Combined BSA-Seq and RNA-Seq Analysis to Identify Candidate Genes Associated with Aluminum Toxicity in Rapeseed (Brassica napus L.)
Abstract
1. Introduction
2. Result
2.1. Statistical Analysis of the Phenotype of the F2:3 Generation Population
2.2. Assessment of Sequencing Quality, SNP Detection, and Annotation
2.3. Indel Detection and Annotation
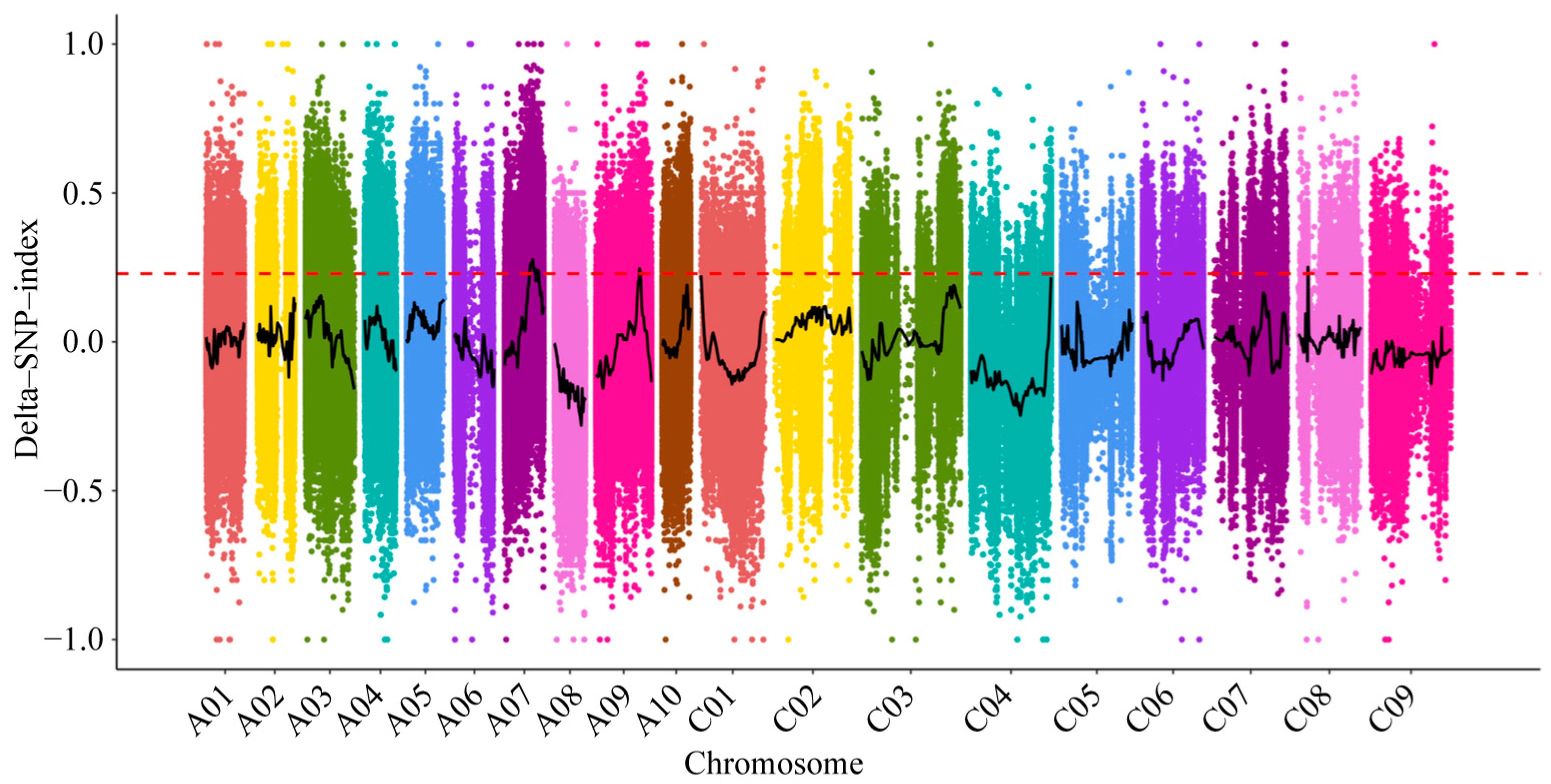
2.4. QTL for Al-Tolerance Identified by SNP and Indel Markers
2.5. Candidate Differentially Expresssed Genes (DEGs) Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Phenotypic Identification of Parents and F2:3 Population
4.3. Construction of DNA Mixed Pool and Re-Sequencing
4.4. Correlation Region Analysis of BSA
4.5. Combined Analysis of BSA-Seq and RNA-Seq
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shetty, R.; Vidya, C.S.; Prakash, N.B.; Lux, A.; Vaculík, M. Aluminum toxicity in plants and its possible mitigation in acid soils by biochar: A review. Sci. Total Environ. 2021, 765, 142744. [Google Scholar] [CrossRef] [PubMed]
- Kochian, L.V.; Piñeros, M.A.; Liu, J.; Magalhaes, J.V. Plant Adaptation to Acid Soils: The Molecular Basis for Crop Aluminum Resistance. Annu. Rev. Plant Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef] [PubMed]
- von Uexküll, H.; Mutert, E. Global extent, development and economic impact of acid soils. Plant Soil 1995, 171, 1–15. [Google Scholar] [CrossRef]
- Zhu, X.F.; Shen, R.F. Towards sustainable use of acidic soils: Deciphering aluminum-resistant mechanisms in plants. Fundam. Res. 2023, 14, 41. [Google Scholar] [CrossRef]
- Mejia-Alvarado, F.S.; Botero-Rozo, D.; Araque, L.; Bayona, C.; Herrera-Corzo, M.; Montoya, C.; Ayala-Díaz, I.; Romero, H.M. Molecular network of the oil palm root response to aluminum stress. BMC Plant Biol. 2023, 23, 346. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, R.; Shu, K.; Lv, W.; Wang, S.; Wang, C. Aluminum stress signaling, response, and adaptive mechanisms in plants. Plant Signal. Behav. Plant Signal. Behav. 2022, 17, 2057060. [Google Scholar] [CrossRef]
- Tohidi, Z.; Baghizadeh, A.; Enteshari, S. The Effects of Aluminum and Phosphorous on Brassica napus. Am. Eurasian J. Agric. Environ. Sci. 2009, 6, 137–142. [Google Scholar]
- Roshani, M.; Abbaspour, H.; Saeidi-sar, S. Effect of aluminium stress on germination and mineral nutrition of kidney bean cultivars with diferent sensitivity to aluminium. Biosci. Biotechnol. Res. Asia 2014, 11, 545–553. [Google Scholar] [CrossRef]
- Guo, J.H.; Liu, X.J.; Zhang, Y.; Shen, J.L.; Han, W.X.; Zhang, W.F.; Christie, P.; Goulding, K.W.; Vitousek, P.M.; Zhang, F.S. Significant acidification in major Chinese croplands. Science 2010, 327, 1008–1010. [Google Scholar] [CrossRef]
- Ryan, P.R.; Tyerman, S.D.; Sasaki, T.; Furuichi, T.; Yamamoto, Y.; Zhang, W.H.; Delhaize, E. The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J. Exp. Bot. 2011, 62, 9–20. [Google Scholar] [CrossRef]
