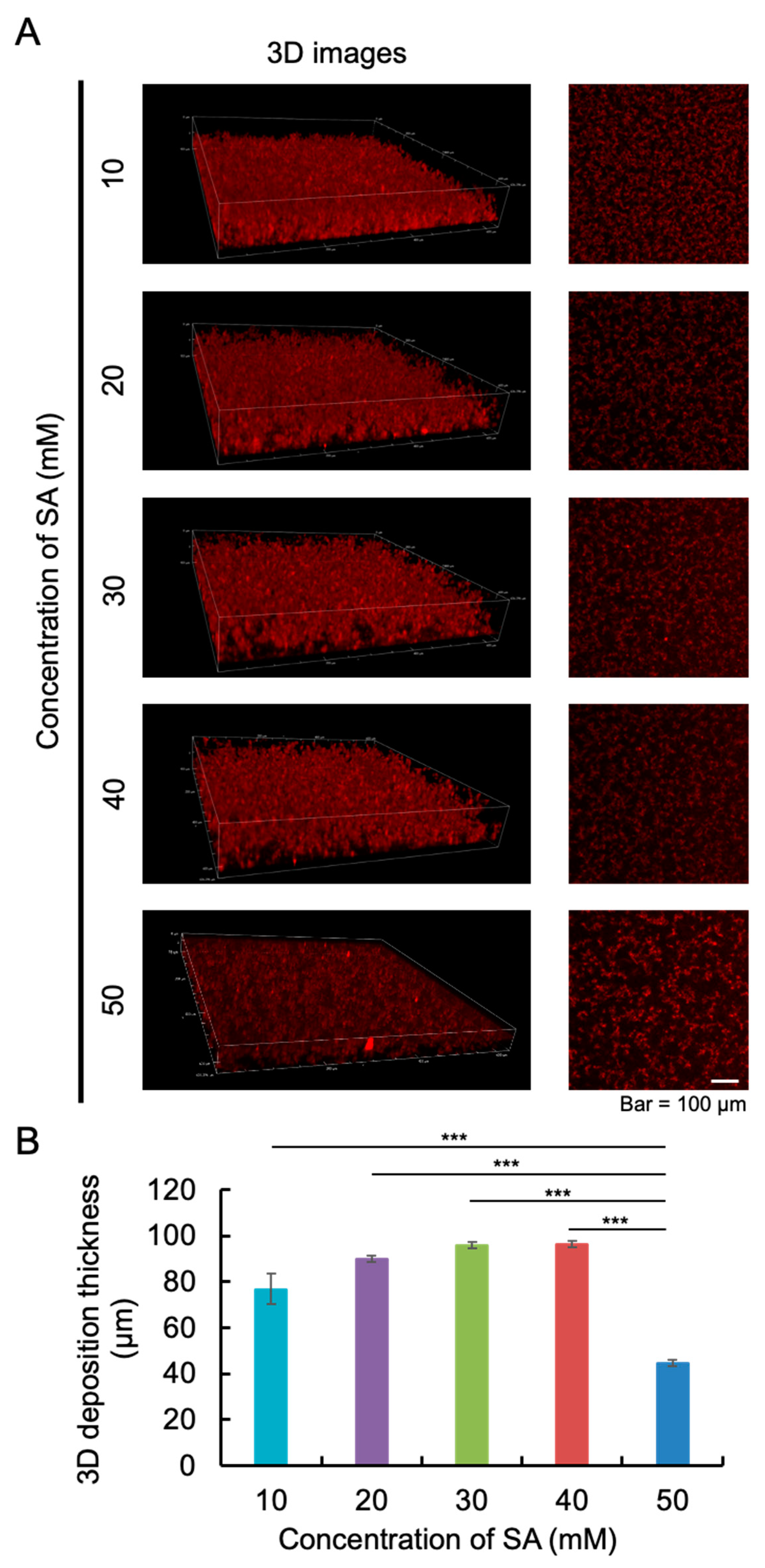
2.1. The Appropriate Concentration of Sodium Acetate (SA) Buffer
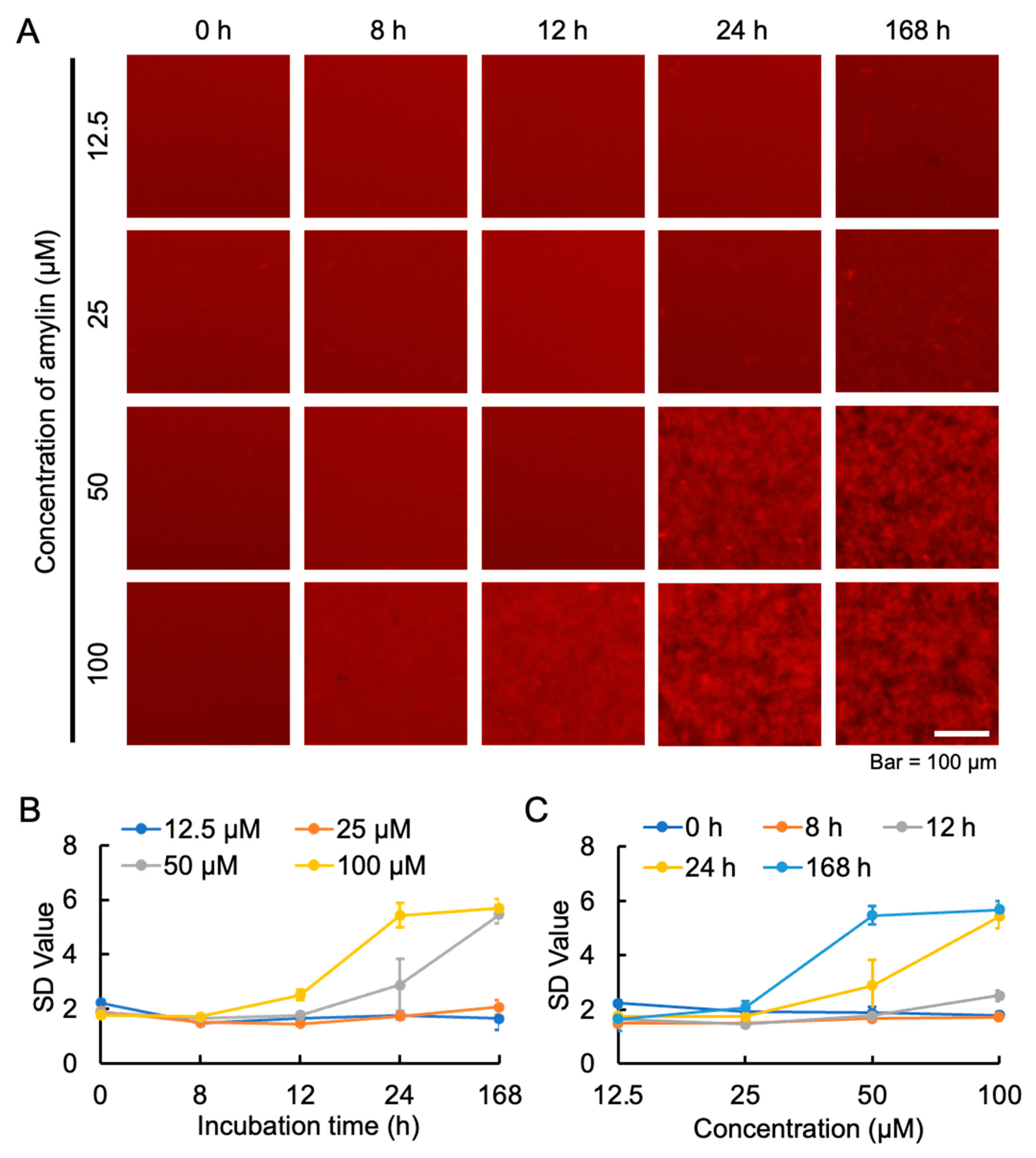
2.2. The Appropriate Concentration of Amylin for Aggregation
2.3. Transmission Electron Microscopy Observations of Amylin Fibrils
2.4. Effect of RA on the Aggregation of Amylin
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation of Amylin
4.3. MSHTS System
4.4. The Appropriate Concentration of SA Buffer for Amylin Aggregation
4.5. The Appropriate Concentration of Amylin for Aggregation
4.6. TEM Observation
4.7. Inhibition of Amylin Aggregation by RA
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, L.; Velander, P.; Brown, A.M.; Wang, Y.; Liu, D.; Bevan, D.R.; Zhang, S.; Xu, B. Rosmarinic acid potently detoxifies amylin amyloid and ameliorates diabetic pathology in a transgenic rat model of type 2 diabetes. ACS Pharmacol. Transl. Sci. 2021, 4, 1322–1337. [Google Scholar] [CrossRef]
- Eisenberg, D.S.; Sawaya, M.R. Structural studies of amyloid proteins at the molecular level. Annu. Rev. Biochem. 2017, 86, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Ali, M.; Zehra, T.; Haider Zaidi, S.A.; Tariq, R. Intermittent fasting: A user-friendly method for type 2 diabetes mellitus. Cureus 2021, 13, e19348. [Google Scholar] [CrossRef]
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.N.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.C.; Liu, Y.; Yu, Z.; Xu, B. Gut-targeted therapies for type 2 diabetes mellitus: A review. World J. Clin. Cases. 2024, 12, 1–8. [Google Scholar] [CrossRef]
- Kanatsuka, A.; Kou, S.; Makino, H. IAPP/amylin and β-cell failure: Implication of the risk factors of type 2 diabetes. Diabetol. Int. 2018, 9, 143–157. [Google Scholar] [CrossRef]
- Zhang, X.X.; Pan, Y.H.; Huang, Y.M.; Zhao, H.L. Neuroendocrine hormone amylin in diabetes. World J. Diabetes 2016, 7, 189–197. [Google Scholar] [CrossRef]
- Hay, D.L.; Chen, S.; Lutz, T.A.; Parkes, D.G.; Roth, J.D. Amylin: Pharmacology, Physiology, and Clinical Potential. Pharmacol. Rev. 2015, 67, 564–600. [Google Scholar] [CrossRef]
- Westermark, P.; Andersson, A.; Westermark, G.T. Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiol. Rev. 2011, 91, 795–826. [Google Scholar] [CrossRef]
- Cooper, G.J.; Willis, A.C.; Clark, A.; Turner, R.C.; Sim, R.B.; Reid, K.B. Purification and characterization of a peptide from amyloid-rich pancreases of type 2 diabetic patients. Proc. Natl. Acad. Sci. USA 1987, 84, 8628–8632. [Google Scholar] [CrossRef]
- Westermark, P.; Wernstedt, C.; Wilander, E.; Hayden, D.W.; O’Brien, T.D.; Johnson, K.H. Amyloid fibrils in human insulinoma and islets of Langerhans of the diabetic cat are derived from a neuropeptide-like protein also present in normal islet cells. Proc. Natl. Acad. Sci. USA 1987, 84, 3881–3885. [Google Scholar] [CrossRef] [PubMed]
- Saafi, E.L.; Konarkowska, B.; Zhang, S.; Kistler, J.; Cooper, G.J. Ultrastructural evidence that apoptosis is the mechanism by which human amylin evokes death in RINm5F pancreatic islet beta-cells. Cell Biol. Int. 2001, 25, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Butler, A.E.; Jang, J.; Gurlo, T.; Carty, M.D.; Soeller, W.C.; Butler, P.C. Diabetes due to a progressive defect in beta-cell mass in rats transgenic for human islet amyloid polypeptide (HIP Rat): A new model for type 2 diabetes. Diabetes 2004, 53, 1509–1516. [Google Scholar] [CrossRef] [PubMed]
- Ritzel, R.A.; Butler, P.C. Replication increases beta-cell vulnerability to human islet amyloid polypeptide-induced apoptosis. Diabetes 2003, 52, 1701–1708. [Google Scholar] [CrossRef]
- Tavana, O.; Puebla-Osorio, N.; Sang, M.; Zhu, C. Absence of p53-dependent apoptosis combined with nonhomologous end-joining deficiency leads to a severe diabetic phenotype in mice. Diabetes 2010, 59, 135–142. [Google Scholar] [CrossRef]
- Bruchez, M., Jr.; Moronne, M.; Gin, P.; Weiss, S.; Alivisatos, A.P. Semiconductor nanocrystals as fluorescent biological labels. Science 1998, 281, 2013–2016. [Google Scholar] [CrossRef]
- Chan, W.C.; Nie, S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 1998, 281, 2016–2018. [Google Scholar] [CrossRef]