- Wu, J.; Guo, X.S.; Wang, W.J.; Zhu, H.B. Effect of dolomite application on soil acidity and yield of rapeseed on yellow-red soil. Chin. J. Oil Crop Sci. 2006, 28, 55–58. [Google Scholar]
- Rahman, R.; Upadhyaya, H. Aluminium Toxicity and Its Tolerance in Plant: A Review. J. Plant Biol. Singmul Hakhoe Chi 2021, 64, 101–121. [Google Scholar] [CrossRef]
- Yang, J.L.; Fan, W.; Zheng, S.J. Mechanisms and regulation of aluminum-induced secretion of organic acid anions from plant roots. J. Zhejiang Univ. Sci. B 2019, 20, 513–527. [Google Scholar] [CrossRef] [PubMed]
- Shweta, S.; Durgesh, K.T.; Swati, S.; Shivesh, S.; Nawal Kishore, D.; Devendra, K.C.; Marek, V. Toxicity of aluminium on various levels of plant cells and organism: A review. Environ. Exp. Bot. 2017, 137, 177–193. [Google Scholar] [CrossRef]
- Li, S.; Yang, J.B.; Li, J.Q.; Huang, J.; Shen, R.F.; Zeng, D.L.; Zhu, X.F. A NAC transcription factor represses a module associated with xyloglucan content and regulates aluminum tolerance. Plant Physiol. 2024, 196, 564–578. [Google Scholar] [CrossRef] [PubMed]
- Zhan, M.; Li, H.; You, J.; Meng, X.; Guan, K.; Zheng, M.; Gao, J.; Yang, Z. SbXTH7, acting downstream of transcription factor SbHY5, regulates aluminum tolerance by modulating cell wall hemicellulose content and aluminum accumulation in sweet sorghum [Sorghum bicolor (L.)]. Ind. Crop. Prod. 2024, 214, 118485. [Google Scholar] [CrossRef]
- Ding, Z.J.; Xu, C.; Yan, J.Y.; Wang, Y.X.; Cui, M.Q.; Yuan, J.J.; Wang, Y.N.; Li, G.X.; Wu, J.X.; Wu, Y.R.; et al. The LRR receptor-like kinase ALR1 is a plant aluminum ion sensor. Cell Res. 2024, 34, 281. [Google Scholar] [CrossRef]
- Fang, Q.; Zhou, F.; Zhang, Y.; Singh, S.; Huang, C.F. Degradation of STOP1 mediated by the F-box proteins RAH1 and RAE1 balances aluminum resistance and plant growth in Arabidopsis thaliana. Plant J. 2021, 106, 493–506. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Guo, J.; Zhou, F.; Singh, S.; Xu, X.; Xie, Q.; Yang, Z.; Huang, C. F-box protein RAE1 regulates the stability of the aluminum-resistance transcription factor STOP1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 319–327. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, H.; Kou, J.; Shi, L.; Zhang, C.; Xu, F. Dual effects of transgenic Brassica napus overexpressing CS gene on tolerances to aluminum toxicity and phosphorus deficiency. Plant Soil 2013, 362, 231–246. [Google Scholar] [CrossRef]
- Ligaba, A.; Katsuhara, M.; Ryan, P.R.; Shibasaka, M.; Matsumoto, H. The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol. 2006, 142, 1294–1303. [Google Scholar] [CrossRef] [PubMed]