- Tokuraku, K.; Marquardt, M.; Ikezu, T. Real-time imaging and quantification of amyloid-beta peptide aggregates by novel quantum-dot nanoprobes. PLoS ONE 2009, 4, e8492. [Google Scholar] [CrossRef]
- Kuragano, M.; Yamashita, R.; Chikai, Y.; Kitamura, R.; Tokuraku, K. Three-dimensional real time imaging of amyloid β aggregation on living cells. Sci. Rep. 2020, 10, 9742. [Google Scholar] [CrossRef]
- Ishigaki, Y.; Tanaka, H.; Akama, H.; Ogara, T.; Uwai, K.; Tokuraku, K. A microliter-scale high-throughput screening system with quantum-dot nanoprobes for amyloid-β aggregation inhibitors. PLoS ONE 2013, 8, e72992. [Google Scholar] [CrossRef]
- Mirza, F.J.; Amber, S.; Sumera Hassan, D.; Ahmed, T.; Zahid, S. Rosmarinic acid and ursolic acid alleviate deficits in cognition, synaptic regulation and adult hippocampal neurogenesis in an Aβ1-42-induced mouse model of Alzheimer’s disease. Phytomedicine 2021, 83, 153490. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G.; Godos, J.; Privitera, A.; Lanza, G.; Castellano, S.; Chillemi, A.; Bruni, O.; Ferri, R.; Caraci, F.; Grosso, G. Phenolic acids and prevention of cognitive decline: Polyphenols with a neuroprotective role in cognitive disorders and Alzheimer’s disease. Nutrients 2022, 14, 819. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, R.; Hatayama, K.; Takahashi, T.; Hayashi, T.; Sato, Y.; Sato, D.; Ohta, K.; Nakano, H.; Seki, C.; Endo, Y.; et al. Structure-activity relations of rosmarinic acid derivatives for the amyloid β aggregation inhibition and antioxidant properties. Eur. J. Med. Chem. 2017, 138, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Hase, T.; Shishido, S.; Yamamoto, S.; Yamashita, R.; Nukima, H.; Taira, S.; Toyoda, T.; Abe, K.; Hamaguchi, T.; Ono, K.; et al. Rosmarinic acid suppresses Alzheimer’s disease development by reducing amyloid β aggregation by increasing monoamine secretion. Sci. Rep. 2019, 9, 8711. [Google Scholar] [CrossRef] [PubMed]
- López, L.C.; Varea, O.; Navarro, S.; Carrodeguas, J.A.; Sanchez de Groot, N.; Ventura, S.; Sancho, J. Benzbromarone, quercetin, and folic acid inhibit amylin aggregation. Int. J. Mol. Sci. 2016, 17, 964. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, L.; Zheng, W.; Chen, T. Inhibition of islet amyloid polypeptide fibril formation by selenium-containing phycocyanin and prevention of beta cell apoptosis. Biomaterials 2014, 35, 8596–8604. [Google Scholar] [CrossRef]
- Singh, S.; Trikha, S.; Bhowmick, D.C.; Sarkar, A.A.; Jeremic, A.M. Role of cholesterol and phospholipids in amylin misfolding, aggregation and etiology of islet amyloidosis. Adv. Exp. Med. Biol. 2015, 855, 95–116. [Google Scholar] [CrossRef]
- Azzam, S.K.; Jang, H.; Choi, M.C.; Alsafar, H.; Lukman, S.; Lee, S. Inhibition of human amylin aggregation and cellular toxicity by lipoic acid and ascorbic acid. Mol. Pharm. 2018, 15, 2098–2106. [Google Scholar] [CrossRef]
- Powell, D.S.; Maksoud, H.; Chargé, S.B.; Moffitt, J.H.; Desai, M.; Da Silva Filho, R.L.; Hattersley, A.T.; Stratton, I.M.; Matthews, D.R.; Levy, J.C.; et al. Apolipoprotein E genotype, islet amyloid deposition and severity of Type 2 diabetes. Diabetes Res. Clin. Pract. 2003, 60, 105–110. [Google Scholar] [CrossRef]
- Kim, S.H.; Yoo, H.J.; Park, E.J.; Na, D.H. Nano differential scanning fluorimetry-based thermal stability screening and optimal buffer selection for immunoglobulin G. Pharmaceuticals 2021, 15, 29. [Google Scholar] [CrossRef]
- da Silva, D.C.; Lima, L.M.T.R. Physico-chemical properties of co-formulated fast-acting insulin with pramlintide. Int. J. Pharm. 2018, 547, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, R.; Tainaka, R.; Ando, Y.; Hashi, Y.; Deepak, H.V.; Suga, Y.; Murai, Y.; Anetai, M.; Monde, K.; Ohta, K.; et al. An automated microliter-scale high-throughput screening system (MSHTS) for real-time monitoring of protein aggregation using quantum-dot nanoprobes. Sci. Rep. 2019, 9, 2587. [Google Scholar] [CrossRef] [PubMed]
- Goldsbury, C.; Baxa, U.; Simon, M.N.; Steven, A.C.; Engel, A.; Wall, J.S.; Aebi, U.; Müller, S.A. Amyloid structure and assembly: Insights from scanning transmission electron microscopy. J. Struct. Biol. 2011, 173, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Galaqin, N.; Tainaka, R.; Shimamori, K.; Kuragano, M.; Noguchi, T.Q.P.; Tokuraku, K. Real-Time 3D Imaging and Inhibition Analysis of Various Amyloid Aggregations Using Quantum Dots. Int. J. Mol. Sci. 2020, 21, 1978. [Google Scholar] [CrossRef]