- Basu, U.; Good, A.G.; Taylor, G.J. Transgenic Brassica napus plants overexpressing aluminium—Induced mitochondrial manganese superoxide dismutase cDNA are resistant to aluminium. Plant Cell Environ. 2001, 24, 1269–1278. [Google Scholar] [CrossRef]
- Zhou, H.; Xiao, X.; Asjad, A.; Han, D.; Zheng, W.; Xiao, G.; Huang, Y.; Zhou, Q. Integration of GWAS and transcriptome analyses to identify SNPs and candidate genes for aluminum tolerance in rapeseed (Brassica napus L.). BMC Plant Biol. 2022, 22, 130. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Chen, L.; Liang, F.; Zhang, Y.; Zhang, C.; Xiao, H.; Tang, R.; Yang, B.; Wang, L.; Jiang, H. Identification of a major QTL and candidate genes analysis for branch angle in rapeseed (Brassica napus L.) using QTL-seq and RNA-seq. Front. Plant Sci. 2024, 15, 1340892. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.; Xie, Y.; He, H.; Wu, H.; Gao, L.; Atif, A.; Zhang, Y.; Tian, H.; Hui, J.; Gao, Y. Integrated BSA-seq and RNA-seq analysis to identify candidate genes associated with nitrogen utilization efficiency (NUtE) in rapeseed (Brassica napus L.). Int. J. Biol. Macromol. 2024, 254, 127771. [Google Scholar] [CrossRef]
- Zheng, R.; Deng, M.; Lv, D.; Tong, B.; Liu, Y.; Luo, H. Combined BSA-Seq and RNA-Seq Reveal Genes Associated with the Visual Stay-Green of Maize (Zea mays L.). Int. J. Mol. Sci. 2023, 24, 17617. [Google Scholar] [CrossRef]
- Majeed, A.; Johar, P.; Raina, A.; Salgotra, R.K.; Feng, X.; Bhat, J.A. Harnessing the potential of bulk segregant analysis sequencing and its related approaches in crop breeding. Front. Genet. 2022, 13, 944501. [Google Scholar] [CrossRef]
- Wang, H.; Liu, J.; Huang, J.; Xiao, Q.; Hayward, A.; Li, F.; Gong, Y.; Liu, Q.; Ma, M.; Fu, D.; et al. Mapping and identifying candidate genes enabling cadmium accumulation in brassica napus revealed by combined BSA-Seq and RNA-Seq Analysis. Int. J. Mol. Sci. 2023, 24, 10163. [Google Scholar] [CrossRef]
- Li, Y.; Ye, H.; Song, L.; Vuong, T.D.; Song, Q.; Zhao, L.; Shannon, J.G.; Li, Y.; Nguyen, H.T. Identification and characterization of novel QTL conferring internal detoxification of aluminium in soybean. J. Exp. Bot. 2021, 72, 4993–5009. [Google Scholar] [CrossRef]
- Zhang, P.; Zhong, K.; Zhong, Z.; Tong, H. Mining candidate gene for rice aluminum tolerance through genome wide association study and transcriptomic analysis. BMC Plant Biol. 2019, 19, 490. [Google Scholar] [CrossRef]
- Gao, H.; Ye, S.; Wu, J.; Wang, L.; Wang, R.; Lei, W.; Meng, L.; Yuan, F.; Zhou, Q.; Cui, C. Genome-wide association analysis of aluminum tolerance related traits in rapeseed (Brassica napus L.) during germination. Genet. Resour. Crop Evol. 2021, 68, 335–357. [Google Scholar] [CrossRef]
- Wang, R.L.; Wang, L.Y.; Ye, S.; Gao, H.H.; Lei, W.; Wu, J.Y.; Yuan, F.; Meng, L.J.; Tang, Z.L.; Li, J.N.; et al. QTL mapping of seed germination-related traits in Brassica napus L. under aluminum toxicity stress. Acta Agron. Sin. 2020, 46, 832–843. [Google Scholar]