- Abioye, R.O.; Okagu, O.D.; Udenigwe, C.C. Disaggregation of islet amyloid polypeptide fibrils as a potential anti-fibrillation mechanism of tetrapeptide TNGQ. Int. J. Mol. Sci. 2022, 23, 1972. [Google Scholar] [CrossRef]
- Khemtemourian, L.; Fatafta, H.; Davion, B.; Lecomte, S.; Castano, S.; Strodel, B. Structural dissection of the first events following membrane binding of the islet amyloid polypeptide. Front. Mol. Biosci. 2022, 9, 849979. [Google Scholar] [CrossRef]
- Jackson, K.; Barisone, G.A.; Diaz, E.; Jin, L.W.; DeCarli, C.; Despa, F. Amylin deposition in the brain: A second amyloid in Alzheimer disease? Ann. Neurol. 2013, 74, 517–526. [Google Scholar] [CrossRef]
- Ly, H.; Despa, F. Hyperamylinemia as a risk factor for accelerated cognitive decline in diabetes. Expert Rev. Proteom. 2015, 12, 575–577. [Google Scholar] [CrossRef][Green Version]
- Charalambous, A.; Andreou, M.; Antoniades, I.; Christodoulou, N.; Skourides, P.A. In vivo, site-specific, covalent conjugation of quantum dots to proteins via split-intein splicing. Methods Mol. Biol. 2012, 906, 157–169. [Google Scholar] [CrossRef]
- Marín-Peñalver, J.J.; Martín-Timón, I.; Sevillano-Collantes, C.; Del Cañizo-Gómez, F.J. Update on the treatment of type 2 diabetes mellitus. World J. Diabetes 2016, 7, 354–395. [Google Scholar] [CrossRef]
- Rochester, C.D.; Akiyode, O. Novel and emerging diabetes mellitus drug therapies for the type 2 diabetes patient. World J. Diabetes 2014, 5, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Jaikaran, E.T.; Clark, A. Islet amyloid and type 2 diabetes: From molecular misfolding to islet pathophysiology. Biochim. Biophys. Acta 2001, 1537, 179–203. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanouil, C.; Chatziathanasiadou, M.V.; Chatzigiannis, C.; Chontzopoulou, E.; Mavromoustakos, T.; Grdadolnik, S.G.; Tzakos, A.G. Unveiling the interaction profile of rosmarinic acid and its bioactive substructures with serum albumin. J. Enzyme Inhib. Med. Chem. 2020, 35, 786–804. [Google Scholar] [CrossRef] [PubMed]