- Wang, R.L.; Wang, L.Y.; Lei, W.; Wu, J.Y.; Shi, H.S.; Li, C.Y.; Tang, Z.L.; Li, J.N.; Zhou, Q.Y.; Cui, C. Screening candidate genes related to aluminum toxicity stress at germination stage via RNA-seq and QTL mapping in Brassica napus L. Acta Agron. Sin. 2021, 47, 2407–2422. [Google Scholar]
- Su, N.; Gong, Y.; Hou, X.; Liu, X.; Shabala, S.; Demidchik, V.; Yu, M.; Jiang, M.; Huang, L. Zinc finger protein ZFP36 and pyruvate dehydrogenase kinase PDK1 function in ABA-mediated aluminum tolerance in rice. Crop J. 2024. [Google Scholar] [CrossRef]
- Liu, L.; Cheng, L.; Liu, K.; Yu, T.; Liu, Q.; Gong, Z.; Cai, Z.; Liu, J.; Zhao, X.; Nian, H.; et al. Transgenic soybean of GsMYB10 shapes rhizosphere microbes to promote resistance to aluminum (Al) toxicity. J. Hazard. Mater. 2023, 455, 131621. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, P.; Bai, Z.; Herde, M.; Ma, Y.; Li, N.; Liu, S.; Huang, C.F.; Cui, R.; Ma, H.; et al. Calmodulin—Like protein CML24 interacts with CAMTA2 and WRKY46 to regulate ALMT1—Dependent Al resistance in Arabidopsis thaliana. New Phytol. 2022, 233, 2471–2487. [Google Scholar] [CrossRef]
- Zhang, L.; Dong, D.; Wang, J.; Wang, Z.; Zhang, J.; Bai, R.Y.; Wang, X.; Rubio, W.M.; Blumwald, E.; Zhang, N.; et al. A zinc finger protein SlSZP1 protects SlSTOP1 from SlRAE1-mediated degradation to modulate aluminum resistance. New Phytol. 2022, 236, 165–181. [Google Scholar] [CrossRef]
- Huang, C. Activation and activity of STOP1 in aluminium resistance. J. Exp. Bot. 2021, 72, 2269–2272. [Google Scholar] [CrossRef]
- Tokizawa, M.; Enomoto, T.; Ito, H.; Wu, L.; Kobayashi, Y.; Mora-Macías, J.; Armenta-Medina, D.; Iuchi, S.; Kobayashi, M.; Nomoto, M.; et al. High affinity promoter binding of STOP1 is essential for the early aluminum-inducible expression of novel Al resistance genes GDH1 and GDH2 in Arabidopsis. J. Exp. Bot. 2021, 72, 2769–2789. [Google Scholar] [CrossRef]
- Li, G.Z.; Wang, Z.Q.; Yokosho, K.; Ding, B.; Fan, W.; Gong, Q.Q.; Li, G.X.; Wu, Y.R.; Yang, J.L.; Ma, J.F.; et al. Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4 expression and enhancement of citrate secretion in rice (Oryza sativa). New Phytol. 2018, 219, 149–162. [Google Scholar] [CrossRef]
- Tao, Y.; Wan, J.X.; Liu, Y.S.; Yang, X.Z.; Shen, R.F.; Zhu, X.F. The NAC transcription factor ANAC017 regulates aluminum tolerance by regulating the cell wall-modifying genes. Plant Physiol. 2022, 189, 2517–2534. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.Q.; Fan, W.; Jin, J.F.; Xu, J.M.; Chen, W.W.; Yang, J.L.; Zheng, S.J. A NAC-type transcription factor confers aluminum resistance by regulating cell wall-associated receptor kinase 1 and cell wall pectin. Plant Cell Environ. 2020, 43, 463–478. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, C.; Wang, L.; Zhong, H.; Xu, X.; Cheng, Y.; Nian, H.; Liu, W.; Chen, P.; Zhang, A.; et al. GmABR1 encoding an ERF transcription factor enhances the tolerance to aluminum stress in Arabidopsis thaliana. Front. Plant Sci. 2023, 14, 1125245. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, X.; Yang, C.; Cheng, Y.; Cai, Z.; Nian, H.; Ma, Q. GsERF1 enhances Arabidopsis thaliana aluminum tolerance through an ethylene-mediated pathway. BMC Plant Biol. 2022, 22, 258. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Z.; Gong, L.; Chen, A.; Liu, N.; Li, S.; Sun, H.; Yang, Z.; You, J. Functional characterization of three MATE genes in relation to aluminum-induced citrate efflux from soybean root. Plant Soil 2019, 443, 121–138. [Google Scholar] [CrossRef]
- Maron, L.G.; Pineros, M.A.; Guimaraes, C.T.; Magalhaes, J.V.; Pleiman, J.K.; Mao, C.; Shaff, J.; Belicuas, S.N.; Kochian, L.V. Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J. 2010, 61, 728–740. [Google Scholar] [CrossRef]
- Miao, Y.; Hu, X.; Wang, L.; Schultze-Kraft, R.; Wang, W.; Chen, Z. Characterization of SgALMT genes reveals the function of SgALMT2 in conferring aluminum tolerance in Stylosanthes guianensis through the mediation of malate exudation. Plant Physiol. Biochem. 2024, 208, 108535. [Google Scholar] [CrossRef]
- Huang, J.; Li, X.; Chen, X.; Guo, Y.; Liang, W.; Wang, H. Genome-Wide Identification of Soybean ABC Transporters Relate to Aluminum Toxicity. Int. J. Mol. Sci. 2021, 22, 6556. [Google Scholar] [CrossRef]
- Kuromori, T.; Miyaji, T.; Yabuuchi, H.; Shimizu, H.; Sugimoto, E.; Kamiya, A.; Moriyama, Y.; Shinozaki, K. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. USA 2010, 107, 2361–2366. [Google Scholar] [CrossRef]
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef]
- Fu, S.; Lu, Y.; Zhang, X.; Yang, G.; Chao, D.; Wang, Z.; Shi, M.; Chen, J.; Chao, D.; Li, R.; et al. The ABC transporter ABCG36 is required for cadmium tolerance in rice. J. Exp. Bot. 2019, 70, 5909–5918. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Xu, J.M.; Wu, P.; Yang, Z.X.; Lou, H.Q.; Chen, W.W.; Jin, J.F.; Zheng, S.J.; Yang, J.L. Alleviation by abscisic acid of Al toxicity in rice bean is not associated with citrate efflux but depends on ABI5-mediated signal transduction pathways. J. Integr. Plant Biol. 2019, 61, 140–154. [Google Scholar] [CrossRef]
- Hou, N.; You, J.; Pang, J.; Xu, M.; Chen, G.; Yang, Z. The accumulation and transport of abscisic acid insoybean (Glycine max L.) under aluminum stress. Plant Soil 2010, 330, 127–137. [Google Scholar] [CrossRef]
- Li, C.; Liu, G.; Geng, X.; He, C.; Quan, T.; Hayashi, K.I.; De Smet, I.; Robert, H.S.; Ding, Z.; Yang, Z.B. Local regulation of auxin transport in root—Apex transition zone mediates aluminium—Induced Arabidopsis root—Growth inhibition. Plant J. 2021, 108, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Geng, X.; He, C.; Zhang, F.; Wang, R.; Horst, W.J.; Ding, Z. TAA1-regulated local auxin biosynthesis in the root-apex transition zone mediates the aluminum-induced inhibition of root growth in Arabidopsis. Plant Cell 2014, 26, 2889–2904. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Tian, Q.Y.; Chen, J.; Zhang, W.H. Aluminium-induced inhibition of root elongation in Arabidopsis is mediated by ethylene and auxin. J. Exp. Bot. 2010, 61, 347–356. [Google Scholar] [CrossRef]
- Zou, Y.; Zhang, X.; Tan, Y.; Huang, J.B.; Zheng, Z.; Tao, L.Z. Phosphoethanolamine N-methyltransferase 1 contributes to maintenance of root apical meristem by affecting ROS and auxin-regulated cell differentiation in Arabidopsis. New Phytol. 2019, 224, 258–273. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]

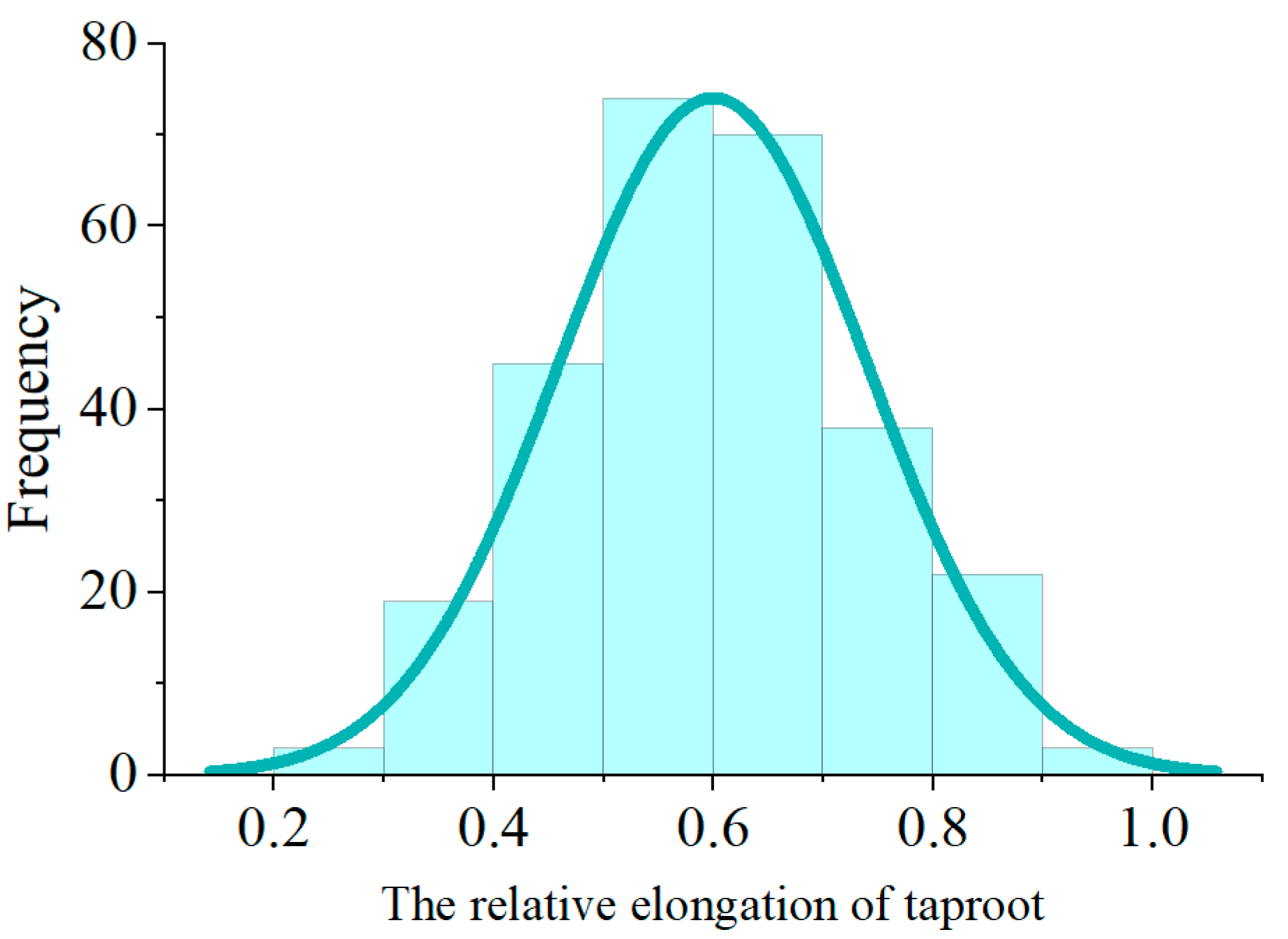

| Indexes | F2 Number | Average | Minimum | Median | Maximum | Standard Deviation | CV/% | Kurtosis | Shapiro–Wilk | Kolmogorov Smirnov |
|---|---|---|---|---|---|---|---|---|---|---|
| Value | 274 | 0.600 | 0.281 | 0.594 | 0.930 | 0.141 | 23.49 | −0.304 | p = 0.144 | 1.000 |
| Chromosome | QTL | Start | End | Size/Mb | Genes |
|---|---|---|---|---|---|
| chrA07 | qAT-A07-1 | 15,712,254 | 18,799,754 | 3.09 | 542 |
| chrA07 | qAT-A07-2 | 20,680,931 | 20,755,053 | 0.07 | 12 |
| chrA09 | qAT-A09-1 | 26,393,183 | 27,036,238 | 0.64 | 102 |
| Gene ID | QTL | Function Description | R178 | S169 | R178 vs. S169 | |||
|---|---|---|---|---|---|---|---|---|
| 6 h vs. 0 h | 24 h vs. 0 h | 6 h vs. 0 h | 24 h vs. 0 h | 6 h | 24 h | |||
| BnaA07g20010D | qAT-A07-1 | DeSI-like protein At4g17486 | −1.43 | - | −1.59 | - | 1.59 | - |
| BnaA07g20030D | qAT-A07-1 | Zinc finger protein 1 | −1.15 | −1.42 | −1.08 | - | - | - |
| BnaA07g20140D | qAT-A07-1 | Vacuolar fusion protein CCZ1 homolog B | - | - | - | - | −1.67 | −1.48 |
| BnaA07g20150D | qAT-A07-1 | Chaperone protein dnaJ 8, chloroplastic | - | - | - | - | −2.52 | −2.49 |
| BnaA07g20260D | qAT-A07-1 | Protein ACTIVITY OF BC1 COMPLEX KINASE 3, chloroplastic | - | - | - | - | 3.15 | 1.46 |
| BnaA07g20290D | qAT-A07-1 | Phosphoglycerate kinase 3, cytosolic | - | - | - | - | −1.71 | −1.48 |
| BnaA07g20500D | qAT-A07-1 | DNA repair protein recA homolog 1, chloroplastic | - | - | - | - | 1.69 | 1.90 |
| BnaA07g20510D | qAT-A07-1 | Photosystem II 10 kDa polypeptide, chloroplastic | - | - | −1.89 | - | 3.05 | 1.17 |
| BnaA07g20530D | qAT-A07-1 | Interactor of constitutive active ROPs 4 | 1.08 | - | 1.28 | - | - | - |
| BnaA07g20740D | qAT-A07-1 | Protein COFACTOR ASSEMBLY OF COMPLEX C SUBUNIT B CCB4, chloroplastic | - | - | - | - | −10.51 | −8.94 |
| BnaA07g20870D | qAT-A07-1 | Thaumatin-like protein | - | - | - | - | 1.76 | 1.69 |
| BnaA07g20960D | qAT-A07-1 | Indole glucosinolate O-methyltransferase 1 | - | 1.22 | - | 1.19 | 1.89 | 1.53 |