- Velander, P.; Wu, L.; Hildreth, S.B.; Vogelaar, N.J.; Mukhopadhyay, B.; Helm, R.F.; Zhang, S.; Xu, B. Catechol-containing compounds are a broad class of protein aggregation inhibitors: Redox state is a key determinant of the inhibitory activities. Pharmacol. Res. 2022, 184, 106409. [Google Scholar] [CrossRef]
- Kao, P.Y.; Green, E.; Pereira, C.; Ekimura, S.; Juarez, D.; Whyte, T.; Arhar, T.; Malaspina, B.; Nogaj, L.A.; Moffet, D.A. Inhibition of Toxic IAPP Amyloid by Extracts of Common Fruits. J. Funct. Foods 2015, 12, 450–457. [Google Scholar] [CrossRef]
- Fuentes, A.L.; Hennessy, K.; Pascual, J.; Pepe, N.; Wang, I.; Santiago, A.; Chaggan, C.; Martinez, J.; Rivera, E.; Cota, P.; et al. Identification of Plant Extracts that Inhibit the Formation of Diabetes-Linked IAPP Amyloid. J. Herb. Med. 2016, 6, 37–41. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, X.; Liu, Z.; Huanood, G.; Sawatari, H.; Shimamori, K.; Kuragano, M.; Tokuraku, K. Analyzing Amylin Aggregation Inhibition Through Quantum Dot Fluorescence Imaging. Int. J. Mol. Sci. 2024, 25, 11132. https://doi.org/10.3390/ijms252011132
Yin X, Liu Z, Huanood G, Sawatari H, Shimamori K, Kuragano M, Tokuraku K. Analyzing Amylin Aggregation Inhibition Through Quantum Dot Fluorescence Imaging. International Journal of Molecular Sciences. 2024; 25(20):11132. https://doi.org/10.3390/ijms252011132
Chicago/Turabian StyleYin, Xiaoyu, Ziwei Liu, Gegentuya Huanood, Hayate Sawatari, Keiya Shimamori, Masahiro Kuragano, and Kiyotaka Tokuraku. 2024. "Analyzing Amylin Aggregation Inhibition Through Quantum Dot Fluorescence Imaging" International Journal of Molecular Sciences 25, no. 20: 11132. https://doi.org/10.3390/ijms252011132
APA StyleYin, X., Liu, Z., Huanood, G., Sawatari, H., Shimamori, K., Kuragano, M., & Tokuraku, K. (2024). Analyzing Amylin Aggregation Inhibition Through Quantum Dot Fluorescence Imaging. International Journal of Molecular Sciences, 25(20), 11132. https://doi.org/10.3390/ijms252011132