| BnaA07g21010D | qAT-A07-1 | Ubinuclein-2 | - | - | - | - | −1.82 | −2.43 |
| BnaA07g21360D | qAT-A07-1 | Increased DNA methylation 3 | - | - | - | - | −1.15 | −1.22 |
| BnaA07g21490D | qAT-A07-1 | Dehydrin ERD14 | - | −2.26 | - | −2.19 | −1.10 | - |
| BnaA07g21500D | qAT-A07-1 | PREDICTED: pro-resilin-like [Brassica rapa] | - | - | - | - | −1.16 | −1.08 |
| BnaA07g21560D | qAT-A07-1 | Rac-like GTP-binding protein ARAC5 | - | - | - | - | 2.72 | 2.71 |
| BnaA07g21640D | qAT-A07-1 | B-box zinc finger protein 21 | - | - | - | - | −1.47 | −1.70 |
| BnaA07g21850D | qAT-A07-1 | Glycine--tRNA ligase, chloroplastic/mitochondrial 2 | - | - | - | - | −1.52 | −1.33 |
| BnaA07g21970D | qAT-A07-1 | Protein MARD1 | 2.01 | - | 2.13 | - | - | - |
| BnaA07g22050D | qAT-A07-1 | Protein NUCLEAR FUSION DEFECTIVE 4 | - | - | 1.35 | - | −1.59 | −1.28 |
| BnaA07g22100D | qAT-A07-1 | Probable arabinosyltransferase ARAD1 | - | - | - | - | −1.58 | −1.77 |
| BnaA07g22220D | qAT-A07-1 | GDSL esterase/lipase At1g74460 | - | - | - | - | 1.48 | 1.79 |
| BnaA07g22620D | qAT-A07-1 | Phosphoethanolamine N-methyltransferase 3 | 1.63 | - | - | 4.73 | 5.71 | - |
| BnaA07g22680D | qAT-A07-1 | Sucrose synthase 6 | - | - | −3.04 | - | 3.61 | 2.44 |
| BnaA07g22710D | qAT-A07-1 | protein-lysine methyltransferase METTL21D [Brassica napus] | - | - | - | - | −3.16 | −5.01 |
| BnaA07g22740D | qAT-A07-1 | Serine carboxypeptidase-like 2 | - | - | −1.40 | - | −2.22 | −3.67 |
| BnaA07g22770D | qAT-A07-1 | F-box/LRR-repeat protein At5g02910 | - | - | - | - | −3.62 | −2.69 |
| BnaA07g22810D | qAT-A07-1 | Anaphase-promoting complex subunit 13 | - | - | - | - | 2.52 | 2.25 |
| BnaA07g22890D | qAT-A07-1 | Beta-galactosidase 17 | - | - | - | 2.94 | 3.79 | 2.07 |
| BnaA07g23200D | qAT-A07-1 | Protein NRT1/ PTR FAMILY 5.11 | - | - | - | - | 2.25 | 2.37 |
| BnaA07g23230D | qAT-A07-1 | Protein NRT1/ PTR FAMILY 5.11 | 1.85 | - | 1.81 | - | 1.35 | 2.23 |
| BnaA07g23320D | qAT-A07-1 | ABC transporter G family member 25 | −1.59 | - | −1.59 | - | −2.57 | −2.51 |
| BnaA07g23350D | qAT-A07-1 | Sucrose transport protein SUC1 | −2.21 | - | −2.07 | −1.15 | −4.03 | −3.87 |
| BnaA07g23650D | qAT-A07-1 | Ethylene-responsive transcription factor ERF070 | - | - | - | - | 1.05 | 1.58 |
| BnaA07g23760D | qAT-A07-1 | Probable peroxygenase 4 | - | - | - | - | −2.39 | −2.67 |
| BnaA07g24010D | qAT-A07-1 | Transcription factor KUA1 | - | −1.58 | −1.14 | - | - | −1.44 |
| BnaA07g24230D | qAT-A07-1 | Cyclic dof factor 5 | −5.40 | −1.09 | −6.03 | - | - | - |
| BnaA07g24270D | qAT-A07-1 | NAC transcription factor 29 | - | - | −1.05 | −1.53 | −1.20 | −4.43 |
| BnaA07g24310D | qAT-A07-1 | Probable WRKY transcription factor 57 | −1.16 | −1.48 | −2.11 | - | - | - |
| BnaA07g28730D | qAT-A07-2 | Protein ALTERED XYL | - | - | - | - | 1.08 | 1.02 |
| BnaA07g28790D | qAT-A07-2 | Probable peroxygenase 4 | - | - | - | - | −3.11 | −3.13 |
| BnaA09g36740D | qAT-A09-1 | Pentatricopeptide repeat-containing protein At3g57430, chloroplastic | - | - | - | - | −1.30 | −1.27 |
| BnaA09g36900D | qAT-A09-1 | protein enabled homolog [Brassica napus] | - | - | - | - | 1.10 | 1.19 |
| BnaA09g36940D | qAT-A09-1 | Probable glucuronoxylan glucuronosyltransferase F8H | - | - | −1.79 | - | 2.29 | 1.48 |
| BnaA09g37240D | qAT-A09-1 | E3 ubiquitin-protein ligase SINAT2 | - | - | - | - | 1.13 | 1.08 |
| BnaA09g37360D | qAT-A09-1 | MATH domain and coiled-coil domain-containing protein At3g58340 | - | - | - | - | 2.71 | 2.18 |
| BnaA09g37400D | qAT-A09-1 | Rhomboid-like protein 15 | - | - | - | - | 1.66 | 1.40 |
| BnaA07g20780D | qAT-A07-1 | PREDICTED: uncharacterized protein LOC103830546 | 1.38 | - | 1.14 | - | - | - |
| BnaA07g21140D | qAT-A07-1 | uncharacterized protein | - | - | - | - | −2.03 | −1.75 |
| BnaA07g21190D | qAT-A07-1 | uncharacterized protein | - | - | - | - | −9.48 | −8.96 |
| BnaA07g23870D | qAT-A07-1 | PREDICTED: uncharacterized protein LOC103830885 [Brassica rapa] | - | - | - | - | 1.72 | 1.37 |
| BnaA07g24050D | qAT-A07-1 | uncharacterized protein | −1.62 | −2.01 | - | - | −1.01 | −1.24 |
| BnaA07g24110D | qAT-A07-1 | PREDICTED: uncharacterized protein LOC103830913 [Brassica rapa] | −2.36 | - | - | - | −2.76 | 2.66 |
| BnaA09g36790D | qAT-A09-1 | uncharacterized protein BNAA09G36790D [Brassica napus] | - | - | - | - | 3.05 | 1.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Yu, P.; Wu, L.; Han, D.; Wu, Y.; Zheng, W.; Zhou, Q.; Xiao, X. Combined BSA-Seq and RNA-Seq Analysis to Identify Candidate Genes Associated with Aluminum Toxicity in Rapeseed (Brassica napus L.). Int. J. Mol. Sci. 2024, 25, 11190. https://doi.org/10.3390/ijms252011190
Zhou H, Yu P, Wu L, Han D, Wu Y, Zheng W, Zhou Q, Xiao X. Combined BSA-Seq and RNA-Seq Analysis to Identify Candidate Genes Associated with Aluminum Toxicity in Rapeseed (Brassica napus L.). International Journal of Molecular Sciences. 2024; 25(20):11190. https://doi.org/10.3390/ijms252011190
Chicago/Turabian StyleZhou, Huiwen, Paolan Yu, Lanhua Wu, Depeng Han, Yang Wu, Wei Zheng, Qinghong Zhou, and Xiaojun Xiao. 2024. "Combined BSA-Seq and RNA-Seq Analysis to Identify Candidate Genes Associated with Aluminum Toxicity in Rapeseed (Brassica napus L.)" International Journal of Molecular Sciences 25, no. 20: 11190. https://doi.org/10.3390/ijms252011190
APA StyleZhou, H., Yu, P., Wu, L., Han, D., Wu, Y., Zheng, W., Zhou, Q., & Xiao, X. (2024). Combined BSA-Seq and RNA-Seq Analysis to Identify Candidate Genes Associated with Aluminum Toxicity in Rapeseed (Brassica napus L.). International Journal of Molecular Sciences, 25(20), 11190. https://doi.org/10.3390/ijms252011190





